SUMMARY
Defective DNA repair by homologous recombination (HR) is thought to be a major contributor to tumorigenesis in individuals carrying Brca1 mutations. Here we show that DNA breaks in Brca1-deficient cells are aberrantly joined into complex chromosome rearrangements by a process dependent on the non-homologous end joining (NHEJ) factors, 53BP1 and DNA Ligase 4. Loss of 53BP1 alleviates hypersensitivity of Brca1 mutant cells to PARP inhibition and restores error-free repair by HR. Mechanistically, 53BP1 deletion promotes ATM-dependent processing of broken DNA ends to produce recombinogenic single-stranded DNA competent for HR. In contrast, Lig4 deficiency does not rescue the HR defect in Brca1 mutant cells, but prevents the joining of chromatid breaks into chromosome rearrangements. Our results illustrate that HR and NHEJ compete to process DNA breaks that arise during DNA replication, and that shifting the balance between these pathways can be exploited to selectively protect or kill cells harboring Brca1 mutations.
INTRODUCTION
Mutations in the Brca1 gene predispose carriers to a high incidence of breast and ovarian cancer (Venkitaraman, 2004). In the absence of Brca1, Xrcc2 or other HR proteins, Rad51 foci formation and homology dependent repair are impaired (Moynahan et al., 1999; Scully et al., 1999). Since the HR pathway is required for repair of spontaneous DSBs that arise during DNA replication, defects in HR result in an accumulation of chromatid breaks (Andreassen et al., 2006; Sonoda et al., 1998). Cells that cannot repair chromatid breaks by HR become more reliant on other poorly-defined alternative repair pathways. These pathways are not template-based like HR and therefore have the propensity to join together DSBs on different chromatids to produce complex chromosomal rearrangements, which promote genomic instability and/or trigger loss in viability (Bryant et al., 2005; Farmer et al., 2005; Sonoda et al., 1998). Genomic instability following loss-of-function of Brca1 is hypothesized to be a key factor leading to tumorigenesis in individuals with the Brca1 mutation; nevertheless, additional mutations are required to enable survival and outgrowth of tumor cells (Deng, 2006; Venkitaraman, 2004).
HR-deficient cells exhibit an acute sensitivity to killing by inhibitors of the single strand DNA (ssDNA) repair protein poly(ADP-ribose) polymerase (PARP) (Bryant et al., 2005; Farmer et al., 2005; Jackson and Bartek, 2009). Mechanistically, loss of PARP activity prevents repair of ssDNA breaks, which are then converted into DSBs during DNA replication. These breaks are normally repaired by Rad51-dependent HR using the sister chromatid as a template, hence PARP inhibition is particularly toxic in Brca1- or Brca2-deficient cells which are HR-defective. The ability of PARP inhibitors to selectively kill HR-deficient cells is currently being used in clinical trials for treatment of breast and ovarian cancers where Brca1 or Brca2 is mutated (Fong et al., 2009; Jackson and Bartek, 2009). More recently, it has been observed that Brca2-deficient tumors are capable of acquiring reversion mutations that enable resistance to chemotherapeutic agents (Edwards et al., 2008; Sakai et al., 2008). This observation raises the possibility that additional secondary mutations could mediate resistance of Brca-deficient tumors to the toxic effects of PARP inhibitors.
Mice homozygous for the exon 11 deletion (Δ11) isoform of Brca1 (Brca1Δ11/Δ11) die in utero (Xu et al., 2001). Embryonic cell death is associated with extensive apoptosis and activation of the ATM-Chk2-p53 arm of the DNA damage response (Cao et al., 2006). Indeed, embryonic lethality can be rescued by complete or heterozygous loss of p53 (Xu et al., 2001), or deletion of ATM or Chk2 (Cao et al., 2006). Deletion of 53BP1 also rescues the viability of Brca1Δ11/Δ11 mice (Cao et al., 2009). However, in contrast to rescue by loss of p53, Brca1Δ11/Δ11 53BP1-/- mice exhibit a low incidence of tumor formation and near-normal lifespan (Cao et al., 2009). Nevertheless, Brca1Δ11/Δ11 53BP1-/- cells showed elevated levels of DSBs, intact ATM-Chk2-p53 signaling and IR induced apoptosis (Cao et al., 2009).
