SUMMARY
TopBP1 has important roles in both DNA replication and checkpoint regulation in vertebrates. We have identified a protein called Treslin that associates with TopBP1 in Xenopus egg extracts. Depletion of Treslin from egg extracts strongly inhibits chromosomal DNA replication. Binding of Treslin to chromatin in egg extracts occurs independently of TopBP1. However, loading of the initiator protein Cdc45 onto chromatin cannot take place in the absence of Treslin. Prior to the initiation of DNA replication, Treslin associates with TopBP1 in a Cdk2-dependent manner. Ablation of Treslin from human cells also strongly inhibits DNA replication. Taken together, these results indicate that Treslin and TopBP1 collaborate in the Cdk2-mediated loading of Cdc45 onto replication origins. Thus, Treslin regulates a pivotal step in the initiation of DNA replication in vertebrates.
INTRODUCTION
In eukaryotic cells, the duplication of the genome involves the strictly ordered assembly of various proteins onto regions of DNA that are destined to serve as origins of replication (Bell and Dutta, 2002; Méndez and Stillman, 2003; Sclafani and Holzen, 2007). First, the Origin Recognition Complex (ORC) associates with prospective sites for the initiation of DNA synthesis. Thereupon, ORC functions essentially as a platform for the binding of Cdc6 and Cdt1. These two proteins subsequently facilitate the loading of the MCM complex onto the DNA to form the prereplication complex (pre-RC) (Diffley, 2004). The MCM proteins are key components of the replicative helicase that unwinds DNA around the origins to create a template for the DNA polymerases (see Pacek and Walter, 2004).
The initiation of DNA replication involves the binding of additional proteins to origins as well as regulation by two conserved kinases. Currently, this process is best understood in budding yeast (Sclafani and Holzen, 2007). Besides the components of the pre-RC, other proteins such as Dpb11, Sld2, Sld3, Mcm10, the GINS complex, and Cdc45 also associate with DNA replication origins. Concomitantly, phosphorylation by the Dbf4-dependent (DDK) and cyclin-dependent kinases (CDK) promotes formation of the preinitiation complex (pre-IC) (Jares and Blow, 2000; Mimura and Takisawa, 1998; Pacek and Walter, 2004; Tercero et al., 2000; Zou and Stillman, 1998). A hallmark of this transformation is the functional incorporation of Cdc45 into the pre-IC. These steps ultimately result in the manifestation of replicative helicase activity.
It has been crucial to understand how these kinases regulate the proteins that carry out DNA replication. In budding yeast, the role of S-phase CDK activity (S-CDK) in controlling the initiation of replication is now understood in some detail (Botchan, 2007; Tanaka et al., 2007a; Tanaka et al., 2007b; Zegerman and Diffley, 2007). It has been shown that Sld2 and Sld3 serve as the minimal CDK targets in the replicative machinery whose phosphorylation is necessary for DNA synthesis. These regulatory steps involve the docking of CDK-phosphorylated forms of Sld2 and Sld3 onto Dpb11. Dpb11 and its homologues in other species contain multiple pairs of BRCT repeats, which form polypeptide domains that recognize phosphopeptide targets (Caldecott, 2003; Garcia et al., 2005). Sld2 and Sld3 latch onto distinct pairs of BRCT repeats within Dpb11. Hence, Dpb11 appears to be acting, at least in part, as a scaffolding protein that helps to position other replication proteins for initiation. For example, these associations are necessary for the functional integration of Cdc45 into the replication-initiating apparatus. Dpb11 and its homologue in fission yeast (Cut5) also play a crucial role in checkpoint responses to damaged DNA (Garcia et al., 2005).
Our understanding of the initiation of replication in vertebrates is less advanced, in part because Sld2 and Sld3 have not been strictly conserved in these organisms. RecQ4 has been proposed as a vertebrate homologue of Sld2, but this protein is quite different from Sld2 in several respects (Matsuno et al., 2006; Sangrithi et al., 2005). Moreover, there has not been a good candidate for a vertebrate form of Sld3. In vertebrates, the functional analogue of Dpb11 is a protein called TopBP1 (Garcia et al., 2005). TopBP1 is a larger and more complex protein that contains eight BRCT repeats. Nonetheless, like its counterparts in yeast, TopBP1 is necessary for both initiation of DNA replication and checkpoint control. In the case of DNA replication, TopBP1 is necessary for the loading of Cdc45 onto replication origins (Hashimoto and Takisawa, 2003; Van Hatten et al., 2002). During checkpoint responses, TopBP1 serves as a direct activator of the ATR-ATRIP complex (Kumagai et al., 2006; Mordes et al., 2008). However, the precise role that TopBP1 fulfills in DNA replication has been unclear.
In this report, we have identified a TopBP1-interacting protein, named Treslin, that is necessary for DNA replication in Xenopus egg extracts. In further analyses, we found that Treslin acts in collaboration with TopBP1 at an early step to facilitate the loading of Cdc45 onto chromatin. We have also established that Treslin possesses a similar function in human cells. Treslin bears no readily discernible sequence homology with yeast Sld2 and Sld3, but it does share certain characteristics with these proteins. In addition to the fact that Treslin associates with the vertebrate homologue of Dpb11, this interaction in egg extracts depends upon Cdk2, the principal S-CDK in this system. This dependency may help to explain how S-CDK activity triggers the initiation of DNA replication in vertebrates. Overall, these studies provide insights into early crucial events that lead to faithful replication of the genome in vertebrates, including humans.
RESULTS
Identification of Treslin
We sought to understand how TopBP1 controls the initiation of DNA replication. Toward this end, we carried out pull-down experiments to search for proteins that might interact with TopBP1 at replication forks in Xenopus egg extracts. In these experiments, we utilized a FLAG-tagged, truncated version of TopBP1 that contains BRCT domains I-VI. We chose this construct because it lacks a key region required for activation of ATR, but nonetheless contains all of the regions necessary to support DNA replication (Hashimoto et al., 2006; Kumagai et al., 2006; Yan et al., 2006). We incubated this fragment in lysates of replicating nuclei from Xenopus egg extracts. These lysates were obtained from nuclei that had been extracted with 300 mM NaCl. This salt concentration is high enough to dislodge all of the TopBP1 from replicating chromatin (data not shown), but low enough that it might plausibly not perturb the binding of TopBP1 to at least some interacting factors.
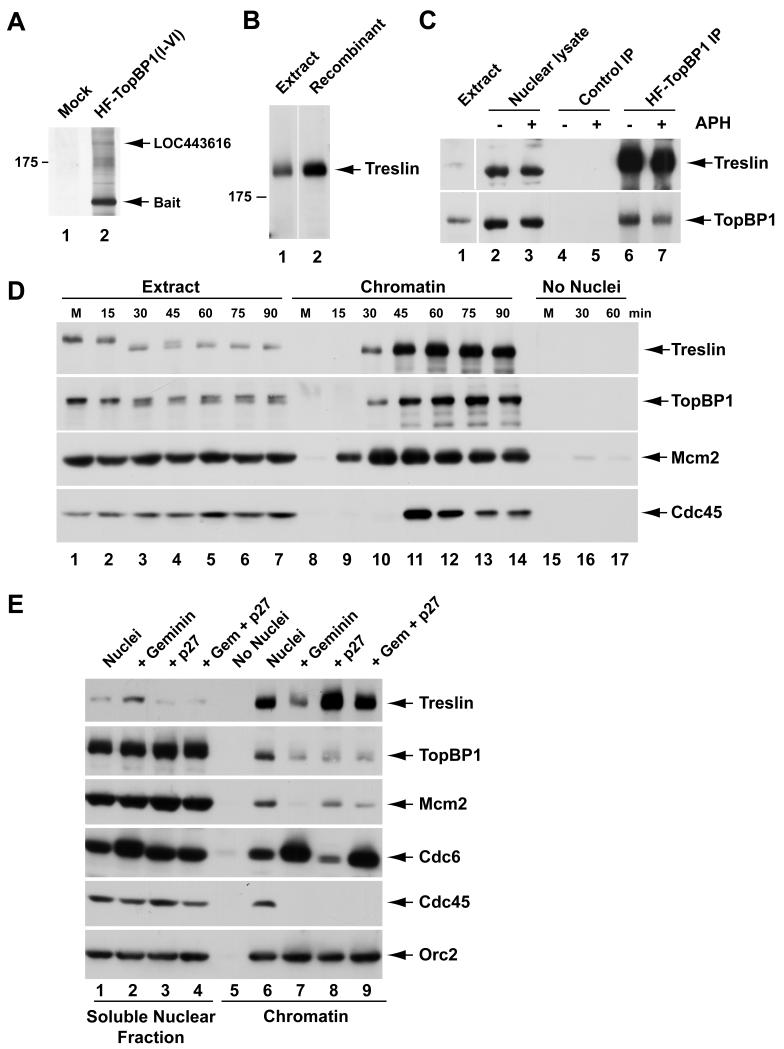
As shown in Figure 1A, we identified a large protein with a molecular mass of approximately 220 kD that associated specifically with the BRCT I-VI fragment of TopBP1. This band contained sequences from a polypeptide (LOC443616) that represents a portion of a Xenopus hypothetical protein of unknown function (see Experimental Procedures). We also identified in the database a full-length version of a human homologue of this protein that is encoded by a functionally uncharacterized open reading frame on chromosome 15 (C15orf42). With this information, we were able to obtain the remaining sequence of the Xenopus protein (see Experimental Procedures). This putative full-length Xenopus cDNA encodes a 1,986-amino acid protein with 42% identity and 50% similarity to the human protein (Table S1). We decided to name this protein Treslin (TopBP1-interacting, replication-stimulating protein) for reasons that are justified below. Treslin does not possess any sequence motifs that would indicate its function. However, one significant feature of the protein is that it contains 67 (Ser/Thr)-Pro motifs that comprise the core of the consensus site for CDK-catalyzed phosphorylation. Among these sites, 35 also contain either an arginine or a lysine two positions downstream of the proline. This feature is characteristic of an optimal site for phosphorylation by a CDK such as Cdk2.
