Abstract
Notch signaling pathways are known to regulate many cellular processes, including cell proliferation, apoptosis, migration, invasion, and angiogenesis, and is one of the most important signaling pathway during normal development. Recently, emerging evidences suggest that microRNAs (miRNAs) can function as key regulators of various biological and pathologic processes during tumor development and progression. Notch signaling has also been reported to be regulated through crosstalk with many pathways and factors where miRNAs appears to play a major role. This article will provide a brief overview of the published evidences for the crosstalks between Notch and miRNAs. Further, we summarize how targeting miRNAs by natural agents could become a novel and safer approach for the prevention of tumor progression and treatment.
Keywords: Notch, miRNA, cancer, signaling, therapy, nutrition
1. Notch signaling
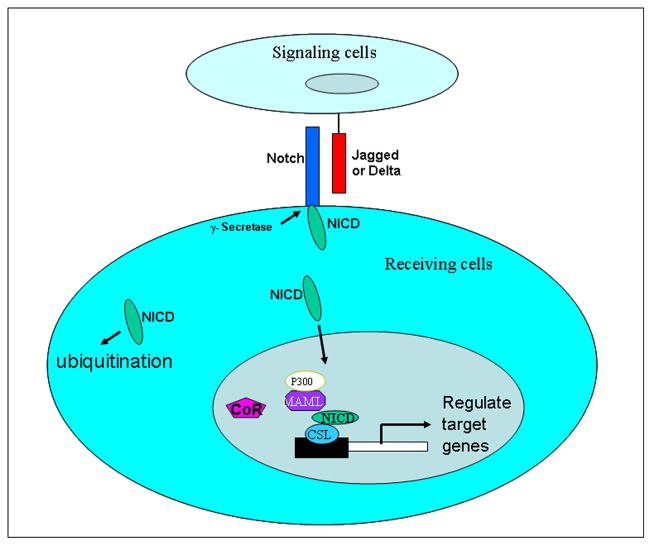
In recent years, we have witnessed the sudden explosion in the literature regarding the role of Notch signaling in tumor progression. It has become clear that Notch signaling is involved in cell proliferation, survival, apoptosis, and differentiation which affects the development and function of many organs [1]. Notch genes encode proteins which can be activated by binding of a family of its ligands. The four members of Notch receptors have been identified to date in mammals, including Notch-1-4. Five Notch ligands have been found in mammals: Dll-1 (Delta-like 1), Dll-3 (Delta-like 3), Dll-4 (Delta-like 4), Jagged-1 and Jagged-2 [1]. Notch signaling is initiated by binding of the Notch transmembrance receptors with their specific ligands between two neighboring cells. Upon activation, Notch is cleaved, releasing the Notch intracellular domain (NICD) through a cascade of proteolytic cleavages by the metalloprotease tumor necrosis factor-α-converting enzyme (TACE) and γ-secretase complex. The NICD can subsequently translocate into the nucleus for transcriptional activation of Notch target genes [2] (Figure-1). Inhibiting γ-secretase function would prevent the cleavage of the Notch receptor, resulting in blocking the Notch signal transduction signaling [3]. Therefore, γ-secretase inhibitors could be useful for the treatment of human malignancies, which are being tested in clinical trials (see website: clinicaltrials.gov). In the absence of NICD, transcription of Notch target genes is inhibited by a repressor complex mediated by the CSL (C protein binding factor 1/Suppressor of Hairless/Lag-1). When NICD enters the nucleus, it binds to CSL and recruits transcription activators to the CSL complex, leading to convert it from a transcriptional repressor into a transcription activator complex [3]. A few Notch target genes have been identified, some of which are dependent on Notch signaling in multiple tissues, while others are tissue specific. Notch target genes include Hes (Hairy enhance of split) family, Hey (Hairy/enhancer of spit related with YRPW motif), nuclear factor-kappa B (NF-κB), vascular growth factor receptor (VEGF), mammalian target of rapamycin (mTOR), cyclin D1, c-myc, p21, p27, Akt, etc. [4–7].
Figure 1.
Schematic of Notch signaling. Notch signaling is initiated by binding of the Notch transmembrance receptors with their specific ligands between two neighboring cells. Upon activation, Notch is cleaved, releasing the Notch intracellular domain (NICD) through cleavage by γ-secretase complex. The NICD can subsequently translocate into the nucleus for transcriptional activation of Notch target genes. When NICD enters the nucleus, co-repressors associated with CSL (CBF1, Suppressor of Hairless, Lag-1) are displaced and a transcriptionally active complex consisting of CSL, NICD, Mastermind, and other co-activators is formed, which converts CSL from a transcriptional repressor into an activator, leading to activation of Notch target genes.
