Abstract
Immunotherapy by cocaine-binding monoclonal antibodies (mAbs) has emerged as a promising strategy for the treatment of cocaine addiction. The human (γ1 heavy chain)/murine (λ light chain) chimeric mAb 2E2 has excellent affinity and specificity for cocaine and recent animal studies have demonstrated 2E2’s ability in vivo to reduce cocaine levels in the brain as well as alter cocaine self-administration behavior in rats. In this study, we used mAb 2E2 amino acid sequence information to create a homology model for the 3-D structure of its Fv fragment. Subsequent computational docking studies revealed the intermolecular interactions potentially responsible for mAb 2E2’s cocaine binding properties. The driving force of cocaine binding was identified as a combination of hydrophobic interactions and a single hydrogen bond between a light chain tyrosine residue and a carbonyl oxygen atom of cocaine. The model also allowed for an in silico evaluation of single/double residue mutations in the heavy and light chain variable regions that might further enhance mAb 2E2’s cocaine binding properties.
Keywords: homology modeling, drug addiction, immunotherapy, molecular docking, antibody, cocaine
1. Introduction
Cocaine abuse has been and remains a serious public health and societal/economic problem in the U.S. and many other countries. Currently available estimates from National Survey on Drug Use and Health reports [1–3] indicate that in the U.S. over 2 million Americans are addicts or heavy users of cocaine with 800,000–900,000 individuals entering treatment at least once in a given year. It is important to note that current treatments of cocaine abuse largely consist of behavioral modification, use of antidepressants, and other indirect approaches [4]. Decades of research have established cocaine’s role as an indirect agonist, raising dopamine levels by blocking the dopamine transporters in defined regions of the brain. While this pharmacologic action contributes to cocaine’s addictive properties, no pharmacological intervention, either by an agonist or an antagonist, has been shown to be an effective intervention treatment [5, 6]. Currently, no pharmacological therapeutics are approved for cocaine abuse treatment.
An alternative approach to targeting neurotransmitter receptors, which is fraught with the potential for untoward central nervous system (CNS)1 side-effects, is the use of anti-cocaine polyclonal or monoclonal antibodies (mAbs) with high affinity and selectivity for cocaine and its active metabolites (e.g. cocaethylene and norcocaine) but not its inactive metabolites (benzoylecgonine, benzoylnorecgonine, ecgoninemethylester and ecgonine). Mechanistically, this immunotherapeutic approach works through the antibody’s sequestering of cocaine in the peripheral circulatory system, preventing it from crossing the blood-brain barrier and being psychoactive, yet allowing naturally occurring cholinesterases to rapidly convert unbound cocaine to its inactive metabolites [7–9]. The anti-cocaine antibodies can be either generated in vivo by a cocaine vaccine consisting of an immunogenic hapten-carrier conjugate or administered through passive immunization with a selected/manufactured “humanized” mAb. Recent clinical trials have demonstrated the safety and potential of a vaccine to generate levels of cocaine-directed polyclonal antibodies capable of lowering the use of cocaine in a subset of vaccinated drug abusers [8, 10] as well as an anti-nicotine vaccine for smoking cessation intervention [11]. As a complementary approach to vaccination, our laboratory has generated a partially human sequence anti-cocaine mAb, designated as mAb 2E2 (a human γ1 heavy (H) chain and a murine λ light (L) chain) that was elicited against the hapten benzoylecgonine coupled to 1,4-butanediamine-derivatived keyhole limpet hemocyanin (KLH). mAb 2E2 has been shown to have a high affinity (~ 4 nM) and specificity for cocaine, norcocaine, and cocaethylene over that of inactive cocaine metabolites [12]. mAb 2E2’s high affinity for cocaethylene is fortuitous since this metabolite is an active derivative that is formed when alcohol is ingested while taking cocaine. More recently, this mAb has been determined to have dramatic in vivo efficacy in mice, raising plasma concentrations of cocaine 10- to 20-fold above control levels while decreasing brain levels of cocaine without altering cocaine’s rate of elimination or metabolism to inactive products [13]. Further, in recent studies of rat self-administration of cocaine, a model of drug abuse, mAb 2E2 has been demonstrated to have significant effects on the levels of cocaine required to re-initiate drug administering behavior in rats trained to self-administer cocaine [14]. Therefore, given that the expected in vivo elimination rate t1/2 value for human IgG1 is approximately 30 days, mAb 2E2 should have the physicochemical properties that may be expected to confer relatively long-term efficacy as a passive immunotherapeutic agent, especially as compared to the short-term action of low molecular weight drugs.
In this study, we aimed to answer the underlying question of how mAb 2E2’s high affinity and specificity for cocaine over inactive metabolites is achieved on the molecular level, given the limitations imposed by the small size of the benzoylecgonine amide (~ 300 Da) that served as the immunizing antigen. The study started with the generation of a homology model of the Fv region (variable region) based on the known sequences of mAb 2E2, which, as a “chimeric” mAb, is comprised of the human γ1 H and the murine λ L chain. By computationally docking cocaine and its metabolites into the model, their intermolecular interactions with mAb 2E2 could be identified. The accuracy of the computational approach was assessed by a comparison of the results with the findings of an earlier 3D quantitative structure-activity relationship model (3D-QSAR) that correlated the structural properties of cocaine and analogues with their experimentally determined binding affinities via comparative molecular similarity index analysis (CoMSIA) [12]. The modeling presented here was also undertaken to reveal possible amino acid mutations in the H and L chain variable region fragments that might improve 2E2’s cocaine binding specificity or be required to be retained in order to maintain its affinity should re-engineering its light chain to generate a more fully human sequence mAb be required. Finally, the model provided a means of investigating how mAb 2E2’s binding of cocaine may differ from that of other cocaine binding and/or catalytic mAbs whose Fab fragment crystal structures have been determined. A comparison of the cocaine binding modes employed by these different mAbs allowed a critical test of Pozharski’s earlier supposition that even for a very small antigen, highly specific recognition by an antibody can be achieved in a variety of ways [15].