Figure 1. Identification of Treslin as a TopBP1-interacting Protein in Xenopus Egg Extracts.
(A) Nuclear lysates were incubated without (lane 1) or with HF-TopBP1(I-VI) (lane 2) in the presence of anti-FLAG antibody beads. Beads were retrieved and processed for silver staining.
(B) Interphase egg extract (lane 1) and recombinant Xenopus Treslin from insect cells (lane 2) were immunoblotted with anti-Xenopus Treslin antibodies. Lanes are aligned from the same gel.
(C) Nuclear lysates from egg extracts lacking (lanes 2, 4, and 6) or containing aphidicolin (APH) (lanes 3, 5, and 7) were incubated with anti-FLAG beads containing no recombinant protein (lanes 4 and 5) or full-length HF-TopBP1 (lanes 6 and 7). Beads were reisolated and immunoblotted for Treslin and TopBP1. Lane 1 shows an aliquot of initial egg extract.
(D) M-phase extracts (M) from unactivated eggs were left unactivated (lanes 1, 8, and 15) or activated with calcium (lanes 2-7, 9-14, and 16-17). Extracts were incubated with (lanes 1-14) or without sperm nuclei (lanes 15-17). Chromatin fractions from activated extracts were isolated at indicated times. Chromatin from M-phase extracts was incubated for 30 min (lanes 8 and 15). Mock chromatin fractions were isolated from extracts lacking sperm nuclei (lanes 15-17) at indicated times. Samples were immunoblotted for indicated proteins.
(E) Egg extracts were incubated in the presence (lanes 1-4 and 6-9) or absence of sperm nuclei (lane 5) with no inhibitor (lanes 1, 5, and 6), geminin (lanes 2 and 7), p27 (lanes 3 and 8), or both geminin and p27 (lanes 4 and 9). Soluble nuclear fractions (lanes 1-4) and chromatin fractions (lanes 5-9) were immunoblotted for indicated proteins. See also Table S1.
In general, Treslin appears to be well conserved in vertebrates, including zebrafish (Table S1). We could also find limited stretches of homology with proteins from animals as distant in evolution as the lower marine organisms Ciona intestinalis and Trichoplax adhaerens. However, we could not find any obvious sequence similarity with proteins in budding or fission yeast, Caenorhabditis elegans, and Drosophila melanogaster.
In order to validate the specificity of the interaction between TopBP1 and Treslin, we performed various immunoprecipitation experiments with Xenopus egg extracts. First, we prepared polyclonal antibodies that recognize the endogenous Treslin in egg extracts (Figure 1B). Next, we added a FLAG-tagged version of full-length TopBP1 to interphase extracts that lacked or contained aphidicolin, a DNA polymerase inhibitor that elicits the formation of stalled DNA replication forks. Finally, we prepared nuclear extracts from the incubations and reisolated the tagged TopBP1 with anti-FLAG antibody beads. We could readily detect binding of endogenous Treslin to beads containing the tagged TopBP1 but not to control beads (Figure 1C). Blockage of replication with aphidicolin appeared not to affect the interaction between Treslin and TopBP1. Finally, we were unable to detect association between Treslin and TopBP1 under any conditions in whole egg extracts lacking nuclei (data not shown). These observations suggest that Treslin and TopBP1 interact only in the nucleus.
Treslin Associates with Replicating Chromatin
To characterize the basic properties of Treslin, we examined its binding to chromatin in comparison with proteins that act at discrete stages of DNA replication. For this purpose, we added demembranated Xenopus sperm nuclei to M-phase egg extracts, added calcium to activate the extracts, and then examined the binding of Treslin and other proteins to chromatin as a function of time. Under these conditions, DNA replication typically commences at approximately 45 min after addition of calcium. By this time, intact nuclear envelopes have formed around the chromatin, which enables the accumulation of various key replication proteins in the nuclei.
We observed that binding of Treslin to chromatin began at 30 min and increased thereafter (Figure 1D). Mcm2 (a marker for the MCM complex) preceded Treslin in associating with chromatin, whereas Cdc45 commenced binding just after Treslin. Moreover, Treslin bound to chromatin with a generally similar timing as TopBP1. We also noticed that Treslin from M-phase egg extracts showed a reduced electrophoretic mobility indicative of a modification such as phosphorylation (Figure 1D). The mobility of Treslin increased within 15-30 min after addition of calcium upon exit from M-phase, but then decreased again at around 45 min during interphase.
We also examined the effect of some well-established inhibitors of DNA replication on the binding of Treslin to chromatin. We observed that geminin, which inhibits the loading of the MCM complex onto DNA (McGarry and Kirschner, 1998), greatly reduced, but did not abolish, the binding of Treslin to chromatin (Figure 1E). As expected, geminin compromised the binding of both Mcm2 and Cdc45 to chromatin, but did not inhibit the binding of Cdc6 (a loading factor for the MCM complex). In fact, as reported, geminin elicited increased binding of Cdc6 (McGarry and Kirschner, 1998). Overall, we conclude that the binding of a substantial fraction of Treslin to chromatin depends on the pre-RC, but some Treslin can associate with chromatin by a mechanism that does not absolutely require the pre-RC.
We also examined the response of Treslin to p27, which blocks DNA replication in egg extracts through inhibition of the Cdk2-cyclin E complex (the major S-CDK activity in egg extracts) (Walter and Newport, 2000). We found that p27 did not block the binding of Treslin to chromatin (Figure 1E). Indeed, this inhibitor elicited a significantly elevated association of Treslin with the DNA. There was also elevated binding of Treslin to chromatin in the presence of both p27 and geminin, although this binding was less than with p27 alone. These experiments indicate that phosphorylation by Cdk2 is not necessary for either the pre-RC-dependent or pre-RC-independent association of Treslin with chromatin. We have not investigated why p27 elicits increased binding of Treslin, but this observation suggests that Cdk2 may somehow regulate dynamic association of Treslin with the DNA. As expected, p27 did inhibit the recruitment of Cdc45 under the same conditions, but it did not affect the binding of Mcm2 (Mimura and Takisawa, 1998; Walter and Newport, 2000). Altogether, these experiments suggest that the initial association of Treslin with chromatin occurs upstream of phosphorylation by Cdk2-cyclin E in the series of events leading to DNA replication.