It has been well documented that Notch signaling maintains the balance between cell proliferation, differentiation and apoptosis. Therefore, alterations in Notch signaling are considered to be associated with tumorigenesis. Indeed, it has been reported that Notch genes are abnormally regulated in many human malignancies [8–10]. These observations suggest that dysfunction of NICD prevents differentiation, ultimately guiding undifferentiated cells toward malignant transformation. Interestingly, it has been shown that the function of Notch signaling in tumorigenesis could be either oncogenic or anti-proliferative, and the function could be context dependent [11]. Notch signaling has been shown to be anti-proliferative in a limited number of tumor types, including skin cancer, human hepatocellular carcinoma and small cell lung cancer [11–13]. However, most of the studies have shown oncogenic function of Notch in many human carcinomas. In summary, emerging evidence suggest that the Notch signaling network is frequently deregulated in human malignancies with up-regulated expression of Notch receptors and their ligands in cervical, lung, colon, head and neck, renal carcinoma, acute myeloid, Hodgkin and large-cell lymphomas and pancreatic cancer [6;14–17].
Moreover, patients with tumors expressing high levels of Jagged-1 or Notch-1 had a significantly poorer overall survival compared with patients expressing low levels of these genes [18–20]. Jagged-1 was also found to be highly expressed in metastatic prostate cancer compared to localized prostate cancer or benign prostatic tissues [18]. Furthermore, high Jagged-1 expression in a subset of clinically localized tumors was significantly associated with recurrence, suggesting that Jagged-1 may be a useful marker in distinguishing indolent vs. aggressive prostate carcinomas [18]. Notch signaling pathway has also been reported to cross-talk with multiple oncogenic signaling pathways, such as NF-κB, Akt, Sonic hedgehog (Shh), mTOR, Ras, Wnt, estrogen receptor (ER), androgen receptor (AR), epidermal growth factor receptor (EGFR) and platelet-derived growth factor (PDGF) [14;21–25], and thus it is believed that the cross-talk between Notch and other signaling pathways may play critical roles in tumor aggressiveness. The main features of these pathways and cross-talk with Notch signaling have recently been reviewed, and thus the readers who are interested in learning more on the cross-talk between these pathways and Notch pathway are referred published review articles [1;6;7;24;25]. Recently, microRNAs (miRNAs) have been reported to cross-talk with Notch pathway for its regulation [26–30], suggesting that the post-transcriptional and/or translational regulation of genes by miRNAs are becoming critically important. Therefore, in the following sections, we have attempted to summarize the functional role of miRNAs in Notch signaling pathway.
2. miRNAs
In recent years, a large body of literature has emerged documenting the biological significance of miRNAs in tumor progression [31–33]. Over 4500 miRNAs have been identified in vertebrates, flies, worms, plants and viruses after the first miRNA, which was discovered in 1993 while studying Caenorhabditis elegans [34]. It is well known that miRNAs work as integral players in cancer biology. The miRNAs elicit their regulatory effects in post-transcriptional regulation by binding to the 3′ untranslated region (3′UTR) of target messenger RNA (mRNA). Either perfect or near perfect complimentary base pairing results in the degradation of the mRNA, while partial base pairing leads to translational inhibition to functional proteins [35]. The miRNAs have been implicated in a wide array of cell functions in many normal biological processes, including cell proliferation, differentiation, apoptosis, and stress resistance [36]. It has also been shown that miRNAs are key players in human cancer. The reason why miRNAs are connected with cancer is that miRNAs are involved in the biological processes of cell proliferation and apoptosis, the two intimately linked processes that are critically involved in the development and progression of human malignancies. It has been reported that there are aberrant expression of miRNAs when comparing various types of cancer with normal tissues [37]. It is very important to note that some miRNAs are thought to have oncogenic activity while others have tumor suppressor activity as indicated earlier. Oncogenic miRNAs are up-regulated in cancer and contribute to its pathology through various mechanisms such as targeting tumor suppressor genes. In contrast to the oncogenic miRNAs, other miRNAs are considered to have tumor suppressor activity and are down-regulated in cancer [38;39]. However, these distinctions may not be so strict, suggesting that some miRNAs may express either activity, depending on the biological context and tissue type.
