Abstract
Mps1, a dual-specificity kinase, is required for the proper functioning of the spindle assembly checkpoint and the maintenance of chromosomal stability. As Mps1 function has been implicated in numerous phases of the cell cycle, it is expected the development of a potent, selective small molecule inhibitor of Mps1 would greatly facilitate dissection of Mps1-related biology. We describe the cellular effects and Mps1 co-crystal structures of novel, selective small molecule inhibitors of Mps1. Consistent with RNAi studies, chemical inhibition of Mps1 leads to defects in Mad1 and Mad2 establishment at unattached kinetochores, decreased Aurora B kinase activity, premature mitotic exit, and gross aneuploidy, without any evidence of centrosome duplication defects. However, in U2OS cells possessing extra centrosomes, an abnormality found in some cancers, Mps1 inhibition increases the frequency of multipolar mitoses. Lastly, Mps1 inhibitor treatment resulted in a decrease in cancer cell viability.
Introduction
The successful segregation of chromosomes in mitosis requires the timely coordination of cell cycle events to ensure the bipolar attachment of sister chromatids via their kinetochores to the mitotic spindle prior to the initiation of anaphase. Deregulation of this process or uncoupling of its component parts can lead to aneuploidy and chromosomal instability (CIN), recognized hallmarks of cancer. Mps1, a dual-specificity kinase 1, was first identified in Saccharomyces cerevisiae (Mps1p) where it was shown to function in multiple pathways critical to the maintenance of genomic integrity, including spindle pole body (SPB) duplication 2, 3, mitotic spindle assembly 4, and the spindle assembly checkpoint 2. The spindle assembly checkpoint (SAC), a conserved pathway in eukaryotes, is responsible for monitoring mitotic spindle attachment at kinetochores. In response to a lack of microtubule occupancy at kinetochores or a lack of tension between sister kinetochores the checkpoint prevents the early onset of anaphase until all chromosomes make stable bipolar attachments to the mitotic spindle (reviewed in 5). Evidence from functional and localization experiments in mammalian cells have demonstrated that Mps1 is required for the maintenance of the mammalian SAC 6–9. In contrast to its unequivocal role in the mammalian SAC, its purported role in centrosome (mammalian equivalent of SPB) duplication in S phase and subsequent bipolar spindle assembly is a topic of considerable debate 10–13. Nevertheless, the necessity of Mps1 kinase activity for the fidelity of the cell cycle and genomic stability is well established.
Investigating how Mps1 kinase activity and its dynamic localization during the cell cycle participate in the coordination of multiple cell cycle processes requires the ability to rapidly inhibit Mps1 kinase activity at specific phases of the cell cycle; a level of temporal control that cannot be attained using RNAi or other common genetic methods. Small molecules that are cell permeable and can inhibit Mps1 kinase activity with rapid and reversible kinetics may provide a powerful tool to probe cell cycle-related Mps1 functions. ATP-competitive inhibitors of mitotic kinases Aurora A/B, cyclin-dependent kinase 1 (Cdk1), and Polo-like kinase 1 (Plk1) have proven invaluable to elucidate the temporal function and potential therapeutic relevance of these proteins due to their ability to inhibit kinase activity in a dose-dependent and rapid fashion (reviewed in 14).
In contrast to partial inactivation of Mps1 kinase activity, complete depletion of Mps1 or replacement of wild-type Mps1 activity with a kinase-dead Mps1 D664A allele results in cell death 15–18. Similar findings with checkpoint components Mad2 19–22 and BubR1 20, 23 support the view that complete spindle checkpoint abrogation is lethal to cells, while decreased checkpoint stability results in non-lethal chromosomal instability (reviewed in 24). Targeted chemical inhibition of Mps1 may therefore prove to be an efficient means of pharmacologically evaluating the consequences of inactivating the checkpoint as 1) epistasis experiments suggest Mps1 functions near the apex of the SAC signaling cascade 8, 9, 25, 2) Mps1 kinase activity is essential to checkpoint function 26, and 3) kinases make excellent targets for inhibitor development. A selective Mps1 kinase inhibitor will also address the question of whether targeted ablation of the mitotic checkpoint in rapidly proliferating tumor cells is a potential therapeutic approach.
Previously, three Mps1 inhibitors have been reported in the literature while other potential inhibitors have been discovered by high-throughput in vitro screens but lack characterization of cellular activity27, 28). The first, cincreasin, was shown to be effective at inhibiting Mps1 kinase activity in S. cerevisiae but was reported to be ineffective in mammalian cells 29. The second inhibitor, 1NM-PP1, was designed to inhibit Mps1 kinase function in yeast but only in the context of a sensitizing mutation in Mps14. The third, a dual JNK, Mps1 inhibitor SP600125 was reported but it exhibited very poor selectivity (Supplementary Fig. S1a, Supplementary Table S1), which would likely limit its utility as a selective probe of Mps1 function 30.
Here we describe the discovery and characterization of two new classes of potent and selective ATP-competitive Mps1 kinase inhibitors and their co-crystal structures with Mps1 kinase domain. Mps1-IN-1 (1) and Mps1-IN-2 (2) (Mps1 Inhibitor 1 and 2) inhibit Mps1 with moderate potency, exhibiting half-maximal inhibitory concentrations (IC50) of 367 nM and 145 nM respectively. Consistent with RNAi studies, chemical inhibition of Mps1 leads to defects in Mad1 and Mad2 establishment at unattached kinetochores, premature mitotic exit and reduced Aurora B kinase activity leading to the manifestation of gross aneuploidy. We found no evidence of centrosome duplication defects upon inhibition of Mps1 kinase activity. However, in U2OS cells possessing extra centrosomes Mps1 inhibition results in a catastrophic mitosis causing massive chromosome mis-segregation. Finally, we demonstrate that SAC silencing by Mps1 inhibition results in aneuploidy and decreases the viability of both cancer and ‘normal’ cells.
Results
Identification of Lead Compounds for Mps1 inhibition
In an effort to discover novel classes of selective small molecule inhibitors of Mps1 and other kinases we screened a diverse library of heterocyclic ATP-site directed kinase scaffolds using an in vitro ATP-site competition binding assay 27, 31. Approximately 400 compounds were profiled at a concentration of 10 µM against a panel of 352 diverse kinases. By screening this ‘library’ of inhibitors versus this large panel of kinases we were able to generate a selectivity-annotated library (SAL) which allowed us to rapidly identify compounds capable of selectively inhibiting Mps1 as well as other kinases of interest. Several different scaffolds including 2,6-disubstituted purines, 2,4-disubstituted pyrrolopyridines, and dihydropyrimidodiazepinones emerged as potential Mps1 inhibitor scaffolds. We focused our attention primarily on the latter two scaffolds as they demonstrated the highest degree of selectivity among available lead scaffolds. The 2,6-disubstituted purine series, though potent for its Mps1 target proved to be a more promiscuous scaffold class. Several iterative rounds of synthesis and biochemical and cellular kinase profiling resulted in the discovery of the two inhibitor series which we named Mps1-IN-1 and Mps1-IN-2 (Mps1 Inhibitor 1 and 2) (Fig. 1a, synthetic chemistry routes available in Supplementary Fig. S2).
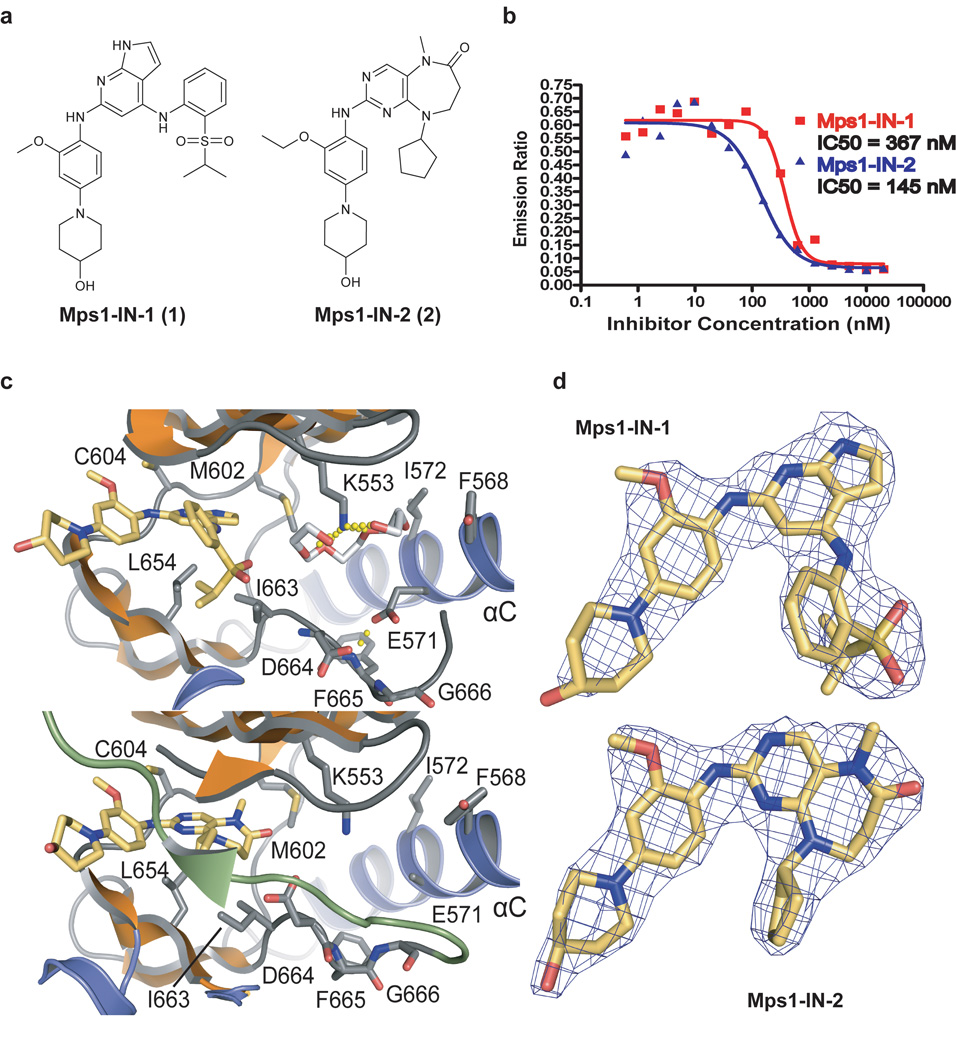
Figure 1. Mps1-IN-1 and Mps1-IN-2 inhibit Mps1 kinase activity and bind Mps1 in the ATP-binding site.