Treslin Interacts with a Region of TopBP1 that Is Critical for DNA Replication
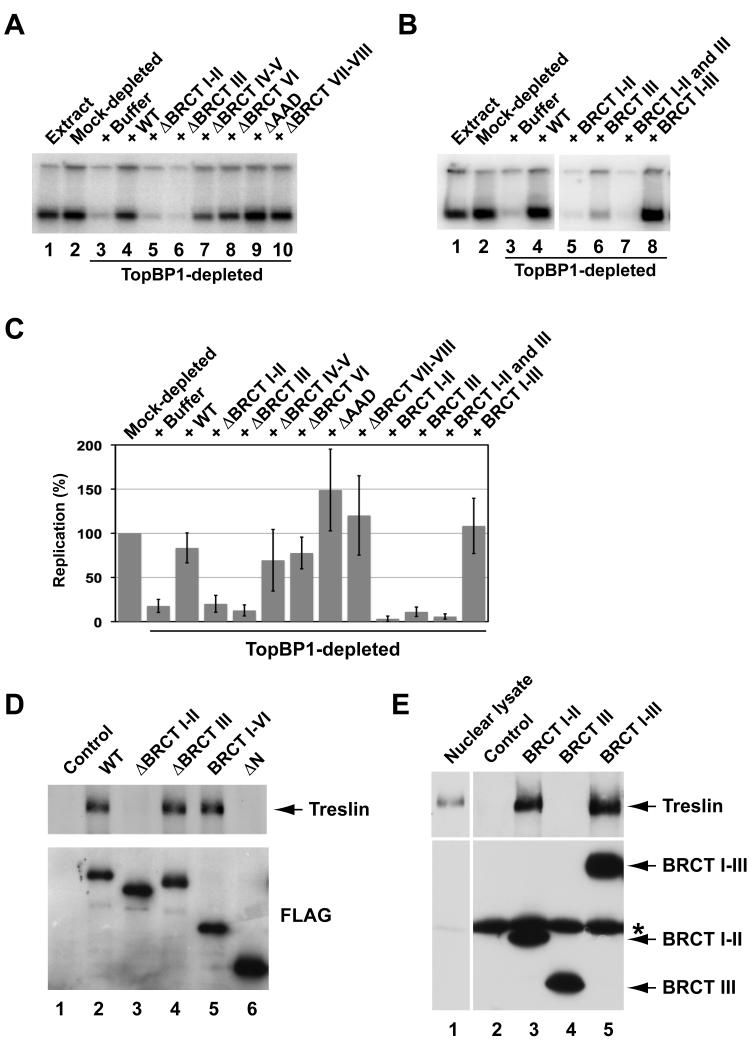
As another means to explore the function of Treslin, we examined which part of TopBP1 is involved in the association with Treslin. Prior to these experiments, we initially performed some structure-function analyses of TopBP1 in order to dissect further its role in DNA replication. Toward this end, we sought to define more precisely the region of TopBP1 that is necessary for replication. For this purpose, we immunodepleted TopBP1 from egg extracts and replaced it with either recombinant full-length TopBP1 or various deletion mutants (Figure 2A and C; Figure S1). As expected, TopBP1-depleted extracts were deficient for DNA replication, and recombinant full-length TopBP1 could reverse this defect (Hashimoto and Takisawa, 2003; Van Hatten et al., 2002). Deletion of BRCT repeats IV-V, VI, or VII-VIII or removal of the ATR-activating domain (AAD) from TopBP1 did not block its activity in promoting replication. These findings are consistent with earlier studies indicating that approximately the N-terminal half of TopBP1 (residues 1-758) is required for replication (Hashimoto et al., 2006; Yan et al., 2006). Conversely, deletion of either BRCT motifs I-II or III abolished the replication-inducing function of TopBP1. Thus, the entire BRCT I-III region appears to be necessary for DNA replication.
Figure 2. Treslin Interacts with a Region of TopBP1 that Is Essential for DNA Replication.
(A) Regions of TopBP1 necessary for DNA replication. Egg extracts (lane 1) were mock-depleted with control antibodies (lane 2) or immunodepleted with anti-TopBP1 antibodies (lanes 3-10). TopBP1-depleted extracts were supplemented with no recombinant protein (lane 3), full-length (WT) HF-TopBP1 (lane 4), or deletion mutants of TopBP1 lacking the indicated BRCT domains or the ATR-activating domain (AAD) (lanes 5-10). DNA replication was determined by incorporation of 32P into chromosomal DNA after 120 min. Labeling of both the upper and lower bands was quantitated.
(B) Egg extracts (lane 1) were subjected to mock depletion (lane 2) or immunodepletion of TopBP1 (lanes 3-8). TopBP1-depleted extracts were supplemented with no recombinant protein (lane 3), full-length HF-TopBP1 (lane 4), or fragments containing BRCT domains I-II (lane 5), III (lane 6), or I-III (lane 8). For lane 7, two fragments containing domains I-II or III were added together.
(C) Quantitation of the data from A and B. Results were compiled from seven independent experiments. Values are from at least two independent experiments (mean ± standard deviation). Replication in mock-depleted extracts is denoted as 100%.
(D) Nuclear lysates from egg extracts containing no tagged protein (lane 1) or the indicated versions of HF-TopBP1 were immunoprecipitated with anti-FLAG beads. Samples were processed for immunoblotting with anti-Treslin (top) and anti-FLAG antibodies (bottom).
(E) Nuclear lysates (lane 1) from egg extracts containing no tagged protein (lane 2) or indicated fragments of TopBP1 (lanes 3-5) were incubated with anti-FLAG beads. Beads were retrieved and immunoblotted with anti-Treslin (top) and anti-FLAG antibodies (bottom). Asterisk denotes cross-reacting IgG heavy chain. See also Figure S1.
To complement these studies, we asked whether the BRCT I-III region is sufficient for supporting DNA replication. For these studies, we utilized recombinant fragments of TopBP1 containing BRCT repeats I-II, I-III, or III. We found that the I-III fragment restored DNA replication in TopBP1-depleted extracts quite well (Figure 2B and C). On the other hand, neither the I-II nor the III fragment alone was able to rescue DNA replication. A mixture of the I-II and III fragments was also ineffective. Interestingly, we observed that the I-II fragment, either alone or in combination with the III fragment, eradicated the small amount of residual replication that occurred in TopBP1-depleted extracts, suggesting that this construct might interfere with replication in some manner in this context. Taken together, these experiments indicate that the BRCT I-III region is both necessary and sufficient for TopBP1 to carry out its function in DNA replication.
We pursued these studies by examining which part of TopBP1 interacts with Treslin. For this purpose, we also first examined deletion mutants of TopBP1 that lack one or more BRCT repeats. We found that the TopBP1(ΔN) construct, which lacks BRCT repeats I-V, could not bind to Treslin (Figure 2D). After examining smaller deletions, we observed that Treslin could not bind to a mutant of TopBP1 lacking BRCT repeats I-II, whereas it bound very well to a mutant missing only the BRCT III region. In conjunction with these experiments, we examined what fragments of TopBP1 might be sufficient for binding of Treslin. We found that Treslin could bind quite well to a fragment of TopBP1 containing only BRCT repeats I-II (Figure 2E). In the same assays, Treslin also bound efficiently to the somewhat larger, replication-competent BRCT I-III fragment, but it could not interact with a fragment containing only the BRCT III region. Altogether, these experiments indicate that Treslin binds to the same general region of TopBP1 that is necessary for DNA replication. However, the region of TopBP1 that associates with Treslin (BRCT I-II) is somewhat less extensive than the area that is necessary for replication (BRCT I-III).
