Abstract
BACKGROUND
We previously demonstrated that a putative anti-tumor gene, peroxisomal membrane protein 4, 24 kDa (PMP24 or PXMP4), is silenced via DNA methylation of a CpG island in its 5′ flanking region (5′-CGI) in prostate cancer (PCa) cells.
METHODS
To identify demethylation hypersensitive site(s) in PMP24 5′-CGI, PC-3 cells with methylated 5′-CGI were treated with a low-dose of 5-aza-2′-deoxycytidine (5-aza-dC) just sufficient to reactivate gene expression, referred as the limited demethylation approach. Gel shift assays and promoter analyses were performed to demonstrate the role of the hypersensitive site in PMP24 gene regulation. Transfection of a methylated oligonucleotide corresponding to the hypersensitive site was conducted to determine the effect of site-specific methylation on the gene expression. Bisulfite sequencing analysis was performed to reveal the methylation status of PMP24 promoter in cultured cells and microdissected samples. In situ hybridization was applied to determine expression positivity of PMP24 mRNA.
RESULTS
A 5-aza-dC hypersensitive site encompasses two CpG dinucleotides in intron 1 was identified. Methylation of the first, but not the second, CpG dinucleotide of this site disrupted DNA-protein interactions and suppressed the gene expression. Using archival specimens, we found the first CpG dinucleotide of the hypersensitive site is hypermethylated with a loss of PMP24 mRNA expression in microdissected PCa cells when compared to normal prostatic epithelial cells.
CONCLUSIONS
These findings support a critical role for a single intronic CpG dinucleotide in PMP24 gene regulation through DNA methylation. The data suggest that methylation-mediated silencing of PMP24 is a molecular event associated with prostate carcinogenesis.
Keywords: PXMP4, intronic regulatory sequence, limited demethylation approach, methylation-demethylation hypersensitive site, 5-aza-2′-deoxycytidine
INTRODUCTION
DNA methylation is widely involved in tumorigenesis, and changes in both its level and pattern have been reported in virtually all types of cancers, including prostate cancer (PCa) [1–3]. Aberrant hypermethylation of promoter CpG islands (CGIs) in cancer is of particular interest because of its close association with gene silencing, especially when the silencing occurs to tumor suppressor genes such as BRCA1 [4], MLH1 [5], and p16 [6]. Tumor suppressor genes often can be reactivated by reducing the extent of methylation in their regulatory CGIs through treatment with epigenetic modifiers such as 5-aza-2′-deoxycytidine (5-aza-dC), offering an attractive option for cancer treatment [7, 8]. In this regard, 5-aza-dC and other DNA methyltransferase inhibitors have been tested in multiple clinical trials for the treatment of patients with hematologic malignancies [9]. However, the efficacy of these drugs in activating tumor suppressor genes is dependent on both the cell type and the target gene [10, 11], and their anti-cancer activity may involve both DNA methylation-dependent and -independent pathways [12]. Therefore, the complex relationship between aberrant promoter CGI methylation and gene silencing requires further exploration, with an anticipatory outcome of improving the clinical potential of demethylating agents as cancer therapies.
CGIs located in the 5′ regulatory region of the genes usually encompass the promoter, the first exon, and occasionally the first intron [13]. Hypermethylation of 5′-CGIs is thought to repress gene transcription by interfering with transcription initiation. However, other studies have shown that exons and introns further downstream can contribute to gene regulation via DNA methylation [14, 15]. Studies have linked DNA methylation-regulatory proteins (eg, DNA methyltransferases and methyl CpG binding-domain proteins) and histone modification proteins (eg, histone deacetylases and acetylases) to higher-order chromatin remodeling events (eg, nucleosome destabilization) as a mechanism of epigenetic regulation of gene expression [2, 16]. These proteins, working in concert, facilitate the assembly of a repressive chromatin that is inaccessible to gene-specific transcription factors (TFs) and/or the general transcriptional machinery involved in the initiation of transcription. DNA methylation also directly affects gene transcription by steric interference of TF binding to cis-elements in the promoter region [17]. We previously demonstrated the correlation between the methylation of three CpG clusters (methylation hotspots) in the estrogen receptor β (ERβ) promoter-CGI and the silencing of the gene in clinical PCa specimens [18]. More recently, we found that the activator protein 2 (AP-2) interacts with the most 5′ methylation hotspot (16-mer) and elicits the transcription of ERβ [19], hence establishing this methylation hotspot as a regulatory cis-element. These findings illustrate how methylation of the cis-acting element of a key TF of a gene conveys epigenetic suppression.
In the prostate, androgen signaling plays a critical role in cancer development [20–22]. In a previous study, using methylation-sensitive restriction fingerprinting [23], we identified the peroxisomal membrane protein 4, 24 kDa (PMP24 or PXMP4) [24], as a gene that undergoes DNA hypermethylation-mediated transcriptional silencing during the transition of LNCaP, a PCa cell line, from androgen dependence to androgen independence [25]. PMP24 is a 24 kDa peroxisomal intrinsic membrane protein [24] with unknown function although recent research has established an indispensable role of peroxisomes in catalyzing a number of essential metabolic functions including fatty acid beta-oxidation, ether phospholipid biosythesis, fatty acid alpha-oxidation and glyoxylate detoxification [26]. We found that PMP24 has a 5′-CGI with 43 CpG dinucleotides encompassing the proximal promoter, exon 1, and part of intron 1 [25]. The gene is actively transcribed in LNCaP and immortalized human normal epithelial cells (NPrEC) [27] and its 5′-CGI is unmethylated in these cells. In the androgen-independent subline, LNCaPCS, generated by maintaining LNCaP in medium with charcoal-stripped (CS) serum for over 30 passages, PMP24 is silenced and exhibits a densely methylated 5′-CGI. PMP24 is also silenced in PC-3, an androgen-independent PCa cell line. Treatment of LNCaPCS and PC-3 with 5-aza-dC readily re-activates expression of PMP24 and induces demethylation of its 5′-CGI. Ectopic expression of PMP24 in LNCaPCS and PC-3 induces a significant reduction in cell growth and colony-formation potential on soft agar, suggesting a tumor-suppressing role for PMP24 [25].
In this study, we aimed to identify specific CpGs in the 5′-CGI of PMP24, whose methylation status is essential for gene regulation. A limited demethylation approach with fine-tuned, low concentrations of 5-aza-dC was used to identify hypersensitive CpG dinucleotides. In parallel, methylated sense oligonucleotides targeting specific CpGs was used to achieve in cellulo sequence-specific methylation [18]. Deletion- and site-directed mutants were then used to validate the transcriptional activities of the putative regulatory CpGs. Laser capture microdissection (LCM) was used to acquire pure populations of normal and malignant prostatic epithelial cells from clinical specimens for subsequent assessment of their PMP24 5′-CGI methylation status in vivo. The expression of PMP24 transcript in PCa foci and benign glands were evaluated by in situ hybridization. These studies conjointly identified a single CpG dinucleotide in the intronic region of the 5′-CGI whose methylation status is critical to the transcription of PMP24 in vitro and in vivo. Our findings implicate an involvement of methylation-mediated silencing of PMP24 in prostate carcinogenesis.
METHODS AND METHODS
Prostate Samples and Laser Capture Microdissection
Formalin-fixed, paraffin-embedded sections were obtained from archival collections of radical prostatectomy specimens [18]. Seven cases of specimens were used to acquire seven LCM samples of benign prostate epithelial cells. Fourteen cases of PCa tissues were used for LCM to obtain 16 samples of PCa cells. For in situ hybridization, sections from another seven cases of prostate specimens which contained both benign glands and PCas were used. The use of these samples was reviewed and approved by the respective institutional review boards at the University of Cincinnati and University of Massachusetts Medical School.