(a) The chemical structures of two ATP-competitive Mps1 kinase inhibitors, Mps1-IN-1 and Mps1-IN-2, are derived from two scaffold series: pyrrolopyridine and pyrimidodiazepinone respectively. (b) In vitro kinase assays using the Lanthascreen technology were performed to assess the in vitro activity of compounds 1 and 2. 5 µg/mL Mps1 (∼ 40 nM) kinase were used in each reaction with 1 µM ATP (Km,app < 1 uM) and 200 nM AF-647 E4Y substrate. (c) Active site of Mps1 in complex with Mps1-IN-1 (upper panel) and the methoxy derivative of Mps1-IN-2 (methoxy-Mps1-IN-2 - lower panel). The most important residues interacting with the inhibitor are shown in stick representation and are labeled. The polyethylene glycol molecule coordinating the conserved active site lysine (K553) in the Mps1-IN-1 complex is also shown. Carbon atoms of Mps1-IN-1 and Mps1-IN-2 are shown in yellow to discriminate from Mps1 kinase residue side chains. (d) 2FoFc electron density map contoured at 2σ around the inhibitor molecules.
Mps1-IN-1 and 2 inhibited Mps1 kinase activity with half-maximal inhibitory concentrations (IC50) of 367 nM and 145 nM respectively when screened at 1 µM ATP (apparent Km for ATP < 1 µM) (Fig. 1b). Both compounds demonstrated greater than 1000-fold selectivity relative to the 352 member kinase panel with the major exceptions of Alk and Ltk for Mps1-IN-1 and Gak and Plk1 for Mps1-IN-2 (Supplementary Fig. S1b, Supplementary Table S1 and Supplementary Table S2). The Alk activity of Mps-IN-1 is not unexpected as this series of pyrrolopyridines is structurally similar to TAE684, a known potent Alk inhibitor 32. Given the restricted tissue expression of both Alk 33 and Ltk 34 these off-target interactions would not be expected to significantly interfere with the use of Mps1-IN-1 in cell types typically used to study the spindle checkpoint. Mps1-IN-2 was overall more selective than Mps1-IN-1, but possesses significant activity against Plk1 which is consistent with this compound being a ring-expanded version of a highly potent Plk1 (and Plk2, Plk3) inhibitor: BI-2536 35. Although the Plk1 activity of Mps1-IN-2 limits its use as a selective Mps1 inhibitor, the compound does provide a unique tool to investigate the combined inhibition of Plk1 and Mps1. In addition because these two compounds do not share any common-off targets with the exception of Mps1, the phenotypes they have in common are likely to result from Mps1 inhibition. The majority of cellular experiments in this study were performed with Mps1-IN-1.
Crystal structure of Mps1 Kinase Domain
Mps1 shares only weak sequence homology (∼20% in kinase domain) with kinases of known structure. To this end we determined the crystal structure of the catalytic domain (F515-Q794) of Mps1 at 2.3 Å resolution (Supplementary Table S3). The structure of Mps1 revealed the characteristic bilobal domain architecture and secondary structure elements of protein kinases (Supplementary Fig. S3a). Following deposition of the coordinates into the protein data base (http://www.rcsb.org/pdb/home/home.do) two more apo-structures were published 36, 37. Our structural model superimposed with an r.m.s.d of 0.78 Å and 0.5 Å with these models that have been refined at 2.7 Å and 3.17 Å, respectively.
Mps1 adopts an inactive conformation as indicated by incorrect positioning of the regulatory helix αC, the lack of a salt bridge between the conserved αC glutamate (E571) and the active site lysine (K553), and an unstructured activation loop (M671-V684). In the lower kinase lobe the loop region between residues S699-K708 was also disordered. Interestingly, a polyethylene glycol molecule present in the crystallization solution was visible in the electron density as a ring around the catalytic lysine (K553).
The observed inactive conformation is surprising considering that nine phosphorylation sites were detected by ESI-MS after expression of Mps1 in bacteria. However, none of these sites were visible in the electron density in the apo-crystal structure suggesting that the location of the auto-phosphorylation sites is confined to the unstructured regions of the protein. Homogeneously de-phosphorylated protein was obtained by co-expressing λ-phosphatase. However, no crystals were obtained using the unphosphorylated protein.
Binding modes of Mps1-IN-1 and methoxy-Mps1-IN-2
Mps1-IN-1 was co-crystallized and structural models were refined at 2.74 Å resolution (Supplementary Table S3). As expected, the inhibitor bound to the ATP binding pocket of Mps1 forming a hydrogen bond with the hinge backbone (E603). The structure of the complex superimposed well with the apo-structure and main structural differences of the protein backbone were confined to the phosphate-binding loop (P-loop) region (Supplementary Fig. S3b). The inhibitor was well defined by electron density (Fig. 1d). The binding mode was stabilized by a number of hydrophobic interactions involving the gatekeeper residue M602 as well as I663, L654 and the P-loop residues V539 and I531 (Fig. 1c, Supplementary Fig. S3b). Both Mps1-IN-1 and the polyethylene glycol molecule occupied the ATP binding sites simultaneously.
We co-crystallized a close analog of Mps1-IN-2, methoxy-Mps1-IN-2 (3), which contains a methoxy instead of an ethoxy substituent at the ortho-position of the C2-aniline. Though a strong selectivity determinant, the ethoxy group of Mps1-IN-2 would not be expected to alter the overall binding mode of the molecule from that of what is seen for methoxy-Mps1-IN-2. Methoxy-Mps1-IN-2 forms a hydrogen bond with the hinge residue G605 and forms a tight contact with the same hydrophobic residues as Mps1-IN-1 (Fig. 1c, Supplementary Fig. S3b). The polyethylene glycol molecule was however not present in the methoxy-Mps1-IN-2 complex structure. The most significant structural change relative to the Mps1-IN-1 complex was that the activation segment became ordered revealing three phosphorylated amino acid residues (T675, T676 and S677) and formed an antiparallel β-sheet interaction with the P-loop (Fig. 1c). The high charge density of the three consecutive phosphorylated residues is compensated by 2 Mg2+ ions that are coordinated by the phosphate oxygens. However, this conformation of the activation segment may be a consequence of crystal contacts. Auto-phosphorylation on T676 has been shown to activate Mps1 18, 38 but the functional role of the other two phosphorylation sites is unknown.
Mps1-IN-1 and Mps1-IN-2 abrogate SAC function
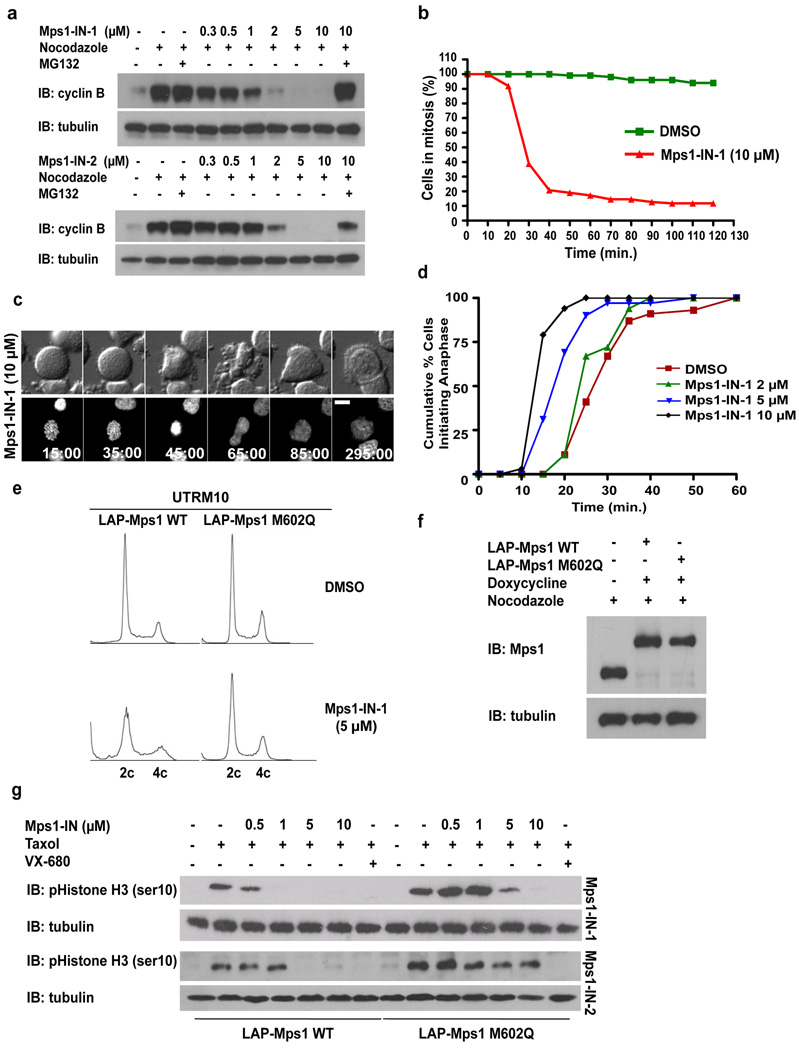
We first investigated whether chemical inhibition of Mps1 would elicit similar phenotypes to that of Mps1 RNAi-mediated knockdown, including silencing of the SAC in mammalian cells 13. Silencing of the SAC, a surrogate readout of Mps1 activity, can be assessed by monitoring events that signal that the cell has progressed through mitosis such as disappearance of phosphorylated histone H3 pSer10 (pHistone H3) or degradation of cyclin B. As assessed by flow cytometry, administration of Mps1-IN-1 to U2OS cells arrested in mitosis using nocodazole, resulted in a dose-dependent accumulation of 4c pHistone H3 negative cells (Supplementary Fig. S4a). Likewise, levels of cyclin B protein, which accumulate in G2 and are sustained during an activated spindle checkpoint 13, dropped with increasing concentration of Mps1-IN-1 or 2, but could be reversed by addition of the proteasome inhibitor, MG132 (Fig. 2a). Downregulation of these mitotic markers indicate that both compounds cause a dose-dependent escape from a checkpoint-mediated mitotic arrest. Mitotic escape induced by Mps1-IN-1 was rapid with DNA de-condensation and nuclear envelope reassembly apparent in as early as 20 minutes with the majority of cells escaping within 1 hour, as compared to DMSO-treated control cells that exhibited sustained arrest (Fig. 2b and 2c). Similar results were obtained when the SAC was activated using other spindle perturbing agents such as the tubulin stabilizer taxol and the kinesin-5 inhibitor s-trityl cysteine (Supplementary Fig. S4b). These results suggest that Mps1 activity is required to maintain mitotic arrest under conditions affecting both attachment and tension.