Treslin Is Necessary for DNA Replication in Egg Extracts
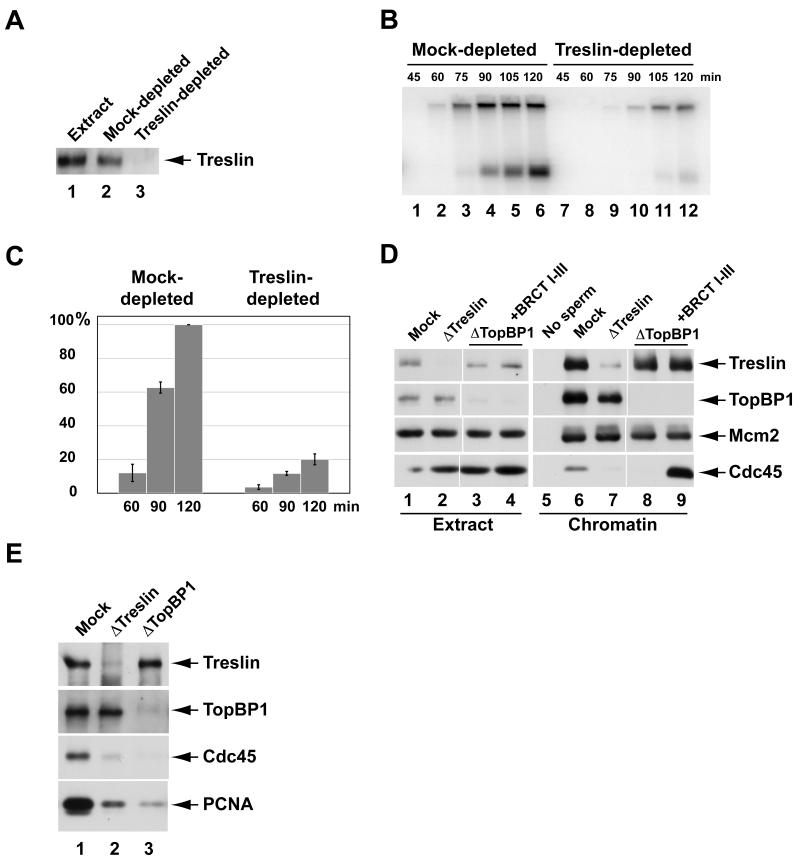
We proceeded to ask explicitly whether Treslin has a role in chromosomal DNA replication in egg extracts. For this purpose, we sought to immunodeplete Treslin from the extracts. As shown in Figure 3A, we could achieve an essentially complete depletion of Treslin with anti-Treslin antibodies. Next, we added sperm chromatin and [α-32P]dATP to the extracts and examined the rate and extent of chromosomal DNA synthesis. We observed that incorporation of 32P into chromosomal DNA was severely diminished in Treslin-depleted extracts in comparison with mock-depleted extracts (Figure 3B and C).
Figure 3. Treslin Is Required for Chromosomal DNA Replication in Xenopus Egg Extracts.
(A) Egg extracts (lane 1) were mock-depleted with control antibodies (lane 2) or immunodepleted with anti-Treslin antibodies (lane 3) and immunoblotted for Treslin.
(B) Mock-depleted (lanes 1-6) and Treslin-depleted extracts (lanes 7-12) were assayed for chromosomal DNA replication at indicated times.
(C) Compilation of results (mean ± standard deviation) from four independent experiments on the extent of replication in mock-depleted (left) and Treslin-depleted extracts (right) at indicated times. Replication at 120 min in mock-depleted extract is denoted as 100%.
(D) Mock-depleted extracts (lanes 1, 5, and 6), Treslin-depleted extracts (lanes 2 and 7), TopBP1-depleted extracts (lanes 3 and 8), and TopBP1-depleted extracts supplemented with HF-TopBP1(I-III) (lanes 4 and 9) were incubated for 90 min in the absence (lane 5) or presence of sperm nuclei (lanes 6-9). Extracts (lanes 1-4) and chromatin fractions (lanes 5-9) were immunoblotted for indicated proteins.
(E) Chromatin fractions from mock-depleted (lane 1), Treslin-depleted (lane 2), and TopBP1-depleted extracts (lane 3) were immunoblotted for indicated proteins. See also Figure S2.
Quantitation indicated that replication was reduced to approximately 20% of the normal level in the absence of Treslin. This decrease is comparable to what we typically observe in TopBP1-depleted extracts (see Figure 2A-C). In the absence of Treslin, assembly of reconstituted nuclei around sperm chromatin in egg extracts occurred normally. For example, nuclear morphology and mean nuclear diameter were very similar in mock-depleted and Treslin-depleted extracts (Figure S2). Proper nuclear assembly is a prerequisite for DNA replication. Therefore, removal of Treslin does not interfere indirectly with DNA replication by perturbing nuclear envelope assembly. We were not able to rescue replication in the Treslin-depleted extracts with recombinant Treslin. However, owing to the fact that Treslin is quite a large protein, the expression and purification of a recombinant version of this protein proved difficult. Therefore, it is not clear that this recombinant protein is functional. As another approach, we attempted to rescue the Treslin-depleted extracts with salt extracts from replicating nuclei. We prepared these salt extracts under essentially the same conditions that we used for the initial identification of Treslin. As shown in Figure S2, Treslin-containing salt extracts efficiently rescued DNA replication in Treslin-depleted egg extracts, which indicates that these extracts had not undergone irreversible inactivation because of the immunodepletion procedure.
It is possible that Treslin exists in a multi-protein complex that is necessary for replication, which also might explain why recombinant Treslin cannot rescue the depleted extracts. In this regard, however, we observed that depletion of Treslin from egg extracts did not appreciably diminish the levels of other key replication proteins, such as Mcm2, TopBP1, and Cdc45 (see Figure 3D). Please note that, owing to the fact that Treslin and TopBP1 do not interact in whole egg extracts, one would not expect a reduction in the amount of TopBP1 in Treslin-depleted extracts. Taken together, these experiments indicate that Treslin, either alone or possibly in a multi-protein complex, plays an important role in DNA replication in the Xenopus egg extract system.
Treslin Is Essential for the Loading of Cdc45 onto Replication Origins
We pursued these observations by attempting to pinpoint the step at which Treslin functions in DNA replication. Toward this end, we assessed the binding of other key replication proteins to chromatin in Treslin-depleted egg extracts. We observed that both Mcm2 and TopBP1 bound well to chromatin in the absence of Treslin (Figure 3D). Thus, some early key events leading to DNA replication can occur normally following removal of Treslin. These observations validate further that immunodepletion of Treslin does not cause non-specific inactivation of egg extracts.
Significantly, however, we could not detect the recruitment of Cdc45 to chromatin in Treslin-depleted extracts. It is well established that Cdc45 is required for the initiation of DNA replication in this system (Mimura et al., 2000; Mimura and Takisawa, 1998; Walter and Newport, 2000). Thus, Treslin appears to function at or before the initiation of replication. Consistent with this observation, there was also dramatically reduced binding of PCNA to chromatin in the absence of Treslin (Figure 3E). PCNA acts as a processivity factor for the elongation of nascent DNA strands at replication forks (Bell and Dutta, 2002). The absence of PCNA on Treslin-depleted chromatin reinforces the concept that replication forks cannot assemble without Treslin.
In conjunction with these experiments, we examined the chromatin-binding behavior of Treslin in the absence of TopBP1, which ultimately forms a complex with Treslin in replicating nuclei (Figure 3D). Consistent with previous studies, the loading of Cdc45 onto chromatin did not occur in the absence of TopBP1 (Hashimoto and Takisawa, 2003; Van Hatten et al., 2002). Addition of the replication-competent BRCT I-III fragment of TopBP1 to the depleted extracts restored the binding of Cdc45 to chromatin. By contrast, the association of Treslin with chromatin took place normally without TopBP1. This observation, taken together with the finding that TopBP1 binds normally to chromatin lacking Treslin, indicates that Treslin and TopBP1 bind independently to future sites for initiation of replication. Therefore, it appears that distinct pathways containing Treslin and TopBP1 ultimately converge to promote the loading of Cdc45.