2. Results and Discussion
2.1. Quality assessment of a three-dimensional homology model for the Fv region of mAb 2E2
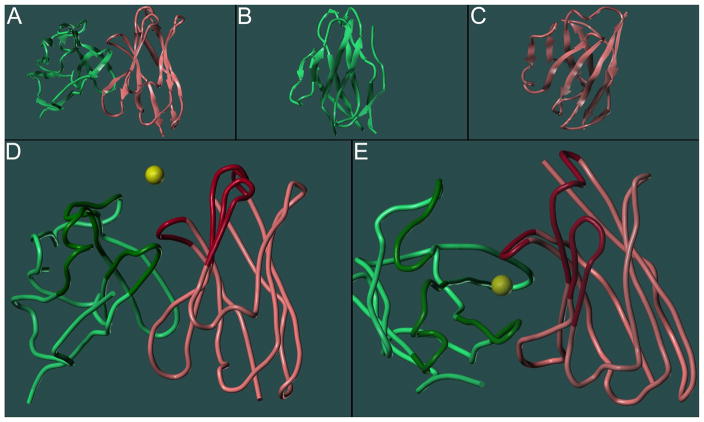
After the determination of the amino acid sequences of both chains of mAb 2E2 (Fig. 1), a structural model of the Fv domain (for a schematic showing the various domains of a mAb, see Supplementary Materials) of the antibody was developed using the antibody modeling software WAM (Web Antibody Modelling). Visual inspection revealed that the homology model was in good agreement with the characteristic immunoglobulin fold adopted by antibody Fv regions, which is composed exclusively of anti-parallel β-sheets connected by loops, including those that form the CDRs [16, 17] (Fig. 2A–C). The secondary structure of the light chain, for example, folded into the typical double sandwich structure composed of three- and four-stranded anti-parallel β-sheets, respectively (Fig. 2B). As revealed by a backbone alignment of the model with three anti-cocaine antibodies with known crystal structures (Supplementary Materials), the model’s contained all the expected characteristic structural features observed in Fv regions of immunoglobulins.
Figure 1.
Sequence of the mouse light and human heavy chain of the chimera mAb 2E2 according to the WAM numbering system. The CDRs as defined by WAM are shown in bold letters.
Figure 2.
Model-predicted immunoglobulin fold of the Fv region of mAb 2E2. β-sheets are depicted as arrows and the CDR regions are located on the top (except for E). A: both chains. B: light chain only (green). C: heavy chain only (red). Close-up views of the position of the docking sphere center in relation to the CDRs of the mAb 2E2 model. D: side view. E: top view. The six CDRs are displayed in darker colors and the yellow sphere represents the center of the docking sphere.
For a more rigorous evaluation, the fully prepared 2E2 Fv model was submitted to the MolProbity website for creation of Ramachandran (ϕ,Ψ) plots [18] (Supplementary Materials) and rotamer analysis. Ramachandran analysis showed that the backbone torsion angles of 200 residues (90.9%) resided in favored regions. Fourteen residues (6.4%) were in allowed regions and six residues (2.7%) were found in the outlier regions of the plot. Of the six outliers, three were part of the complementarity determining regions (CDRs) - Thr32(L3), Glu211(H3) and Pro214(H3). Among these three residues, only Glu211 was predicted to be involved in direct interactions with cocaine and its analogues. As anticipated for a homology model, the overall percentage of outliers was slightly larger than that observed for most high resolution Fv X-ray crystal or NMR structures. Nevertheless, there are a number of cases in which homology models of antibodies proved to be sufficiently accurate for an analysis of ligand binding on the molecular level. Examples include antibodies with affinity for cardiac glycosides [19], biaryl ligands [20], the Fas ligand protein [21], and others.
Rotamer analysis of the side chain conformations showed that 159 of 177 rotamers displayed torsion angles commonly observed in protein structures. Among the remaining 18 (10.2%) rotamers, eight were part of the CDRs: Thr31(L1), Thr32(L1), Thr54(L2), Trp94(L3), Thr97(L3), Tyr99(L3), Asp144(H1), and Trp145(H1). Only two - Trp94(L3) and Trp145(H1) - of these eight residues were predicted to be involved in cocaine/mAb interactions.
2.2. Development and validation of a molecular docking protocol
For the study of the intermolecular interactions responsible for mAb 2E2’s affinity and specificity for cocaine, the modeled structures of cocaine and its metabolites were computationally docked into the homology model using GOLD (Genetic Optimisation for Ligand Docking), a powerful docking tool that operates with a genetic search algorithm and allows for full conformational flexibility of the ligand and partial flexibility of the binding site. As outlined in “Materials and Methods”, the most suitable choice of the user-defined docking parameters (choice between ChemScore or GoldScore fitness functions as well as selection of the docking sphere center) was determined by systematic variation of these parameters and observing their effects on the success of the docking results. The success of a docking experiment was judged by its reproducibility, which was measured by the number of repeats that generated essentially indistinguishable docking solutions, called a consensus. Based on this approach, ChemScore [22] was the chosen scoring function and the docking sphere had a radius of 15 Å centered at the location specified in Table 1. As expected, the docking sphere center was surrounded by the six CDRs of mAb 2E2 (Fig. 2D–E).
Table 1.