The only known functions of 53BP1 are in transducing a subset of ATM dependent cell cycle checkpoints (DiTullio et al., 2002; Fernandez-Capetillo et al., 2002; Lee et al., 2009; Wang et al., 2002) and facilitating the joining of distal DSBs formed at dysfunctional telomeres and during lymphocyte antigen receptor recombination (Difilippantonio et al., 2008; Dimitrova et al., 2008; Manis et al., 2004; Reina-San-Martin et al., 2007; Ward et al., 2004). None of these activities would appear to account for the survival of Brca1Δ11/Δ11 mice in the absence of 53BP1. Thus, the underlying mechanism by which loss of 53BP1 rescues cell death and prevents tumorigenesis in Brca1 mutant mice remains unclear.
Here we show that the presence of 53BP1 limits the capacity of Brca1-deficient cells to accurately repair DSBs. Importantly, the high levels of genomic instability and cell death induced in Brca1-deficient cells by treatment with an inhibitor of PARP are not present in Brca1/53BP1 double-deficient cells. Genomic stability is rescued because the HR pathway is largely restored in cells lacking Brca1 and 53BP1. In contrast, Brca1 deficient repair is not normalized by deletion of the NHEJ factor, DNA Ligase IV (Lig4), although deletion of Lig4 does prevent accumulation of chromosomal fusions. Our results indicate a new role for 53BP1 and Brca1 in regulating the choice between NHEJ and HR pathways, which has implications for anti-cancer therapies using PARP inhibitors.
RESULTS
53BP1 promotes genomic instability and mammary tumorigenesis in Brca1-mutant mice
Brca1Δ11/Δ11 mice rescued by loss of one or both copies of p53 (Brca1Δ11/Δ11 p53+/- or Brca1Δ11/Δ11 p53-/-) develop multiple types of tumors (Cao et al., 2006; Xu et al., 1999), whereas rescue by deletion of 53BP1 (Brca1Δ11/Δ11 53BP1-/-) results in near normal lifespan with significantly reduced tumorigenesis. To further investigate the requirement for 53BP1 in tumorigenesis in Brca1-deficient animals, we followed the onset of development of mammary tumors in cohorts of female mice with deletion mutations in Brca1 exon 11 (Fig. 1A). As expected, mice with breast-specific deletion of Brca1 exon 11 succumbed to tumors of the mammary tissue (Brodie et al., 2001; Xu et al., 1999), with 12 out of 27 animals affected by 18 months of age (Fig. 1A). Mice that were doubly deficient for Brca1 exon 11 and 53BP1, by contrast, developed almost no breast tumors, with just one animal affected at the age of 22 months (Fig. 1A; not shown).
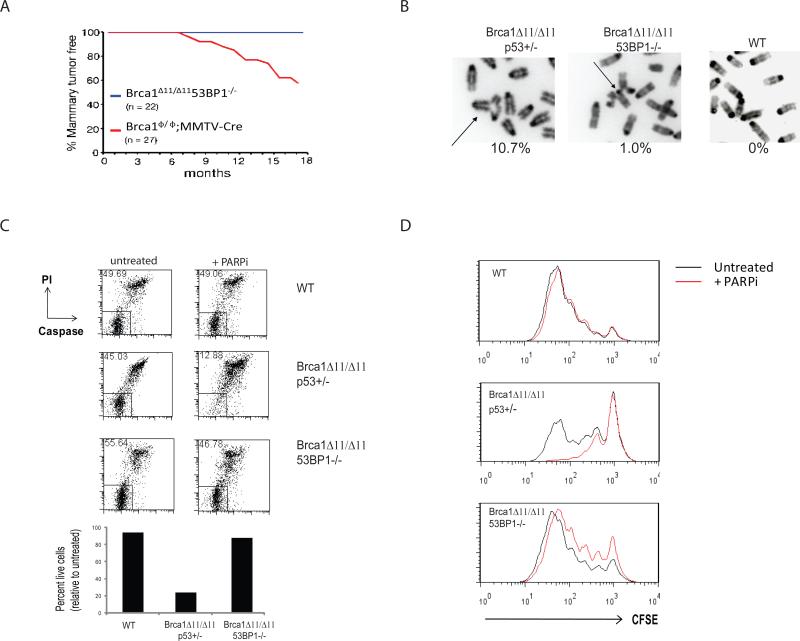
Figure 1. Deletion of 53BP1 reduces mammary tumorigenesis, radial chromosome formation and cellular proliferation defects in Brca1Δ11/Δ11 cells.