Treslin Associates with TopBP1 in a Cdk2-dependent Manner
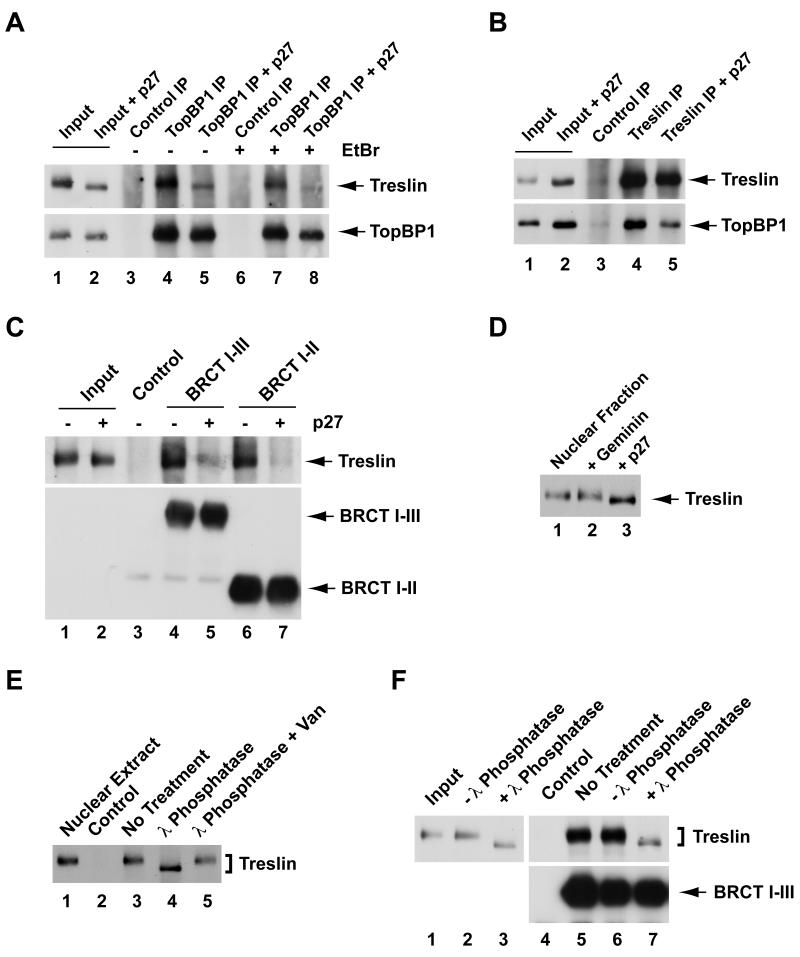
In view of the observations that both Treslin and Cdk2 activity are necessary for the loading of Cdc45 onto replication origins, we examined whether Cdk2 regulates the functional properties of Treslin. For these studies, we utilized p27 to inhibit the Cdk2-cyclin E complex in egg extracts. First, we immunoprecipitated TopBP1 from lysates of nuclei that had been isolated from egg extracts lacking or containing p27 (Figure 4A). In the absence of p27, we could efficiently coimmunoprecipitate endogenous TopBP1 and Treslin with anti-TopBP1 antibodies. This interaction was resistant to treatment with ethidium bromide, an agent that disrupts interactions between proteins and DNA. Thus, the association between Treslin and TopBP1 is dependent upon protein-protein interactions. By contrast, there was greatly reduced binding of TopBP1 to Treslin in nuclear lysates from p27-treated extracts. We obtained similar results by immunoprecipitating Treslin and subsequently immunoblotting for TopBP1 (Figure 4B).
Figure 4. Association of Treslin with TopBP1 Depends Upon Cdk2.
(A) Nuclear lysates were prepared from egg extracts incubated without (lanes 1, 3, 4, 6, and 7) or with p27 (lanes 2, 5, and 8). Input lysates (lanes 1 and 2), control immunoprecipitates (lanes 3 and 6), and anti-TopBP1 immunoprecipitates (lanes 4, 5, 7, and 8) were immunoblotted for Treslin and TopBP1. Immunoprecipitations were carried out in the absence (lanes 3-5) or presence of 50 μg/ml ethidium bromide (EtBr) (lanes 6-8).
(B) Nuclear lysates were prepared from egg extracts incubated without (lanes 1, 3, and 4) or with p27 (lanes 2 and 5). Input lysates (lanes 1 and 2), control immunoprecipitates (lanes 3), and anti-Treslin immunoprecipitates (lanes 4 and 5) were immunoblotted for Treslin and TopBP1.
(C) Nuclear lysates from egg extracts lacking (lanes 1, 3, 4, and 6) or containing p27 (lanes 2, 5, and 7) were incubated with anti-FLAG antibody beads containing no tagged protein (lane 3) or indicated fragments of TopBP1 (lanes 4-7). Beads were retrieved and immunoblotted with anti-Treslin and anti-FLAG antibodies. Lanes 1 and 2 depict input nuclear lysates.
(D) Nuclear fractions were prepared from extracts that were mock-treated (lane 1) or treated with geminin (lane 2) or p27 (lane 3). Samples were immunoblotted for Treslin.
(E) Nuclear lysates (lane 1) containing no tagged protein (lane 2) or the HF-TopBP1(I-III) fragment (lanes 3-5) were incubated with anti-FLAG beads. Beads were retrieved and incubated without (lanes 2 and 3) or with 20 U/μl lambda phosphatase (lanes 4 and 5) in the absence (lane 4) or presence of 10 mM sodium vanadate (lane 5) for 30 min at room temperature. Vanadate is an inhibitor of lambda phosphatase. Samples were immunoblotted for Treslin.
(F) Eluates of chromatin treated with 0.45 M NaCl (lane 1) were incubated in the absence (lanes 2 and 6) or presence of lambda phosphatase (lanes 3 and 7) for 30 min at room temperature or left on ice (lanes 4 and 5). Subsequently, eluates were mixed with anti-FLAG beads in the absence (lane 4) or presence of HF-TopBP1(I-III) (lanes 5-7). Beads were retrieved and immunoblotted with anti-Treslin (top) or anti-FLAG antibodies (bottom). See also Figure S3.
We also explored this issue by using recombinant fragments from the N-terminal end of TopBP1 that bind very efficiently to Treslin. In particular, we incubated the FLAG-tagged BRCT I-II and I-III fragments of TopBP1 in nuclear lysates in the absence and presence of p27 and then recovered these polypeptides with anti-FLAG antibodies. We observed that there was greatly reduced binding of Treslin to both fragments of TopBP1 in the presence of p27 (Figure 4C).
We noticed that treatment with p27 caused an increased electrophoretic mobility of Treslin in nuclei from interphase extracts (see Figure 4D). By contrast, geminin had no obvious effect on the mobility of Treslin. In order to address explicitly the issue of whether the interphase form of Treslin is phosphorylated, we incubated this protein in the absence and presence of lambda phosphatase. We observed that treatment of Treslin with lambda phosphatase resulted in a clear increase in electrophoretic mobility due to dephosphorylation (Figure 4E). Moreover, treatment of chromatin eluates with lambda phosphatase abolished the binding of Treslin to TopBP1 (Figure 4F). Taken together, these observations indicate that Treslin is phosphorylated during interphase and that Cdk2 promotes both phosphorylation of Treslin and formation of a Treslin-TopBP1 complex. Consistent with this scenario, binding of Treslin to TopBP1 increases substantially near the commencement of DNA replication (Figure S3).
Treslin-depleted Human Cells Display Defects in S Phase
As another means to assess the function of Treslin, we sought to ablate the human version of this protein from tissue culture cells by using small interfering RNA (siRNA) technology. To pursue these studies, we first raised anti-human Treslin antibodies. These antibodies specifically recognize a 220 kD polypeptide in human U2OS cells (Figure 5A). As shown in Figure 5A, treatment with Treslin siRNA elicited the essentially complete ablation of Treslin within 48 hr. We first noted that the Treslin-depleted cells displayed a pronounced defect in proliferation. There was no increase in cell number in Treslin-depleted cultures from 24-96 hr after treatment with the siRNA (Figure 5B). However, the cells did not undergo cell death and otherwise appeared to remain healthy. On the other hand, depletion of Treslin from HeLa cells resulted in extensive cell death within 72 hr (data not shown). We also noted that the Treslin-depleted U2OS cells had grown to be noticeably larger than the control cells by 72 hr after transfection (data not shown).
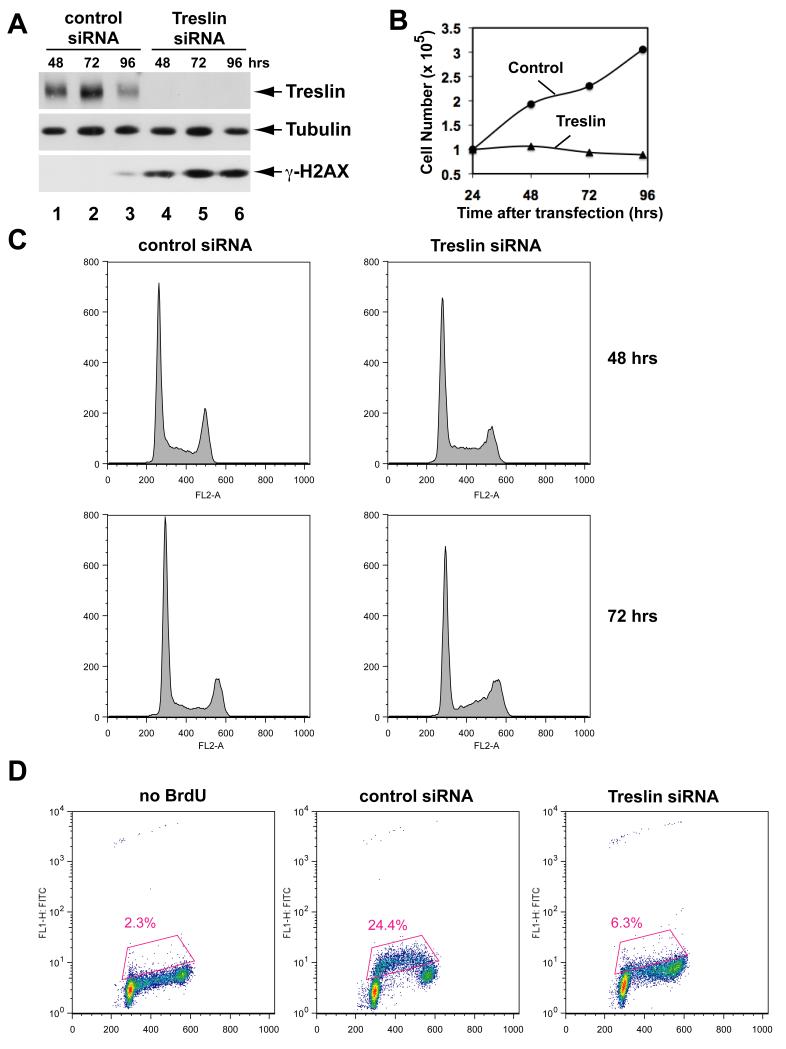
Figure 5. Depletion of Treslin from Human Cells Results in Defective S-Phase.