Location of docking sphere center as defined by its distance to four residues of the mAb 2E2 model.
| atom | distance/Å |
|---|---|
| Tyr34 (L) backbone nitrogen | 16.68 |
| Thr97 (L) backbone nitrogen | 10.85 |
| Ser33 (L) backbone nitrogen | 14.21 |
| Leu212 (H) backbone nitrogen | 17.85 |
Next, the docking protocol was employed to dock modeled structures of cocaine analogues into the homology model. Based largely on their physiological significance, the analogues were grouped into two sets. The “focused set” was comprised of cocaine as well as its metabolites cocaethylene, norcocaine, benzoylecgonine, benzoylnorecgonine, ecgonine, and ecgoninemethylester. A larger set of 31 compounds, designated the “extended set”, contained the “focused set” plus a series of 24 cocaine analogues that were not cocaine metabolites but whose affinity to mAb 2E2 had been measured in an earlier QSAR study [12]. The purpose of the “extended set” was to provide a structurally diverse set of compounds to be used for a thorough test of the predictive capabilities of the docking protocol.
When docking the “focused set” into the homology model, a consensus orientation was obtained for each molecule. The majority of the metabolites were predicted to bind in a similar pose, meaning that most of their shared structural features overlapped (Fig. 3A). The only exceptions were the two smaller and low affinity metabolites ecgonine and ecgoninemethylester, whose tropane rings displayed a minor tilt compared to the orientation of the larger compounds (Fig. 3B).
Figure 3.
Binding poses obtained for the “focused set” of cocaine metabolites (top view, cocaine is displayed in yellow). For clarity, the left panel contains the larger metabolites whereas the right panel shows the smaller compounds ecgonine and ecgoninemethylester alone. The surface of the antibody model is colored according to hydrophobicity (brown: more hydrophobic; green: less hydrophobic).
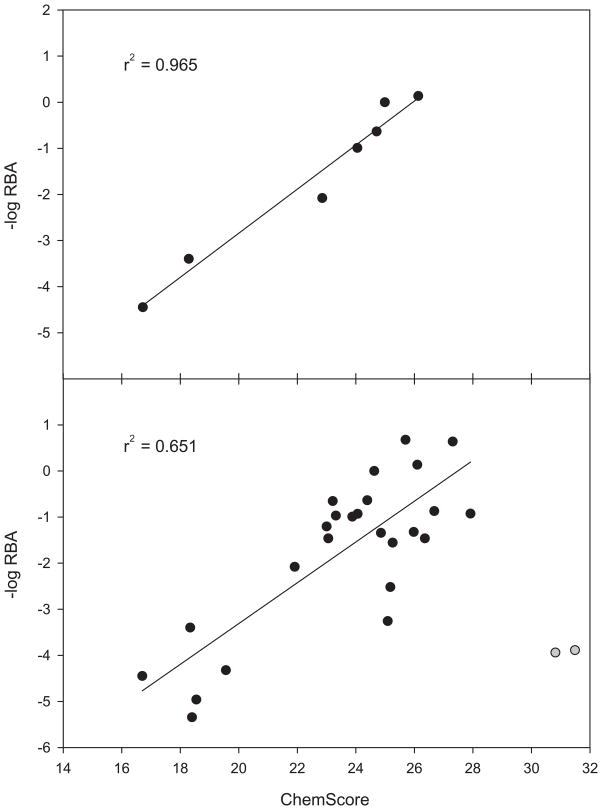
An additional test of the docking protocol’s predictive powers was a comparison of experimentally determined binding affinities to the obtained docking scores. Since the ChemScore scoring function was developed by regression analysis against a set of experimentally determined binding affinities of ligand/protein complexes [22], the ChemScore values of docked cocaine analogues ought to correlate with their previously determined binding affinities to mAb 2E2 (Supplementary Materials) [12]. As illustrated in Fig. 4A, such a correlation was clearly present for cocaine and its immediate metabolites. It should be noted that separate docking runs were executed with the tropane nitrogen in both a protonated and deprotonated state, as either form might exist at physiological pH. Although slightly higher for the deprotonated form, the correlation coefficients did not differ significantly (r2 = 0.96 versus r2 = 0.91) and were therefore not a suitable criterion for the determination of the protonation state of the ligands in the binding pocket.
Figure 4.
ChemScore values compared to experimentally observed relative binding affinities (RBA). Upper panel: “focused set”, lower panel: “extended set”. The two outliers that were excluded from linear regression are shown as grey circles.
To further test the robustness of the protocol model, the “extended” set of cocaine analogues was docked into the homology model. Consensus orientations were obtained for 27 of the 31 compounds (Supplementary Materials). The initially obtained poor correlation coefficient (r2 = 0.19) was significantly improved (r2 = 0.64; Fig. 4B) by the omission of the two outliers benzatropine and 4-4-difluoro-3α-diphenylmethoxytropane, both of which were low affinity binders (~8,000-fold lower than cocaine). A common structural feature of the two outliers was the presence of two instead of one phenyl group, which greatly increased their hydrophobic surface area in comparison to the other compounds. Fig. 4B shows that ChemScore greatly overestimated the binding affinities of these two compounds, an observation that agrees with the previously noted tendency of ChemScore to overrate the contribution of hydrophobic contacts to the binding energy. For one of the outliers, the cause of the binding affinity/ChemScore mismatch could also have originated from the presence of a fluorine atom. In ligand/protein complexes, fluorine is not a frequently encountered element and it might therefore not have been sufficiently accounted for during the calibration of ChemScore [22]. However, the overall correlation obtained between the scoring function and binding affinities was strong and suggested a general accuracy and predictiveness of the developed docking protocol.
