Abstract
Cellular processes propelled by actin polymerization require rapid disassembly of filaments, and then efficient recycling of ADF/cofilin-bound ADP-actin monomers back to an assembly-competent ATP-bound state. How monomer recharging is regulated in vivo is still not well understood, but recent work suggests the involvement of the ubiquitous actin-monomer binding protein Srv2/CAP. To better understand Srv2/CAP mechanism, we explored the contribution of its WH2 domain, the function of which has remained highly elusive. We found that the WH2 domain binds to actin monomers and, unlike most other WH2 domains, exhibits similar binding affinity for ATP-actin and ADP-actin (Kd ~1.5μM). Mutations in the WH2 domain that impair actin binding disrupt the ability of purified full-length Srv2/CAP to catalyze nucleotide exchange on ADF/cofilin-bound actin monomers and accelerate actin turnover in vitro. The same mutations impair Srv2/CAP function in vivo in regulating actin organization, cell growth, and cell morphogenesis. Thus, normal cell growth and organization depend on the ability of Srv2/CAP to recharge actin monomers, and the WH2 domain plays a central role in this process. Our data also reveal that while most isolated WH2 domains inhibit nucleotide exchange on actin, WH2 domains in the context of intact proteins can help promote nucleotide exchange.
Keywords: actin, yeast, turnover, Srv2/CAP, ADF/cofilin, profilin, WH2 domain
INTRODUCTION
The dynamic turnover of actin networks is vital for maintenance of cell shape and structure, and for such processes as endocytosis, cell motility and cell division (Ono, 2007). Continuous subunit turnover provides cells with the plasticity to rapidly remodel their actin cytoskeletons in response to internal and external signals. Further, this process ensures rapid replenishment of the assembly-competent pool of ATP-actin monomers required for new growth. The major rate-limiting steps in actin turnover are filament disassembly and monomer recycling, which includes nucleotide exchange (ATP for ADP) on actin monomers. Filament disassembly is catalyzed by the combined actions of ADF/cofilin, Aip1, coronin, and other proteins (Insall and Machesky, 2009), and leads to the rapid accumulation of ADP-actin monomers bound to ADF/cofilin. ADF/cofilin preferentially binds to ADP-actin over ATP-actin (Blanchoin and Pollard, 1998; Maciver and Weeds, 1994) and strongly inhibits nucleotide exchange on actin (Hawkins et al., 1993; Hayden et al., 1993; Nishida, 1985). Since ATP-actin is far more competent for polymerization compared to ADP-actin (Pollard, 1986), recharging of actin monomers to an ATP-bound state represents a critical step in actin turnover (Carlier, 1989; Korn et al., 1987).
Despite its central importance in the actin turnover process, the mechanism for catalyzing monomer recharging has remained poorly understood. Early observations suggested that profilin plays a major role in this function, based on the observation that purified profilin increases the rate of exchange of radioactively- or chemically-labeled ATP for unlabeled ATP on actin (Goldschmidt-Clermont et al., 1992; Goldschmidt-Clermont et al., 1991; Mockrin and Korn, 1980; Nishida, 1985). However, more recently it was shown that profilin can only weakly promote nucleotide exchange on ADF/cofilin-bound ADP-actin monomers, the physiological substrate (Balcer et al., 2003). This is also consistent with the known (and opposite) binding preferences of profilin and ADF/cofilin, for ATP-actin and ADP-actin, respectively (Blanchoin and Pollard, 1998; Vinson et al., 1998). These observations call into question whether profilin has a major role in recharging ADF/cofilin-bound ADP-actin monomers, and suggests that other cellular factors may be involved.
Mounting evidence has pointed to the involvement of the ubiquitous actin-binding protein Suppressor of RasV19/ Cyclase-Associated Protein (Srv2/CAP) in monomer recycling in the presence of ADF/cofilin. Srv2/CAP is widely expressed in plant, animal, and fungal cells, where its genetic disruption causes severe defects in actin organization and actin-based processes including cell division, cell motility, cell polarization and endocytosis (Deeks et al., 2007; Hubberstey and Mottillo, 2002). Biochemical analyses show that Srv2/CAP accelerates ADF/cofilin-dependent actin turnover in vitro (Balcer et al., 2003; Chaudhry et al., 2007; Mattila et al., 2004; Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009), and genetic interactions of SRV2 with COF1 and PFY1 are consistent with this role (Balcer et al., 2003; Quintero-Monzon et al., 2009; Vojtek et al., 1991). Further, the depletion of CAP in mammalian cells slows actin turnover and leads to actin and ADF/cofilin accumulating in abnormal cytoplasmic aggregates (Bertling et al., 2004).
How does Srv2/CAP promote monomer recycling in the presence of ADF/cofilin? Recent work suggests a multi-step mechanism that involves the coordinated effects of multiple domains in Srv2/CAP (Quintero-Monzon et al., 2009). This includes an N-terminal helical folded domain (HFD) (Yusof et al., 2005), which binds specifically to ADF/cofilin-bound ADP-actin monomers, but not to uncomplexed ADF/cofilin or actin monomers (Moriyama and Yahara, 2002; Quintero-Monzon et al., 2009), and a C-terminal β-sheet domain (Dodatko et al., 2004), which binds to ADP-actin (Mattila et al., 2004). Native Srv2/CAP is a higher order oligomer (Balcer et al., 2003; Gieselmann and Mann, 1992; Wang et al., 1992). The oligomer is maintained by the N-terminal coiled-coil domain in Srv2/CAP, which has only an ancillary role in optimizing actin turnover (Quintero-Monzon et al., 2009). The central region of Srv2/CAP consists of a proline-rich motif (P1) that binds to profilin (Bertling et al., 2007; Drees et al., 2001; Lambrechts et al., 1997), a predicted WASp Homology-2 (WH2) domain, and a second proline-rich motif (P2) that binds to the SH3 domain of Abp1 to regulate Srv2/CAP localization (Balcer et al., 2003; Lila and Drubin, 1997). To date, the only conserved region of Srv2/CAP that has not been assigned a function is the WH2 domain. WH2 domains are found in a variety of actin-associated proteins (e.g. β-thymosins, WASP, WIP, MIM, and IRSp53), where they bind to actin monomers and assist in either promoting actin assembly or sequestering actin (Paunola et al., 2001). However, there has been no experimental evidence to support a role for the WH2 domain of Srv2/CAP in binding actin or in contributing to Srv2/CAP function. One previous study found that mutation of the two conserved lysine residues in the LKKV motif of the WH2 domain had little effect on Srv2/CAP function in vitro or in vivo (Mattila et al., 2004). Further, a purified fragment of Srv2/CAP containing the WH2 domain (a.a. 253-373) was reported to have no detectable binding affinity for NBD-labeled G-actin. These observations have left the function of the Srv2/CAP WH2 domain uncertain.
Here, we show that the WH2 domain of yeast Srv2/CAP binds directly to ATP-actin and ADP-actin monomers with similar affinity, and makes a fundamental contribution to Srv2/CAP biochemical and genetic functions in catalyzing nucleotide exchange on actin monomers and accelerating actin turnover. This defines the WH2 domain as a critical element in mediating the actin-binding interactions of Srv2/CAP, and extends the known utilities of WH2 domains in actin-binding proteins to include roles in promoting nucleotide exchange and actin turnover.