(A) Human U2OS cells were treated with control siRNA (lanes 1-3) or Treslin siRNA #1 (lanes 4-6) for 48, 72, and 96 hrs. Cell lysates were immunoblotted for Treslin, α-tubulin, and γ-H2AX.
(B) Cell numbers in control and Treslin siRNA-treated cultures were counted at indicated times after transfection.
(C) DNA content of cells treated with siRNA was analyzed by flow cytometry. Cells were stained with propidium iodide. DNA content is shown on x-axis. Y-axis indicates relative number of cells.
(D) Cells treated with control or Treslin siRNA were incubated in the absence or presence of BrdU for 60 min at 72 hr post-transfection. Intensity of BrdU incorporation is indicated on logarithmic y-axis; DNA content is shown on x-axis. Boxed area indicates cells incorporating BrdU. See also Figure S4.
We proceeded to examine cell cycle progression and DNA replication in the siRNA-treated cells. First, we performed FACS analysis to examine the cell cycle distribution of the ablated cells as a function of time (Figure 5C). By 48 hr, there was an accumulation of cells in the Treslin-depleted culture with a content of DNA between 2N and 4N (S-phase population), suggesting that the cells might have some difficulty in replicating their DNA. At 72 hr, there was a significant decrease in the early S-phase population, which suggests that by this time the cells were suffering a defect in the entry into S-phase.
Next, we directly examined DNA replication by two different methods. First, we monitored incorporation of BrdU into the cells by staining with anti-BrdU antibodies. In the culture treated with the control siRNA, we observed strong incorporation of BrdU at 72 hr post-transfection (Figure 5D). By contrast, only a very small portion of the Treslin-depleted cells showed incorporation of BrdU at this time.
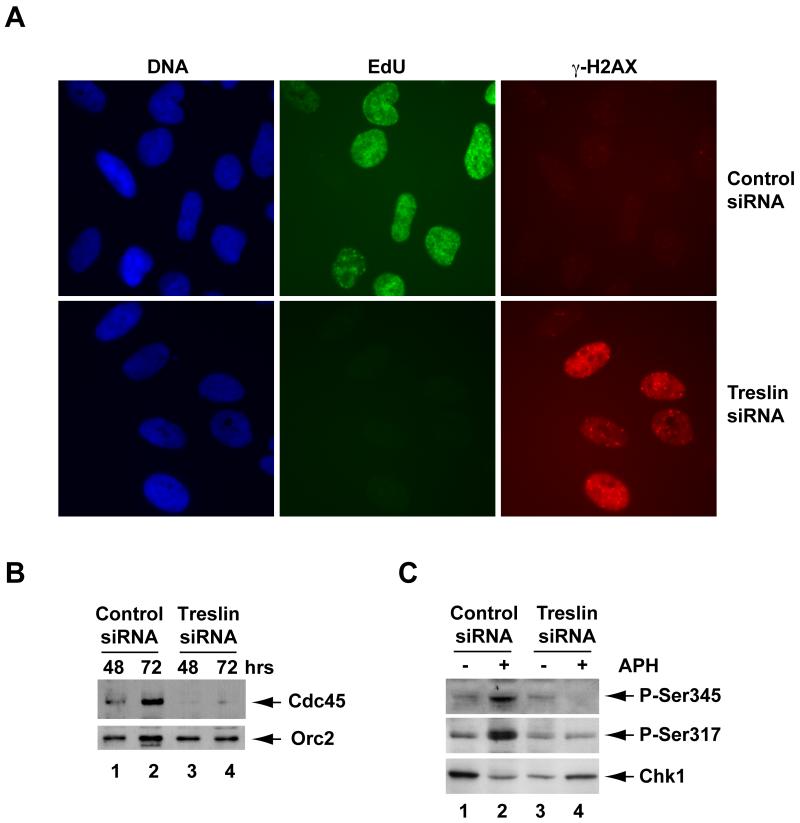
We also visualized DNA replication in the siRNA-treated cells by using a derivative of deoxyuridine (5-ethynyl-2′-deoxyuridine) that reacts directly with a modified fluorescent dye. These experiments also indicated that there is a very pronounced reduction in DNA replication in the absence of Treslin (Figure 6A). We observed a very similar reduction in DNA replication in cells treated with two additional siRNAs (Figure S4). As another means to validate the specificity of the siRNA-mediated knockdown of Treslin, we also performed rescue experiments with recombinant human Treslin. For this purpose, we generated a line of U2OS T-Rex cells (Monroe et al., 2003) in which the expression of an siRNA-resistant version of Treslin was under the control of a doxycycline-inducible promoter. Consistent with the results described above, treatment of these cells with Treslin siRNA in the absence of doxycycline elicited a marked reduction of DNA replication, while there was an effective rescue of DNA replication in the presence of doxycycline (Figure S5). Finally, we observed that ablation of Treslin virtually eliminated the binding of Cdc45 to chromatin fractions from U2OS cells (Figure 6B), which suggests that Treslin acts at a similar point in DNA replication in both Xenopus and humans. Taken together, these experiments indicate that the inhibition of replication in human cells is due to the absence of Treslin and not to some non-specific effect of the siRNA treatment. Moreover, the findings that removal of Treslin has similar consequences in both human cells and Xenopus egg extracts mutually reinforce the concept that Treslin is an important, functionally conserved replication protein in vertebrates.
Figure 6. Treslin-depleted Cells Defective for DNA Replication Also Accumulate Damaged DNA.
(A) Cells treated with control siRNA or Treslin siRNA #1 for 72 hr were stained with Hoechst dye to detect DNA, Click-iT reagent to detect incorporated EdU, and antibodies against γ-H2AX.
(B) Chromatin fractions were prepared at 48 and 72 hrs from U2OS cells transfected with control siRNA (lanes 1 and 2) or Treslin siRNA #1 (lanes 3 and 4). Fractions were immunoblotted for Cdc45 (top) and Orc2 (bottom).
(C) Control (lanes 1 and 2) and Treslin siRNA-treated cultures (lanes 3 and 4) were incubated for 15 min in the absence (lanes 1 and 3) or presence of 10 μg/ml aphidicolin (lanes 2 and 4). Cultures were processed for immunoblotting with antibodies that detect Chk1 or phosphorylation of Chk1 on Ser345 or Ser317. See also Figure S5.
In the course of performing the siRNA experiments, we also noticed significantly elevated phosphorylation of H2AX in Treslin-depleted cells by immunoblotting with antibodies that recognize the Ser139-phosphorylated form of this protein, known as γ-H2AX (Figure 5A). This phosphorylation is a well-established indicator for the presence of damaged DNA. We confirmed this finding by performing indirect immunofluorescence on the siRNA-treated cells with these antibodies (see Figure 6A). Moreover, in these experiments, we ascertained that damaged DNA, as indicated by the punctate staining pattern of γ-H2AX, had accumulated in 50.6% of the Treslin-depleted cells at 72 hr. By contrast, only 3.7% of the cells treated with control siRNA displayed γ-H2AX staining, and this staining was much weaker than in the Treslin-ablated cells. Interestingly, we could not detect any increased phosphorylation of Chk1 on either S317 or S345 in Treslin-depleted cultures. Also, there was no increase in these phosphorylations upon treatment of the Treslin-ablated cells with aphidicolin (Figure 6C). These observations suggest that Treslin might be necessary directly or indirectly for the activation of Chk1 (see Discussion). In either case, these experiments provide further evidence for the functional importance of Treslin in the replicative machinery of the cell.