2.3. Molecular interactions between cocaine analogues and mAb 2E2
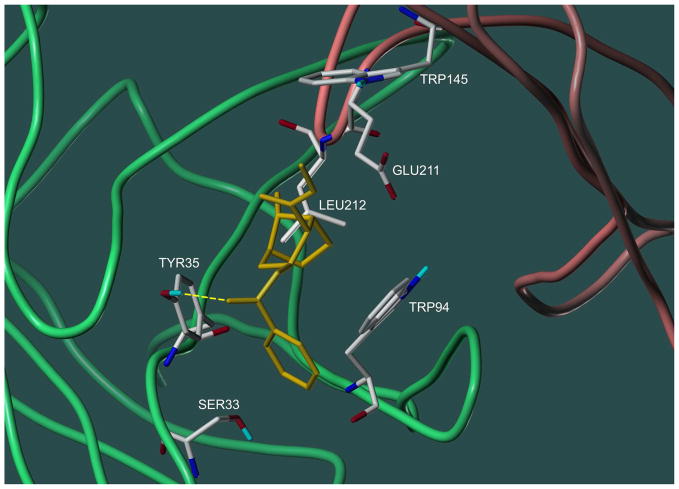
The discussion of the intermolecular interactions responsible for ligand binding by mAb 2E2 will be restricted to cocaine and the six metabolites contained in the “focused set” because of their physiological importance for immunotherapy and the good correlation between their binding affinities and ChemScore docking values. An inspection of the GOLD-generated output identified hydrophobic interactions and a single hydrogen bond as the primary forces responsible for cocaine binding to mAb 2E2. The hydrogen bond involved the carbonyl oxygen atom of the ester bond between ecgonine and the benzoyl group and the hydroxyl group of Tyr35, which was part of the CDR L1 (Fig. 5). Without exception, this hydrogen bond was predicted for all other cocaine metabolites as well. The bulk of the ligand binding energy, however, was provided by hydrophobic interactions that occurred between non-polar parts of the ligand and hydrophobic sections of the L chain amino acids Ser33(L1), Try35(L1), and Trp94(L3) and the H chain amino acids Trp145(H1), Glu211(H3), and Leu212(H3) (Fig. 5). Therefore, the binding specificity of mAb 2E2 for larger ligands such as cocaine, cocaethylene, and norcocaine could be attributed to the large size of their hydrophobic surface areas. Smaller compounds such as ecgoninemethylester or ecgonine were not bound as well by the mAb because of their lack of a large hydrophobic area, which would also explain the slightly altered binding pose for some of them (Fig. 3B). Other contributions to the overall docking score of the metabolites were either similar for all compounds and did therefore not contribute to binding specificity (loss in conformational entropy upon binding) or were negligible (steric clash penalties). Interestingly, cocaine’s amino group, which had been postulated previously to be involved in interactions with the binding site [12], was not predicted to engage in any binding interactions–an observation in agreement with its modeled partially solvent-exposed position.
Figure 5.
Docking predicted orientation of cocaine (yellow) in the binding site of mAb 2E2 (top view). Amino acid residues engaged in interactions with the ligand are displayed as capped sticks. The backbones of the light and heavy chains are shown in green and red, respectively.
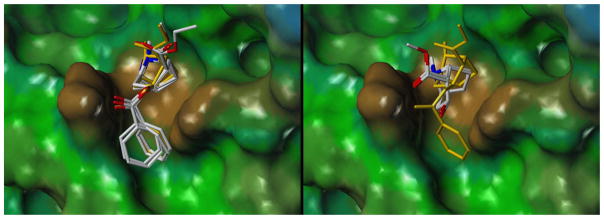
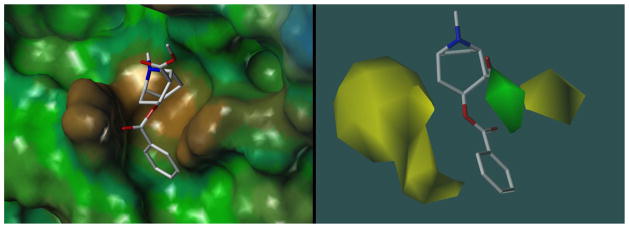
The overall shape of the modeled cocaine binding site and the position of cocaine in mAb 2E2 was consistent with the aforementioned CoMSIA model [12]. The tropane ring of cocaine was buried facing towards the back of the binding cavity whereas the ester group of the benzoyl moiety was solvent-exposed and engaged in a hydrogen bond with Tyr35(L). Fig. 6 compares the surface of the antibody model to the CoMSIA steric contour plot. The binding site is defined by a cavity lined by two protruding bulges formed by Ser33(L) and Tyr35(L) and Trp145(H) that coincide with the sterically restricted areas of the contour plot. Furthermore, the sterically favored region (green) represents unoccupied space in which the ethyl and propyl groups of cocaethylene and cocainepropylester can be favorably accommodated.
Figure 6.
Side-by-side comparison of cocaine docked into the mAb 2E2 model (left panel, hydrophobic surface) and the CoMSIA steric contour plot for cocaine binding (right panel, sterically restricted yellow areas reduce binding affinity, green areas favor binding). The CoMSIA model was generated as described previously [12].
2.4. Predicted effects of point mutations on cocaine binding to mAb 2E2
To be effective for immunotherapy, an antibody should have a clear binding preference for cocaine and any major psychoactive metabolites over major inactive ones, in particular, in humans, benzoylecgonine. While mAb 2E2 has an approximate 10-fold specificity for cocaine over that of benzoylecgonine, benzoylecgonine in humans has a t1/2 of elimination that is about 6-times slower than that for cocaine and, as a result, its plasma concentrations can easily exceed those of cocaine by several fold [23]. Thus, increasing mAb 2E2’s specificity may prove important. In addition to its specificity, the antibody should also exhibit a good overall binding affinity. Whereas extremely tight antigen binding (Kd values in nanomolar range) may not be a requirement for pharmacological efficacy [13, 23, 24], it could allow antibody treatment levels to be maintained at minimum concentrations. Typical cocaine blood concentrations after smoking or injection have been reported to be as low as in the submicromolar range [24], implying that only a mAb with a Kd in the nanomolar range would be fully saturated under these conditions. Minimizing the dose of mAb could reduce the likelihood for undesired side effects and adverse events–in the case of 2E2 with its murine light chain–this is somewhat of a concern.