Recent studies also suggest that miRNAs could have diagnostic, prognostic, and therapeutic value. For example, up-regulation of miR-21 is strongly associated with both a high Ki-67 proliferative index and the presence of liver metastasis [40]. High expression of miR-196a-2 had a median survival of 14.3 months compared with a median of 26.5 months for those with low expression in pancreatic cancer [41], suggesting that miR-196a-2 could be important predictor of survival. Moreover, high expression of miR-15b was significantly associated with poor prognosis and tumorigenesis in melanoma [42]. Furthermore, Patients whose liver tumors had low miR-26 expression had shorter overall survival [43]. Many other published papers showed that miRNAs expression profiling not only can be used in diagnosis, but can also be used as prognostic markers in cancer [37]. Although the research studies for the role of miRNAs in cancer have exploded in recent years, the question remains whether the alteration in miRNAs expression could be ascertained as the cause or the consequence of cancer development [37]. It is not clear for the specific targets and functions of miRNAs although there are several excellent review articles published documenting the role of miRNAs in human cancers [31–37;44], and thus we will not discuss the functions of miRNAs in cancers in this article rather we will present evidence regarding the cross-talk regulation of Notch and miRNAs in cancer development and progression.
3. Cross talk between Notch and miRNAs
Recently, it has been reported that miRNAs play critical roles in Notch signaling pathway. Several miRNAs have been shown to crosstalk with Notch pathway. However, the role of miRNAs in the Notch pathway remains unclear. Therefore, in this article, we will discuss the effect of miRNAs in the Notch signaling pathway and their cross-talk in tumor development and progression.
3.1. miR-1
It has been well known that some miRNAs have tumor suppressor activity and are down-regulated in cancer. One such miRNA which belongs to tumor suppressor group is the miR-1. In several studies investigating the expression levels of miR-1, the authors have found that the miR-1 was markedly reduced in primary human hepatocellular carcinoma (HCC), prostate cancer, head and neck, and lung cancer [45–49]. Datta et al have shown that ectopic expression of miR-1 inhibited HCC cell growth and reduced clonogenic survival [47]. In prostate cancer cell lines, transfection with miR-1 represses the expression of its target genes exportin-6 and protein tyrosine kinase 9 [45]. Nasser et al reported that reexpression of miR-1 in lung cancer cells reversed their tumorigenic properties, including growth, migration, clonogenic survival, and tumor formation in nude mice. The anti-tumor effect of miR-1 in lung cancer may be mediated through down-regulation of oncogenic targets, such as MET, Pim-1, FoxP1, and HDAC-4. Further, ectopic miR-1 expression was found to induce apoptosis in lung cancer cells in response to the potent anticancer drug doxorubicin, suggesting that miR-1 has potential therapeutic application against lung cancers [48]. Interestingly, the exon 1 and intron 1 of miR-1-1 was methylated in HCC cell lines and in primary human HCC [47]. Recently, it has been reported that miR-1 regulated Notch signaling pathway. Kwon et al reported that miR-1 directly targets the Notch ligand delta in Drosophila for repression [50]. Recently, it has also been found that Dll-1 protein levels are negatively regulated by miR-1 in mouse embryonic stem cells [51]. These results suggest that miR-1 could regulate the Notch signaling pathway; however further in-depth research is needed in order to fully understanding how miR-1 regulate the Notch pathway.
3.2. miR-34
Another important miRNA is miR-34, which has been found to participate in p53 and Notch pathways regulation consistent with tumor suppressor activity [29;52]. In mammalians, the miR-34 family is composed of three processed miRNAs: miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript. It has been reported that the expression of miR-34a was lower or undetectable in pancreatic cancer, osteosarcoma, breast cancer and non-small cell lung cancer [53–56]. Recently, the inactivation of miR-34a was identified in cell lines derived from some tumors including lung, breast, colon, kidney, bladder, pancreas and melanoma [57]. More recently, the inactivation of miR-34b/c due to CpG methylation was found in malignant melanoma, colorectal cancer, and oral squamous cell carcinoma [58–60]. Moreover, lower levels of miR-34a expression was correlated with higher probability of relapse in non-small cell lung cancer (NSCLC), suggesting that miR-34a could work as a novel prognostic marker in NSCLC patients [61]. All published data to-date suggests that the inactivation of the miR-34 is a common event in human malignancies.