Figure 2. Mps1-IN-1 and Mps1-IN-2 Induce Bypass of a Checkpoint-mediated Mitotic Arrest.
(a) Immunoblot of cyclin B from U2OS cells treated with Mps1 inhibitors. U2OS cells were arrested in mitosis by combination treatment of thymidine and nocodazole prior to treatment with nocodazole or co-administration with Mps1 inhibitor ± MG132 for 4 hrs. (b) Hela S3 cells expressing histone H2B-GFP were treated with nocodazole ± Mps1-IN-1. Cells were followed by immunofluorescence to determine exit from mitosis via nuclear envelope reassembly. 150 cells of each cell population were counted. (c) Selected frames from a time-lapse series of cells after treatment as in (b). Time is given in minutes:seconds. (d) Cumulative percentage of cells initiating anaphase. U2OS H2B–GFP cells were treated with DMSO or Mps1-IN-1 and imaged using fluorescence time-lapse microscopy. Time refers to the duration between nuclear envelope breakdown (NEBD) and anaphase initiation. 100 cells of each cell population were analyzed. (e) FACS analysis of UTRM10 cells expressing LAP-Mps1 WT or M602Q treated with Mps1-IN-1 or DMSO for 48 hrs. (f) Immunoblot showing induction of the RNAi-resistant LAP-tagged proteins. Clones were harvested and the relative levels of LAP-Mps1 in mitosis were compared to the UTRM10 parental cell line 17, 18. (g) Immunoblot of pHistone H3 in UTRM10 cells expressing LAP-Mps1 WT or M602Q after treatment with Mps1 inhibitors. UTRM10 cell lines arrested in mitosis by combination treatment of thymidine and taxol were treated with taxol ± inhibitor. All graphics were obtained from three independent experiments. Scale bars in C are 10 µm.
To probe the effects of Mps1-IN-1 on an unperturbed mitosis, U2OS cells expressing fluorescently-tagged histone H2B (H2B–GFP) cells were treated with the inhibitor and the time spent in mitosis, from nuclear envelope breakdown (NEBD) to anaphase initiation, was assessed. Mps1-IN-1 administration resulted in a dose-dependent decrease in the time spent in mitosis with nearly 100% U2OS cells initiating anaphase within 20 minutes (10 µM Mps1-IN-1) as compared to roughly 10% in DMSO-treated cells (Fig. 2d, Supplementary Fig. S5a and Movie S1 and Movie S2). Acceleration of mitosis kinetics in Mps1-IN-1-treated cells had direct consequences on genomic stability with cells exhibiting significant signs of chromosome mis-alignment and chromosome mis-segregation (Supplementary Fig. S5a–c) manifesting aneuploidy (Fig. 2e, left two panels and Supplementary Fig. S5d, S5e), phenomena previously reported for RNAi-mediated knockdown of Mps1 and other checkpoint components 17–20, 30
Mutation of the kinase active site gatekeeper residue from methionine (M) to glutamine (Q) in Mps1 (M602Q) was previously shown reduce the potency of inhibition by the ATP-competitive small molecule inhibitor, SP600125 without significantly disrupting Mps1 kinase activity or proper kinetochore localization 30. To investigate whether the M602Q mutation would also confer resistance to the new inhibitors, a radioenzymatic immunoprecipitation kinase assay was performed using WT or M602Q LAP-Mps1. The M602Q Mps1 mutant was 5 and 19-fold less sensitive to Mps1-IN-1 and Mps1-IN-2 respectively (Supplementary Fig. S6a, S6b).
To investigate the effect of this mutation on intracellular inhibitor activity, endogenous Mps1 was replaced with the putative inhibitor resistant allele of Mps1 (M602Q). This was achieved by simultaneous expression of a plasmid-based Mps1 shRNA and an RNAi-insensitive LAP-tagged Mps1 M602Q allele to generate a stable U2OS cell line expressing LAP-Mps1 M602Q (UTRM10 Mps1 M602Q) (Fig. 2f) 17, 18. Expression of LAP-Mps1 M602Q restored pHistone H3 positivity to wild-type levels in the presence of Mps1-IN-1 and Mps1-IN-2 up to concentrations of 1 and 10 µM respectively (Fig. 2g). In contrast, treatment with VX-680 39, a potent pan-Aurora kinase inhibitor that causes a similar mitotic arrest override, was still capable of executing an efficient mitotic escape. Additionally, expression of LAP-Mps1 M602Q reduced the aneuploidy and chromosome mis-segregation phenotypes caused by Mps1-IN-1 (Supplementary Fig. S7, Fig. 2e, compare left and right panels). Finally, Mps1 has been reported to reside in a hyper-phosphorylated and activated form during checkpoint activation due in large part to autophosphorylation 18. Addition of Mps1-IN-1 to UTRM10 LAP-Mps1 WT cells causes a dose-dependent reduction in hyper-phosphorylated Mps1 as demonstrated by a decrease in phosphorylation-induced mobility shift (Supplementary Fig. S6c, compare lanes 2 −5). In contrast, UTRM10 LAP-Mps1 M602Q cells maintained high levels of hyper-phosphorylated Mps1 in the presence of Mps1-IN-1 (Supplementary Fig. S6c, compare lanes 7–10). Together these findings support the assertion that Mps1-IN-1 and 2 override the checkpoint through direct inhibition of Mps1.
Mps1-IN-1 disrupts recruitment of Mad2 to kinetochores
The effect of chemical inhibition of Mps1 kinase activity on the localization of Mad2, an essential component of the mitotic checkpoint, was examined next. Previous reports examining the kinetochore localization of checkpoint components, particularly Mad1 and Mad2, after RNAi-mediated knockdown of Mps1 have yielded conflicting results 8, 15, 30. In this study, the use of time-lapse fluorescence microscopy allowed us to determine the effect of inhibition of Mps1 kinase activity on the establishment of Mad2 at unattached kinetochores in the presence or absence of nocodazole.
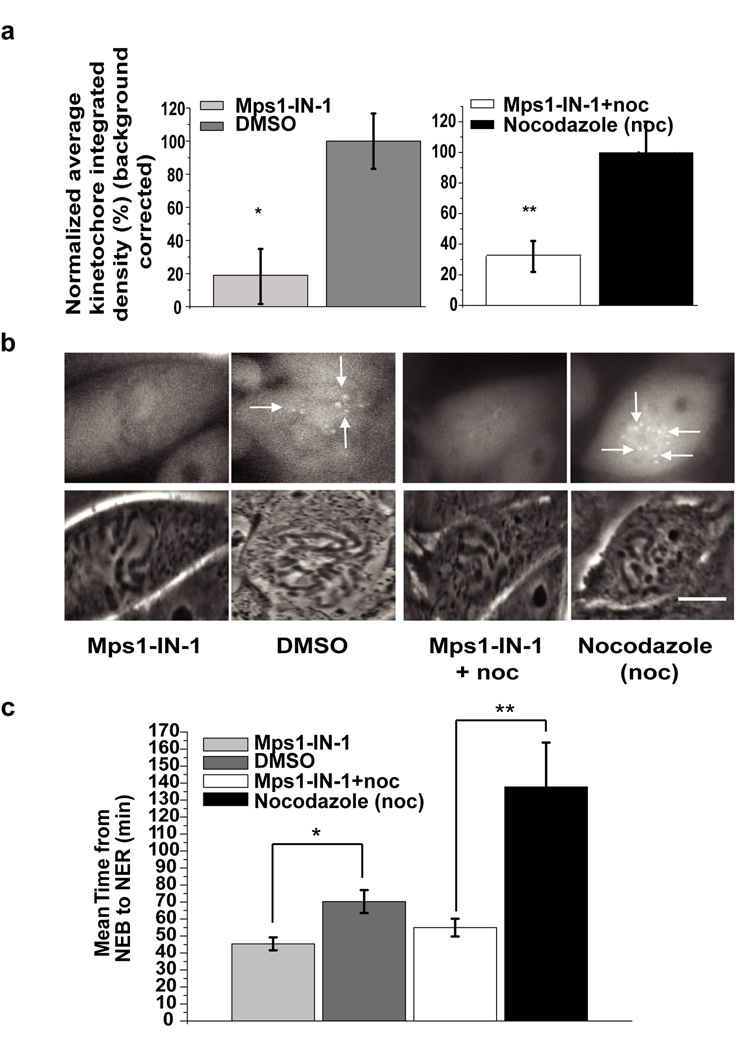
To this end, a PtK2 cell line stably expressing hsMad2-EYFP (Supplementary Method) was used to determine whether Mps1 kinase activity is required for the checkpoint activation and initial establishment of Mad2 at unattached kinetochores. Mps1-IN-1-treated hsMad2-EYFP PtK2 cells were followed as they entered mitosis and the levels of Mad2 at kinetochores were quantified. As compared to DMSO-treated control cells, Mps1-IN-1-treated cells exhibited an 80% decrease in kinetochore-bound Mad2 (Fig. 3a and 3b, Supplementary Movie S3 and Supplementary Movie S4). As a result, Mps1-IN-1-treated cells spent roughly 40% less time in mitosis as compared to DMSO-treated cells (Fig. 3c). Similar results were obtained when cells entered mitosis in the presence of both Mps1-IN-1 and nocodazole to stimulate checkpoint response. Mps1-IN-1 and nocodazole co-treated cells displayed a 70% reduction in the amount of kinetochore-bound Mad2 as compared to nocodazole-treated cells (Fig. 3a and 3b, Supplementary Movie S5 and Supplementary Movie S6). Mps1-IN-1-treated cells were unable to establish a proficient checkpoint in the presence of nocodazole and exited mitosis prematurely (Fig. 3c). The disappearance of hsMad2 (and hsMad1) from kinetochores following Mps1-IN-1 treatment was corroborated in Hela cells using indirect immunofluorescence (Supplementary Fig. S8). These results clearly demonstrate that Mps1 kinase activity is required for the recruitment of Mad2 to kinetochores and SAC activation.