In an effort to guide future modifications and potential improvements of mAb 2E2 by genetic engineering, we introduced several mutations into the homology model of the 2E2 Fv and studied their effects on binding properties in silico. Computational mAb modeling using the online modeling software WAM in conjunction with genetic engineering of antibodies has been employed successfully in several cases in the past, both in the absence of [25, 26] and with known crystal structures [27].
Due to their modeled proximity to the bound cocaine ligand, residues Ser33, Asn34, Tyr35, Asn37, and Trp94, and Tyr99 on the light chain and residues Trp145, Glu211, Leu212 on the heavy chain were subjected to mutation. A total of 57 single point mutations were modeled, corresponding to about six mutations per amino acid. Each one of the targeted residues was replaced by a randomly selected representative from the following amino acid classes: basic, acidic, small polar, large polar, small non-polar, and large non-polar. A second mutation was introduced in those mutants in which the first mutation had been predicted to enhance the binding properties, yielding a total of 36 double mutants. Subsequently, the “focused” subset containing cocaine and its metabolites was docked into all 93 mutant structures.
For the quantification of a mutant’s binding properties, two metrics were defined. Using the wild-type antibody as a reference, specificity was considered to be the relative difference in the ChemScore value of the psychoactive compound (cocaine, norcocaine, and cocaethylene) with the lowest predicted affinity and the ChemScore value of the non-psychoactive metabolite (all others) with the highest affinity. In other words, the specificity parameter numerically defined the gap between the predicted affinities of the psychoactive and non-psychoactive compounds and an improvement in specificity - an increase in the size of the gap - was reflected in a positive numerical value for this parameter. Second, the change in overall binding affinity for cocaine was defined as the difference in the ChemScore values between a given mutant and the wild-type model. Consequently, an improvement in affinity for cocaine resulted in a positive value for this parameter (Table 2).
Table 2.
Effect of mutations on binding properties of mAb 2E2. See text for definition of metrics, which are derived from ChemScore values expressed in kJ/mol.
| Mutation | specificity change | cocaine affinity change |
|---|---|---|
| wild-type | 0.0 | 0.0 |
| Glu211Ala | −0.2 | 1.6 |
| Leu212Asp | 0.1 | −1.8 |
| Tyr99Glu/Leu212Asp | 0.3 | −2.2 |
| Tyr99Ser/Leu212Ser | 0.3 | −2.2 |
| Tyr99Glu | 0.6 | −0.3 |
Analysis of all 93 mutations showed that none increased both specificity and affinity simultaneously. However, some mutants displayed increased affinity at the expense of decreased specificity or vice versa. Based on the quality of the docking results (existence of a consensus for all docked compounds) and the obtained predictions of binding properties, five mutant structures were considered further (Table 2). Among them, Glu211(H)Ala was the only one that had a significantly increased affinity for cocaine, primarily caused by an altered binding pose that strengthened hydrogen bonding. However, the gain in cocaine affinity was accompanied by a slight increase in the affinity for the inactive compounds, thereby reducing specificity. Among the remaining four mutants were two single and two double mutants, all of which had a predicted increase in specificity accompanied by an overall drop in cocaine affinity. For the mutant Tyr99(L)Glu, the gain in specificity was most pronounced whereas the loss in affinity was only minor. Overall, the two mutants Glu211(H)Ala and Tyr99(L)Glu appear to be the most promising starting points for future genetic engineering work aimed at a further improvement of the binding properties of mAb 2E2 for immunotherapy of cocaine addiction.
2.5. Comparison of mAb 2E2’s cocaine binding mode to that of other mAbs
An interesting aspect of molecular recognition relates to the question of whether protein receptors that recognize and bind the same ligand accomplish this task by utilizing similar or fundamentally different binding strategies, as defined by the type relevant intermolecular interactions and the pose of the ligand in the binding site. In the case of cocaine, a comparison of the binding modes observed for different anti-cocaine mAbs can reveal whether the employed binding modes share similarities that might be necessary for high affinity and specificity for cocaine, or whether each mAbs is unique in this regard.
The availability of several high resolution X-ray crystal structures of Fabs or entire antibodies in complex with cocaine permitted a direct comparison of their binding modes and a search for common strategies that might be required for effective cocaine binding. Examples for cocaine-binding Fabs with known crystal structures include the anti-cocaine murine/human chimeric Fab fragment of mAb GNC92H2 [28] and the murine mAb M82G2 [15]. Both mAbs have significantly lower cocaine affinities than mAb 2E2 with reported Kd values of ~ 400 nM and ~ 140 μM, respectively. Furthermore, the structure of cocaine in complex with mAb 7A1 fragment–a cocaine-hydrolyzing murine mAb with a KM value of ~ 2 mM–has been reported [29]. One binding feature shared by all mAbs related to the dominance of hydrophobic interactions and the role of Trp and Tyr residues for cocaine binding. After superimposition of the mAbs protein backbones, however, a comparison of the cocaine molecules relative positions revealed no general similarities with regard to their position in relation to the antibody scaffold (see Supplementary Materials). For example, cocaine binds mAb 2E2 with the benzoyl group extended and oriented towards the L chain loops while it has the opposite orientation when binding to mAbs M82G2 and 7A1. mAb GNC92H2 forms a deep binding pocket that buries the tropane ring but leaves the benzoyl group solvent exposed without any contact residues. Overall, other than significant cocaine/light chain interactions, we were unable to identify a specific common feature amongst these four binding modes that would suggest a shared ligand structural conformation or orientation of cocaine in the binding site. Likewise, no specific drug/mAb topology that might be required for the generation of a small molecule binding site could be observed. Finally, no specific differences between the modeling results for mAb 2E2 and the crystallographic data for the other three mAbs were recognized that would have accounted for the unusually high cocaine affinity of mAb 2E2 compared to the other mAbs. In conclusion, these findings seem to support the general hypothesis by Pozharski and coworkers [15] that highly specific recognition of a small antigen by an antibody can be accomplished in a variety of ways.