RESULTS
Effects of WH2 domain mutations on cell growth and genetic interactions
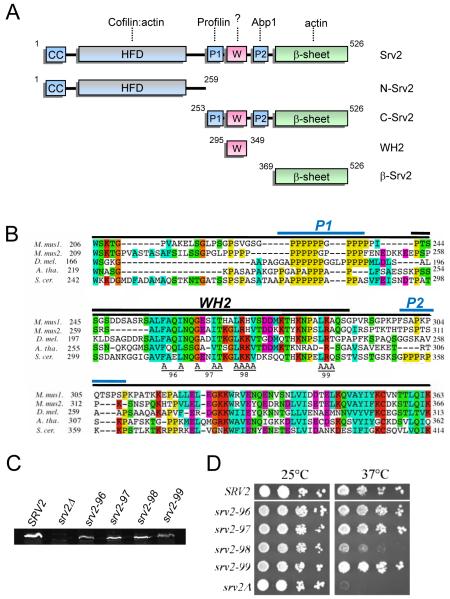
The WH2 domain is positioned in the C-terminal half of Srv2/CAP, flanked by proline-rich motifs P1 and P2 (Fig. 1A). Based on WH2 sequence alignments and known structural and functional boundaries (Paunola et al., 2001), the WH2 domain of S. cerevisiae Srv2/CAP is predicted to span 35 residues (Fig. 1B). To explore the in vivo importance of the WH2 domain for Srv2/CAP function, we designed four alanine substitution alleles (srv2-96, srv2-97, srv2-98, and srv2-99), targeting clusters of residues that are conserved in Srv2/CAP across distant species (Fig. 1B). The alleles were integrated at the SRV2 locus in haploid yeast, replacing the wild type copy of SRV2. The resulting haploid strains were found to express the mutant Srv2 proteins at levels comparable to wild type Srv2 protein (Fig. 1C). Mutant strains were compared to isogenic wild type SRV2 and srv2Δ strains for cell growth at 25 and 37°C after serial dilution. One mutant (srv2-98) showed highly impaired growth at 37°C, almost as defective as srv2Δ (Fig. 1D). The other three mutant strains showed growth comparable to wild type SRV2.
Figure 1. Mutational analysis of the WH2 domain of Srv2/CAP.
(A) Schematic of Srv2/CAP domain organization and fragments purified for biochemical analysis in this study. Relevant binding partners are shown above each domain. (B) Alignment of P1-WH2-P2 domain sequences from diverse Srv2/CAP homologues using ClustalW. M. mus1, mouse CAP1; M. mus2, mouse CAP2; D. mel., Drosophila melanogaster CAP; A. tha, Arabidopsis thaliana CAP; and S. cer., S. cerevisiae Srv2. Specific residues changed to alanine are marked A for each allele (srv2-96 through srv2-99). A black bar denotes the WH2 domain of S. cerevisiae Srv2 (residues 295-349). Blue bars denote the two proline-rich regions (P1 and P2). (C) Immunoblot of whole cell extracts from wild type SRV2 and srv2 mutant strains probed with anti-Srv2 antibodies. D) SRV2 and srv2 mutant strains were grown to log phase, serially diluted, plated on YPD plates, and grown at 25°C and 37°C.
Interestingly, the defects in cell growth observed for srv2-98 were more severe than any other srv2 alleles generated in previous mutational studies targeting the HFD and β-sheet domains (Mattila et al., 2004; Quintero-Monzon et al., 2009). Thus, srv2-98 has the strongest growth defects of any point mutant introduced into SRV2 to date. This result was somewhat unexpected, because the WH2 domain has not previously been implicated in any study as making a contribution to Srv2/CAP function.
Effects of WH2 domain mutations on cell morphology and actin organization
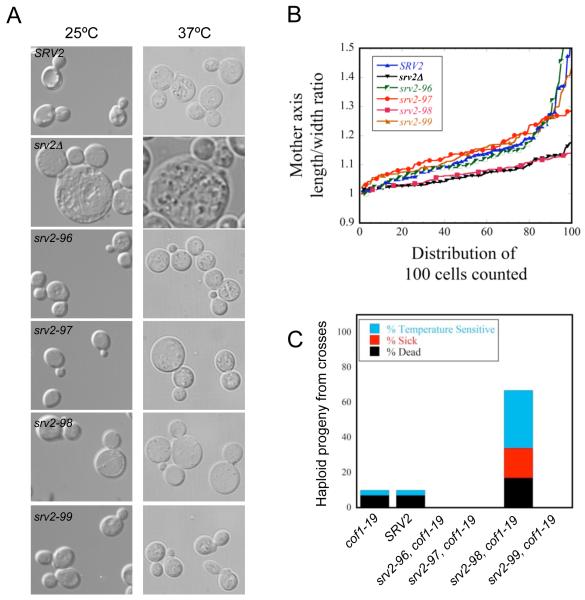
We next examined the morphology of srv2 mutant cells. DIC imaging showed that srv2-98 cells were noticeably larger and rounder than wild-type cells (Fig. 2A), albeit not as large as the srv2Δ cells. Further analysis of length/width axis ratio in mother cell compartments revealed that srv2Δ and srv2-98 mutants had lost their ellipsoid shape (indicated by a decrease in the length/width ratio), which is a strong indication of polarity defects (Fig. 2B). The remaining srv2 mutants grown at 25°C were indistinguishable from wild-type SRV2. However, srv2-97 cells were noticeably larger and rounder when grown at 37°C (Fig. 2A), suggesting that this mutation may cause a partial loss of function.
Figure 2. Cell morphology defects and genetic interactions of srv2 mutants.
(A) DIC imaging of wild type SRV2 and srv2 mutant cells. Cells were grown to mid-log phase at 25°C or 37°C and fixed. (B) Mother cell length/width ratio of wild type SRV2, srv2Δ, and mutant srv2 cells grown at 25°C were determined using CalMorph freeware and averaged (n=100 cells each). (C) Genetic interactions of srv2 alleles with cof1-19. Haploid yeast strains carrying integrated srv2 alleles were crossed to the haploid cof1-19 strain. Diploids were sporulated and tetrads dissected (minimum 20 tetrads, 80 spores), and haploid progeny were compared for cell growth after serial dilution on YPD plates and growth at 25, 30, 34, and 37°C. For each cross, we determined the percentage of haploid progeny compared to a wild type strain that exhibited impaired growth at 37°C (TS), impaired growth at all temperatures (‘Sick’), or were dead at 25°C (‘Dead’).
As an independent test of in vivo function, we crossed each of the srv2 alleles into the cof1-19 background, as this mutation is synthetic lethal with srv2Δ (Balcer et al., 2003). Haploid single and double mutant progeny from these genetic crosses were analyzed for defects in cell growth at a range of temperatures. Only one allele, srv2-98, displayed strong synthetic growth defects with cof1-19 (Fig. 2C), in close agreement with the single mutant analysis above. Taken together, these results demonstrate a dramatic loss of in vivo function for the srv2-98 allele. In this mutation, all four residues of the conserved LKKV motif of the WH2 domain are replaced by alanines. Interestingly, an allele (srv2-101) generated in a previous study targeted only the two lysine residues in the LKKV motif and showed little if any defects in Srv2 cellular or biochemical function (Mattila et al., 2004). Thus, the residues at positions 1 and 4 in the LKKV motif appear to be important for function in vivo. We also note that srv2-96, srv2-97, and srv2-99 appeared to suppress cof1-19; however, presently we do not have a clear understanding or explanation for this effect.