Figure 3. Mps1-IN-1 treatment causes disruption in recruitment of Mad2 to kinetochores.
(a) Establishment of Mad2 at the kinetochores is dependent on Mps1 kinase activity when cells enter mitosis. PtK2 cells stably expressing HsMad2-EYFP were treated with either Mps1-IN-1 (10 µM) (N=29) or Mps1-IN-1 (10µM) and nocodazole (N=25) and then imaged entering mitosis. Corresponding controls were PtK2 cells stably expressing HsMad2-EYFP treated with DMSO (N=26) or treated with nocodazole only (N=20) and then imaged entering mitosis. Normalized average kinetochore density representing Mad2-EYFP at kinetochores was measured and background corrected for each experimental case and control. Error bars represent the 95% confidence interval of each data set (* p-value = 5.63e−09, ** p-value = 7.39−07, Student’s t test). (b) Representative images of PtK2 cells stably expressing HsMad2-EYFP that were treated as in (a). The top panel are images of HsMad2-EYFP fluorescence and the bottom panel are corresponding phase images. Scale bar is equal to 10 µm. (c) Inhibition of Mps1 kinase activity affects mitotic timing. Randomly cycling PtK2 cells stably expressing HsMad2-EYFP were treated with either Mps1-IN-1 (10 µM) (N=29) alone or with Mps1-IN-1 (10 µM) and nocodazole (N=25) and then imaged entering mitosis from nuclear envelope breakdown (NEB) through to nuclear envelope reassembly (NER). Untreated PtK2 cells (N=26) or those treated with nocodazole only (N=20) were also imaged. Average time spent in mitosis (from NEB to NER) is shown. Error bars represent the 95% confidence interval of each data set (* p< 2.23e−08, ** p<3.08−08, Student’s t test).
Mps1-IN-1 affects the kinase activity of Aurora B
Previous evidence has suggested that an epistatic signaling relationship exists between Mps1 and Aurora B, whereby Mps1 modulates the cellular kinase activity of Aurora B 17. This finding suggested a plausible mechanism whereby an activated checkpoint could serve to initiate or modulate the tension-sensing process of attachment error correction with the Mps1-Aurora B interaction acting as a bridge between the two processes.
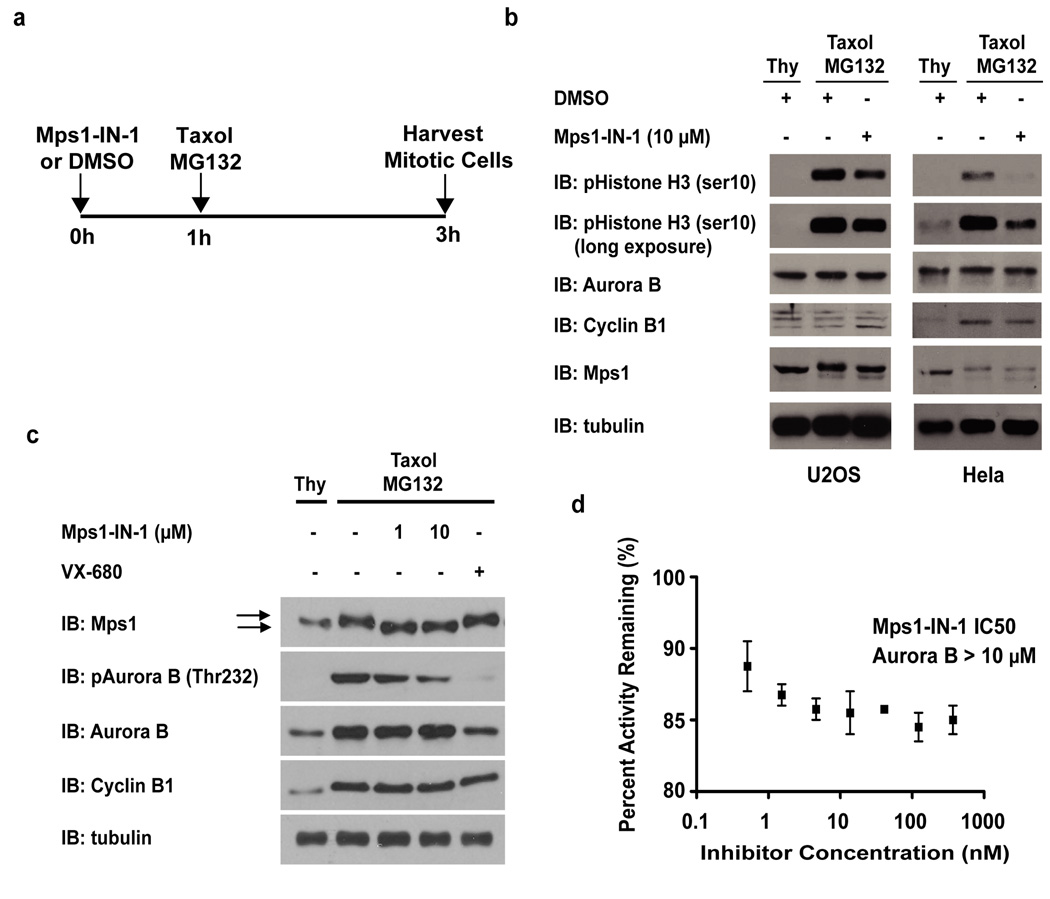
We sought to use Mps1-IN-1 as a means to test this epistatic relationship. Both Hela and U2OS cells were released from a thymidine block and prior to mitotic entry were treated with Mps1-IN-1 or DMSO vehicle for 1 hr. Cells were subsequently treated with taxol and MG132 (Fig. 4a) and mitotic cells were collected by shake-off. The sustained cyclin B levels corroborated that the collected cells were mitotic, yet a reduction in pHistone H3 (ser10) signal (a direct Aurora B substrate) upon Mps1-IN-1 treatment was apparent in both cell lines (Fig. 4b). Additionally, Mps1-IN-1 treatment caused a dose-dependent reduction in the phosphorylation status of Aurora B at threonine-232 (Thr232), an activation loop phosphorylation of the kinase that is responsible for increased kinase activity 40, further suggesting that inhibition of Mps1 reduces Aurora B kinase activity (Fig. 4c, lanes 2–4). The reciprocal experiment showed that treatment with VX-680 inhibited the activation and phosphorylation of Aurora B (Thr232), but did not adversely affect the hyperphosphorylation status of Mps1, as evidenced by the maintained phosphorylation-induced mobility shift in the Mps1 protein band (Fig. 4c). Direct inhibition of Aurora B by Mps1-IN-1 is unlikely as in vitro assays indicate that Mps1-IN-1 does not exhibit activity against Aurora B (Fig. 4d, Supplementary Table S1). These results support the hypothesis that Mps1 kinase activity regulates Aurora B intracellular kinase activity.
Figure 4. Mps1-IN-1 treatment decreases intracellular Aurora B kinase activity.
(a) Schematic of the treatment regimen used to assess the effect of Mps1-IN-1 treatment on intracellular Aurora B kinase activity during taxol treatment in Hela and U2OS cells. (b) Mps1-IN-1 treatment decreases the phosphorylation of pHistone H3 (ser10), a direct substrate of Aurora B. Immunoblot of Hela and U2OS cells treated as described in (a). An antibody to pHistone H3 (ser10) was used to assess the kinase activity of Aurora B, while the mobility shift in Mps1 was used to assess the relative amount of Mps1 auto-phosphorylation. Cyclin B levels served as a mitotic marker. (c) Mps1-IN-1 treatment decreases the phosphorylation of Aurora B in its activation loop (Thr232). U2OS cells were treated as described in (a). An antibody to pAurora B (Thr232) was used to assess the phosphorylation of Aurora B, while the mobility shift in Mps1 was used to assess the relative amount of Mps1 auto-phosphorylation. Cyclin B levels served as a mitotic marker. Arrows indicate phosphorylation-induced shift of Mps1 total protein band. (d) In vitro kinase assay using the Lanthascreen technology was performed to assess the in vitro activity of Mps1-IN-1 on Aurora B. Aurora B kinase (5 nM) was used in each reaction with ATP at the apparent Km (63.7 nM) and the substrate, 30 nM Kinase Tracer 236.
Mps1-IN-1 does not affect centrosome duplication
The founding member of the Mps1 kinase family (Mps1p of S. cerevisiae) has been implicated in the duplication of the yeast spindle pole body (SPB) due to the phenotypic manifestation of monopolar spindles after Mps1p inactivation 2, 3. Additional evidence with murine Mps1 (mMps1) suggested mMps1 to be required for centrosome duplication supporting the notion that this function was evolutionary conserved in mammalian cells 41. However, conflicting reports in human cells have raised questions as to whether human Mps1 (hMps1) is required for centrosome (re)duplication 10–13.