4. Conclusions
In this study, we have demonstrated the value of homology modeling and computational docking methodologies for the study of antibody/antigen interactions in mAb 2E2, a chimeric human/mouse mAb of considerable medicinal interest because of its potential for immunotherapy of cocaine addiction and relapse. The developed models account accurately for the experimentally determined cocaine binding properties of mAb 2E2 and can provide guidance for future modifications of the antibody by genetic engineering. Since the modeling methodologies employed in our study are fast, user-friendly, and readily available, they should be of general value for a broad variety of other antibody/antigen systems with known antibody amino acid sequences.
5. Experimental and Computational Protocols
5.1. Sequencing of the variable regions of mAb 2E2
The hybridoma cell line secreting anti-cocaine mAb 2E2 was generated using standard hybridoma technology by fusing splenocytes obtained from a human antibody producing transgenic mouse, strain HCo7/Ko5 [30], immunized with a hapten (benzoylecgonine)-derivatized carrier protein KLH conjugate as described previously [12]. After separation of purified mAb 2E2’s H and L chains by polyacrylamide gel electrophoresis, significant portions of the amino acid sequences were determined by liquid chromatography (LC)/tandem mass spectrometry (MS) analysis of their tryptic fragments by MALDI-time-of flight (TOF) mass spectrometry. Sequences were obtained both by direct sequence matching of the LC/MS results and by calculating theoretical peptide fragment masses as per initial cDNA sequencing of mAb 2E2’s variable H and L regions and matching these to MALDI-TOF fragments. The H chain was identified as a γ1 protein of the human VH3 family gene DP-50 whereas the L chain was a murine type λ V-IIB. Subsequently, Vybion Inc. (Ithaca, NY) generated cDNAs from the cloned full-length H and L chain mRNAs and confirmed the MS results through nucleotide sequencing, corrected and filled in a few missing sequences, and determined the leader sequences of the two immunoglobulin chains. Confirmation of the H and L cloning was achieved by transiently co-transfecting human embryonic kidney cells (HEK293T) with expression vector constructs that separately contained the full length H and L chain cDNAs and growing the cells for 24–48 hr post transfection. Culture media samples were collected which tested positive in competition binding enzyme-linked immunosorbent assays (ELISA) for production of an mAb with a high affinity and specificity for cocaine binding that was indistinguishable from that of the hybridoma cell produced mAb 2E2 (unpublished results).
5.2. Development of a three-dimensional model for the Fv region of mAb 2E2
Molecular modeling of the three-dimensional structure of the Fv region of mAb 2E2 was performed using the online modeling software WAM [19]. The largely automated modeling process starts with sequence alignment by AUTOALIGN, which detects certain key residues in the proximity of the CDRs in the template sequence and estimates the bounds of the CDRs from those. After sequence alignment of the mAb 2E2 murine λ light and human γ1 heavy chain Fv domains, the backbone and the side chains of the framework sections (light and heavy chains are treated separately) were built using corresponding parts of antibodies with the largest sequence homology as templates. Next, the CDRs of the target antibody sequence were classified into canonical and noncanonical classes. The CDRs from the L2, H1, and H2 chain domains were classified as canonical whereas the remaining three CDRs (L1, L3 and H3) could not be assigned to any group of frequently occurring conformations. Interestingly, the H3 loop was comprised of only four residues. For canonical loops, the coordinates of backbone atoms and side chains of conserved amino acids were assigned using as templates loops (within the same canonical class) that possessed the largest degree of homology. The backbones of noncanonical loops were built by a combination search (CAMAL), first completing a PDB database search and then a conformational search with CONGEN [31]. CONGEN is an iterative algorithm that sequentially builds each side chain, starting from the lowest energy conformation of a given residue and then iteratively adjusting each conformation taking into account the changes in the new environment. In the last step, several conformations of the H3 loop were modeled and a root-mean square deviation screen determined the most likely conformation of the target sequence.
Atom coordinates for the model were downloaded from the WAM website and converted into PDB files using the program DeepView, version 4.0 [32]. The following manipulations of the model structure were performed with SYBYL 8.0 (Tripos, St. Louis, MO): hydrogen atoms were added, end groups were fixed using the charged option, and charges were assigned to the models according to the Amber7FF99 library. While keeping the heavy atoms static, the positions of the added hydrogen atoms were subjected to energy minimization using the Powell method, the Amber7FF99 force field with the previously loaded charges, and a distance dependent dielectric constant of 4. Minimization was terminated when the energy gradient dropped below 0.05 kcal/(mol Å). For validation and quality assessment, the generated model was submitted to the online tool MolProbity at http://molprobity.biochem.duke.edu [33, 34] for the creation of Ramachandran plots and rotamer analysis. Mutations of selected amino acids were performed with the “Mutate Monomer” tool in the “Composition” menu of SYBYL’s Biopolymer module by replacing a targeted residue by another amino acid and then conducting a local energy minimization (using the same parameters as above) that was restricted to the conformation of the new residue’s side chain.
For structural comparisons, the coordinates of the mAb 2E2 model and of the X-ray crystal structures of the cocaine-binding mAbs GNC92H2 (PDB code: 1I7Z) and M82G2 (PDB code: 1Q72) were imported into SYBYL. The alignment of the crystal structures to the model was performed with the “Fit Monomers” option in SYBYL’s “Biopolymer” module. The fit was completed using the backbone atoms of amino acids that were not part of the CDR loops, as defined by WAM for 2E2 and as specified by Pozharski et al. for mAbs GNC92H2 and M82G2 [15].