Cell Culture and 5-aza-dC Treatment
Prostate cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA) and maintained according to the supplier’s suggestions. An immortalized human normal prostate epithelial cell line (NPrEC) was generated in our laboratory and maintained as previously described [27]. For the demethylation assay, PC-3 cells were seeded at a density of 1 × 105 cells per T25 flask at day 0, one day before treatment with 5-aza-dC (Sigma, St. Louis, MO). The cells were treated for a total of 5 days with medium containing fresh 5-aza-dC changed every two days. The cells were collected at day 6 for bisulfite sequencing and real-time RT-PCR analyses.
Bisulfite Sequencing Analyses and Real-time RT-PCR
The LCM bisulfite-sequencing analysis was targeted on a 372 bp region within the CGI encompassing 37 CpG dinucleotides. The primers P24-bisF1/P24-bisR1 and P24-bisF2/P24-bisR2 (Table I) were used in the first round and nested PCR, respectively. The amplified region was from −151 to +221 (Figure 1A), with the translation start site designated as +1. The real-time RT-PCR primers for PMP24 transcript were PMP24f and PMP24r (Table I). GAPDH and 18S RNA was used as the controls [19].
Table I.
Oligonucleotide sequences used in the study.
| Assay | Name | Primer sequences (5′➔3′) |
|---|---|---|
| Bisulfite sequencing | P24-bisF1 | GAG TGT TAT TGT TTT ATG AGA TAA GTA |
| P24-bisR1 | AAC TCC CAT TAC ACA AAT AAT AAC | |
| P24-bisF2 | TGA TTG GTT TTT TAT GTA TGA GT | |
| P24-bisR2 | AAA ACT ACT CCA AAA CAA ACA A | |
| Real-time RT-PCR | PMP24f | AGG GCT CTG CTC GTA GTC GTC |
| PMP24r | GCC ATT CCG GAA GAG AAA GGT | |
| Gel shift | WT | CTG TGT GAG TGG GGC CGT CGG GCC GGG CTG |
| MutCG1 | CTG TGT GAG TGG GAT TAT CGG GCC GGG CTG | |
| MutCG2 | CTG TGT GAG TGG GGC CGT TAA ACC GGG CTG | |
| MutCG1-2 | CTG TGT GAG TGG GGT TAT TAA GCC GGG CTG | |
| In cellulo methylation | metODN | G*G*G* mCGT mCGG GCmC GG*G* C*T |
| In situ hybridization | T7 | CAG TAA TAC GAC TCA CTA TA |
| P24probe | TGG TAG CGG CGC TTG CGC AGC AGT GCG TTG ACG ACT ACG AGC AGA CCC TAT AGT GAG TCG TAT TAC TG |
|
| Scramble probe |
GTG TAA CAC GTC TAT ACG CCC A |
Note: Primers for the real-time RT-PCR are located at exon 1 and 2, with the PCR size 153 bp. In the gel-shift assay, CpG dinucleotides in the WT probe and mutated nucleotides at the first and second CpG dinucleotides (mutCG1 and mutCG2, respectively) are underlined. mC indicates methylated C.
indicates nucleotide phosphorothioated bonds. For in situ hybridization, the underlined is the consensus promoter sequence of T7 RNA polymerase.
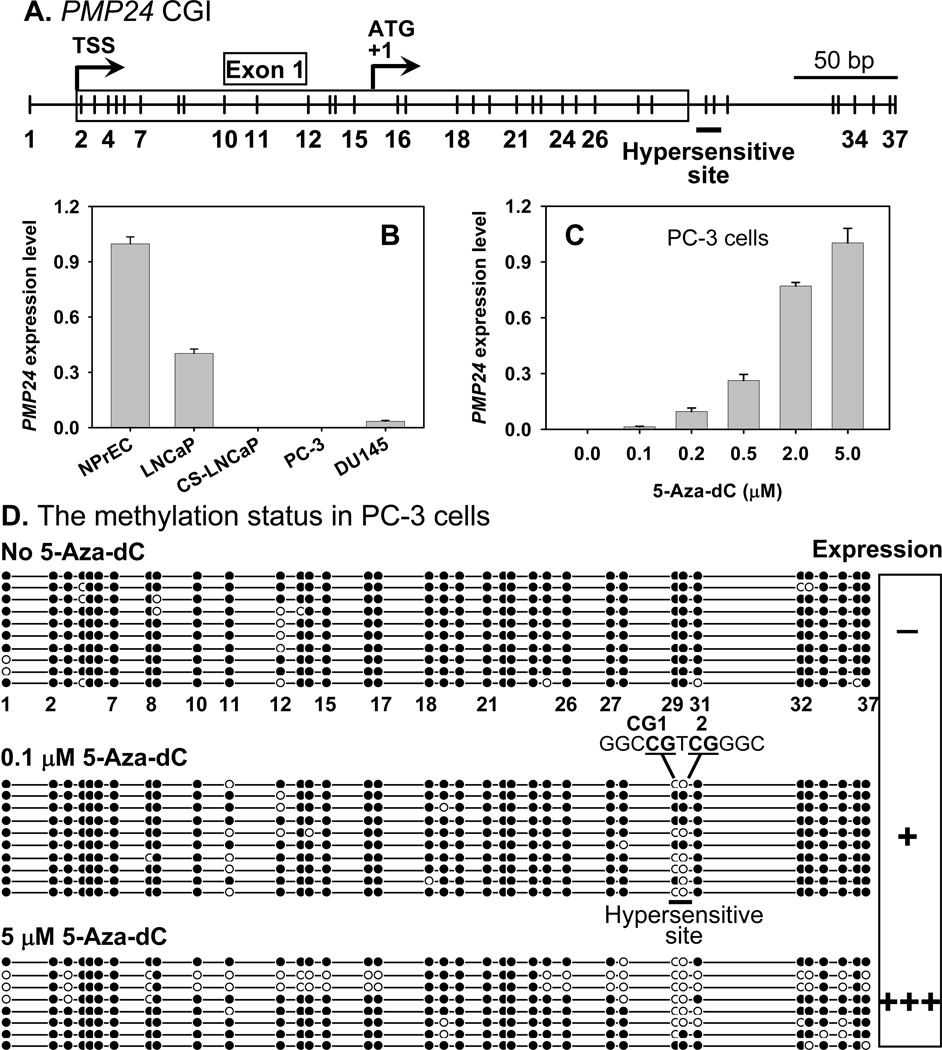
Fig. 1. Treatment of 5-aza-dC by limited demethylation approach preferentially demethylated specific CpG dinucleotide in PMP24 promoter CGI.
A: The organization of PMP24 promoter CGI. The major 5-aza-dC-hypersensitive site at intronic CpG 1 and 2 that corresponding to CG29 and CG30 is marked with a line. B: Relative PMP24 expression levels, as determined by real-time RT-PCR, among the normal and malignant prostate cell lines, which correlated with the extent of promoter methylation [25]. C: Treatment with 5-aza-dC reactivated PMP24 gene expression in a dose-dependent manner, with 0.1 µM the lowest concentration causing gene reactivation (n=4). D: Using the limited demethylation approach, we found one major region in PMP24 CGI that is hypersensitive to 5-aza-dC demethylation after PC-3 cells were treated briefly with a very low dose of 5-aza-dC. Figure 1A and 1D represent the same region with the same scale.
Gel Shift Assay
A 30-bp putative cis-element encompassing the hypersensitive site was used for the gel shift assay (Table I). Fluorescence dye IRDye 800-labeled probe (Li-Cor, Lincoln, NB), together with the unlabeled (cold) wild-type or mutated oligonucleotides (Table I), were used in the assay to determine the binding specificity. Methylated probe was generated by in vitro methylation of oligonucleotides with M. SssI CpG methyltransferase (NEB, Beverly, MA) and purified with the QIAquick nucleotide removal kit (Qiagen, Valencia, CA). Nuclear proteins were extracted by Nuclear Extract Kit from Active Motif (Carlsbad, CA). DNA-protein binding reactions were set up with 5 µg of NPrEC nuclear extract in 1× binding buffer (20 mM HEPES, pH 7.9, 100 mM KC1, 2.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT) containing 5% glycerol and 0.1 mg/ml poly d(I-C) with or without unlabeled competitive oligonucleotides. After 10 min of incubation at 22°C, 0.05 pmol of IRDye-labeled probe was added to each reaction and incubated for another 20 min before separation on 6% DNA Retardation Gels (Invitrogen, Carlsbad, CA) and detected by the Odyssey Infrared Imaging System (Li-Cor).