The reports from several groups have shown that the members of the miR-34 family could direct p53 signaling. Expectedly, ectopic miR-34 inhibited cell proliferation, colony formation, and caused a cell cycle arrest in the G1 phase [53;62]. Moreover, re-expression of miR-34a induced apoptotic cell death [52]. It has been suggested that miR-34-mediated apoptosis could be suppressed by inactivation of p53 gene. It was also documented that miR-34a could target several mRNAs, such as SIRT1, Bcl-2, N-myc, cyclin D1, leading to translational repression of these genes [53;63;64]. Recently, Li et al reported that transfection of miR-34a to glioma cells down-regulated the protein expression of Notch-1, Notch-2, and CDK6 [26]. More recently, Ji et al reported that human gastric cancer cells with miR-34 restoration reduced the expression of target gene Notch [28]. Very recently, the same group reported that Notch-1, Notch-2 is downstream genes of miR-34 in pancreatic cancer cells. They found that restoration of miR-34 expression in the pancreatic cancer cells down-regulated Notch-1 and Notch-2 [29]. They also reported that pancreatic cancer stem cells are enriched with tumor-initiating cells or cancer stem cells with high levels of Notch-1/2 and loss of miR-34. These results suggested that miR-34 may be involved in pancreatic cancer stem cell self-renewal, potentially via the direct modulation of downstream target Notch [29]. Taken together, it may be possible to restore miR-34 function for cancer therapeutic for which novel and innovative research is warranted.
3.3. miR-146
The miR-146 was previously reported to function as novel negative regulators that help to fine-tune the immune response. Konstantin et al described the role for miR-146 in the control of Toll-like receptor and cytokine signaling through a negative feedback regulation loop involving inhibition of TNF receptor-associated factor 6 protein and IL-1 receptor-associated kinase 1 levels [65]. Recently, it has been found that miR-146a/b acts as terminal transducers of TLR4 signaling by targeting NF-κB activation by TLR4 [66]. They also demonstrated a decrease in miR-146b in adult T-cell Leukemia cells. The decrease in miR-146b may lead to increased inflammation and decreased T-reg functions, resulting in leukemia [66]. Very recently, miR-146a has been found to have the strongest predictive accuracy for stratifying prognostic groups and have also shown superiority in predicting overall survival in lung squamous cell carcinoma [67]. The miR-146 has been reported to crosstalk with breast cancer metastasis suppressor 1 (BRMS1), a predominantly nuclear protein that inhibits metastasis without blocking orthotopic tumor growth. Specifically, BRMS1 significantly up-regulates miR-146a and miR-146b in breast cancer cells. Transduction of miR-146a or miR-146b into breast cancer cells decreased expression of epidermal growth factor receptor, down-regulated NF-κB activity, inhibited migration and invasion in vitro, and suppressed lung metastasis in experimental xenograft models [68;69]. These provided experimental support suggesting that the modulation in the levels of miR-146 could have therapeutic value in inhibiting breast cancer metastasis. Very recently, miR-146a was found to regulate Numb in C2C12 cells [27], which is interesting because Numb is known to regulate Notch signaling negatively through interaction with Notch and the subsequent ubiquitin-mediated protein degradation. Indeed, Notch activation and the loss of Numb expression were found in a large proportion of breast carcinomas [70;71]. It has been reported that over-expression of Notch-1 stimulates NF-κB activity in several cancer cell lines [72] and since miR-146 also regulate NF-κB activity, it clearly suggest that miR-146 could regulate NF-κB through Notch mediated signaling pathway. However, the role of miR-146 in Notch signaling pathway need further innovative investigations.
3.4. miR-199
It has been reported that miR-199a was down-modulated in ovarian cancer [73]. Murakami et al also found that miR-199a was down-regulated in hepatocellular cancer. Moreover, they found that over-expression of miR-199a can introduce cell cycle arrest in G2/M phase [74]. Recently, It was reported that miR-199a and miR-199b were down-regulated after 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), a potent tobacco-specific carcinogen, treated rats up to 20 weeks [75]. Very recently, miR-199b-5p was seen to be a regulator of the Notch pathway through its targeting of the transcription factor Hes-1 in medulloblastoma (MB) tumors. Inhibition of Hes-1 by miR-199b-5p negatively regulated the MB cell growth. Moreover, over-expression of miR-199b-5p decreased the MB stem-like cells (CD133+) and also blocked expression of several cancer stem-cell genes. Further, the expression of miR-199b-5p in the non-metastatic cases was significantly higher than in the metastatic cases. The patients with high levels of miR-199b expression showed a better overall survival [30]. These results clearly suggest that miR-199 family could be very important in the regulation of multiple signaling pathways including Notch, and thus further in-depth studies are needed in order to clarify the biological significance and mechanisms on how miR-199 can regulate the Notch signaling pathway in human cancers.