(A) Breast cancer incidence in mice deficient in Brca1. The red line indicates tumor incidence in mice with deletion of Brca1 exon 11 targeted to the mammary cells using a Cre transgene under control of the MMTV LTR promoter (Brca1ϕ/ϕ;MMTV-Cre). The blue line shows mammary tumor incidence in Brca1Δ11/Δ11 53BP1-/- mice, in which Brca1 exon 11 and 53BP1 are deleted in all cells. (B) Radial chromosome structures characteristic of metaphases from Brca1Δ11/Δ11 p53+/- B cells. The percentage of metaphases containing radial chromosomes in Brca1Δ11/Δ11 p53+/- (n=300), Brca1Δ11/Δ11 53BP1-/- (n=300) and WT (n=100) cells is indicated. Note that equivalent results were seen with Brca1Δ11/Δ11 p53+/- and Brca1Δ11/Δ11 p53-/-cells. (C) Flow cytometry analysis of B cells to measure cell survival. The labeled populations in the dot plots are viable cells, identified by their ability to exclude PI and their lack of caspase 3 activation. The chart shows the frequency of live cells treated with PARP inhibitor normalized to the untreated population. (D) Proliferation of B cells pulsed with CFSE and cultured with and without PARP inhibitor. CFSE signal diminishes with increasing cell division, so that cells that do not divide have the highest CFSE signal. See also Fig. S1.
Brca1 is thought to suppress malignancy by promoting HR (Moynahan et al., 1999; Scully et al., 1999; Venkitaraman, 2004). In light of the dramatic reduction in the frequency of mammary tumors in Brca1Δ11/Δ11 53BP1-/- animals, we hypothesized that loss of 53BP1 might specifically affect the ability of Brca1-deficient cells to repair replication-associated DNA damage. To test this, we examined chromosomal aberrations in metaphase spreads from WT, Brca1Δ11/Δ11 p53+/- and Brca1Δ11/Δ11 53BP1-/- B cells that were stimulated to divide ex vivo for three days. 10.7 % of Brca1Δ11/Δ11p53+/- B cells (n=300) carried one or more asymmetric radial chromosome structures, a type of chromatid exchange characteristic of HR deficiency (Venkitaraman, 2004) (Fig. 1B). Strikingly, these chromosome aberrations were present in just 1.0% of Brca1Δ11/Δ11 53BP1-/- cells, equivalent to a 10-fold reduction relative to Brca1Δ11/Δ11 p53+/- (n=300). Radial chromosomes were undetectable in WT and 53BP1-/- cells (n=100 cells analyzed for each genotype; Fig. 1B and Fig. 2A).