Construction of Promoter-Luciferase Plasmids
Luciferase reporter vector pGL3 basic (Promega, Madison, WI) was used in the promoter-reporter analysis. As the 5′ UTR and a part of the open reading frame of PMP24 gene was included, the vector was modified with an insertion of a 0.6-kb internal ribosomal entry site (IRES) [28] immediately upstream of the luciferase gene and named pGL3/IRES. The IRES element eliminates the potential interference from the open reading frame of PMP24 on the translation of the luciferase reporter gene. The promoter region from −415 to +330 of the PMP24 gene was inserted upstream of IRES in pGL3/IRES vector and designated as pGL3/IRES/(−415/+330). Additional constructs with nested 3′-end deletions of the promoter including pGL3/IRES/(−415/+194) and pGL3/IRES/(−415/+93) were generated using the Delete-A-Base kit (Promega). Plasmids with mutations of the two intronic CpG dinucleotides in the promoter-hypersensitive region cis-element (Figure 1 and Table I) were generated from pGL3/IRES/(−415/+330) using the GeneTailer Site-Directed Mutagenesis System (Invitrogen) and referred to as pGL3/IRES/(−415/+330)/mutCG1 and pGL3/IRES/(−415/+330)/mutCG2, respectively. The sequence of the mutated region is the same as the corresponding mutated probe sequence used in the gel shift assay (Table I). The promoter sequence was confirmed by sequencing (Macrogen Inc, Korea).
Promoter Luciferase Assay
Transient transfection of the promoter-reporter plasmid into LNCaP cells was performed with Lipofectamine and Plus Reagents (Invitrogen) according to the manufacturer’s instructions. In brief, 6 × 104 cells were plated in each well of a 24-well plate for 48 h before transfection with 0.2 µg of promoter-luciferase plasmid and 40 ng of CMV promoter-driven LacZ gene plasmid which expresses β-galactosidase as the internal control. The cells were then incubated with complete medium for 24 h before luciferase assay (Bright-Glo Luciferase Assay Kit, Promega). Luciferase and β-galactosidase assays were performed as previously described [29]. Relative luciferase activity of each sample was normalized by its β-galactosidase activity.
Delivery of Methylated Oligonucleotides to NPrEC Cells
A 16-bp phosphorothioated oligonucleotide (Proligo, Boulder, CO) with 5′-methylcytosines (mC) in CpG dinucleotide, named metODN (Table I), was used for targeting the 5-aza-dC-hypersentive site (Figure 4). Unmethylated phosphorothioated oligonucleotide was used as a control. NPrEC cells that express PMP24 with an unmethylated promoter background were seeded at 3 × 105 cells per well in a 6-well plate for 24 h before transfection. Two rounds of transfection of the oligonucleotides at 2 µM were carried out using Lipofectamine and Plus reagents. The experiment was repeated once to generate four samples for each regimen. Transfected cells were harvested after 96 h for real-time RT-PCR and bisulfite sequencing analyses.
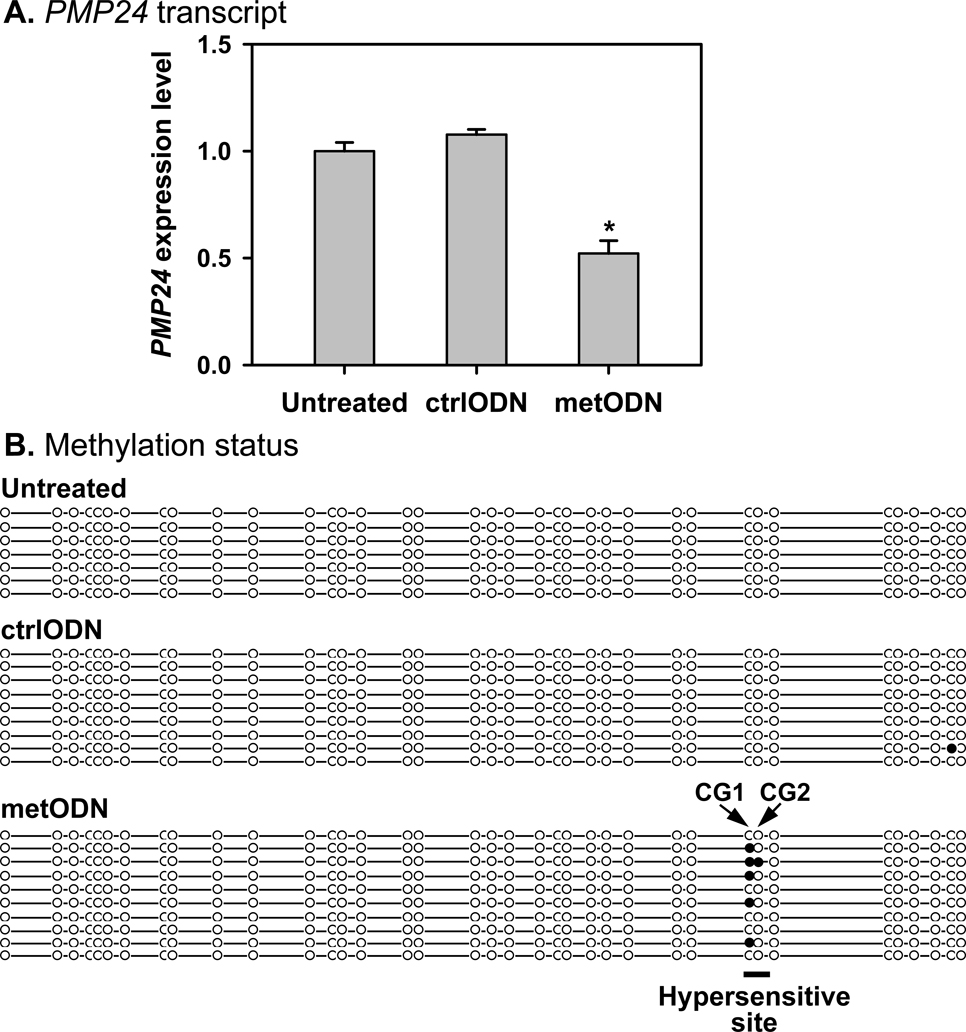
Fig. 4. Methylation of the first CpG in the 5-aza-dC-hypersensitive site repressed PMP24 gene expression in NPrEC cells.
A: Compared with non-methylated oligonucleotide (ctrlODN)-transfected control, real-time RT-PCR analysis showed significant downregulation of PMP24 transcript in the cells transfected with a methylated oligonucleotide (metODN) targeting the 5-aza-dC-hypersensitive site (*, n=3, p<0.001). Transfection with ctrlODN did not significantly change PMP24 expression as compared with the untreated cells. B: The metODN induced ~50% methylation at the first CpG dinucleotide but almost no methylation at the second CpG dinucleotide of the hypersensitive site.
SiRNA-mediated Knockdown and Ectopic Expression in NPrEC Cells
SiRNA knockdown of AP-2α, -2γ and Sp1 were performed as previously described [19, 29]. TransIT TKO kit (Mirus, Madison, WI) was used according to the manufacturer’s recommendation for siRNA transfection to NPrEC cells. The ectopic expression of AP-2 isoforms in NPrEC cells was achieved by transfection of expression plasmids [19] into these cells using the human PrEC Nucleofector kit purchased from Lonza (Valais, Switzerland).