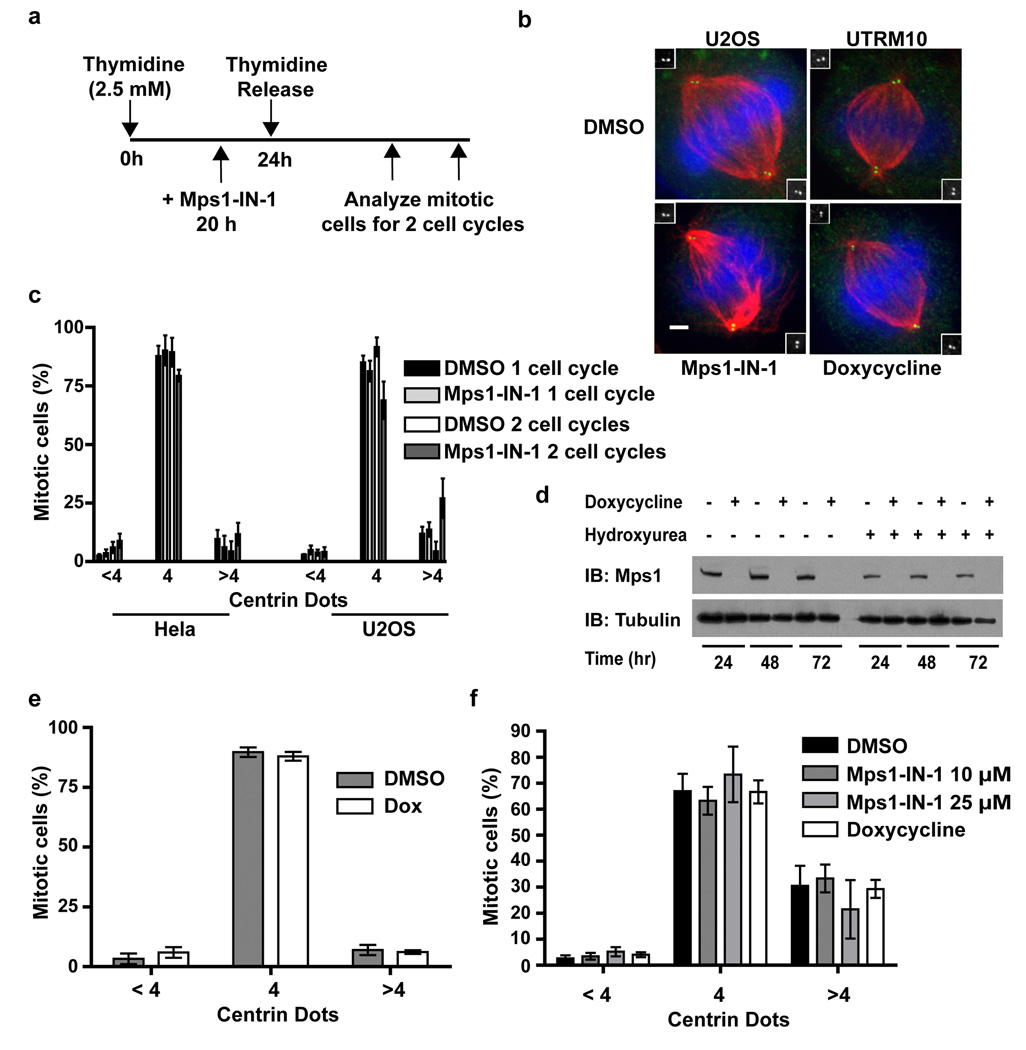
To determine if Mps1 kinase activity is indeed necessary for centrosome duplication we treated synchronized cells with 10 µM Mps1-IN-1 (Fig. 5a), a concentration known to inhibit all checkpoint-associated Mps1 kinase activity (Fig. 2a and Supplementary Fig. S4a), for two cell cycles. Mitotic cells were then scored for the number of centrioles they contained. Un-treated mitotic cells have 2 centrioles per centrosome (4 per cell), whereas mitotic cells exhibiting centrosome duplication defects would have fewer. Analysis of cells after one cell cycle ensures that all mitotic cells scored had passed through only one S phase in the presence of Mps1-IN-1; a level of time resolution that can only be attained with small molecule intervention. Analysis of cells after two cell cycles allows one to visualize the appearance of monopolar spindles that would arise by dilution of centrioles by passage through successive rounds of cell division without concomitant centriole duplication. Using this scheme we found no significant difference between vehicle- and compound-treated cell populations after 1 or 2 cell doublings as the mean percentage of mitotic cells containing less than 4 centrioles (centrin dots) remained unchanged within one standard deviation (Fig. 5b and 5c).
Figure 5. Mps1-IN-1 compound treatment does not affect centrosome duplication.
(a) Schematic of the treatment regimen used to assess the effect of Mps1-IN-1 treatment on centrosome duplication in Hela and U2OS cells. (b) Immunofluorescence images of U2OS cells treated with Mps1-IN-1 or doxycycline to induce Mps1 shRNA expression. Centrin, α-tubulin and DNA are shown in green, red and blue, respectively. Insets show high magnification images of centrin staining. (c) Mps1-IN-1 treatment does not affect centrosome number in cycling Hela S3 or U2OS cells. The centriole number of mitotic spindles from synchronized cycling cells treated with Mps1-IN-1 or DMSO was quantitated using centrin staining. Cells counted (1 and 2 cell cycles): Hela DMSO - 288 and 260 cells, Hela Mps1-IN-1 – 296 and 260 cells, U2OS DMSO - 300 and 234 cells, U2OS Mps1-IN-1 – 300 and 135 cells. (d) Immunoblot showing shRNA-mediated depletion of endogenous Mps1 in UTRM10 cells by doxycycline treatment. (e) Depletion of Mps1 does not affect centrosome number in UTRM10 cells. The centriole number of mitotic spindles from cycling cells treated with doxycycline for 72 hrs. was scored as in (c). 304 DMSO-treated mitotic cells and 267 doxycycline-treated mitotic cells were scored. (f) UTRM10 cells co-treated with hydroxyurea and either Mps1-IN-1, doxycycline, or DMSO vehicle for 48 hours were scored using centrin staining. Cells counted: DMSO – 443, Mps1-IN-1 10 µM/25 µM - 433/433, doxycycline - 485. All graphics represent mean ±SD and were obtained from three independent experiments. Scale bars in B are 10 µm.
Hydroxyurea treatment of U2OS has been shown to result in overduplication of centrioles. To determine whether Mps1-IN-1 treatment disrupts centriole overduplication during a prolonged S phase arrest, UTRM10 cells were co-treated with hydroxyurea and Mps1-IN-1 (10 or 25 µM) or DMSO vehicle for 48 hours. Similar to the results obtained with cycling cells, no effect of Mps1 inhibition on centriole overduplication was seen with the fraction of each cell population remaining statistically unchanged (Fig. 5f). In accordance with compound treatment, shRNA-mediated ablation of Mps1 in UTRM10 cells displayed no significant difference in centrosome duplication from DMSO control despite the lack of detectable Mps1 protein (Fig. 5d, 5e). Therefore, we conclude that in the presence of Mps1-IN-1 or shRNA-mediated knockdown of Mps1 we find no evidence for Mps1-dependent effects on centrosome duplication.
Mps1-IN-1 increases multipolar cell divisions in extra centrosomal cells
Centrosome amplification is a common feature of many cancer cells. Recently, several groups have reported that cells with extra centrosomes activate the spindle assembly checkpoint 42–44. SAC-dependent delay of anaphase onset is an important mechanism to protect against the harmful consequences of extra centrosomes by providing time for cancer cells to cluster extra centrosomes, enabling bipolar spindle assembly and normal cell division 42, 44. A natural hypothesis is whether Mps1-IN-1-induced checkpoint abrogation would be preferentially cytotoxic to cancer cells with extra centrosomes.
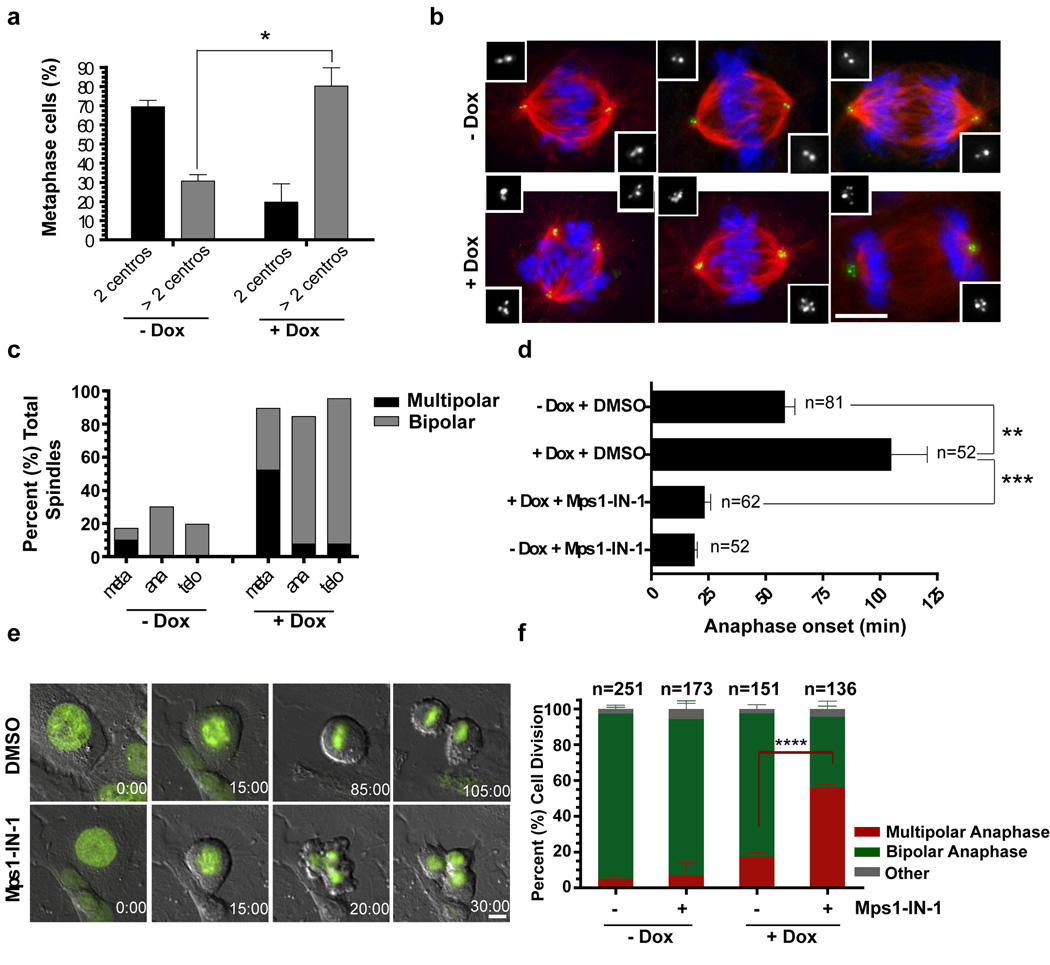
To determine if Mps1-dependent SAC activation promotes bipolar divisions in mammalian cells harboring extra centrosomes we characterized mitosis in U2OS cells where centrosome number can be controlled by the inducible expression of Plk4. Plk4 kinase is the ‘master regulator’ of centriole duplication and its overexpression induces centrosome amplification 45. As expected, doxycycline treatment of cells rapidly induced extra centrosomes, generating populations of cells where ∼80% contain extra centrosomes (Fig. 6a). Despite the presence of extra centrosomes, the majority of extra centrosomal cells (∼80%) successfully divide in a bipolar manner because of efficient clustering of the extra centrosomes (Fig. 6b) and multipolar metaphase figures are resolved into bipolar spindles prior to anaphase onset (Fig. 6c). To directly determine if SAC inhibition by Mps1-IN-1 induces multipolar anaphases in extracentrosomal cells, U2OS cells expressing H2B–GFP were synchronized with a double thymidine block, released for 6–7 hrs and treated with Mps1-IN-1, with or without Plk4 overexpression. Note that this protocol excludes the possible consequences of inhibiting Mps1 on centriole duplication, a controversial point in mammalian cells 10, 13. Time lapse imaging to monitor time from NEBD to anaphase onset revealed that the induction of extra centrosomes (+Dox) led to a ∼ 2 fold delay in anaphase onset (Fig. 6d and 6e, Supplementary Movie S7). In these cells, Mps1-IN-1 treatment abolished the anaphase delay (Fig. 6d and 6e, Supplementary Movie S8). Moreover, Mps1-IN-1 treatment resulted in marked increase in multipolar anaphases (Fig. 6f, Supplementary Movie S8). Approximately 60% of Mps1-IN-1–treated cells enter anaphase prematurely and undergo fragmentary divisions into multiple daughter cells. By contrast, without Mps1-IN-1 treatment cells spent more time in metaphase, eventually achieving bipolar metaphase plates, and dividing normally into two daughters (Fig. 6f). Thus, Mps1-IN-1 blocks the SAC dependent delay in anaphase onset that occurs in cancer cells with extra centrosomes. Surprisingly, despite the increased frequency of fragmentary cell divisions, no detectable additional cytotoxicity as assessed by MTS assay of Mps1-IN-1 was attained as compared to the parental U2OS cell lines (Supplementary Fig. S9a).