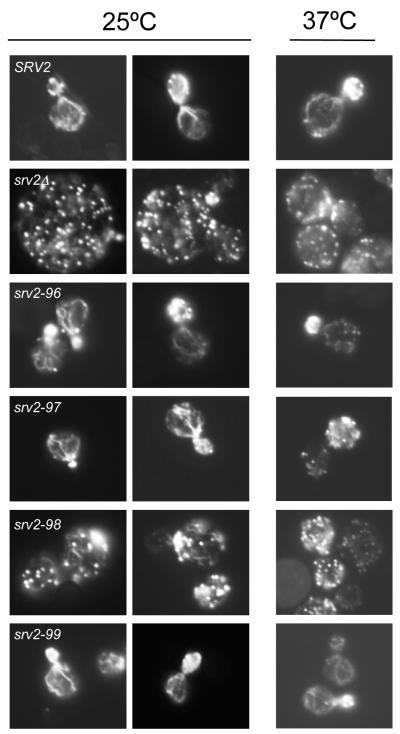
Next, we compared wild type and mutant strains for actin organization after growth at 25°C and 37°C (Fig. 3). Wild type SRV2 cells had polarized actin patches, concentrated in the bud, and actin cables running along the mother-bud axis. srv2-96 and srv2-99 cells resembled wild type cells. However, the majority of srv2-98 cells showed highly depolarized actin patches, distributed throughout mother and daughter cells, and disorganized and partially diminished actin cables. Defects in the srv2Δ strain were similar, but exhibited a more extreme loss of cables. Interestingly, srv2-97 cells grown at 37°C showed a modest defect modest loss of actin patch polarization, consistent with their temperature sensitive morphological defects (Fig. 2A).
Figure 3. Cellular actin organization defects in srv2 mutants.
Wild type SRV2 and mutant strains were grown in YPD medium to log phase at 25°C and 37°C, fixed, and stained with Alexa488-phalloidin to visualize filamentous actin.
In vitro actin binding by purified wild type and mutant Srv2 proteins
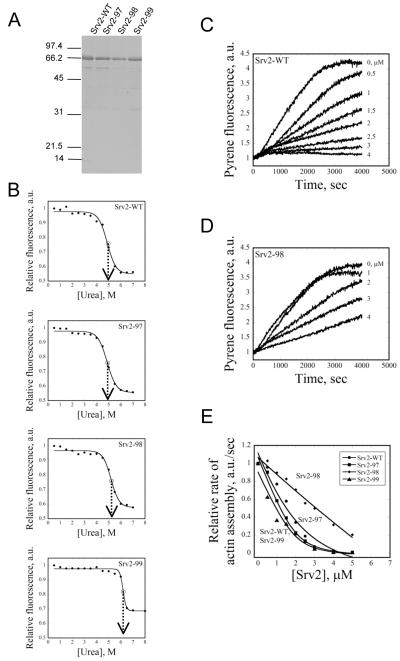
To better understand the mechanistic basis of the phenotypes described above, we purified full-length wild type and mutant Srv2 proteins from E. coli (Fig. 4A) to compare their binding interactions with G-actin. Our analysis included Srv2-97 and Srv2-98, since they showed clear defects in vivo, and Srv2-99 as one pseudo-wild type control. Urea denaturation tests suggested that the purified Srv2-WT, Srv2-97, and Srv2-98 proteins were folded correctly (Fig. 4B). Interestingly, Srv2-99 showed a slight shift in its melting curve in the direction of increased protein stability; the significance of this observation is not yet clear.
Figure 4. G-actin binding activities of wild type and mutant Srv2 proteins.
(A) Coomassie stained gel of full-length wild type and mutant Srv2 proteins. (B) The stability of Srv2 proteins was compared using a fluorescence-monitored urea denaturation assay. The normalized fluorescence is shown on the y-axis and urea concentration on the x-axis. Srv2-WT, Srv2-97, and Srv2-98 unfold at ~5 M urea, while Srv2-99 unfolds at ~6 M urea. (C and D) Monomeric actin (3 μM, 5% pyrene labeled) was polymerized in the presence of different concentrations of wild type and mutant Srv2 proteins (μM indicated by each curve). (E) Rates of inhibition of actin polymerization were plotted against concentration of wild type or mutant Srv2 protein.
To assess G-actin binding, we compared wild type and mutant Srv2 proteins for their ability to suppress the kinetics of spontaneous actin assembly (Fig. 4C). Srv2-WT inhibited spontaneous actin assembly in a concentration-dependent manner, reaching full inhibition at a saturating ratio of Srv2 to G-actin (Fig. 4C). Srv2-98 was noticeably impaired in suppressing actin assembly (Fig. 4D and 4E), Srv2-97 showed only slight defects, and Srv2-99 was similar to Srv2-WT (Fig. 4E). These data are highly consistent with our in vivo analysis above, and indicate that the srv2-98 mutations in the WH2 domain weaken the ability of Srv2/CAP to associate with G-actin.
The WH2 domain of Srv2/CAP binds directly to G-actin
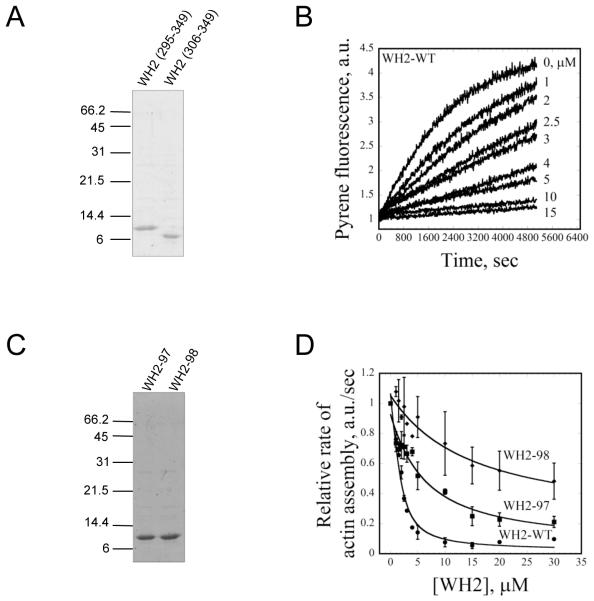
To test directly whether the WH2 domain of Srv2/CAP is capable of binding to G-actin, we purified two different WH2 domain-containing peptides (a.a. 295-349 and 306-349) (Fig. 5A). Neither WH2 peptide showed any detectable binding affinity for F-actin in co-sedimentation assays (data not shown). Since both WH2 peptides interacted with G-actin, we arbitrarily selected the longer peptide (a.a. 295-349) for all further analysis. As an independent test of G-actin binding, we asked whether the WH2 peptide could suppress spontaneous actin assembly similar to full-length Srv2 protein (Fig. 5C). Indeed, the WH2 peptide alone inhibited actin assembly in a concentration-dependent manner (Fig. 5C). Its potency of inhibition was not as strong as that of full-length Srv2-WT, but this was expected since full-length Srv2 contains additional G-actin-binding domains. Although the WH2 domain suppressed spontaneous actin nucleation, it did not alter the steady state critical concentration for actin assembly (actin alone Cc = 0.21; actin + WH2 Cc = 0.26 μM). This was is in contrast to full-length Srv2 protein, which increased critical concentration 10-fold (Cc = 2.4 μM) (data not shown). Thus, the WH2 domain alone is not sufficient to sequester monomers like full-length Srv2, which in turn suggests that sequestering by Srv2 may depend on the combined effects of the WH2 and β-sheet domains.