In Situ Hybridization (ISH) of PMP24 Transcript and Evaluation of Expression
ISH was performed on sections of formalin-fixed paraffin-embedded clinical specimens containing benign glands and cancer foci. The criteria for the probe design were based on the previously described approach [30, 31] and a scrambled probe served as a negative control (Exiqon, Denmark). The sequences of the probes are listed in Table I. The oligo DNA template for the cRNA probe was generated as reported [29]. The DIG-labeled cRNA probes were synthesized according to the protocol (Roche Applied Science, Indianapolis, IN). The ISH for PMP24 transcript was performed based on a published method [32] with the following modifications: acetylation with 0.1 M triethanolamine and 0.25% acetic anhydride was performed for 10 min prior to prehybridization. Hybridization was carried out at 55°C for 16 hr. The signals were developed with BCIP/NBT substrate (Millipore, Berillica, MA), and amplified by the TSA DNP (AP) System (Perkin Elmer, Waltham, MA). The sections were counterstained with methyl green for nuclear staining.
In each sample, 3–5 representative foci of benign glands and PCa were selected to evaluate PMP24 transcript expression (positive or negative), and were represented as the percentage of cells expressing the transcript. Evaluation was conducted by two individuals (XZ and MTL) under light microscopy and the percentage of positivity in each focus was averaged.
Bioinformatics and Statistical Analyses
Promoter CpG island was analyzed by Methprimer [33]. Putative TF binding sites in the PMP24 5′-CGI was predicted with the MatInspector (www.genomatix.de) based on the TRANSFAC database.
In the promoter-reporter assays, a one-way analysis of variance (ANOVA), followed by Tukey's HSD post hoc test (SigmaPlot version 9.01; Systat Software Inc., Chicago, IL), was used for the comparisons of the promoter activity between the mutated or truncated promoter constructs with the wild-type promoter of pGL3/IRES(−415/+330). In the real-time RT-PCR, a two-tailed, unpaired t-test was performed between two groups. The statistical significance of the differences of methylation percents between microdissected benign (n=7) and cancer foci (n=16) was evaluated. Logistic regression was performed to compare methylation percents for each CpG dinucleotide. In order that inferences may be applied to any set of samples and not just the samples that were observed, a random sample effect was modeled. Probabilities (p-values) were calculated testing the difference between mean methylation percents in the two groups for each CpG dinucleotide. The analysis was preformed using the GLIMMIX procedure in the Statistical Analysis System (SAS, version 9.2; SAS Institute Inc., Cary, NC). Unless otherwise stated, p<0.05 was considered as statistically significant and error bars represented standard deviation.
RESULTS
Identification of a Single Intronic CpG Cluster Exhibiting Hypersensitivity to 5-aza-dC-demethylationand Its Correlation with Gene Re-activation
The PMP24 5′-CGI encompasses the promoter, first exon, and a part of intron 1 (Figure 1A). PMP24 expression, quantified by real-time RT-PCR, was highest in NPrEC, followed by LNCaP and DU145 (Figure 1B). LNCaPCS and PC-3 cells are devoid of expression. The degree of methylation in the PMP24 5′-CGI was inversely related to gene expression [25]. To identify the CpG dinucleotide whose methylation status is most important for gene regulation, we first used the PC-3 cell as a model. This cell line lacks PMP24 expression, and the 5′-CGI of the gene is heavily methylated. By exposing PC-3 cells to various concentrations of 5-aza-dC, we found the minimal concentration of the drug (0.1 µM) that reactivated expression of the gene (Figure 1C). The gene expression level in response to 5-aza-dC treatment exhibited a concentration-dependent manner. We reasoned that, if only a few CpG dinucleotides were demethylated at this low concentration of 5-aza-dC and the gene expression was reactivated, these dinucleotides must play a pivotal role in gene regulation. Because of the lack of standard terminology to describe our approach, we arbitrarily referred to it as a “limited demethylation approach” and to these sites as “hypersensitive sites” (to demethylating agents). Using this approach, we found only one CpG cluster comprising two CpGs (CpG 29 and 30) that exhibited hypersensitivity to 5-aza-dC demethylation, along with reactivation of gene expression (a 5-aza-dC hypersensitive site, demethylated at 0.1 µM, Figure 1D). Higher concentrations of 5-aza-dC (5 µM), however, extended demethylation to other CpGs flanking this site and to multiple regions of the CGI with less site-specificity. These results, taken together, suggested that the methylation status of these two CpG dinucleotides in the PMP24 5′-CGI may play a pivotal role in the regulation of gene expression.
The 5-aza-dC-Hypersensitive Site Binds Nuclear Protein(s) in a Sequence- and Methylation Status-specific Manner
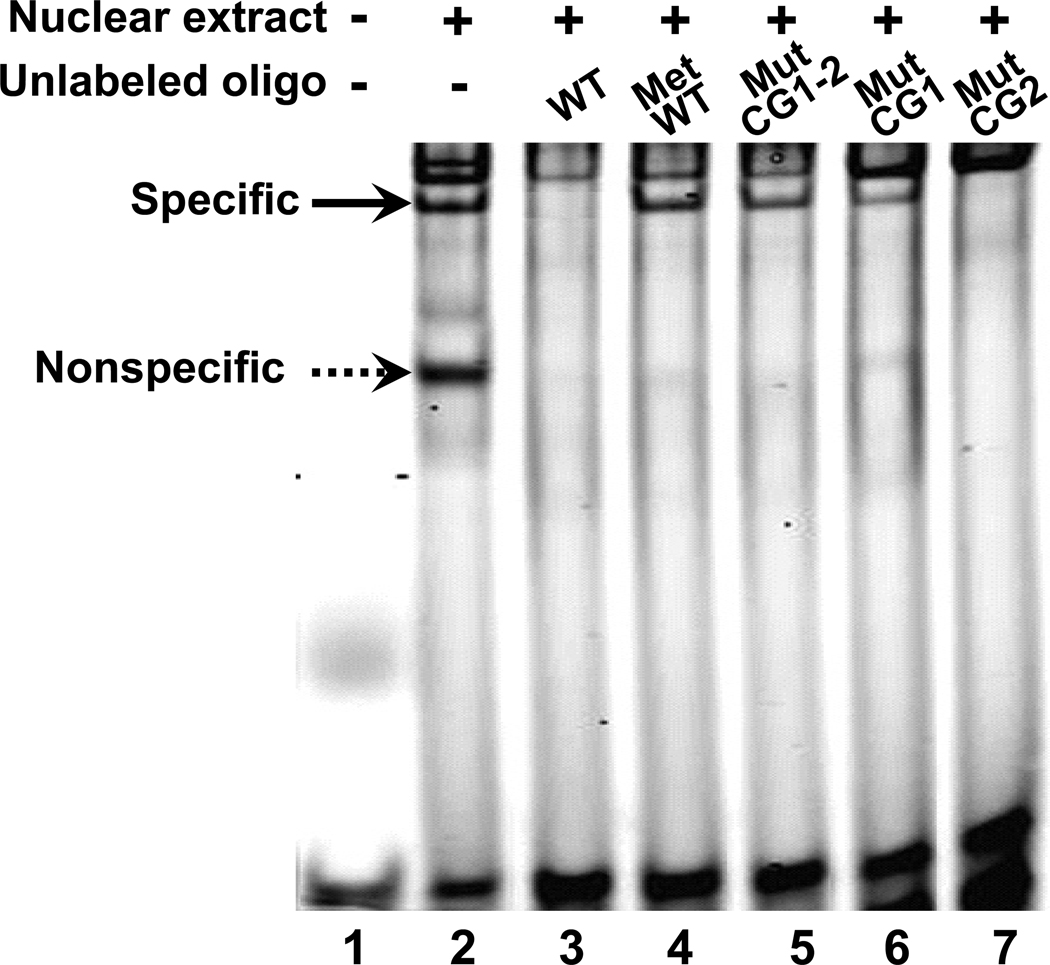
We then performed a gel-shift assay to determine if one or both of the CpGs (CpG 29 and 30, Figure 1A) in the 5-aza-dC-hypersensitive site was essential for protein interaction. Nuclear extracts were prepared from NPrEC cells because they have high levels of PMP24 expression and thus should contain all essential proteins supporting transcription. A 30-mer oligonucleotide corresponding to the hypersensitive site was synthesized and labeled as the probe (Table I). As shown in Figure 2, following incubation of the labeled probe with NPrEC nuclear extracts, a sequence-specific DNA-protein complex (indicated by the solid arrow; lane 2) was formed; this complex could be completely blocked by co-incubation with an excess of unlabeled wild-type oligonucleotide (WT, lane 3). We then investigated the effect of cytosine methylation on this DNA-protein interaction. When the oligonucleotide was methylated, it completely lost its ability to compete with the labeled probe, indicating that methylation interfered with DNA-protein interaction (MetWT, lane 4). Mutation at both CpGs (MutCG1-2, lane 5) or only at the first CpG site (MutCG1, lane 6) in the oligonucleotide resulted in the loss of their ability to compete with the labeled probe. In contrast, a mutation at the second CpG dinucleotide of the oligonucleotide alone did not abolish its ability to block the DNA-protein complex formation (MutCG2, lane 7), suggesting that the first, but not the second, CpG dinucleotide was crucial for DNA-protein-interaction. Collectively, these data indicated that the 5-aza-dC-hypersensitive site interacts with nuclear protein(s) in a sequence-specific manner and that the interaction is sensitive to cytosine methylation, with the first intronic CpG dinucleotide being crucial to the interaction.