Figure 6. Mps1-IN-1 drives cancer cells with extra centrosomes into a catastrophic anaphase.
(a) Doxycycline (Dox) inducible Plk4 overexpression induces extra centrosomes in U2OS cells. Metaphase spindles with 2 or more centrosomes (centros.) were quantified after treatment with doxycycline or DMSO by centrin staining. ∼300 spindles were scored. (b) Cells cluster extra centrosomes, passing through a multipolar intermediate before undergoing bipolar divisions. Immunofluorescence images of cells from (a). Centrin, α-tubulin and DNA are shown in green, red and blue, respectively. Insets show high magnification images of centrin staining. (c) Resolution of metaphase multipolar spindles into bipolar spindles in anaphase in cells with extra centrosomes. Quantitation of bipolar or multipolar spindles during metaphase, anaphase and telophases in cells with extra centrosomes. ∼300 spindles were scored. (d) Extra centrosomes lead to delays in anaphase onset in a Mps1 (SAC)-dependent manner. (e) In extra centrosomal cells, the abrogation of SAC by Mps1-IN-1 treatment induces multipolar anaphases/telophases (bottom panels), whereas SAC-dependent delay enables bipolar division in the absence of Mps1-IN-1 (upper panels). Timelapse still images taken from movies of U2OS H2B–GFP after Plk4 overexpression. Time is shown in minutes after NEBD. Cells were synchronized with double thymidine blocks and released for 6 hrs. prior to Mps1-IN-1 treatment. (f) Quantitation of bipolar or multipolar anaphases/telophases in (e). All graphics represent mean ±SD (*p-value = 7.11e−05, **p-value = 0.0011, *** p-value = 2.46e−07, **** p-value = 0.0024, Student’s t test, 3 independent experiments). Scale bars in B, E are equal to 10 µm.
Mps1-IN-1 treatment decreases cell viability
Recent experimental evidence has given rise to the idea that complete spindle checkpoint inactivation is lethal to cells, while partial inactivation is associated with non-lethal chromosomal instability 18, 20–22 (reviewed in 24). Cells in which endogenous Mps1 was removed and reconstituted with LAP-Mps1 T676A, a de-activating mutation in the activation loop of Mps1, suffered no detectable decrease in cell viability despite exhibiting a weakened checkpoint and errors in chromosome segregation 18. In contrast, cells depleted of endogenous Mps1 and reconstituted with LAP-Mps1 KD exhibited no cell growth. These genetic results suggest that complete and selective inhibition of Mps1 kinase activity represents another way to inhibit cell growth but it remains unclear whether selective cytotoxicity can be achieved towards cancer cells.
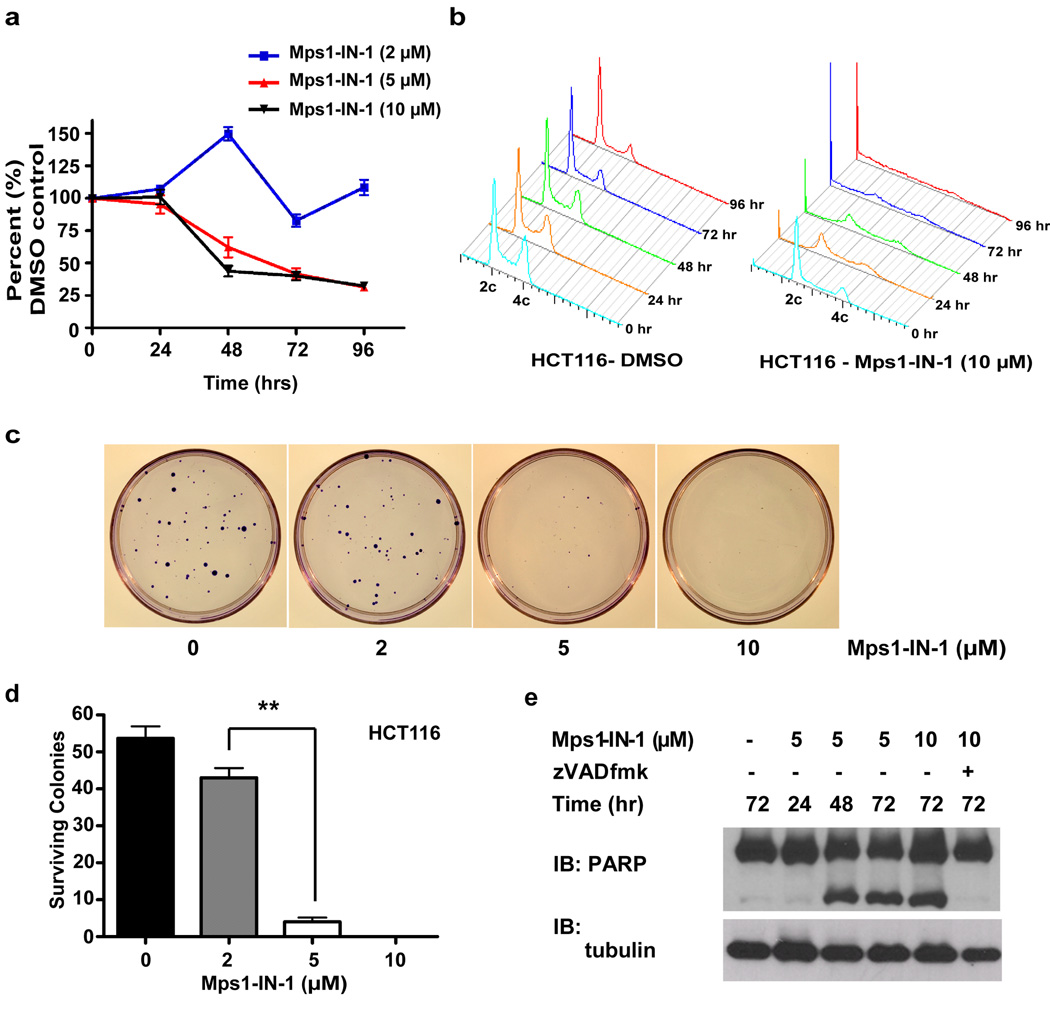
First, to examine the effect of chemical inhibition of Mps1 on cell proliferation, HCT116 colorectal cells, demonstrated to have a competent spindle checkpoint 46, were treated with Mps1-IN-1 at various concentrations and analyzed for cell number over a 4-day period. After 96 hour treatment with 5–10 µM Mps1-IN-1, the proliferative capacity of HCT116 cells was reduced to 33% that of DMSO control (Fig. 7a), during which time this cell population exhibited gross signs of aneuploidy and accumulation of cells with <2c DNA content (Fig. 7b). The cytotoxic concentrations of 5–10 µM Mps1-IN-1 are consistent with concentrations previously shown to inhibit all Mps1-dependent SAC activity. In congruence with proliferation results, concentrations of 5–10 µM Mps1-IN-1 caused severe loss of cell viability and clonal survival in a colony outgrowth assay (Fig. 7c, 7d). Similar concentrations of Mps1-IN-1 were found to have anti-proliferative effects when tested against a small panel of tumor cell lines and non-cancerous ‘normal’ cell lines 47, 48 (Supplementary Fig. S9b and Supplementary Fig. S10a). Consistent with previous reports 18, shRNA- (Supplementary Fig. S10b and S10c) and siRNA- (Supplementary Fig. S11) mediated silencing of endogenous Mps1 also resulted in decreased cell viability 18.
Figure 7. Mps1 is required for cell viability.