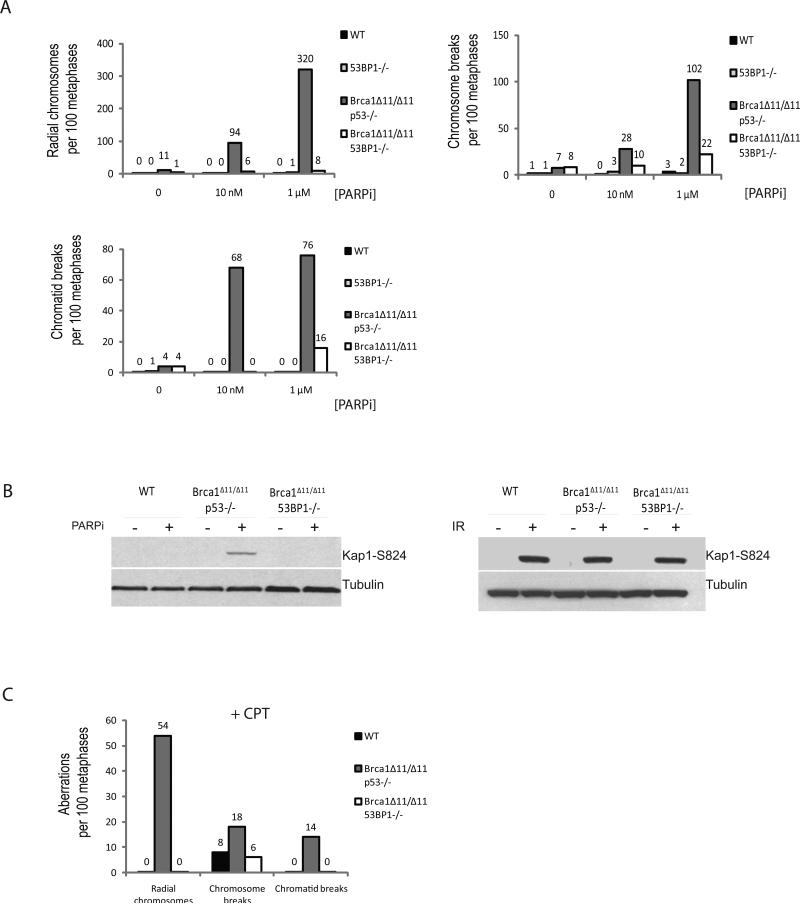
Figure 2. Deletion of 53BP1 reverses sensitivity of Brca1Δ11/Δ11 cells to PARPi and camptothecin.
(A) Analysis of genomic instability in metaphases from B cells treated with 0, 10 nM and 1μM PARP inhibitor. Charts show the number of radial chromosomes, chromatid breaks and chromosome breaks per 100 metaphases (n=50 metaphases analyzed in each case). Note that genomic instability in Brca1Δ11/Δ11 cells is independent of p53 status and equivalent results were seen in Brca1Δ11/Δ11 p53+/- and Brca1Δ11/Δ11 p53-/- cells. (B) Western blot showing Kap1 phosphorylation in B cells from the indicated genotypes treated with PARP inhibitor or 5Gy ionizing radiation. (C) Analysis of genomic instability in metaphases from B cells treated with 4nM camptothecin (CPT). B cells were cultured overnight with CPT prior to fixation and preparation of metaphase slides.
To exclude the possibility that p53 heterozygosity provides a survival advantage to genomically unstable Brca1 deficient cells by allowing aberrant chromosomes to persist (Callen et al., 2007; Difilippantonio et al., 2008), we quantified the incidence of radial chromosomes in Brca1Δ11/Δ11 conditional B cells. By infecting these cells with a virus expressing Cre recombinase, we were able to specifically delete Brca1 exon 11 in p53-sufficient cells. We found that these Brca1ϕ/ϕ cells still showed a significant increase (8-fold) in the number of radial chromosomes relative to Cre-infected Brca1ϕ/ϕ 53BP1-/- cells (Fig. S1A). Radial chromosome formation in Brca1ϕ/ϕ cells is therefore independent of p53 status but dependent on 53BP1.
To investigate further the effect of 53BP1 in regulating repair of DNA breaks occurring during replication, we challenged Brca1 mutant cells with a chemical inhibitor of PARP (PARPi; KU58948) (Fig.1 C and D). This drug has been shown to cause cell death in Brca1-deficient cells, associated with high levels of genomic instability (Farmer et al., 2005). In the presence of PARPi, Brca1Δ11/Δ11 p53+/- cells showed much lower levels of survival relative to either WT or Brca1Δ11/Δ11 53BP1-/- cells (Fig. 1C). Equivalent PARPi-induced cell death was seen with Brca1Δ11/Δ11 p53-/- cells, which were nonetheless resistant to IR-induced apoptosis (Fig. S1B). Thus, PARPi-mediated apoptosis in Brca1Δ11/Δ11 cells, in contrast to IR-induced apoptosis, occurs independently of p53 status.