Fig. 2. The first CpG site in the 5-aza-dC-hypersensitive region is essential for DNA-protein interaction.
A 30-bp labeled oligonucleotide (WT) corresponding to the hypersensitive site was incubated with a nuclear extract from NPrEC cells. A gel shift assay demonstrated the formation of a sequence-specific DNA-protein complex, as indicated by the solid arrow. The presence of 100-fold molar excess of unlabeled WT probe (lane 3) or mutCG2 (mutation at the second CpG dinucleotide, lane 7) diminished the shifted band (DNA-protein complex), while the same excess amount of MetWT (methylated WT sequence, lane 4), mut1-2CGs (mutations at both CpG1 and 2, lane 5), and mutCG1 (mutation at the first CpG dinucleotide, lane 6) showed no competition with the labeled probe. Dotted arrow indicates nonspecific DNA-protein interaction as any excessive unlabeled oligos can compete for the protein binding regardless of their sequences and methylation status (lane 3 to 7). Using nuclear extract from different passage numbers of NPrEC cells, three independent experiments were done and one representative figure is shown.
The First CpG in the Hypersensitive Site Is Crucial for PMP24 Promoter Activity
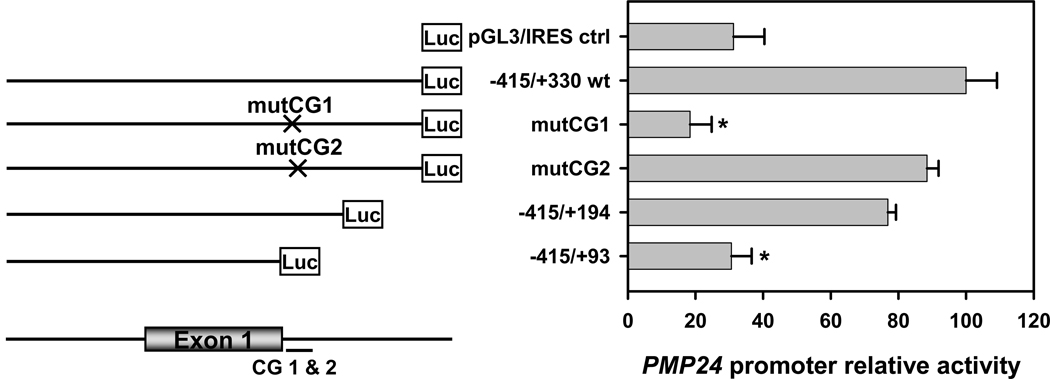
To evaluate the individual contribution of the first and the second CpGs in the 5-aza-dC-hypersensitive site of the PMP24 5′-CGI on promoter activity, we generated site-specific mutants and deletion mutants from a minimal proximal promoter (−415/+330 wt). Mutation of CpG1 (mutCG1) significantly decreased the promoter activity by 82% (p<0.001) in LNCaP cells as compared with that of the wild-type promoter in the pGL3/IRES/(−415/+330) plasmid (Figure 3). In comparison, mutation of CpG2 (mutCG2) showed activity comparable to that of the wild-type promoter (p>0.05). Nested deletion at the 3′ flanking region did not significantly affect the promoter activity (pGL3/IRES/(−415/+194), p>0.05) until the entire 5-aza-dC-hypersensitive region was removed (pGL3/IRES/(−415/+93), 69% decrease, p<0.001). These results demonstrated that the promoter activity depends critically on the first CpG located at the 5-aza-dC-hypersensitive site (corresponding to CpG 29 in the CGI depicted in Figure 1A).
Fig. 3. PMP24 promoter analysis.
Promoter luciferase reporter pGL3/IRES/(−415/+330)mutCG1 and pGL3/IRES/(−415/+330)/mutCG2 that have mutations at the first or second CpG dinucleotide respectively, were generated from the WT promoter reporter pGL3/IRES/(−415/+330). Nested 3′ deletion of the promoter generated two additional plasmids of pGL3/IRES/(−415/+194) and pGL3/IRES/(−415/+93) with or without the hypersensitive site (+128), respectively. Deletion of the hypersensitive site (intronic CG1 &2) or the first CpG dinucleotide but not deletion of the second significantly decreased the promoter activity (*p<0.001) as compared with the activity of the WT promoter (n=3). The position of the translation start site was designated as +1. The IRES element immediately upstream of the luciferase gene is not shown.
Site-specific Methylation of the Hypersensitive Region Downregulated PMP24 Expression
To directly demonstrate that methylation of the hypersensitive site can impede PMP24 expression, we used methylated oligonucleotides (MetODN, Table I). MetODNs are 18–25-bp oligonucleotides designed to be corresponding to the targeted sequence at which one wants to induce in cellulo methylation [34, 35]; each has at least three methylated CpG dinucleotides. It is believed that, following transfection, the MetODNs will form hemi-methylated DNA intermediates with the targeted sequence during cell replication. Since hemi-methylated DNAs are preferred substrates for DNA methyltransferase I, site-specific methylation could be attained after multiple rounds of cell proliferation. MetODNs targeting the 5-aza-dC-hypersensitive site and its unmethylated counterpart (CtrlODN) were transfected into NPrEC cells, which have a completely unmethylated PMP24 5′-CGI [25]. The expression level of PMP24 decreased by 44% (p<0.001) in the MetODN-treated cells, but that of the CtrlODN-transfected cells showed no change as compared with the untransfected cells (p>0.05, Figure 4A). In concordance with the PMP24 expression level, bisulfite sequencing analysis of the MetODN-transfected cells confirmed selective methylation of in the hypersensitive site, with methylation occurring predominately at the first CpG dinucleotide of the hypersensitive site (Figure 4B). In contrast, transfection of the unmethylated CtrlODN did not elicit any methylation and had no effect on gene expression.