Figure 5. G-actin binding affinities of wild type and mutant WH2 peptides.
(A) Coomassie stained gel of two WH2 domain peptides (a.a. 295-349 and 306-349). (B) Monomeric actin (3 μM, 5% pyrene labeled) was polymerized in the presence of different concentrations of wild type and mutant WH2 peptides (μM indicated by each curve). (C) Coomassie stained gel of WH2-97 and WH2-98 peptides (a.a. 295-349) (D) Comparison of wild type and mutant WH2 peptide effects on rate of actin polymerization (conditions as in B).
To better understand the defects caused by the srv2-97 and srv2-98 alleles, we purified WH2 peptides (a.a. 295-349) carrying these mutations (Fig. 5D). The WH2-98 peptide showed greatly reduced binding to G-actin compared to wild type WH2 peptide, and the WH2-97 peptide showed intermediate defects (Fig. 5E). These data support the biochemical analyses above using full-length Srv2 proteins.
We next addressed whether the WH2 domain of Srv2/CAP shows binding preference for ATP-actin or ADP-actin. Almost all WH2 domains that have been characterized show much stronger binding affinity for ATP-actin compared to ADP-actin (Chereau et al., 2005). To derive the dissociation constants for Srv2/CAP WH2 peptide interactions with ATP-actin and ADP-actin, we analyzed data from multiple experiments comparing WH2 peptide inhibition of nucleotide exchange as described (Kovar et al., 2001). Our analysis revealed that the WH2 peptide from Srv2/CAP binds with similar affinity to ATP-actin (Kd = 1.4±0.56 μM) (Fig. 6A) and ADP-actin (Kd = 1.5±0.28 μM) (Fig. 6B). In parallel reactions, Cof1 showed much lower affinity for ATP-actin (Kd = 1.8±0.57 μM) (Fig. 6C) compared to ADP-actin (Kd = 0.32±0.17 μM), consistent with its reported binding preference (Blanchoin and Pollard, 1998; Maciver and Weeds, 1994).
Figure 6. WH2 peptide binding affinities for ATP- and ADP-G-actin.
Dissociation constants (Kd shown in each graph) were determined by averaging data from multiple experiments (n=3 each), in which we determined the concentration-dependent effects of WH2 peptides (A and B) or Cof1 (C and D) on rate of nucleotide exchange on ADP-G-actin or ATP-G-actin. Data were fit with a Quadratic Function (see Methods) to derive Kd values.
The WH2 domain is required but not sufficient for recharging actin monomers and accelerating actin turnover in the presence of ADF/cofilin
To investigate the functional contribution of the WH2 domain to Srv2/CAP activity in recharging actin monomers, we measured rate of nucleotide exchange on 2 μM Cof1-bound ADP-actin monomers (Fig. 7). We first directly compared the activities of wild type full-length Srv2 protein and profilin over a range of concentrations (Fig. 7A). Wild type Srv2 increased the rate of nucleotide exchange in a concentration-dependent manner with its effects peaking at 300-400nM Srv2, consistent with previous reports (Quintero-Monzon et al., 2009). At the same concentrations, profilin showed no detectable activity. Next, we directly compared wild type Srv2 and mutant Srv2-97 and Srv2-98 proteins for their effects in this assay (Fig. 7B). In stark contrast to wild type Srv2, Srv2-98 showed almost no activity, and Srv2-97 had impaired activity. These data indicate that WH2 domain interactions with G-actin are essential for the ability of Srv2/CAP to catalyze nucleotide exchange on Cof1-bound ADP-actin monomers.
Figure 7. Role of the WH2 domain in promoting nucleotide exchange on ADF/cofilin-bound ADP-actin monomers.
All assays contained 2 μM ADP-G-actin and 5 μM Cof1, except where noted. (A) Comparison of the effects of full-length wild type Srv2 protein (Srv2-WT) and profilin on rate of nucleotide exchange. (B) Comparison of the effects of Srv2-WT and mutant Srv2-97 and Srv2-98 proteins on rate of nucleotide exchange. (C) Comparison of the effects of different fragments of Srv2 (400 nM) on rate of nucleotide exchange. (D) Comparison of the abilities of wild type and mutant WH2 peptides to restore nucleotide exchange activity to full-length Srv2-98 protein. (E) Effects of different domains of Srv2, alone and in combination (400 nM or 4 μM, as indicated), on the nucleotide exchange rate of Cof1-bound ADP-G-actin. (F) Effects of yeast profilin (5 μM) on the nucleotide exchange rate of ADP-G-actin in the presence and absence of full-length Srv2-WT (400 nM) and/or Cof1 (5 μM).
To dissect Srv2/CAP domain requirements for monomer recharging in the presence of ADF/cofilin, we first compared the activities of different fragments of Srv2/CAP at 400 nM (Fig. 7C). Our analysis showed that no single domain in Srv2/CAP is sufficient for the activity, and that both the N- and C-terminal halves of Srv2/CAP make critical contributions to this function. Given that the WH2 domain is required but not sufficient for this function, we asked whether free WH2 peptide, when combined with full-length mutant Srv2-98 protein, might be capable of restoring activity. Remarkably, the WH2 peptide showed a concentration-dependent ability to function ‘in trans’ with mutant Srv2-98 protein to promote nucleotide exchange on actin (Fig. 7D; circles). Further, mutant WH2-98 peptide showed no activity when combined with Srv2-98 (Fig. 7D; squares). The WH2-WT peptide alone had no effect on nucleotide exchange at any concentrations tested (Fig. 7D; diamonds).
We noted that C-Srv2 is capable of promoting nucleotide exchange on Cof1-bound ADP-G-actin (Fig. 7C), albeit not as effectively as full-length Srv2-WT. Since C-Srv2 contains two actin monomer-binding domains (WH2 and β-sheet), we asked whether the WH2 and β-Srv2 constructs might be capable of functioning in trans to promote nucleotide exchange on actin (Fig. 7E). Neither individual construct alone showed activity at lower (400 nM) or higher (4 μM) concentrations. However, when mixed together at the higher concentrations (4 μM each), WH2 and β-Srv2 constructs promoted nucleotide exchange with a similar ability to 400 nM C-Srv2 (the effects of 4 μM C-Srv2 were too rapid to measure). These data suggest that the WH2 and β-sheet domains are capable of functioning in trans, but require higher concentrations, perhaps due to their actin binding affinities being weaker when the two domains are physically uncoupled. This view is consistent with C-Srv2 having a significantly higher binding affinity for ADP-G-actin compared to the WH2 or β-Srv2 constructs (Mattila et al., 2004; and Fig. 6 in this study). Collectively, these results demonstrate that efficient actin monomer recharging requires both the WH2 and β-sheet domains of Srv2/CAP, and that these domains are capable of functioning together even when physically separated, but are more effective when they are connected.
Since profilin has been hypothesized to promote nucleotide exchange on actin and recharge monomers (see Introduction), we investigated the effects of yeast profilin (5 μM) on the nucleotide exchange rate of Cof1-bound ADP-G-actin, both in the presence and absence of full-length Srv2-WT (400 nM). We found that profilin alone had little if any effect on nucleotide exchange rate, and had no effect on the ability of Srv2-WT to promote nucleotide exchange (Fig. 7F). Thus, profilin does not synergize with Srv2 in this function in vitro. However, these findings do not rule out the possibility that profilin catalyzes nucleotide exchange on ADP-actin monomers in vivo that might be bound to other cellular factors besides Cof1.