(a) HCT116 cells were treated with Mps1-IN-1 (2, 5, 10 µM) for 24, 48, 72, and 96 hours. Effects on cell proliferation were quantified by measuring fluorescence of Syto60 nucleic acid stain emitted at 695 nm (excitation 635 nm). Values were normalized to DMSO control for each time point. All graphics represent mean ± SD and were obtained from 4 independent experiments. (b) FACS profile of HCT116 cells treated with DMSO vehicle or Mps1-IN-1 (10 µM) for the indicated time points. (c) Colony outgrowth assay of HCT116 cells treated with DMSO vehicle or Mps1-IN-1 (2, 5, 10 µM). Cells were plated at a density of 200 cells/60-mm dish and harvested for crystal violet staining after 10 days. (d) Quantitation of colony outgrowth assays for HCT116 cells treated with DMSO vehicle or Mps1-IN-1 (2, 5, 10 µM). All graphics represent mean + SD and were obtained from 3 independent experiments (**p-value = 3.22e−05, Student’s t test). (e) Immunoblot of PARP cleavage of HCT116 cells treated with Mps1-IN-1 at the indicated concentrations. Cell lysates were harvested after 24, 48, and 72 hours for immunoblot detections.
Interestingly, loss of viability induced by compound treatment (or Mps1 siRNA) was associated with induction of apoptosis as exemplified by PARP cleavage beginning after 48 hours (Fig. 7e, Supplementary Fig. S11a and Supplementary Fig. S12a). This cleavage was shown to be specific to caspase activation as the co-administration of Z-VAD-FMK, a pan-caspase inhibitor, prevented PARP cleavage (Supplementary Fig. S12a), but was unable to rescue the anti-proliferative defects elicited by Mps1-IN-1, suggesting a possible caspase-independent cell death also becoming activated (Supplementary Fig. S12b). However, treatment of non-cancerous ‘normal’ cell lines, hTERT-RPE1 and MCF10A cells, with high doses of Mps1-IN-1 (25 µM) (also Mps1 siRNA in hTERT-RPE1) caused moderate aneuploidy and associated decrease in colony survival (Supplementary Fig. S10a and Supplementary Fig. S11), however with little-to-no apoptosis-associated PARP cleavage (Supplementary Fig. S12c). Further research using primary cultures or through in vivo investigations will be required to establish whether a ‘therapeutic window’ with respect to induction of apoptosis or other forms of cell death exists between normal and cancerous cell lines upon inhibition of Mps1 kinase activity.
Discussion
Previous reports have highlighted the utility of inhibiting Mps1 by chemical inhibition. However, these reports used either non-selective inhibitors or inhibitors that were specific to a particular chemical genetics background 4, 29, 30. Here, we described the discovery of a selective Mps1 inhibitor Mps1-IN-1 and a dual Mps1/Plk1 inhibitor Mps1-IN-2 by screening a kinase directed library against a large panel of 352 diverse kinases. The basis for recognition of these new compounds by the ATP-site of Mps1 was demonstrated by co-crystallography with the kinase domain. We demonstrated that these inhibitors recapitulated many of the hallmarks of Mps1 inhibition and SAC abrogation that have been previously demonstrated using RNAi approaches. The cellular specificity for Mps1 inhibition was demonstrated by introduction of an inhibitor-resistant allele of Mps1 (M602Q).
Using the combined approach of a selective Mps1 inhibitor, Mps1-IN-1, to afford rapid inhibition of Mps1 kinase activity and time-lapse microscopy we reported the first direct experimental evidence that shows that Mps1 is required for the checkpoint activation and establishment of Mad2 at kinetochores as cells enter mitosis. Cells that entered mitosis in the presence of Mps1-IN-1 exhibited severe defects in the establishment of kinetochore-bound Mad2, leading to a premature mitotic exit. Overall our results suggest that Mps1 kinase activity is essential for the establishment of and sustained activity of the spindle checkpoint.
Additionally, we used Mps1-IN-1 treatment to test the hypothesis that Mps1 lies upstream of Aurora B in checkpoint signaling. Inhibition of Mps1 kinase activity led to a decrease in phosphorylation of a direct Aurora B substrate (Histone H3) and of Thr232 in the activation loop of Aurora B. Inhibition of Aurora B by VX-680, a potent pan-Aurora inhibitor, had no reciprocal effect on Mps1 activation. These results support the notion that Mps1 activity is necessary for full enzymatic activity of Aurora B and thus serves to regulate attachment error correction, a tension-sensing process.
At concentrations of Mps1-IN-1 known to inhibit Mps1 mitotic kinase activity or using shRNA-mediated depletion of Mps1, we find no quantitative effect on centrosome number in Hela or U2OS cells, consistent with some findings 12, 13 but not with others that have suggested that minimal Mps1 kinase activity is required to fulfill its centrosome-associated function 10, 11, 41.
We demonstrated that Mps1 inhibition results in aneuploidy and a gradual loss of cell viability over several cell doublings as exemplified by decreased proliferative capacity, decreased clonal survival, an increase in the number of cells with sub-2c DNA content and induction of apoptosis. These results as well those conducted using shRNA/siRNA-mediated ablation of Mps1 suggest that massive chromosome loss could serve as a mechanism by which Mps1 inhibition (and checkpoint inactivation) ultimately leads to loss of cell viability. In extra centrosome containing cells, Mps1-IN-1 treatment resulted in catastrophic multipolar anaphase. Interestingly, though these cells were particularly sensitive to Mps1 inhibition, as exemplified by a 4-fold decrease in the time spent in mitosis, the proliferative capacity of these cells was not significantly affected as compared to the parental U2OS cell line (2-fold decrease in time spent in mitosis). This suggests that the effects on mitosis kinetics may not be the sole determining factor affecting cell proliferation and viability. Alternatively, the fold increase in aneuploidy upon compound treatment may represent a saturation point with respect to the induction cell death, beyond which the cell-type, genetic background, and specific chromosomes lost/gained becomes the over-riding factor influencing cell death. A detailed study of chromosome mis-segregation in the presence of compound in different cell types may shed light as to whether the overall rate of chromosome mis-segregation and/or the frequency of specific chromosomal loss correlate with decreased proliferative capacity and viability. Interestingly, recent data suggest that sensitizing tumor cells to checkpoint inhibition by administration of sub-lethal doses of taxol may provide an avenue to augmenting chromosome loss and induction of cell death49.
Finally, high dosage compound treatment in the ‘normal’ cell lines, hTERT RPE1 and MCF10A, did elicit an anti-proliferative effect though not associated with PARP cleavage. Given our current data it remains unclear whether this differential apoptotic induction in ‘normal’ vs. tumor cell lines can form the basis of a therapeutic window for which inhibitors of Mps1 (and more generally against the SAC) can be used as potential anti-cancer agents. Recent evidence showed that administration of sub-lethal doses of taxol (a checkpoint activator) combined with checkpoint inhibition produced differential cell death responses in cancer versus normal cell lines 49. However, more extensive profiling of Mps1-IN-1 and analogs with improved potency for inhibition of Mps1 against a larger panel of ‘normal’ and tumorigenic cell lines and in tumor models will be required to determine if a therapeutic advantage against tumor cells can be obtained.
Methods
Chemistry
Compounds from both scaffold classes were synthesized using existing procedures (WO 03/020722, WO2005080393, WO2009032694). See online Supplementary Methods for synthetic schemes and procedures; additional characterization data available upon request.
Plasmids, Transfections, and Treatments
The pSuper-based shRNA plasmid used in this study: Mps1 (GACAGATGATTCAGTTGTA) was constructed as described previously 17. LAP WT MPS1 cDNA subcloned into pCDNA3.1 was constructed as described previously 17. shRNA-insensitive Mps1 (modified codons 288 and 289) was obtained by site-directed mutagenesis 17. LAP-Mps1 M602Q (shRNA insensitive) was obtained by site directed mutagenesis of codon 602. The PTK2 (Male Potorous tridactylus kidney epithelial cells) cell lines stably expressing Homo sapiens (hs) MAD2 were generated via retroviral plasmids 50. siRNA transient transfections were performed using Dharmacon siGENOME TTK (MPS1) siRNAs (cat#s D-004105–01 and D-004105–03) and control siRNA-A (Santa Cruz Biotechnology, cat# sc-37007) with Lipofectamine RNAiMax (Invitrogen) following the manufacturer’s protocol for reverse transfections. Thymidine (2.5 mM), nocodazole (200 ng·ml−1), taxol (1 µM), MG132 (10 µM), s-trityl cysteine (10 µM), hydroxyurea (4 mM) and doxycycline (2 µg·ml−1) were all from Sigma. Puromycin (2 µg·ml−1) was from Invivogen and Z-VAD-FMK (50 µM) was from BD Biosciences. VX-680 (2 µM) was custom synthesized using published synthetic methods. SP600125 (10 µM) was from Calbiochem.
Cell Culture
Hela S3, U2OS, and A549 cells were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Sigma), and all HCT116 cells were grown in McCoy’s 5A (Invitrogen), and hTERT-RPE1 cells were grown in DMEM and Ham's F-12, 50/50 Mix (Cellgro – Fisher) supplemented with sodium bicarbonate (Invitrogen) and hygromycin B (Invitrogen) to final concentrations of 0.348% and 10 µg·mL−1 respectively. UTRM10 cells were generated and grown as previously described 17, 18. MCF10A cells were cultured as described previously 48. All PTK2 (Male Potorous tridactylus kidney epithelial) cells were cultured in Advanced MEM media (Invitrogen), 2% FBS, penn/strep, and 2mM GlutaMAX ™ (Invitrogen). Doxycycline-inducible PLK4-overexpressing U2OS cells were a gift from Dr. Nigg 45. H2B–GFP expressing cells were a gift from Dr. King. The HCT116 p53+/+, p53−/−, BAX +/+, and BAX−/− cell lines were a gift from Dr. Vogelstein. All cell lines were supplemented with 10% FBS (Sigma) and 100 U·mL−1 penicillin, 100 µg·mL−1 streptomycin (Invitrogen) and cultured at 37°C in a humidified chamber in the presence of 5% CO2, unless otherwise noted
Kinase Screening
The screening method is described in Supplementary Table S4. The data presented in Fig. 1 and Supplementary Fig. S1 were generated at Ambit Biosciences, using binding assays as previously described 27, 31. Kinome trees shown in Fig. 1C were generated using the KinomeScan TreeSpot kinome data visualization tool.