In Silico Predicted Transcription Factors Failed to Regulate PMP24 Expression in NPrEC
We spent much effort to identify the TFs that bind to the 5-aza-dC hypersensitive site and regulate PMP24 gene expression. We used both MatInspector from Genomatix and TRANSFAC to search for putative transcription factor-binding sites in the hypersensitive region. In silico analyses predicted putative AP-2- and Sp1-binding sites at this region. We therefore used NPrEC cells as a model system for targeted gene knockdown experiments since PMP24 5′-CGI is completely unmethylated and the gene is expressed in this cell model. Using real-time RT-PCR, we first established that NPrEC express significant levels of AP-2α and AP-2γ but little or no AP-2β, -2δ and -2ε transcripts. It also expresses high levels of Sp1 mRNA. We then conducted two-round siRNA-mediated knockdown experiments [19] to down-regulate AP-2α, AP-2γ, or Sp1 expression in NPrEC. Although the knockdown experiments effectively repressed the siRNA-targeted genes (>80%), they failed to change the level of PMP24 expression (data not shown). We also overexpressed AP-2α, AP-2γ and their dominant negative mutant in NPrEC, but these efforts failed to change PMP24 transcript levels in the transfected cells (data not shown). These data lead us to conclude that the 5-aza-dC hypersensitive site does not harbor an AP-2- or a Sp1 cis-element; rather an unknown TF may interact with it to regulate PMP24 expression.
Differential Methylation of a Single CpG Dinucleotide in Clinical Prostate Specimens Correlates with PMP24 Transcript Expression
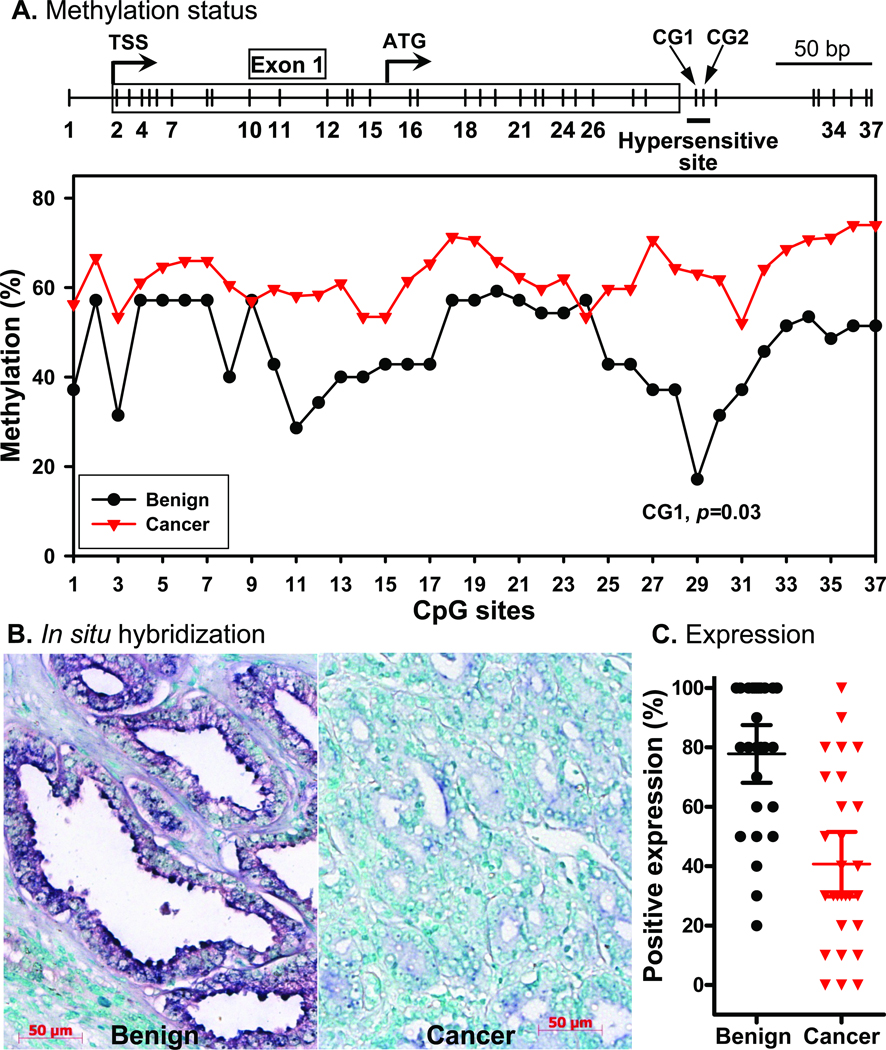
Using cell model experiments, we indentified a single CpG dinucleotide in the intronic region of PMP24 5′-CGI (CpG 29 in Figure 1A) playing a crucial in gene regulation via DNA methylation. To better understand the clinical relevance of our in vitro data, we compared the degree of methylation at each CpG dinucleotide of the PMP24 5′-CGI in microdissected samples of prostate epithelial cells from morphologically normal tissue versus that in microdissected PCa cells. As shown in Figure 5A, a single CpG dinucleotide, CpG29 of the CGI, was found to be significantly hypermethylated (p=0.03) in PCa cells when compared to normal prostate epithelial cells; statistical significance was not observed for any other CpGs within the CGI (error bars not shown). ISH staining showed PMP24 transcript expression in all benign glands foci but marked reduction in expression in many, but not all, PCa foci (Figure 5B). Quantitatively, the levels of transcript expression, represented as percent of positively stained cells (% of positivity), were significantly higher in foci of benign glands than those with PCa (Figure 5C, p<0.01). Thus, an inverse association was observed between methylation of CpG29 in PMP24 5′-CGI and the level of gene expression in clinical samples.
Fig. 5. Methylation status of PMP24 CGI in prostate LCM samples.
A: Bisulfite sequencing result from the benign (n=7) and cancer foci (n=16) of LCM samples. Overall, the CGI is hypermethylated in the cancer group with the average methylation percentage of 63%, whereas in the benign group the average methylation percentage is 46%. One hypomethylated site, CG29 (methylation percentage 17%), was identified in the benign but not in the cancer group (63%). This site exactly matches the 5-aza-dC hypersensitive region shown in Figure 1. CG29 but not other sites exhibited significant difference of the methylation status (p=0.03) between the two groups. B: Representative ISH staining results of benign prostatic glands and PCa foci. C: Comparison of the PMP24 mRNA expression positivity between benign glands and PCa foci (3–5 foci/case) from seven cases of specimens analyzed by ISH (p<0.01). Error bar: 95% confident interval.
DISCUSSION
Our previous study showed that PMP24 in PCa cells is silenced by a mechanism associated with the hypermethylation of its promoter CGI and the silenced gene can be reactivated by treatment with 5-aza-dC [25]. In this study, by carefully varying the concentration of 5-aza-dC until it was just enough to reactivate PMP24 expression in PC-3 cells, we successfully identified a single CpG dinucleotide, within the PMP24 5′-CGI, whose methylation status plays a crucial role in gene regulation. Several features of this CpG dinucleotide are intriguing: 1) it is harbored within the first intron; 2) its methylation status critically controls PMP24 expression; 3) it is one of the many CpG dinucleotides in a relatively large CGI; 4) nuclear proteins bind to it in a sequence-specific manner but it does not match to the cis-regulatory elements predicted by in silico analysis; and 5) it exhibits hypersensitivity to both demethylation by 5-aza-dC and MetODN-mediated methylation in cellulo. Most interestingly, however, we found that this CpG exactly matches a single CpG (CG29) in the PMP24 5′-CGI whose degree of methylation was found to be significant different between normal prostate epithelial cells and PCa cells obtained by LCM of clinical samples. Importantly, ISH data support a regulatory role of this CpG in regulating gene expression via DNA methylation under the in vivo setting. In short, we have identified a single intronic CpG, within a relatively large CGI, that is highly susceptible to methylation-demethylation and thereby can play a dynamic role in the regulating PMP24 expression. Methylation of this CpG appears to be a molecular event associated with PMP24 silencing during prostate carcinogenesis.