3.5. miR-200
The microRNA-200 family has five members: miR-200a, miR-200b, miR-200c, miR-141 and miR-429. The miR-200c was down-regulated in benign or malignant hepatocellular tumors [76]. It has been shown that three miR-200 miRNAs (miR-200a, miR-200b and miR-429) are significantly associated with cancer recurrence and overall survival in ovarian tumors [77]. Recently many studies have shown that the miR-200 family regulates epithelial-mesenchymal transition (EMT) by targeting zinc-finger E-box binding homeobox 1 (ZEB1) and ZEB2 [78–81]. EMT is a process by which epithelial cells undergo remarkable morphological changes characterized by a transition from epithelial cobblestone phenotype to elongated fibroblastic phenotype. We have found that PDGF-D over-expression led to the acquisition of EMT phenotype in PC-3 prostate cells (PC3 PDGF-D cells) consistent with loss of miR-200 expression, and that the re-expression of miR-200b in PC3 PDGF-D cells led to the reversal of the EMT phenotype, which was associated with the down-regulation of ZEB1, ZEB2, and Snail2 expression [82]. Moreover, transfection of PC3 PDGF-D cells with miR-200b inhibited cell migration and invasion with concomitant repression of cell adhesion to the culture surface and cell detachment [82]. We also found that miR-200a, miR-200b, miR-200c, and many members of the tumor suppressor let-7 family were down-regulated in gemcitabine-resistant (GR) pancreatic cancer cells, which show the acquisition of EMT phenotype [83]. Furthermore, we have shown that miR-200 family regulates the expression of ZEB1, slug, E-cadherin, and vimentin, and thus the re-expression of miR-200 could be useful for the reversal of EMT phenotype to mesenchymal-to-epithelial transition [83]. We have found that the expression of both mRNA and protein levels of Notch 1-4, Dll-1, Dll-3, Dll-4, Jagged-2 as well as Notch downstream targets, such as Hes and Hey, were significantly higher in PC3 PDGF-D cells (unpublished data). More importantly, we found that Notch-1 could be one of miR-200b targets because over-expression of miR-200b significantly inhibited Notch-1 expression (unpublished data). However, how the miR-200b regulates Notch gene expression will certainly require further in-depth investigations.
4. miRNA as targets by natural agents
Emerging experimental studies have shown that targeting miRNA could be a novel strategy for cancer prevention and/or treatment. There are several strategies that could be used for targeting the regulation of miRNAs, which could be useful tool for the inhibition of tumor progression and, as such, could be useful for therapy. One potential strategy could be the inactivation of oncogenic miRNAs. It has been found that 2′-O-methyl oligonucleotides or locked nucleic acid-modified oligonucleotides can block miRNA function. For example, using this anti-sense oligonucleotide, one could significantly decrease the activity of miR-21 as compared to control oligonucleotides [84]. Another strategy is to restore down-regulated miRNAs that function as tumor suppressors, such as let-7. It has been shown that over-expression of let-7 by using exogenously transfected pre-let-7 RNAs consistently showed reduction in the number of proliferating cells in lung and liver cancer cell lines [85]. This finding clearly suggests the possibility of restoration of tumor suppressor miRNAs toward cancer therapy. A third possible strategy could be the use of “natural agents” to target miRNAs that are known to contribute in the processes of tumor development and progression.
To that end, recent studies have shown that “natural agents” including curcumin, isoflavone, indole-3-carbinol (I3C), 3,3′-diindolylmethane (DIM), EGCG, and others could alter the expression of specific miRNAs, which may lead to increased sensitivity of cancer cells to conventional therapeutic agents, and thereby may result in the inhibition of tumor growth. We have found that alteration in the expression of miRNAs could be achieved by treating cancer cells with DIM or isoflavone. We have shown that treatment of Panc-1 or Colo-357 cells with B-DIM or genistein (isoflavone) showed decreased expression of the oncogenic miRNA such as miR-17, miR-20a, miR-106a, and increased the expression of the tumor suppressor miRNAs such as let-7, miR-16-1 [86]. Our results clearly suggest that “natural agents” may exhibit their anti-tumor effects through the regulation of miRNAs. Further support to this statement comes from findings reported by Sun et al showing that curcumin could altere specific miRNA expression in human pancreatic cancer cells especially showing up-regulation of miR-22. They also found that up-regulation of miR-22 expression by curcumin in pancreatic cancer cells suppressed the expression of its target genes SP1 transcription factor (SP1) and estrogen receptor 1 (ESR1) [87]. Melkamu et al reported that I3C can inhibit the expression of several oncogenic miRNAs, such as miR-21, miR-31, miR-130a, miR-146b, and miR-377 in vinyl carbamate treated animals. Further investigation showed that I3C up-regulated PTEN tumor suppressor gene though inhibition of miR-21 [49]. Tsang, et al. recently reported that EGCG treatment could up-regulate the expressions of miR-16 in human hepatocellular carcinoma cells [88]. We also found that the expression of miR-200 and let-7 families could be up-regulated in gemcitabine resistant cells by DIM or isoflavone treatment as indicatd above. Our results also showed that DIM treatment cause down-regulation of ZEB1, slug, and vimentin, and the morphologic reversal of EMT to epithelial morphology [83]. Considering the non-toxic characteristics of “natural agents”, one could speculate that targeting miRNAs by “natural agents” could be a novel and safer approach for the prevention of tumor progression and/or treatment of human malignancies in the future.