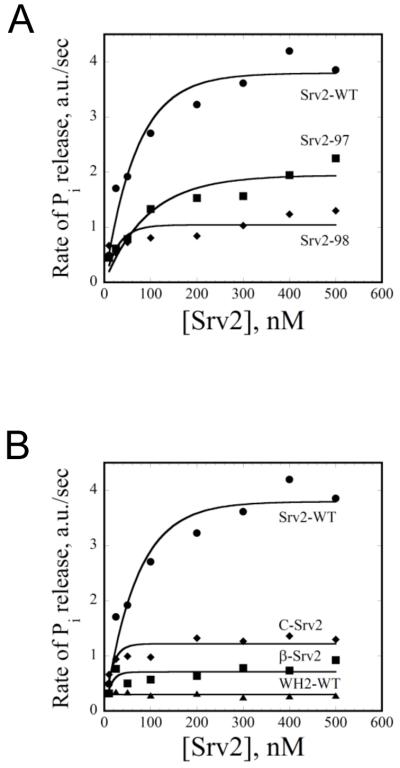
Next, to further assess the contributions of the WH2 domain to actin turnover, we employed an in vitro actin filament turnover assay that measures rate of inorganic phosphate (Pi) release (Wolven et al., 2000). In this assay, actin monomers and polymer are in steady state equilibrium (constant polymer mass), and the rate of ATP hydrolysis and Pi release on actin subunits is directly proportional to rate of filament turnover. Consistent with previous reports, we observed that wild type Srv2 increased the rate of turnover in a concentration-dependent manner in the presence of ADF/cofilin, peaking at a 4-fold rate increase (Fig. 8A, circles) (Quintero-Monzon et al., 2009). In contrast, Srv2-98 had little if any activity, and Srv2-97 showed impaired activity (Fig. 8A, diamonds and squares, respectively). We also dissected Srv2/CAP domain requirements in this assay, testing each fragment of Srv2/CAP for its effects over a range of concentrations (Fig. 8B). As observed for the nucleotide exchange assay, the activity required N- and C-terminal halves of Srv2/CAP, and no single domain was sufficient for function, suggesting that multiple domains in Srv2/CAP function in unison to promote ADF/cofilin-dependent actin turnover. Further, the close correlation between our results in the nucleotide exchange assay (Fig. 7) and the tunover assay (Fig. 8) suggest that the primary mechanistic function of Srv2/CAP in driving turnover is to recharge actin monomers.
Figure 8. Role of the WH2 domain in promoting actin turnover in the presence of ADF/cofilin.
All assays contained 8 μM F-actin and 4 μM Cof1. (A) Comparison of full-length wild type and mutant Srv2 proteins (0-500 nM) effects on rate of steady state actin turnover measured by rate of Pi release. (B) Comparison of effects of different fragments of Srv2 (0-500 nM) on rate of actin turnover.
DISCUSSION
The presence of a WH2 domain sequence in animal and fungal Srv2/CAP homologues has long been recognized (Hubberstey and Mottillo, 2002; Paunola et al., 2001), but its functional relevance has remained uncertain. Here, we have shown that the WH2 domain of yeast Srv2/CAP binds to G-actin, and performs a crucial role in facilitating Srv2/CAP recharging of G-actin in the presence of ADF/cofilin to accelerate actin turnover in vitro and in vivo. We used mutagenesis to disrupt four distinct clusters of conserved residues in the WH2 domain, and identified one mutant (srv2-98) that caused: 1) strong defects in cell growth at elevated temperature similar to an srv2Δ; 2) synthetic lethality with cof1-19 similar to srv2Δ; 3) severe depolarization of the actin cytoskeleton; and 4) corresponding defects in cell morphology (enlarged, rounded cells). Notably, the srv2-98 phenotypes are the strongest observed to date for any point mutant introduced into Srv2/CAP. These data show that the WH2 domain has an indispensable role in Srv2/CAP biochemical and cellular functions.
To explore the mechanistic basis underlying the in vivo observations above, we performed a biochemical analysis of purified wild type and mutant WH2 peptides, as well as purified full-length wild type and mutant Srv2 proteins. In two independent assays, the WH2 peptide bound to actin monomers, and the affinity of the interaction was found to be within a physiologically relevant range (Kd ~ 1.5 μM) (Fig. 6A and B). Further, mutant full-length Srv2-98 protein showed markedly reduced affinity for G-actin. These results resolve a long-standing paradox from an earlier study (Mattila et al., 2004), in which inclusion of the WH2 region increased the affinity of the adjacent β-sheet domain for ADP-G-actin by 20-fold, yet a purified fragment containing the WH2 domain (a.a. 253-373) alone showed no detectable binding to NBD-labeled G-actin. Given our results showing that the WH2 domain binds directly to unlabelled actin (Fig. 5B, and Fig. 6A and B), the negative results from the earlier study may have been due to an inability of the WH2 domain to bind actin covalently modified by NBD, or alternatively, an inability of the WH2 domain to alter NBD-actin fluorescence upon binding.
Our biochemical analysis also shows that the WH2 domain is critical for Srv2/CAP functions in promoting actin turnover. In the presence of ADF/cofilin, full-length mutant Srv2-98 protein showed striking defects in actin monomer recharging (Fig. 7B) and actin turnover (Fig. 8A) compared to wild type Srv2 protein. These observations demonstrate that the WH2 domain has an integral role in enabling Srv2/CAP to catalyze nucleotide exchange on ADF/cofilin-bound actin monomers to promote actin turnover. This function of the WH2 domain is likely to extend to other organisms, as the LKKV motif (disrupted by srv2-98) is conserved in Srv2/CAP homologues from animals, fungi, and plants (Fig. 1B). Together, our data suggest that the ability of Srv2/CAP to catalyze nucleotide exchange on actin is critical in vivo, and that loss of this function leads to drastic defects in cellular actin organization and polarized growth.
Our data also point to interesting similarities and differences in the properties of the WH2 domain of Srv2/CAP compared to the WH2 domains of other proteins. First, we found that the Srv2/CAP WH2 peptide inhibited nucleotide exchange on actin (Fig. 7D), similar to other isolated WH2 domains (Bosch et al., 2007; Chereau et al., 2005; Hertzog et al., 2004). Second, we identified the LKKV motif as crucial for Srv2/CAP WH2 domain interactions with G-actin. This agrees with analyses of other WH2 domains, which show that the analogous LKKT motif is critical for G-actin binding (Carlier et al., 2007; Chereau et al., 2005; Dominguez, 2007; Hertzog et al., 2004; Simenel et al., 2000). In this well-conserved motif, the first amino acid is invariably Leu, the second and third amino acids are usually Lys or Arg, and the fourth amino acid is Thr, Val, Ala, or Ser. Based on WH2-actin co-crystal structures, the most critical residue in the LKKT motif is the invariable Leu, which interacts with the hydrophobic pocket located between actin subdomains I and III (Chereau et al., 2005; Dominguez, 2007). The importance of this Leu may explain why one earlier study (Matilla et al., 2004) found that mutation of the two Lys residues (srv2-101) had little effect on actin binding or Srv2/CAP cellular function. Together, our data suggest that the WH2 domain of Srv2/CAP shares at least some common actin-binding properties with other WH2 domains.