Traditionally, the majority of studies on gene silencing by DNA methylation are focusing on CGIs in the promoter region. It is believed that hypermethylation of a promoter CGI increases the binding of methyl-CpG-binding proteins, along with the recruitment of other chromatin modifiers, and leads to chromatin remodeling and transcriptional repression. Up till recently, not much is known about the relevance of non-promoter CGI(s) or, in the case of our study, a single CpG, in gene regulation via cytosine methylation. Nevertheless, Jones and associates [36, 37] offered an attractive model to explain the role of non-promoter CGI in epigenetic gene regulation. They demonstrated that non-promoter CGIs are more susceptible to de novo methylation than promoter islands and may serve as foci for the seeding of de novo methylation which can then spread into adjacent islands that contain enhancer elements for transcriptional regulation. Whether the reverse scenario, i.e., demethylation, is true remains unknown. Specifically, pertaining to our results, it will be important to determine, in future studies, whether the demethylation of a single CpG (CG29) in the PMP24 CGI can initiate “spreading” of demethylation to adjacent sites, leading to the expansion of chromatin relaxation and increased gene reactivation. In support of this possibility, in this study, we did observe the spreading of demethylation from CG29 to adjacent CGs when PC-3 cells were exposed to higher levels of 5-aza-dC (Figure 1D) along with progressive increases in gene expression (Figure 1C).
Recent reports have broadened our understanding of the importance of intronic CGIs in epigenetic gene regulation. For example, hypermethylation of an intron 1 CGI was shown to silence the EGR2 gene and the CGI was found to harbor cis-elements similar to enhancers commonly found in promoter regions [38]. Similarly, an intron 4 CGI that exhibited all the characteristics of a bona fide cis-element including direct interaction with the 5′ proximal promoter was shown to regulate the expression of IL-10 [39]. In another scenario, transcriptional silencing of the MCJ gene was found to associate with methylation of a CGI in its intron 1 region and reduction of histone acetylation within the CGI as well as in the promoter region of the gene [40]. This finding suggests that methylation of an intronic CGI can alter the chromatin structure of the entire upstream region, leading to the repression of gene expression. These studies provide emerging evidence that intronic CGIs are equally essential in gene regulation as their counterparts in promoter regions.
In the literature there is only a handful of single CpG identified to be essential for gene regulation via methylation. The scarcity of single CpG dinucleotide that plays the most critical role in gene regulation is due to the way 5-aza-dC experiments normally conducted. Most investigations were aimed at achieving maximal gene reactivation and therefore adopted a prolonged treatment protocol using relatively high concentrations of the demethylating agent [41]. Using this traditional approach, it is difficult to ascertain if any of the CpG dinucleotides exhibit differential susceptibility towards methylation-demethylation or to locate the critical regulatory sequences within a CGI. The fact that we have identified CG29 within the PMP24 5′-CGI as the key regulatory site is due to the careful choice of the time and the dose of 5-aza-dC for demethylation treatment. The lowest concentration of 5-aza-dC was used to identify the CpG(s) most sensitive to demethylation along with gene reactivation. Intriguingly, when we used the methylation sense ODN to induce site-specific methylation CG29 is also the most susceptible CpG for methylation with concordant gene silencing of PMP24. Therefore, we can conclude that the limited demethylation approach has proven useful in identifying hypersensitive methylation-demethylatoin CpG(s) essential for gene regulation. These sites may serve as dynamic switches to activate or inactivate of a gene through cytosine methylation.
It is noteworthy to mention that this site may harbor a yet-to-be identified cis-element whose methylation status determines gene expression. Unfortunately, forced expression and siRNA-mediated knockdown experiments failed to identify this site as an AP-2- or a Sp1-cis-element. Regarding to the function of PMP24 in peroxisomes, essential cell organelles involved in lipid metabolism and detoxification [42], however, not much is known about the function of PMP24 except our earlier observations demonstrating that it has an anti-tumor action on PCa cells [25]. Future studies are required to elucidate the TF-hypersensitive site interaction and biological function of this gene.
Finally, using LCM samples from clinical prostate specimens combined with bisulfite sequencing analysis and ISH, we established an inverse correlation between methylation of CG29 in the PMP24 5′-CGI and expression of the gene transcript in clinical specimens. In essence, normal prostate epithelial cells express PMP24 and have a hypomethylated CG29 in the gene’s 5′-CGI whereas PCa cells exhibit significant reduction in PMP24 expression and have a hypermethylated CG29 site. These data strongly suggest that CG29 is essential to PMP24 gene regulation via cytosine methylation in vivo and the DNA methylation-mediated silencing of the gene is a molecular event associated with prostate carcinogenesis. The fact that CG29 identified in clinical samples matches the 5-aza-dC hypersensitive site identified by limited demethylation in cell model systems inspires confidence that this site serves as a versatile switch for PMP24 regulation in cell model systems and in tissue.
CONCLUSION
By using the limited demethylation approach, we identified a single CpG dinucleotide in the PMP24 5′-CGI to be essential for gene regulation via methylation. The importance of this site in mediating gene silencing through cytosine methylation was also illustrated using LCM-sampling of benign glands and PCa cells from clinical specimens along with ISH staining. Collectively, these data provide a strong disease relevance to this unique intronic CpG dinucleotide that plays a pivotal role in PMP24 regulation.
ACKNOWLEDGEMENTS
We thank Ms. Katerina Mardilovich in the Department of Cancer Biology, University of Massachusetts Medical School for her excellent technical support, Dr. Irwin Leav in the Department of Cancer Biology and Pathology, University of Massachusetts Medical School for the dissection of the LCM samples.
Grant sponsor: NIH; Grant numbers: ES006096, ES015584, CA15776, and CA112532 to SMH. Grant sponsor: US Army Prostate Cancer Program; Grant numbers: 81XWH-04-1-0165 to SMH and W81XWH-06-1-0376 to XZ.