DISCUSSION
In this study, we have sought to understand how TopBP1 functions in DNA replication in vertebrates. In budding yeast, it is well established that Dpb11, the functional analogue of TopBP1 in this organism, has a critical role in associating with other replication factors, including Sld2 and Sld3 (Garcia et al., 2005; Sclafani and Holzen, 2007). It has been long known that phosphorylation by a CDK is necessary for the initiation of replication in eukaryotic cells (reviewed in Tanaka et al., 2007a). However, an understanding of the molecular basis for this requirement proved elusive for many years. Recent studies in budding yeast have indicated that CDK-mediated phosphorylation of both Sld2 and Sld3 allows the docking of these proteins onto distinct pairs of BRCT repeats within Dpb11 (Tanaka et al., 2007b; Zegerman and Diffley, 2007). These interactions are necessary for the incorporation of Cdc45 into the pre-IC. These phosphorylations of Sld2 and Sld3 account for the need of S-CDK activity in turning on the activity of the replication apparatus in budding yeast.
An important question is whether similar control mechanisms operate in vertebrates. In this regard, it has been proposed that the RecQ4 protein in vertebrates may be a functional homologue of Sld2. This protein is necessary for DNA replication in Xenopus egg extracts and human tissue culture cells (Matsuno et al., 2006; Sangrithi et al., 2005). However, the homology between Sld2 and RecQ4 is low and limited to a small region of these proteins. In addition, yeast Sld2 does not possess a RecQ helicase domain. Finally, functional analysis of RecQ4 indicated that this protein is not required for the loading of Cdc45 onto chromatin prior to replication in Xenopus egg extracts (Matsuno et al., 2006; Sangrithi et al., 2005). Hence, it seems unlikely that the association of TopBP1 with RecQ4 could explain how TopBP1 promotes the loading of Cdc45 in this system. The situation is even more nebulous in the case of Sld3, which apparently does not have a counterpart in vertebrates with obvious sequence homology.
In this report, we have searched for proteins that associate physically with TopBP1 as a means for uncovering how TopBP1 exerts its replication-promoting function. With this approach, we identified Treslin as a binding partner of TopBP1 in replicating nuclei from Xenopus egg extracts. Our studies indicate that Treslin acts at a crucial step in the commencement of DNA replication in this system. It is well established that the association of Cdc45 with origins is a pivotal event in the initiation of eukaryotic DNA replication. In the Xenopus system, association of Cdc45 with chromatin requires prior loading of TopBP1 onto origins and phosphorylation of some unknown protein(s) by the Cdk2-cyclin E complex (Hashimoto and Takisawa, 2003; Van Hatten et al., 2002). Evidence has also been presented that TopBP1 itself appears not to be one of the direct targets of Cdk2 that is necessary for replication in egg extracts (Hashimoto and Takisawa, 2003).
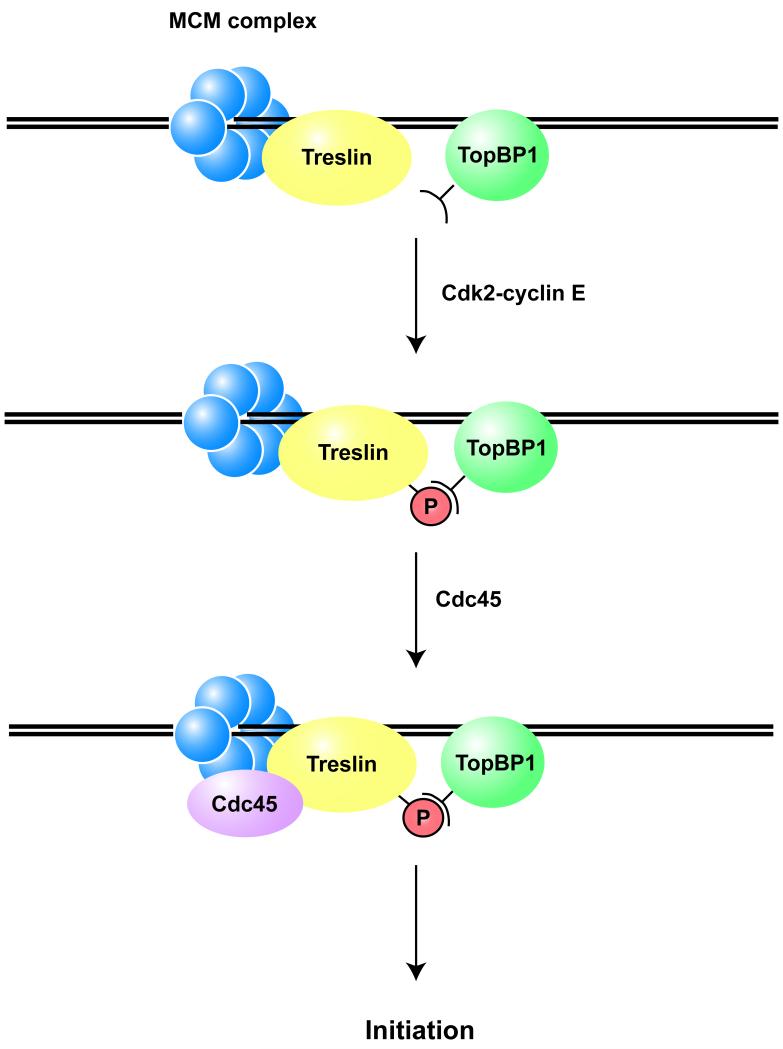
We have found that Treslin is also necessary for the binding of Cdc45 to chromatin. TopBP1 and Treslin each first associate with chromatin independently. However, we can readily detect an association between TopBP1 and Treslin in salt-extracted lysates of replicating nuclei from Xenopus egg extracts. We believe that it is reasonable to infer that this complex formed after these proteins had bound to chromatin. We have also observed that the kinase activity of Cdk2 is necessary for the formation of this complex. On the basis of these observations, a simple model would be that phosphorylation of Treslin by Cdk2 would allow docking with TopBP1 (see Figure 7). This complex would promote the subsequent loading of Cdc45 and the ensuing activation of replicative helicase activity. At this point, we do not have direct evidence that Treslin is a substrate for Cdk2. However, Treslin possesses an unusually large number of potential CDK phosphorylation sites and its electrophoretic mobility increases significantly following treatment of interphase egg extracts with p27. The p27-sensitive mobility shift of Treslin can be reversed by treatment with a protein phosphatase. These observations argue that Cdk2 regulates the phosphorylation of Treslin and thereby promotes the formation of a TopBP1-Treslin complex.
Figure 7. Model for the Role of Treslin in DNA Replication.
See text for details.
Interestingly, Treslin interacts with the same general region of TopBP1 as the Rad9-Hus1-Rad1 (9-1-1) and Mre11-Rad50-Nbs1 (MRN) complexes (Delacroix et al., 2007; Lee et al., 2007; Yoo et al., 2009). An intriguing possibility is that binding of TopBP1 to Treslin versus checkpoint control proteins such as the 9-1-1 complex may be mutually incompatible. Furthermore, termination of checkpoint responses may allow TopBP1 to associate with Treslin again. If so, it is conceivable that Treslin could be important for the re-start of replication at previously stalled replication forks.
Our observations on the consequences of the depletion of Treslin from mammalian cells also strongly support a role for Treslin in S-phase regulation. Treslin-depleted cells displayed pronounced defects in both proliferation and DNA replication. We have observed these defects after treatment with three different siRNAs, and a recombinant form of Treslin impervious to siRNA treatment reverses these effects. Moreover, Treslin appears to regulate DNA replication at a similar juncture in human cells. In particular, Treslin-ablated tissue culture cells are severely deficient in the recruitment of Cdc45 to chromatin. The findings that removal of Treslin has comparable deleterious effects on replication in two completely different systems (Xenopus egg extracts and human tissue culture cells) and blocks replication at a similar point in both systems argue strongly that Treslin is an important replication protein in vertebrates.