On the other hand, we found that the WH2 domain of Srv2/CAP bound to ATP-actin and ADP-actin with similar affinity (Fig. 6A and B), which differs from the observed binding preference of other WH2 domains for ATP-actin (Chereau et al., 2005). This property may be relevant to the functional role of Srv2/CAP as a ‘middleman’ in actin turnover, in which it must interact sequentially with ADP-actin and ATP-actin, promoting a hand off of actin monomers from ADF/cofilin to profilin. This unique ability to bind equally well to ADP-actin and ATP-actin may stem from sequence differences in the WH2 domain of Srv2/CAP compared to other WH2 domains (Paunola et al., 2001). It may be relevant that we observed reduced actin binding affinity for the Srv2-97 mutant, which targets a short inserted sequence (GENIT) in the WH2 domain of diverse Srv2/CAP homologues (Fig. 1B), but is absent from WH2 domains of other proteins. This raises the intriguing possibility that the GENIT insert increases the ADP-actin binding affinity of the WH2 domain of Srv2/CAP, which will require further investigation.
Our observation that the WH2 domain binds indiscriminately to ADP-actin versus ATP-actin in turn raises a new question. What element(s) in the C-terminal half of Srv2/CAP account for its observed 100-fold higher binding affinity for ADP-actin over ATP-actin (Mattila et al., 2004)? It is now clear that there are two distinct actin-binding sites in Srv2/CAP, the WH2 domain and the β-sheet domain, both located in the C-terminal half of Srv2/CAP. Interestingly, the affinity of the WH2 domain for ADP-actin and ATP-actin is 1.5 μM, similar to the affinity of the entire C-terminal half of Srv2 for ATP-actin (Mattila et al., 2004). These observations predict that the β-sheet domain is primarily responsible for endowing the C-terminus of Srv2/CAP with high affinity for ADP-actin, and indeed, we have observed that β-Srv2 construct binds with much higher affinity to ADP-actin compared to ATP-actin (F.C. and B.G., unpublished data).
Despite the progress in elucidating Srv2/CAP mechanism, made here and in other recent studies (Bertling et al., 2007; Mattila et al., 2004; Quintero-Monzon et al., 2009), there are still many fundamental questions left open. For instance, what are the specific roles of the WH2 domain and β-sheet domain in recharging actin monomers? Do they function to displace ADF/cofilin from ADP-actin? Do they promote nucleotide exchange while ADF/cofilin is still bound to actin? Why does the nucleotide exchange function of Srv2/CAP require both the WH2 and the β-sheet domains, and how do they work together? We have shown that the WH2 domain is crucial for the activity, but not sufficient, and that WH2 peptides alone inhibit rather than promote nucleotide exchange (Fig. 6A and B). One explanation for these observations is that the WH2 and β-sheet domains cooperate, through separate interactions with actin, to induce a conformational change in actin that promotes nucleotide exchange. Perhaps this synergy also explains why the WH2-WT peptide can function in trans to restore monomer-recharging activity to full-length Srv2-98 protein (Fig. 7D), and why the WH2 and β-sheet domains can function in trans in these assays (Fig. 7E). Further mechanistic investigations to understand this conserved and central regulator of cellular actin turnover should help answer these and other open questions.
Finally, while our data demonstrate that Srv2 is very efficient at promoting nucleotide exchange on Cof1-bound ADP-G-actin, we found that profilin has little if any activity in this regard and does not enhance Srv2’s effects (Fig. 7F). These observations call into question whether a normal physiological function of profilin is to promote nucleotide exchange on ADF/cofilin-bound ADP-G-actin. On the other hand, this leaves open the possibility that profilin catalyzes nucleotide exchange on actin monomers bound to other cellular factors besides ADF/cofilin. In contrast to profilin, Srv2/CAP acts potently and catalytically to convert ADF/cofilin-bound ADP-actin monomers into ATP-actin monomers, which would replenish the pool of assembly-competent actin monomers. Srv2/CAP also recycles ADF/cofilin from ADP-actin monomers, which indirectly enhances filament disassembly. Thus, through two complementary effects (increasing nucleotide exchange rate on actin monomers and retrieving ADF/cofilin), Srv2/CAP drives the rapid turnover of actin in vivo. These conserved activities may explain why genetic disruption of Srv2/CAP leads to severe actin phenotypes in a variety of plant, animal, and fungal systems.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmid Construction
Standard methods were used for all DNA manipulations and for growth and transformation of yeast strains (Rose et al., 1989). Mutant srv2 strains were generated as described (Quintero-Monzon et al., 2009). A TRP1-marked SRV2 integration plasmid (pSRV2+) was used as the template for PCR-based site directed mutagenesis to produce integration plasmids for srv2-96, srv2-97, srv2-98, and srv2-99. To integrate the alleles, the plasmids were digested with SacII and transformed into the haploid yeast strain BGY330 (srv2Δ::HIS3). Transformants were selected by growth on Trp- media and loss of growth on His- media. Successful integrations were verified by PCR amplification of the coding region of the SRV2 locus from genomic DNA and diagnostic restriction analysis. For purification of full-length 6xHis-tagged mutant Srv2-96, Srv2-97, Srv2-98, and Srv2-99 proteins, ORFs from the integration plasmids above were subcloned into NcoI and NotI sites of the E. coli expression vector pHAT2. A similar strategy was used to construct plasmids for expressing 6xHis-tagged wild type and mutant WH2 domains. All plasmids were verified by DNA sequencing.
Fluorescent microscopy
To visualize actin organization, yeast cells were grown to log phase in YPD medium, fixed in 2% formaldehyde for 30 min, and stained with Alexa-488-phalloidin (Molecular Probes, Eugene, OR). Images were acquired on a Zeiss E600 microscope (Thornwood, NY) equipped with a Hammamatsu Orca ER CCD camera (Bridgewater, NJ) running Openlab software (Improvision Inc., Waltham, MA). To quantify cell size and mother cell length/width axis ratios, cells were stained with calcoflour Fluorescent brightener 28 (Sigma, St. Louis, MO), then imaged as above and processed using the freeware CalMorph.
Protein purification
Rabbit skeletal muscle actin was purified as previously described (Spudich and Watt, 1971), and converted to ADP-actin (Pollard, 1986). Yeast actin was purified as described (Balcer et al., 2003). Yeast profilin (Pfy1) was purified from E. coli as described (Wolven et al., 2000). Yeast cofilin (Cof1) was purified from E. coli and cleaved from its GST tag as described (Quintero-Monzon et al., 2009). β-srv2 and C-Srv2 polypeptides were purified as described (Mattila et al., 2004). Wild type and mutant full-length 6xHis-tagged Srv2 proteins were expressed in E. coli BL21-RP (DE3) cells and purified as described (Quintero-Monzon et al., 2009) with the following modifications. After the cell lysate was clarified by centrifugation, it was loaded onto a 1 ml Bio-Rad Profinity IMAC resin cartridge using the Profinia™ Protein Purification System (Bio-Rad Laboratories, Hercules, CA, U.S.A.). After loading, the cartridge was washed with Wash Buffer A (300 mM KCl, 50 mM KH2PO4, 5 mM imidizole, pH 8.0), then Wash Buffer B (300 mM KCl, 50 mM KH2PO4, 10 mM imidizole, pH 8.0). 6xHis-tagged protein was eluted with Native IMAC elution buffer (300 mM KCl, 50 mM KH2PO4, 250 mM imidazole, pH 8.0). Srv2 proteins were further purified on a Superose 6 gel-filtration column (AP Biotech, Piscataway, NJ) equilibrated in Buffer C (20 mM Tris pH 8.0, 50 mM NaCl and 1 mM DTT). Peak fractions were concentrated using a Centricon-10 device (Millipore, Billerica, MA), aliquoted, snap-frozen in liquid N2, and stored at −80°C. WH2 peptides were purified as above, except that after elution from the IMAC resin cartridge, they were purified further on a Superdex 75 10/30 gel-filtration column (AP Biotech) equilibrated in Buffer C. Peak fractions were concentrated using a Centricon-3 device (Millipore), aliquoted, snap-frozen in liquid N2, and stored at −80°C.