REFERENCES
- 1.Domann FE, Futscher BW. Flipping the epigenetic switch. Am J Pathol. 2004;164:1883–1886. doi: 10.1016/S0002-9440(10)63748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 3.Watts GS, Futscher BW, Holtan N, Degeest K, Domann FE, Rose SL. DNA methylation changes in ovarian cancer are cumulative with disease progression and identify tumor stage. BMC Med Genomics. 2008;1:47. doi: 10.1186/1755-8794-1-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snell C, Krypuy M, Wong EM, Loughrey MB, Dobrovic A. BRCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res. 2008;10:R12. doi: 10.1186/bcr1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hitchins MP, Lin VA, Buckle A, Cheong K, Halani N, Ku S, Kwok CT, Packham D, Suter CM, Meagher A, Stirzaker C, Clark S, Hawkins NJ, Ward RL. Epigenetic inactivation of a cluster of genes flanking MLH1 in microsatellite-unstable colorectal cancer. Cancer Res. 2007;67:9107–9116. doi: 10.1158/0008-5472.CAN-07-0869. [DOI] [PubMed] [Google Scholar]
- 6.Sarbia M, Geddert H, Klump B, Kiel S, Iskender E, Gabbert HE. Hypermethylation of tumor suppressor genes (p16INK4A, p14ARF and APC) in adenocarcinomas of the upper gastrointestinal tract. Int J Cancer. 2004;111:224–228. doi: 10.1002/ijc.20212. [DOI] [PubMed] [Google Scholar]
- 7.Issa JP. DNA methylation as a therapeutic target in cancer. Clin Cancer Res. 2007;13:1634–1637. doi: 10.1158/1078-0432.CCR-06-2076. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z, Karam J, Frenkel E, Sagalowsky A, Hsieh JT. The application of epigenetic modifiers on the treatment of prostate and bladder cancer. Urol Oncol. 2006;24:152–160. doi: 10.1016/j.urolonc.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Kihslinger JE, Godley LA. The use of hypomethylating agents in the treatment of hematologic malignancies. Leuk Lymphoma. 2007;48:1676–1695. doi: 10.1080/10428190701493910. [DOI] [PubMed] [Google Scholar]
- 10.Farinha NJ, Shaker S, Lemaire M, Momparler L, Bernstein M, Momparler RL. Activation of expression of p15, p73 and E-cadherin in leukemic cells by different concentrations of 5-aza-2'-deoxycytidine (Decitabine) Anticancer Res. 2004;24:75–78. [PubMed] [Google Scholar]
- 11.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 12.Ghoshal K, Bai S. DNA methyltransferases as targets for cancer therapy. Drugs Today (Barc ) 2007;43:395–422. doi: 10.1358/dot.2007.43.6.1062666. [DOI] [PubMed] [Google Scholar]
- 13.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog. 2003;38:78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- 15.Ortmann CA, Eisele L, Nuckel H, Klein-Hitpass L, Fuhrer A, Duhrsen U, Zeschnigk M. Aberrant hypomethylation of the cancer-testis antigen PRAME correlates with PRAME expression in acute myeloid leukemia. Ann Hematol. 2008 doi: 10.1007/s00277-008-0514-8. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 17.Muiznieks I, Doerfler W. The topology of the promoter of RNA polymerase II- and III-transcribed genes is modified by the methylation of 5'-CG-3' dinucleotides. Nucleic Acids Res. 1994;22:2568–2575. doi: 10.1093/nar/22.13.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, McNeal JE, Ho SM. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Leung YK, Ho SM. AP-2 regulates the transcription of estrogen receptor (ER)-beta by acting through a methylation hotspot of the 0N promoter in prostate cancer cells. Oncogene. 2007;26:7346–7354. doi: 10.1038/sj.onc.1210537. [DOI] [PubMed] [Google Scholar]
- 20.Tindall D, Horne FM, Hruszkewycz A, Mohla S, Shuman M, Wang Z, Kantoff P. Symposium on androgen action in prostate cancer. Cancer Res. 2004;64:7178–7180. doi: 10.1158/0008-5472.CAN-04-1752. [DOI] [PubMed] [Google Scholar]
- 21.Jiang F, Wang Z. Identification of androgen-responsive genes in the rat ventral prostate by complementary deoxyribonucleic acid subtraction and microarray. Endocrinology. 2003;144:1257–1265. doi: 10.1210/en.2002-220718. [DOI] [PubMed] [Google Scholar]
- 22.Alimirah F, Panchanathan R, Chen J, Zhang X, Ho SM, Choubey D. Expression of androgen receptor is negatively regulated by p53. Neoplasia. 2007;9:1152–1159. doi: 10.1593/neo.07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang TH, Laux DE, Hamlin BC, Tran P, Tran H, Lubahn DB. Identification of DNA methylation markers for human breast carcinomas using the methylation-sensitive restriction fingerprinting technique. Cancer Res. 1997;57:1030–1034. [PubMed] [Google Scholar]
- 24.Reguenga C, Oliveira ME, Gouveia AM, Eckerskorn C, Sa-Miranda C, Azevedo JE. Identification of a 24 kDa intrinsic membrane protein from mammalian peroxisomes. Biochim Biophys Acta. 1999;1445:337–341. doi: 10.1016/s0167-4781(99)00061-5. [DOI] [PubMed] [Google Scholar]
- 25.Wu M, Ho SM. PMP24, a gene identified by MSRF, undergoes DNA hypermethylation-associated gene silencing during cancer progression in an LNCaP model. Oncogene. 2004;23:250–259. doi: 10.1038/sj.onc.1207076. [DOI] [PubMed] [Google Scholar]
- 26.Visser WF, van Roermund CW, Ijlst L, Waterham HR, Wanders RJ. Metabolite transport across the peroxisomal membrane. Biochem J. 2007;401:365–375. doi: 10.1042/BJ20061352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mobley JA, Leav I, Zielie P, Wotkowitz C, Evans J, Lam YW, L'Esperance BS, Jiang Z, Ho SM. Branched fatty acids in dairy and beef products markedly enhance alpha-methylacyl-CoA racemase expression in prostate cancer cells in vitro. Cancer Epidemiol Biomarkers Prev. 2003;12:775–783. [PubMed] [Google Scholar]
- 28.Jan E. Divergent IRES elements in invertebrates. Virus Res. 2006;119:16–28. doi: 10.1016/j.virusres.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Leav I, Revelo MP, Deka R, Medvedovic M, Jiang Z, Ho SM. Deletion hotspots in AMACR promoter CpG island are cis-regulatory elements controlling the gene expression in the colon. PLoS Genet. 2009;5:e1000334. doi: 10.1371/journal.pgen.1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erdtmann-Vourliotis M, Mayer P, Riechert U, Handel M, Kriebitzsch J, Hollt V. Rational design of oligonucleotide probes to avoid optimization steps in in situ hybridization. Brain Res Brain Res Protoc. 1999;4:82–91. doi: 10.1016/s1385-299x(99)00006-9. [DOI] [PubMed] [Google Scholar]
- 31.Kim DH, Longo M, Han Y, Lundberg P, Cantin E, Rossi JJ. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 32.Tsurusaki T, Koji T, Sakai H, Kanetake H, Saito Y. Expression profile of prostate-specific antigen messenger RNA assessed by in situ hybridization is a novel prognostic marker for patients with untreated prostate cancer. Clin Cancer Res. 1998;4:2187–2194. [PubMed] [Google Scholar]
- 33.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim Biophys Acta. 2004;1708:87–102. doi: 10.1016/j.bbcan.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhu Y, Spitz MR, Zhang H, Grossman HB, Frazier ML, Wu X. Methyl-CpG-binding domain 2: a protective role in bladder carcinoma. Cancer. 2004;100:1853–1858. doi: 10.1002/cncr.20199. [DOI] [PubMed] [Google Scholar]
- 35.Yao X, Hu JF, Daniels M, Shiran H, Zhou X, Yan H, Lu H, Zeng Z, Wang Q, Li T, Hoffman AR. A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma. J Clin Invest. 2003;111:265–273. doi: 10.1172/JCI15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen C, Liang G, Nguyen TT, Tsao-Wei D, Groshen S, Lubbert M, Zhou JH, Benedict WF, Jones PA. Susceptibility of nonpromoter CpG islands to de novo methylation in normal and neoplastic cells. J Natl Cancer Inst. 2001;93:1465–1472. doi: 10.1093/jnci/93.19.1465. [DOI] [PubMed] [Google Scholar]
- 37.Nguyen CT, Gonzales FA, Jones PA. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 2001;29:4598–4606. doi: 10.1093/nar/29.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Unoki M, Nakamura Y. Methylation at CpG islands in intron 1 of EGR2 confers enhancer-like activity. FEBS Lett. 2003;554:67–72. doi: 10.1016/s0014-5793(03)01092-5. [DOI] [PubMed] [Google Scholar]
- 39.Tsuji-Takayama K, Suzuki M, Yamamoto M, Harashima A, Okochi A, Otani T, Inoue T, Sugimoto A, Toraya T, Takeuchi M, Yamasaki F, Nakamura S, Kibata M. The production of IL-10 by human regulatory T cells is enhanced by IL-2 through a STAT5-responsive intronic enhancer in the IL-10 locus. J Immunol. 2008;181:3897–3905. doi: 10.4049/jimmunol.181.6.3897. [DOI] [PubMed] [Google Scholar]
- 40.Strathdee G, Davies BR, Vass JK, Siddiqui N, Brown R. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis. 2004;25:693–701. doi: 10.1093/carcin/bgh066. [DOI] [PubMed] [Google Scholar]
- 41.Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, Jones PA, Lubbert M. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2'-deoxycytidine (decitabine) treatment. Blood. 2002;100:2957–2964. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 42.Ma C, Subramani S. Peroxisome matrix and membrane protein biogenesis. IUBMB Life. 2009;61:713–722. doi: 10.1002/iub.196. [DOI] [PMC free article] [PubMed] [Google Scholar]