5. Concluding remarks
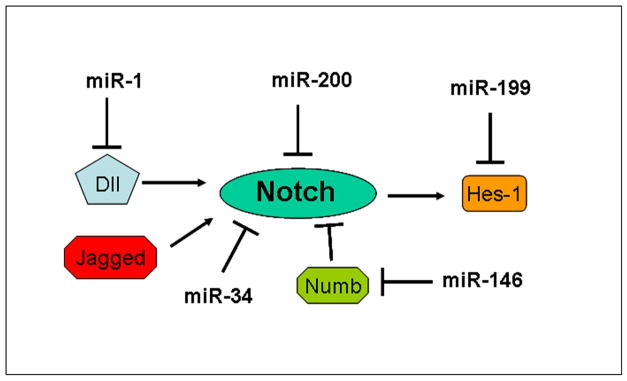
In conclusion, we believe that the deregulation of miRNAs plays important roles in the development and progression of human cancers, and during the acquisition of EMT phenotype that are in part associated with the formation and maintenance of cancer stem cells (CSCs). Importantly, miRNAs have been characterized as biomarkers for diagnosis and prognosis, as well as targets for cancer therapy. Although emerging evidence suggest an interrelationship between miRNAs and Notch signaling pathway (Figure-2), further research is warranted to ascertain the value of specific miRNA in the regulation of Notch signaling in order to exploit preventive and therapeutic strategies. Due to the non-toxic nature of “natural agents”, we believe that targeting miRNAs by “natural agents” combined with conventional chemotherapeutics could be a novel and safer approach for the treatment of cancer. The findings reported in the short review article are very interesting; however, further investigations are needed in order to elucidate the roles of these and numerous other miRNAs that could be mechanistically linked with Notch and other cell signaling, and devising novel approaches on how “natural agents” could be useful in combination therapy for the prevention and/or treatment of human malignancies in the future.
Figure 2.
Diagram of roles of miRNA in the Notch pathway
Acknowledgments
The authors’ work cited in this review was funded by grants from the National Cancer Institute, NIH (5R01CA131151, 5R01CA083695, 1R01CA132794, 1R01CA101870) to F.H.S. and Department of Defense Postdoctoral Training Award W81XWH-08-1-0196 (Zhiwei Wang) and also partly supported by a subcontract award (F.H.S.) from the University of Texas MD Anderson Cancer Center through a SPORE grant (5P20-CA101936) on pancreatic cancer awarded to James Abbruzzese. We also sincerely thank both Puschelberg and Guido foundation for their generous contributions to our research.
Footnotes
Conflict of interest
No.