Inhibition of spontaneous actin assembly assays
To test the ability of purified full-length Srv2 or WH2 domain peptides to suppress spontaneous polymerization of actin monomers, 3 μM gel filtered rabbit muscle actin monomers (5% pyrene-labeled) were mixed with different concentrations of Srv2 proteins and added to 0.05 volume 20x initiation mix. Actin polymerization was monitored over time at 365 nm excitation and 407 nm emission in a Tecan fluorescence multi-well plate reader held at 25°C (Tecan Group Ltd, Männedorf, Switzerland). Rates of actin polymerization were determined from the slopes of the assembly curves.
Nucleotide exchange assays
Rates of nucleotide exchange on monomeric rabbit muscle ATP-actin and ADP-actin were measured by increase in fluorescence upon incorporation of 1,N6-ethenoadenosine 5′-triphosphate (ε-ATP; Sigma) (Goldschmidt-Clermont et al., 1992). The relative activities of wild type and mutant Srv2 proteins observed were similar in assays substituting yeast actin for rabbit muscle actin (Quintero-Monzon et al., 2009); and not shown). Briefly, 2 μM ATP-actin in G-buffer (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.2 mM DTT, 0.2 mM ATP) or 2 μM ADP-actin in G-buffer lacking ATP were mixed with Tris/NaCl buffer (20 mM Tris pH 8.0, 50 mM NaCl) alone or protein mixtures (5 μM Cof1 and/or variable concentrations of Srv2 proteins) in the same buffer, then added to 50 μM ε-ATP (Molecular Probes, Eugene, OR). Reactions were monitored for at least 600 sec at 350 nm excitation and 410 nm emission in a fluorescence spectrophotometer held at 25°C (Photon Technology International; Lawrenceville, NJ). To determine binding affinities, a range of concentrations of WH2 domain and Cof1 were tested for their inhibitory effects on nucleotide exchange rates of ATP-actin and ADP actin. Rates as a function of WH2 or Cof1 concentration were plotted using Kaleidagraph version 4.0 software (Synergy Software, Reading, PA), and the dissociation equilibrium constants (Kd) were calculated using the equation (Kovar et al., 2001):
where ka is the nucleotide exchange rate of free actin, kap is the nucleotide exchange rate of actin bound to ligand (WH2 domain or Cof1), W is the concentration of ligand, and A is the molar concentration of G-actin.
Phosphate release assays
The kinetics of inorganic phosphate (Pi) release during steady state turnover of F-actin mixtures were measured using EnzChek kit (Molecular Probes). 4 μM Cof1 and variable concentrations of Srv2 (0-500 nM) were premixed with polymerization buffer (2 mM MgCl2, 0.5 mM ATP, 50 mM KCl), 0.2 mM MESG (2-amino-mercapto-7 methylpurine ribonucleoside) and 0.1 units of purine nucleoside phosphorilase (PNP). This mixture was added to 8 μM rabbit muscle actin monomers to initiate polymerization. Pi release was monitored at 360 nm in a Tecan fluorescence multi-well plate reader held at 25°C (Tecan Group Ltd). Rates of release were measured at the point where reactions reached steady state. At that point, data were collected for 15 min. After the data were corrected for path length, the slopes at steady state were determined from a linear curve fit.
Miscellaneous
Concentrations of purified Srv2, Cof1, and actin were determined by spectrophotometry using extinction coefficients of ε280nm= 50,100 M−1cm−1 for Srv2, ε280nm=15,930 M−1cm−1 for Cof1, and ε290nm=26,600 M−1cm−1 for actin. Concentrations of mutant Srv2 proteins were determined by comparing band intensities on Coomassie stained gels against a standard curve of wild type Srv2 protein. Western blots were probed sequentially with a 1:5,000 dilution of affinity-purified anti-Srv2 antibody in 1:4 Odyssey Blocking Solution, then a 1:50,000 dilution of goat anti-chicken secondary antibody. Bands were analyzed on a Li-Cor Odyssey Infrared Imaging System (Li-Cor Biosciences, Lincoln, NE). The stability of wild type and mutant full-length Srv2 proteins was measured by fluorescence-monitored urea denaturation as previously described (Lappalainen et al., 1997). Steady state actin critical concentration in the presence and absence of saturating full-length Srv2 or WH2 peptide was measured as described (Brenner and Korn, 1983).
ACKNOWLEDGMENTS
We are extremely grateful to L. Blanchoin for his guidance in processing data to derive Kd values, to C. Gould and other members of the Goode lab for intellectual input and technical assistance throughout the project, and to C. Barber, M. Chesarone, M. Gandhi, E. Jonasson, and A. Rodal for assistance in editing the manuscript. This work was supported by grants from the National Institute of Health (GM63691 and GM083137) to B.G.
REFERENCES
- Balcer HI, Goodman AL, Rodal AA, Smith E, Kugler J, Heuser JE, Goode BL. Coordinated regulation of actin filament turnover by a high-molecular weight Srv2/CAP complex, cofilin, profilin, and Aip1. Current Biology. 2003;13:2159–2169. doi: 10.1016/j.cub.2003.11.051. [DOI] [PubMed] [Google Scholar]
- Bertling E, Hotulainen P, Mattila P, Matilainen T, Salminen M, Lappalainen P. Cyclase-associated protein 1 (CAP1) promotes cofilin-induced actin dynamics in mammalian nonmuscle cells. Molecular Biology of the Cell. 2004;15:2324–2334. doi: 10.1091/mbc.E04-01-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertling E, Quintero-Monzon O, Mattila PK, Goode BL, Lappalainen P. Mechanism and biological role of profilin-Srv2/CAP interaction. Journal of Cell Science. 2007;120:1225–1234. doi: 10.1242/jcs.000158. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem. 1998;273:25106–25111. doi: 10.1074/jbc.273.39.25106. [DOI] [PubMed] [Google Scholar]
- Bosch M, Le K, Bugyi B, Correia J, Renault L, Carlier MF. Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Molecular Cell. 2007;28:555–568. doi: 10.1016/j.molcel.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Brenner S, Korn E. On the mechanism of actin monomer-polymer subunit exchange at steady-state. Journal of Biological Chemistry. 1983;258:5013–2020. [PubMed] [Google Scholar]
- Carlier MF. Role of nucleotide hydrolysis in the dynamics of actin filaments and microtubules. International Review of Cytology. 1989;115:139–170. doi: 10.1016/s0074-7696(08)60629-4. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Hertzog M, Didry D, Renault L, Cantrelle F-X, Van Heijenoort C, Knossow M, Guittet E. Structure, function, and evolution of the β-thymosin/WH2 (WASP-Homology 2) actin-binding module. Annals of the New York Academy of Sciences. 2007;1112:67–75. doi: 10.1196/annals.1415.037. [DOI] [PubMed] [Google Scholar]
- Chaudhry F, Guérin C, von Witsch M, Blanchoin L, Staiger C. Identification of Arabidopsis cyclase-associated protein 1 as the first nucleotide exchange factor for plant actin. Molecular Biology of the Cell. 2007;18:3002–3014. doi: 10.1091/mbc.E06-11-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetma K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proceedings of the National Academy of Science, USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Rodriques C, Dimmock S, Ketelaar T, Maciver SK, Malhó R, Hussey PJ. Arabidopsis CAP1- a key regulator of actin organization and development. Journal of Cell Science. 2007;120:2609–2618. doi: 10.1242/jcs.007302. [DOI] [PubMed] [Google Scholar]