Immunoprecipitations and in vitro kinase assays
Conditions for immunoprecipitations of LAP-Mps1 using S-protein agarose (Novagen) have been described previously 17, 18, with minor modifications in Supplementary Research Data.
Flow Cytometry and Immunoblotting
Cells were released from a 24 hr thymidine-induced block into nocodazole or taxol for 2 hr prior to co-treatment with microtubule poison and test compound for 4 hours and analyzed using immunoblotting and flow cytometry. For immunoblotting standard protocols were followed. Flow cytometric analysis of cells was performed as described in Supplementary Methods using an anti-phospho-Histone H3 (ser10) antibody (Upstate).
Immunofluorescence and Time Lapse Microscopy
Immunofluorescence microscopy was carried out as described in Supplementary Material. For time lapse microscopy, cells were plated in 35 mm glass bottom microwell (14 mm, No 1.5 coverglass) dishes (MatTek Corporation), transfected and/or treated with chemical reagents and imaged in a heated chamber (37°C and 5% CO2). Time lapse images from mitotic timing experiments were captured using a Nikon TE2000E Automated Inverted Microscope (Nikon USA) using a 20X/0.75 NA Plan Apochromat objective lens. Twelve bit phase (25 msec exposure) and green fluorescent (50 msec exposure) images were acquired every 2 minutes using a Hamamatsu Orca AG Cooled CCD Camera and stored on a computer using Metamorph software. Time-lapse images from mitotic escape and chromosome mis-segregation experiments were captured using a Nikon TE2000U Inverted Microscope (Nikon USA) using a 60X/1.4 NA oil objective lens. Twelve bit DIC (200 msec exposure) and green fluorescent (180 msec exposure) images were acquired every 5 minutes (5 z-planes) using a Hamamatsu Orca ER Cooled-CCD camera and analyzed in Metamorph. Images of UTRM10 cells transfected with H2B–EYFP were captured on an Olympus IX-81 Microscope (Olympus) using a 20X/0.5 NA UPLFLN objective lens. Twelve bits yellow fluorescent (20 msec exposure) images were acquired every 3 minutes (5 z-planes) using a Hamamatsu ORCA-ER Cooled-CCD camera and processed using Cell-M software. All images collected to look at chromosome mis-segregation were (H2B–EYFP or H2B–GFP) are maximum intensity projections of all z-planes. For time lapse imaging of doxycycline-inducible PLK4 overexpressing U2OS cells, cells were synchronized with thymine for 18 hrs and released to doxycycline for 10 hrs followed by 2nd thymidine treatment for additional 18 hrs. Cells were then released to medium for 6hrs prior to 5µM of Mps1-IN-1 treatment.
Mad2 Kinetochore Establishment Assay
Ptk2 cells were treated with Mps1-IN-1 (10 µM) or DMSO vehicle for 1 hour before co-administration with either DMSO vehicle or nocodazole. Time-lapse microscopy was used to follow cells as they entered mitosis, as judged by nuclear envelope breakdown (NEBD) and kinetochore-localized HsMad2-EYFP fluorescence was quantified and background corrected as described in Supplementary Methods.
Centrosome Duplication Experiments
Hela S3, U2OS, and UTRM10 cells were plated on 12-mm coverslips. Cells were treated with Mps1-IN-1 with or without hydroxyurea as described and analyzed for centrosome duplication with a centrin antibody (see Supplementary Material). Similarly, UTRM10 cells stably expressing a doxycycline-inducible Mps1 shRNA construct were treated with doxycycline, immunoblotted to assess extent of Mps1 depletion and analyzed for centrosome duplication..
Proliferation, Colony Outgrowth, and Karyotyping Assays
Proliferation and colony formation assays were performed as previously described 16, 20. For karyotyping, asynchronous cells were treated with Mps1-IN-1 (10 µM) for 24 hours, the medium was removed and replaced with medium containing 100 ng/mL colcemid (Irvine Scientific) for 2 hours to arrest cells in metaphase prior to harvesting. Processing of cells was carried out as described in Supplementary Methods.
Supplementary Material
Acknowledgements
The authors thank Drs. R. King, B. Vogelstein, and E. Nigg for reagents and Drs. R. King, T. Mitchison, A. Abrieu, U. Eggert, C. Walsh, F. Sigoillot, and E. Chung for helpful discussions. The authors also thank Ambit Biosciences and Invitrogen Corporation for technical support in initial compound screening and enzymatic activity assays respectively as well as the Nikon Imaging Facility (HMS) and the Dana Farber Flow Cytometry Lab (DFCI) for technical help and instrument use. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, the Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., Inc., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research and the Wellcome Trust.
Footnotes
Supplementary Information. Supplementary Methods and Supplementary Results include expanded protocols, complete protocols for protein expression and crystallography data collection, supplementary references, figures, tables, and movies.
Author Contributions
N.S.G. and T.S. conceived and directed chemistry effort. H.W.C. and N.K. performed chemical synthesis and small molecule structure determination. N.K., N.J., M.S.M, M.K., Q.L.D., S.R., D.P., J.V.S., G.J.P.L.K., and N.S.G. designed biological experimental research. N.K., N.J., M.S.M., and M.K. performed experimental research and analysis. S.K. conceived and directed x-ray crystallography research. P.F. and M.S. performed x-ray crystallography research and analysis. N.K., P.F., M.K., and N.S.G. co-wrote the paper. All authors read and edited the manuscript.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Lauze E, et al. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. Embo J. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991;114:745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schutz AR, Winey M. New alleles of the yeast MPS1 gene reveal multiple requirements in spindle pole body duplication. Mol Biol Cell. 1998;9:759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones MH, et al. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr Biol. 2005;15:160–165. doi: 10.1016/j.cub.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 6.Abrieu A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 7.Vigneron S, et al. Kinetochore localization of spindle checkpoint proteins: who controls whom? Mol Biol Cell. 2004;15:4584–4596. doi: 10.1091/mbc.E04-01-0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu ST, et al. Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol Biol Cell. 2003;14:1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 10.Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci U S A. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasbek C, et al. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18:4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stucke VM, Baumann C, Nigg EA. Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma. 2004;113:1–15. doi: 10.1007/s00412-004-0288-2. [DOI] [PubMed] [Google Scholar]
- 13.Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. Embo J. 2002;21:1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt M, Bastians H. Mitotic drug targets and the development of novel anti-mitotic anticancer drugs. Drug Resist Updat. 2007;10:162–181. doi: 10.1016/j.drup.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Tighe A, Staples O, Taylor S. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J Cell Biol. 2008;181:893–901. doi: 10.1083/jcb.200712028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDermott U, et al. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc Natl Acad Sci U S A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jelluma N, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 18.Jelluma N, et al. Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PLoS ONE. 2008;3:e2415. doi: 10.1371/journal.pone.0002415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci U S A. 2005;102:11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel L, et al. Complete loss of the tumor suppressor MAD2 causes premature cyclin B degradation and mitotic failure in human somatic cells. Proc Natl Acad Sci U S A. 2004;101:4459–4464. doi: 10.1073/pnas.0306069101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michel LS, et al. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- 23.Dai W, et al. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res. 2004;64:440–445. doi: 10.1158/0008-5472.can-03-3119. [DOI] [PubMed] [Google Scholar]
- 24.Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Q, et al. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by Mps1 phosphorylation. Mol Biol Cell. 2009;20:10–20. doi: 10.1091/mbc.E08-03-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell BJ, et al. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- 27.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 28.Chu ML, et al. Biophysical and X-ray Crystallographic Analysis of Mps1 Kinase Inhibitor Complexes. Biochemistry. doi: 10.1021/bi901970c. [Epub - Ahead of print] [DOI] [PubMed] [Google Scholar]
- 29.Dorer RK, et al. A small-molecule inhibitor of Mps1 blocks the spindle-checkpoint response to a lack of tension on mitotic chromosomes. Curr Biol. 2005;15:1070–1076. doi: 10.1016/j.cub.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt M, Budirahardja Y, Klompmaker R, Medema RH. Ablation of the spindle assembly checkpoint by a compound targeting Mps1. EMBO Rep. 2005;6:866–872. doi: 10.1038/sj.embor.7400483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabian MA, et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 32.Galkin AV, et al. Identification of NVP-TAE684, a potent, selective, and efficacious inhibitor of NPM-ALK. Proc Natl Acad Sci U S A. 2007;104:270–275. doi: 10.1073/pnas.0609412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwahara T, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–449. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- 34.Bernards A, de la Monte SM. The ltk receptor tyrosine kinase is expressed in pre-B lymphocytes and cerebral neurons and uses a non-AUG translational initiator. Embo J. 1990;9:2279–2287. doi: 10.1002/j.1460-2075.1990.tb07399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steegmaier M, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 36.Chu ML, Chavas LM, Douglas KT, Eyers PA, Tabernero L. Crystal structure of the catalytic domain of the mitotic checkpoint kinase Mps1 in complex with SP600125. J Biol Chem. 2008;283:21495–21500. doi: 10.1074/jbc.M803026200. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, et al. Structural and Mechanistic Insights into Mps1 Kinase Activation. J Cell Mol Med. 2008 doi: 10.1111/j.1582-4934.2008.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattison CP, et al. Mps1 activation loop autophosphorylation enhances kinase activity. J Biol Chem. 2007;282:30553–30561. doi: 10.1074/jbc.M707063200. [DOI] [PubMed] [Google Scholar]
- 39.Harrington EA, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 40.Yasui Y, et al. Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J Biol Chem. 2004;279:12997–13003. doi: 10.1074/jbc.M311128200. [DOI] [PubMed] [Google Scholar]
- 41.Fisk HA, Winey M. The mouse Mps1p–like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- 42.Basto R, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 46.Cahill DP, et al. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 47.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 48.Soule HD, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 49.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci U S A. 2009;106:19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shah JV, et al. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.