“Conflict of Interest Statement”, all authors disclose no any financial and personal relationships with other people or organisations that could innapropriately influence (bias) this work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Miele L, Miao H, Nickoloff BJ. Notch signaling as a novel cancer therapeutic target. Curr Cancer Drug Targets. 2006;6:313–323. doi: 10.2174/156800906777441771. [DOI] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miele L. Notch signaling. Clin Cancer Res. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- 4.Dufraine J, Funahashi Y, Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene. 2008;27:5132–5137. doi: 10.1038/onc.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li JL, Harris AL. Notch signaling from tumor cells: a new mechanism of angiogenesis. Cancer Cell. 2005;8:1–3. doi: 10.1016/j.ccr.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Li Y, Banerjee S, Sarkar FH. Exploitation of the Notch signaling pathway as a novel target for cancer therapy. Anticancer Res. 2008;28:3621–3630. [PubMed] [Google Scholar]
- 8.Wang Z, Li Y, Banerjee S, Sarkar FH. Emerging role of Notch in stem cells and cancer. Cancer Lett. 2009;279:8–12. doi: 10.1016/j.canlet.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malinge S, Izraeli S, Crispino JD. Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood. 2009;113:2619–2628. doi: 10.1182/blood-2008-11-163501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demarest RM, Ratti F, Capobianco AJ. It’s T-ALL about Notch. Oncogene. 2008;27:5082–5091. doi: 10.1038/onc.2008.222. [DOI] [PubMed] [Google Scholar]
- 11.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sriuranpong V, Borges MW, Ravi RK, Arnold DR, Nelkin BD, Baylin SB, Ball DW. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Research. 2001;61:3200–3205. [PubMed] [Google Scholar]
- 13.Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van NM, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Banerjee S, Li Y, Rahman KM, Zhang Y, Sarkar FH. Down-regulation of Notch-1 inhibits invasion by inactivation of nuclear factor-{kappa}B, vascular endothelial growth factor, and matrix metalloproteinase-9 in pancreatic cancer cells. Cancer Research. 2006;66:2778–2784. doi: 10.1158/0008-5472.CAN-05-4281. [DOI] [PubMed] [Google Scholar]
- 15.Wang Z, Zhang Y, Li Y, Banerjee S, Liao J, Sarkar FH. Down-regulation of Notch-1 contributes to cell growth inhibition and apoptosis in pancreatic cancer cells. Mol Cancer Ther. 2006;5:483–493. doi: 10.1158/1535-7163.MCT-05-0299. [DOI] [PubMed] [Google Scholar]
- 16.Real PJ, Ferrando AA. NOTCH inhibition and glucocorticoid therapy in T-cell acute lymphoblastic leukemia. Leukemia. 2009;23:1374–1377. doi: 10.1038/leu.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strizzi L, Hardy KM, Seftor EA, Costa FF, Kirschmann DA, Seftor RE, Postovit LM, Hendrix MJ. Development and cancer: at the crossroads of Nodal and Notch signaling. Cancer Res. 2009;69:7131–7134. doi: 10.1158/0008-5472.CAN-09-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Santagata S, Demichelis F, Riva A, Varambally S, Hofer MD, Kutok JL, Kim R, Tang J, Montie JE, Chinnaiyan AM, Rubin MA, Aster JC. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 19.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Research. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 20.Reedijk M, Pinnaduwage D, Dickson BC, Mulligan AM, Zhang H, Bull SB, O’Malley FP, Egan SE, Andrulis IL. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res Treat. 2007 doi: 10.1007/s10549-007-9805-3. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, Sengupta R, Banerjee S, Li Y, Zhang Y, Rahman KM, Aboukameel A, Mohammad R, Majumdar AP, Abbruzzese JL, Sarkar FH. Epidermal growth factor receptor-related protein inhibits cell growth and invasion in pancreatic cancer. Cancer Research. 2006;66:7653–7660. doi: 10.1158/0008-5472.CAN-06-1019. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Kong D, Banerjee S, Li Y, Adsay NV, Abbruzzese J, Sarkar FH. Down-regulation of platelet-derived growth factor-D inhibits cell growth and angiogenesis through inactivation of Notch-1 and nuclear factor-kappaB signaling. Cancer Res. 2007;67:11377–11385. doi: 10.1158/0008-5472.CAN-07-2803. [DOI] [PubMed] [Google Scholar]
- 23.Chan SM, Weng AP, Tibshirani R, Aster JC, Utz PJ. Notch signals positively regulate activity of the mTOR pathway in T-cell acute lymphoblastic leukemia. Blood. 2007;110:278–286. doi: 10.1182/blood-2006-08-039883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Wang XF. Signaling cross-talk between TGF-beta/BMP and other pathways. Cell Res. 2009;19:71–88. doi: 10.1038/cr.2008.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poellinger L, Lendahl U. Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr Opin Genet Dev. 2008;18:449–454. doi: 10.1016/j.gde.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuang W, Tan J, Duan Y, Duan J, Wang W, Jin F, Jin Z, Yuan X, Liu Y. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem Biophys Res Commun. 2009;378:259–263. doi: 10.1016/j.bbrc.2008.11.041. [DOI] [PubMed] [Google Scholar]
- 28.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, Xiang D, Desano JT, Bommer GT, Fan D, Fearon ER, Lawrence TS, Xu L. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De MD, Esposito V, Galeone A, Navas L, Esposito S, Gargiulo S, Fattet S, Donofrio V, Cinalli G, Brunetti A, Vecchio LD, Northcott PA, Delattre O, Taylor MD, Iolascon A, Zollo M. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578–585. doi: 10.1038/nrg2628. [DOI] [PubMed] [Google Scholar]