- Dodatko T, AA F, Grynberg M, Patskovsky Y, Rozwarski D, Jaroszewski L, Aronoff-Spencer E, Kondraskina E, Irving T, Godzik A, Almo S. Crystal structure of the actin binding domain of the cyclase-associated protein. Biochemistry. 2004;43:10628–10641. doi: 10.1021/bi049071r. [DOI] [PubMed] [Google Scholar]
- Dominguez R. The β-thymosin/WH2 fold. Annals of the New York Academy of Sciences. 2007;1112:86–94. doi: 10.1196/annals.1415.011. [DOI] [PubMed] [Google Scholar]
- Drees B, Sundin B, Brazeau E, Caviston J, Chen G, Guo W, Kozminski K, Lau M, Moskow J, Tong A, et al. A protein interaction map for cell polarity development. Journal of Cell Biology. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann R, Mann K. ASP-56, a new actin sequestering protein from the pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Letters. 1992;298:149–153. doi: 10.1016/0014-5793(92)80043-g. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont P, Furman M, Wachsstock D, Safer D, Nachmias V, Pollard T. The control of actin nucleotide exchange by thymosin ß4 and profilin. A potential regulatory mechanism for actin polymerization in cells. Molecular Biology of the Cell. 1992;3:1015–1024. doi: 10.1091/mbc.3.9.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont PJ, Machesky LM, Doberstein SK, Pollard TD. Mechanism of the interaction of human platelet profilin with actin. Journal of Cell Biology. 1991;113:1081–1089. doi: 10.1083/jcb.113.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M, Pope B, Maciver SK, Weeds AG. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- Hayden SM, Miller PS, Brauweiler A, Bamburg JR. Analysis of the interaction of actin depolymerizing factor with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier M-F. The β-Thymosin/WH2 domain: structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- Hubberstey AV, Mottillo EP. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. The FASEB Journal. 2002;16:487–499. doi: 10.1096/fj.01-0659rev. [DOI] [PubMed] [Google Scholar]
- Insall R, Machesky L. Actin dynamics at the leading edge: from simple machinery to complex networks. Developmental Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Korn ED, Carlier M-F, Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Yang P, Sale WS, Drobak BK, Staiger CJ. Chlamydomonas reinhardtii produces a profilin with unusual biochemical properties. Journal of Cell Science. 2001;114:4293–4305. doi: 10.1242/jcs.114.23.4293. [DOI] [PubMed] [Google Scholar]
- Lambrechts A, Verschelde JL, Jonckheere V, Goethals M, Vandekerckhove J, Ampe C. The mammalian profilin isoforms display complementary affinities for PIP2 and proline-rich sequences. EMBO J. 1997;16:484–494. doi: 10.1093/emboj/16.3.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen P, Fedorov EV, Fedorov AA, Almo SC, Drubin DG. Essential functions and actin-binding surfaces of yeast cofilin revealed by systematic mutagenesis. EMBO Journal. 1997;16:5520–5530. doi: 10.1093/emboj/16.18.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila T, Drubin D. Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Molecular Biology of the Cell. 1997;8:367–385. doi: 10.1091/mbc.8.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Weeds AG. Actophorin preferentially binds monomeric ADP-actin over ATP-bound actin: consequences for cell locomotion. FEBS Lett. 1994;347:251–256. doi: 10.1016/0014-5793(94)00552-4. [DOI] [PubMed] [Google Scholar]
- Mattila PK, Quintero-Monzon O, Kugler J, Moseley JB, Almo SC, Lappalainen P, Goode BL. A high affinity interaction with ADP-actin monomers underlies the mechanism and in vivo function of Srv2/cyclase-associated protein (CAP) Molecular Biology of the Cell. 2004;15:5158–5171. doi: 10.1091/mbc.E04-06-0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mockrin SC, Korn ED. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1980;19:5359–5362. doi: 10.1021/bi00564a033. [DOI] [PubMed] [Google Scholar]
- Moriyama K, Yahara I. Human CAP1 is a key factor in the recycling of cofilin and actin for rapid actin turnover. Journal of Cell Science. 2002;115:1591–1601. doi: 10.1242/jcs.115.8.1591. [DOI] [PubMed] [Google Scholar]
- Nishida E. Opposite effects of cofilin and profilin from porcine brain on rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry. 1985;24:1160–1164. doi: 10.1021/bi00326a015. [DOI] [PubMed] [Google Scholar]
- Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. International Review of Cytology. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- Paunola E, Mattila P, Lappalainen P. WH2 domain: a small, versatile adapter for actin monomers. FEBS Letters. 2001;513:92–97. doi: 10.1016/s0014-5793(01)03242-2. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. Journal of Cell Biology. 1986;103:2747–2754. doi: 10.1083/jcb.103.6.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero-Monzon O, Jonasson EM, Bertling E, Talarico L, Chaudhry F, Sihvo M, Lappalainen P, Goode BL. Reconstitution and dissection of the 600 kdA SRV2/CAP complex: Roles for oligomerization and cofilin-actin binding in driving actin turnover. Journal of Biological Chemistry. 2009;284:10923–10934. doi: 10.1074/jbc.M808760200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. p. 198. [Google Scholar]
- Simenel C, Van Troys M, Vanderkerckhove J, Ampe C, Delepierre M. Structural requirements for thymosin beta4 in its contact with actin. An NMR-analysis of thymosin beta4 mutants in solution and correlation with their biological activity. European Journal of Biochemistry. 2000;267:3530–3538. doi: 10.1046/j.1432-1327.2000.01380.x. [DOI] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. Journal of Biological Chemistry. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Vinson VK, Delacruz EM, Higgs HN, Pollard TD. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry. 1998;37:10871–10880. doi: 10.1021/bi980093l. [DOI] [PubMed] [Google Scholar]
- Vojtek A, Haarer B, Field J, Gerst J, Pollard TD, Brown S, Wigler M. Evidence for a functional link between profilin and CAP in the yeast S. cerevisiae. Cell. 1991;66:497–505. doi: 10.1016/0092-8674(81)90013-1. [DOI] [PubMed] [Google Scholar]
- Wang J, Suzuki N, Kataoka T. The 70-kilodalton adenyl cyclase-associated protein is not essential for interaction of Saccharomyces cerevisiae adenyl cyclase with RAS proteins. Molecular and Cellular Biology. 1992;12:4937–4945. doi: 10.1128/mcb.12.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven A, Belmont L, Mahoney N, Almo S, Drubin D. In vivo importance of actin nucleotide exchange catalyzed by profilin. Journal of Cell Biology. 2000;150:895–903. doi: 10.1083/jcb.150.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusof A, Hu N-J, Wlodawer A, Hofmann A. Structural evidence for variable oligomerization of the N-terminal domain of cyclase-associated protein (CAP) Proteins: Structure, Function, and Bioinformatics. 2005;58:255–262. doi: 10.1002/prot.20314. [DOI] [PubMed] [Google Scholar]