Interestingly, many cells lacking Treslin accumulated damaged DNA, as indicated by the appearance of phosphorylated H2AX. However, we cannot detect obvious activation of Chk1 in these cells. Furthermore, we do not observe activation of Chk1 in these cells following additional treatment with aphidicolin. It could be that the damaged DNA cannot assemble the appropriate structures necessary for activation of Chk1 unless Treslin is present. Alternatively, Treslin itself might also play a direct role in checkpoint signaling in addition to its role in DNA replication. Further studies will be required to distinguish between these possibilities.
In conclusion, we have identified a replication protein from both Xenopus and humans that collaborates with TopBP1 in facilitating some of the earliest steps in replication of the genome. Further study of Treslin should enhance our understanding of how vertebrates perpetuate their genetic material in a faithful manner.
EXPERIMENTAL PROCEDURES
Xenopus Egg Extracts
Xenopus egg extracts were prepared as described (Kumagai and Dunphy, 2000). Preparation and handling of nuclear lysates and chromatin fractions from egg extracts are described in Supplemental Data. Chromosomal DNA replication assays were performed with [α-32P]dATP as described (Yoo et al., 2004).
Recombinant Proteins
HF-TopBP1 constructs, which contain both hemagglutinin and His6 tags at the N-terminal end and a FLAG tag at the C-terminal end, were produced in baculovirus-infected Sf9 cells (Lee et al., 2007). These constructs are described in detail in Supplemental Data. Xenopus Treslin with a FLAG tag at the C-terminal end was also produced in Sf9 cells.
Identification of TopBP1-interacting Proteins in Nuclear Lysates
Recombinant HF-TopBP1(I-VI) was incubated in nuclear lysates from egg extracts for 1 hr at 4°C in the presence of protein G magnetic beads coated with anti-FLAG antibodies. After retrieval of the beads with a magnet, bound proteins were processed for mass spectrometric analysis as described (Junqueira et al., 2008; Shevchenko et al., 2006; Yoo et al., 2009). Further information is provided in Supplemental Data. The presence of the LOC443616 polypeptide (accession number Q6GPQ1) was established by the presence of 47 individual peptides matching 70 MS/MS spectra and covering 31% of the available sequence.
Cloning of Xenopus and Human Treslin
A 220 kD protein that interacted with HF-TopBP1(I-VI) in egg extracts contained sequences from the GenBank entry for the Xenopus LOC443616 polypeptide. The human homologue of LOC443616 is the protein NP_689472 corresponding to C15orf42. Preparation of full-length cDNA clones encoding these proteins is described in Supplemental Data. GenBank accession numbers for full-length human and Xenopus Treslin are GQ227787 and GQ227788, respectively.
Antibodies and Immunodepletion
Polypeptides containing residues 1-487 and 1566-1909 of Xenopus and human Treslin, respectively, were produced in bacteria using the pET30a expression vector. Antibodies were produced in rabbits and affinity-purified. For immunodepletion of Treslin, egg extract (100 μl) was incubated at 4°C with 75 μg anti-Treslin antibodies bound to Affiprep protein A beads (Bio-Rad) for two rounds of 60 min. Mock depletions were performed with rabbit IgG fraction (Zymed).
Human Tissue Culture Cells
U2OS cells were grown in DMEM containing 10% fetal calf serum. Stealth siRNA duplexes specific for Treslin (with the following sense strand sequences: #1, GACCUGAGAGAAGAUUCAGAAGUUA; #2, AGGACACAUUCUGCCUCCUUCUAUU; and #3, CCUGUUACGCCAAAGAAACUGUUUA) and control siRNA (low GC) were obtained from Invitrogen. The siRNAs (50 nM) were transfected into U2OS cells with Lipofectamine RNAiMAX (Invitrogen). Functional analyses of the siRNA-treated cells are described in Supplemental Data.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to laboratory members for comments on the manuscript. We would also like to thank Rochelle Diamond for assistance with the FACS analyses and Dr. T. C. Spelsberg (Mayo Clinic) for the U2OS T-REx cells. This work was supported by NIH grants GM043974 and GM070891 to W.G.D.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Botchan M. Cell biology: a switch for S phase. Nature. 2007;445:272–274. doi: 10.1038/445272a. [DOI] [PubMed] [Google Scholar]
- Caldecott KW. Cell signaling. The BRCT domain: signaling with friends? Science. 2003;302:579–580. doi: 10.1126/science.1091463. [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev. 2007;21:1472–1477. doi: 10.1101/gad.1547007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF. Regulation of early events in chromosome replication. Curr. Biol. 2004;14:R778–786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Garcia V, Furuya K, Carr AM. Identification and functional analysis of TopBP1 and its homologs. DNA Repair (Amst.) 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Takisawa H. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 2003;22:2526–2535. doi: 10.1093/emboj/cdg238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Tsujimura T, Sugino A, Takisawa H. The phosphorylated C-terminal domain of Xenopus Cut5 directly mediates ATR-dependent activation of Chk1. Genes Cells. 2006;11:993–1007. doi: 10.1111/j.1365-2443.2006.00998.x. [DOI] [PubMed] [Google Scholar]
- Jares P, Blow JJ. Xenopus cdc7 function is dependent on licensing but not on XORC, XCdc6, or CDK activity and is required for XCdc45 loading. Genes Dev. 2000;14:1528–1540. [PMC free article] [PubMed] [Google Scholar]
- Junqueira M, Spirin V, Balbuena T. Santana, Waridel P, Surendranath V, Kryukov G, Adzhubei I, Thomas H, Sunyaev S, Shevchenko A. Separating the wheat from the chaff: unbiased filtering of background tandem mass spectra improves protein identification. J. Proteome Res. 2008;7:3382–3395. doi: 10.1021/pr800140v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol. Cell. 2000;6:839–849. doi: 10.1016/s1097-2765(05)00092-4. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lee J, Kumagai A, Dunphy WG. The Rad9-Hus1-Rad1 checkpoint clamp regulates interaction of TopBP1 with ATR. J. Biol. Chem. 2007;282:28036–28044. doi: 10.1074/jbc.M704635200. [DOI] [PubMed] [Google Scholar]
- Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell. Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Méndez J, Stillman B. Perpetuating the double helix: molecular machines at eukaryotic DNA replication origins. Bioessays. 2003;25:1158–1167. doi: 10.1002/bies.10370. [DOI] [PubMed] [Google Scholar]
- Mimura S, Masuda T, Matsui T, Takisawa H. Central role for Cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells. 2000;5:439–452. doi: 10.1046/j.1365-2443.2000.00340.x. [DOI] [PubMed] [Google Scholar]
- Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe DG, Getz BJ, Johnsen SA, Riggs BL, Khosla S, Spelsberg TC. Estrogen receptor isoform-specific regulation of endogenous gene expression in human osteoblastic cell lines expressing either ERalpha or ERbeta. J. Cell. Biochem. 2003;90:315–326. doi: 10.1002/jcb.10633. [DOI] [PubMed] [Google Scholar]
- Mordes DA, Glick GG, Zhao R, Cortez D. TopBP1 activates ATR through ATRIP and a PIKK regulatory domain. Genes Dev. 2008;22:1478–1489. doi: 10.1101/gad.1666208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrithi MN, Bernal JA, Madine M, Philpott A, Lee J, Dunphy WG, Venkitaraman AR. Initiation of DNA replication requires the RECQL4 protein mutated in Rothmund-Thomson syndrome. Cell. 2005;121:887–898. doi: 10.1016/j.cell.2005.05.015. [DOI] [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM. Cell cycle regulation of DNA replication. Annu. Rev. Genet. 2007;41:237–280. doi: 10.1146/annurev.genet.41.110306.130308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Tak YS, Araki H. The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div. 2007a;2:16. doi: 10.1186/1747-1028-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007b;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Labib K, Diffley JF. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol. Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- Yan S, Lindsay HD, Michael WM. Direct requirement for Xmus101 in ATR-mediated phosphorylation of Claspin bound Chk1 during checkpoint signaling. J. Cell Biol. 2006;173:181–186. doi: 10.1083/jcb.200601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. The Mre11-Rad50-Nbs1 complex mediates activation of TopBP1 by ATM. Mol. Biol. Cell. 2009;20:2351–2360. doi: 10.1091/mbc.E08-12-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Shevchenko A, Shevchenko A, Dunphy WG. Mcm2 is a direct substrate of ATM and ATR during DNA damage and DNA replication checkpoint responses. J. Biol. Chem. 2004;279:53353–53364. doi: 10.1074/jbc.M408026200. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.