5.3. Molecular modeling of cocaine, metabolites, and analogues
The structures of cocaine and its analogues were modeled in SYBYL by building their known structure with the “Sketch Molecule” function. To assure that docking of each individual compound commenced from comparable starting points, the conformational energy of each molecule was minimized prior to docking. For minimization, the conjugate gradient method with a termination criterion of 0.01 kcal/(mol Å) and the MMFF94s force field with MMFF94 charges were used in conjunction with a distance dependent dielectric constant of 4.
5.4. Molecular docking of cocaine analogues into the binding region of the mAb 2E2 model
Docking of cocaine and its analogues was carried out with GOLD 4.0 (CCDC, Cambridge, UK) [35, 36]. GOLD operates with a genetic search algorithm to find the energetically most favorable orientation of a small molecule in a protein binding site. During the search, GOLD allows for complete ligand flexibility and permits terminal carboxyl and amino groups of amino acids to rotate in order to engage in hydrogen bonds. Optimum docking conditions were determined by systematic variation of the user-defined GOLD parameters such as the choice of scoring function (GoldScore versus ChemScore) and the location of the docking sphere center (radius of 15 Å). Since antigens are known to be in close contact with the CDRs or a mAb, initial docking was conducted by placing the sphere’s center at a point that was approximately equidistant from all six CDRs. The center was then systematically moved in 5Å increments in all six directions of the coordinate system and docking was repeated for each new location. The center location giving the largest consensus for cocaine was then used as the starting point for a second search cycle in 3Å increments. 30 independent docking repeats were performed for each compound and all other settings were kept at their default values. For docking results to be considered reproducible and thus usable for further analysis, a requirement for a majority consensus was implemented. A majority consensus was defined as a clustering of at least half of the docking repeats within a RMSD range of 2.0 Å or below.
Supplementary Material
Acknowledgments
This work was supported a summer student fellowship to M.L. from the Greaves foundation at Northern Kentucky University, a grant from the Kentucky Biomedical Research Infrastructure Network (P20RR016481–08), and a grant from the National Institute on Drug Abuse (DA018538, P.I. Dr. Andrew Norman). We thank Christopher Elam for help in the preparation of the mutant models for mAb 2E2. We gratefully acknowledge the mAb 2E2 heavy and light chain cloning and cDNA sequencing work done at Vybion, Inc. (Ithaca, NY) under the direction of Drs. Lee Henderson (CEO), John Noti, and Wally Fish.
Footnotes
Abbreviations: mAb: monoclonal antibody; CNS: central nervous system; H: heavy chain; L: light chain; KLH: keyhole limpet hemocyanin; QSAR: quantitative structure-activity relationship; CoMSIA: comparative molecular similarity index analysis; Fab: fragment, antigen-binding; Fv: fragment, variable; CDR: complementarity determining region; GOLD: genetic optimization for ligand docking; LC: liquid chromatography; MS: mass spectrometry; TOF: time of flight; HEK: human embryonic kidney; ELISA: enzyme-linked immunosorbent assay; WAM: Web Antibody Modeling; RMSD: root-mean square deviation
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.U.S. Dept. H.H.S. The NSDUH Report. Cocaine Use: 2002 and 2003. National Survey on Drug Use and Health. 2005 [Google Scholar]
- 2.U.S. Dept. H.H.S. Trends in Cocaine Treatment Admissions, by State: 1992–2002. The DASIS Report. 2005 [Google Scholar]
- 3.U.S. Dept. H.H.S. Results from 2008 National Survey on Drug Use and Health: Detailed Tables. Substance Abuse and Mental Health Services Administration (SAMHAS); 2008. [Google Scholar]
- 4.Elkashef A, Biswas J, Acri JB, Vocci F. Biotechnology and the treatment of addictive disorders: new opportunities. BioDrugs. 2007;21:259–267. doi: 10.2165/00063030-200721040-00006. [DOI] [PubMed] [Google Scholar]
- 5.Nestler EJ. Historical review: Molecular and cellular mechanisms of opiate and cocaine addiction. Trends in pharmacological sciences. 2004;25:210–218. doi: 10.1016/j.tips.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Preti A. New developments in the pharmacotherapy of cocaine abuse. Addiction biology. 2007;12:133–151. doi: 10.1111/j.1369-1600.2007.00061.x. [DOI] [PubMed] [Google Scholar]
- 7.Kantak KM. Vaccines against drugs of abuse: a viable treatment option? Drugs. 2003;63:341–352. doi: 10.2165/00003495-200363040-00001. [DOI] [PubMed] [Google Scholar]
- 8.Kosten TR, Rosen M, Bond J, Settles M, Roberts JS, Shields J, Jack L, Fox B. Human therapeutic cocaine vaccine: safety and immunogenicity. Vaccine. 2002;20:1196–1204. doi: 10.1016/s0264-410x(01)00425-x. [DOI] [PubMed] [Google Scholar]
- 9.Moreno AY, Janda KD. Immunopharmacotherapy: vaccination strategies as a treatment for drug abuse and dependence. Pharmacology, biochemistry, and behavior. 2009;92:199–205. doi: 10.1016/j.pbb.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martell BA, Mitchell E, Poling J, Gonsai K, Kosten TR. Vaccine pharmacotherapy for the treatment of cocaine dependence. Biological psychiatry. 2005;58:158–164. doi: 10.1016/j.biopsych.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 11.LeSage MG, Keyler DE, Pentel PR. Current status of immunologic approaches to treating tobacco dependence: vaccines and nicotine-specific antibodies. The AAPS journal. 2006;8:E65–75. doi: 10.1208/aapsj080108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ., Jr Three-dimensional quantitative structure-activity relationship modeling of cocaine binding by a novel human monoclonal antibody. Journal of medicinal chemistry. 2004;47:133–142. doi: 10.1021/jm030351z. [DOI] [PubMed] [Google Scholar]