- 32.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 33.Petri A, Lindow M, Kauppinen S. MicroRNA silencing in primates: towards development of novel therapeutics. Cancer Res. 2009;69:393–395. doi: 10.1158/0008-5472.CAN-08-2749. [DOI] [PubMed] [Google Scholar]
- 34.Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson KM, Weiss GJ. MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 36.O’Hara SP, Mott JL, Splinter PL, Gores GJ, LaRusso NF. MicroRNAs: key modulators of posttranscriptional gene expression. Gastroenterology. 2009;136:17–25. doi: 10.1053/j.gastro.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 39.Medina PP, Slack FJ. microRNAs and cancer: an overview. Cell Cycle. 2008;7:2485–2492. doi: 10.4161/cc.7.16.6453. [DOI] [PubMed] [Google Scholar]
- 40.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–4684. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 41.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 42.Satzger I, Mattern A, Kuettler U, Weinspach D, Voelker B, Kapp A, Gutzmer R. MicroRNA-15b represents an independent prognostic parameter and is correlated with tumor cell proliferation and apoptosis in malignant melanoma. Int J Cancer. 2009 doi: 10.1002/ijc.24960. [DOI] [PubMed] [Google Scholar]
- 43.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, Qin LX, Man K, Lo CM, Lee J, Ng IO, Fan J, Tang ZY, Sun HC, Wang XW. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 45.Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, Croce CM. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–6170. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Childs G, Fazzari M, Kung G, Kawachi N, Brandwein-Gensler M, McLemore M, Chen Q, Burk RD, Smith RV, Prystowsky MB, Belbin TJ, Schlecht NF. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, Ghoshal K, Jacob ST. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, Wang B, Suster S, Jacob ST, Ghoshal K. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl-carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- 50.Kwon C, Han Z, Olson EN, Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 53.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, Zhai Y, Giordano TJ, Qin ZS, Moore BB, MacDougald OA, Cho KR, Fearon ER. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 54.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He C, Xiong J, Xu X, Lu W, Liu L, Xiao D, Wang D. Functional elucidation of MiR-34 in osteosarcoma cells and primary tumor samples. Biochem Biophys Res Commun. 2009;388:35–40. doi: 10.1016/j.bbrc.2009.07.101. [DOI] [PubMed] [Google Scholar]
- 56.Kato M, Paranjape T, Muller RU, Nallur S, Gillespie E, Keane K, Esquela-Kerscher A, Weidhaas JB, Slack FJ. The mir-34 microRNA is required for the DNA damage response in vivo in C. elegans and in vitro in human breast cancer cells. Oncogene. 2009;28:2419–2424. doi: 10.1038/onc.2009.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Korner H, Knyazev P, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 58.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 59.Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, Tokino T. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 61.Gallardo E, Navarro A, Vinolas N, Marrades RM, Diaz T, Gel B, Quera A, Bandres E, Garcia-Foncillas J, Ramirez J, Monzo M. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- 62.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 63.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci U S A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 65.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bellon M, Lepelletier Y, Hermine O, Nicot C. Deregulation of microRNA involved in hematopoiesis and the immune response in HTLV-I adult T-cell leukemia. Blood. 2009;113:4914–4917. doi: 10.1182/blood-2008-11-189845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, Beer DG. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 68.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappaB activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008 doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 71.Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167:215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osipo C, Golde TE, Osborne BA, Miele LA. Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest. 2008;88:11–17. doi: 10.1038/labinvest.3700700. [DOI] [PubMed] [Google Scholar]
- 73.Iorio MV, Visone R, Di LG, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 74.Murakami Y, Yasuda T, Saigo K, Urashima T, Toyoda H, Okanoue T, Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 75.Kalscheuer S, Zhang X, Zeng Y, Upadhyaya P. Differential expression of microRNAs in early-stage neoplastic transformation in the lungs of F344 rats chronically treated with the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis. 2008;29:2394–2399. doi: 10.1093/carcin/bgn209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 77.Hu X, Macdonald DM, Huettner PC, Feng Z, El NI, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 78.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 80.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, Sarkar FH. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–6712. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–550. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack FJ. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 86.Vandenboom Ii TG, Li Y, Philip PA, Sarkar FH. MicroRNA and Cancer: Tiny Molecules with Major Implications. Curr Genomics. 2008;9:97–109. doi: 10.2174/138920208784139555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 88.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2009 doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]