- 13.Norman AB, Tabet MR, Norman MK, Buesing WR, Pesce AJ, Ball WJ. A chimeric human/murine anticocaine monoclonal antibody inhibits the distribution of cocaine to the brain in mice. The Journal of pharmacology and experimental therapeutics. 2007;320:145–153. doi: 10.1124/jpet.106.111781. [DOI] [PubMed] [Google Scholar]
- 14.Norman AB, Norman MK, Buesing WR, Tabet MR, Tsibulsky VL, Ball WJ. The effect of a chimeric human/murine anti-cocaine monoclonal antibody on cocaine self-administration in rats. The Journal of pharmacology and experimental therapeutics. 2009;328:873–881. doi: 10.1124/jpet.108.146407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozharski E, Moulin A, Hewagama A, Shanafelt AB, Petsko GA, Ringe D. Diversity in hapten recognition: structural study of an anti-cocaine antibody M82G2. Journal of molecular biology. 2005;349:570–582. doi: 10.1016/j.jmb.2005.03.080. [DOI] [PubMed] [Google Scholar]
- 16.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. Journal of molecular biology. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 17.Schiffer M, Girling RL, Ely KR, Edmundson AB. Structure of a lambda-type Bence-Jones protein at 3.5-A resolution. Biochemistry. 1973;12:4620–4631. doi: 10.1021/bi00747a013. [DOI] [PubMed] [Google Scholar]
- 18.Ramachandran GN, Sasisekharan V. Conformation of polypeptides and proteins. Advances in protein chemistry. 1968;23:283–438. doi: 10.1016/s0065-3233(08)60402-7. [DOI] [PubMed] [Google Scholar]
- 19.Paula S, Monson N, Ball WJ., Jr Molecular modeling of cardiac glycoside binding by the human sequence monoclonal antibody 1B3. Proteins. 2005;60:382–391. doi: 10.1002/prot.20484. [DOI] [PubMed] [Google Scholar]
- 20.Rasmussen BS, Pedersen JM, Sorensen J, Egebjerg J, Schiott B, Mortensen KK, Skrydstrup T. Enantioselective proteins: selection, binding studies and molecular modeling of antibodies with affinity towards hydrophobic BINOL derivatives. Chembiochem. 2007;8:1974–1980. doi: 10.1002/cbic.200700295. [DOI] [PubMed] [Google Scholar]
- 21.Obungu VH, Gelfanova V, Rathnachalam R, Bailey A, Sloan-Lancaster J, Huang L. Determination of the mechanism of action of anti-FasL antibody by epitope mapping and homology modeling. Biochemistry. 2009;48:7251–7260. doi: 10.1021/bi900296g. [DOI] [PubMed] [Google Scholar]
- 22.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J Comput Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 23.Karch SB. The Pathology of Drug Abuse. 2. CRC Press; 1996. [Google Scholar]
- 24.Jenkins AJ, Oyler JM, Cone EJ. Comparison of heroin and cocaine concentrations in saliva with concentrations in blood and plasma. Journal of analytical toxicology. 1995;19:359–374. doi: 10.1093/jat/19.6.359. [DOI] [PubMed] [Google Scholar]
- 25.Kamei H, Shimazaki K, Nishi Y. Computational 3-D modeling and site-directed mutation of an antibody that binds Neu2en5Ac, a transition state analogue of a sialic acid. Proteins. 2001;45:285–296. doi: 10.1002/prot.1149. [DOI] [PubMed] [Google Scholar]
- 26.Sivasubramanian A, Chao G, Pressler HM, Wittrup KD, Gray JJ. Structural model of the mAb 806-EGFR complex using computational docking followed by computational and experimental mutagenesis. Structure. 2006;14:401–414. doi: 10.1016/j.str.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Sivasubramanian A, Maynard JA, Gray JJ. Modeling the structure of mAb 14B7 bound to the anthrax protective antigen. Proteins. 2008;70:218–230. doi: 10.1002/prot.21595. [DOI] [PubMed] [Google Scholar]
- 28.Larsen NA, Zhou B, Heine A, Wirsching P, Janda KD, Wilson IA. Crystal structure of a cocaine-binding antibody. Journal of molecular biology. 2001;311:9–15. doi: 10.1006/jmbi.2001.4839. [DOI] [PubMed] [Google Scholar]
- 29.Zhu X, Dickerson TJ, Rogers CJ, Kaufmann GF, Mee JM, McKenzie KM, Janda KD, Wilson IA. Complete reaction cycle of a cocaine catalytic antibody at atomic resolution. Structure. 2006;14:205–216. doi: 10.1016/j.str.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Fishwild DM, O’Donnell SL, Bengoechea T, Hudson DV, Harding F, Bernhard SL, Jones D, Kay RM, Higgins KM, Schramm SR, Lonberg N. High-avidity human IgG kappa monoclonal antibodies from a novel strain of minilocus transgenic mice. Nature biotechnology. 1996;14:845–851. doi: 10.1038/nbt0796-845. [DOI] [PubMed] [Google Scholar]
- 31.Bruccoleri RE, Karplus M. Prediction of the folding of short polypeptide segments by uniform conformational sampling. Biopolymers. 1987;26:137–168. doi: 10.1002/bip.360260114. [DOI] [PubMed] [Google Scholar]
- 32.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 33.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 34.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic acids research. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 36.Cole JWMNJC, Taylor R. Protein-Ligand Docking and Virtual Screening with GOLD. In: Shoichet JAB, editor. Virtual Screening in Drug Discovery. CRC Press; Boca Raton, Florida, USA: 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.