The ability of PARPi to inhibit cell proliferation in culture was measured by pulsing the cells with the fluorescent dye, CFSE (carboxyfluorescein succinimidyl ester). In this assay, cellular proliferation is revealed by stepwise loss of CFSE during the culture period. PARPi inhibited growth in Brca1Δ11/Δ11 p53+/- cells (Fig. 1D) as well as in Brca1Δ11/Δ11 p53-/- B cells (Fig. 5F). Brca1Δ11/Δ11 53BP1-/- cells were distinct in that they were practically insensitive to PARPi-induced apoptosis and divided almost normally whether or not PARPi was present (Fig.1C and Fig. 1D). Consistent with these findings in B cells, Brca1Δ11/Δ11 53BP1-/- mouse embryonic fibroblasts (MEFs) were also insensitive to cell killing with PARPi, whereas Brca1Δ11/Δ11 MEFs were hypersensitive to the drug (Fig. S1C). We conclude that 53BP1 is required for the toxic effect of PARP inhibition on Brca1Δ11/Δ11 cells.
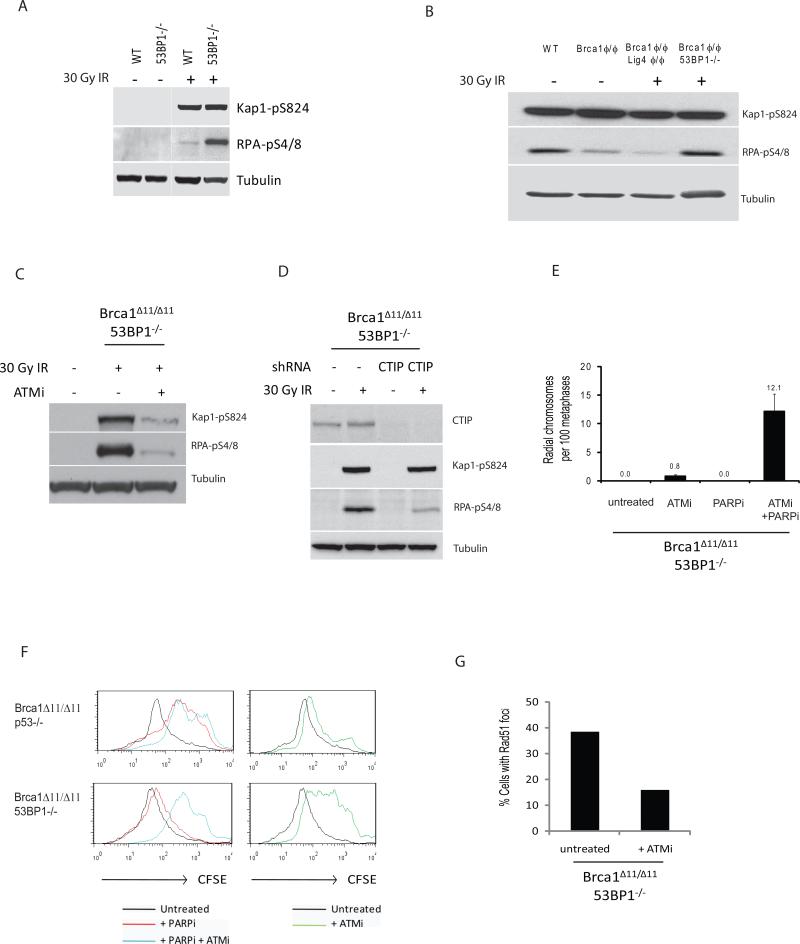
Figure 5. Rescue of homologous recombination in Brca1Δ11/Δ11 cells by 53BP1 deletion correlates with increased phosphorylation of RPA and is dependent on ATM and CtIP.
(A) Western blot analysis showing Kap1 and RPA phosphorylation in WT and 53BP1-/- cells in response to 30Gy ionizing radiation. (B) RPA and Kap1 phosphorylation after 30Gy ionizing radiation in Brca1- and Lig4-deficient cells. Knockout Brca1ϕ/ϕ and Lig4ϕ/ϕ cells were prepared by sorting B cells homozygous for the conditional alleles after infection with pMX-Cre-GFP retrovirus. (C) Western blot showing Kap1 and RPA phosphorylation in Brca1Δ11/Δ11 53BP1-/- cells after 30Gy ionizing radiation with and without ATM inhibitor (KU55993). ATMi (5 μM) was added 2 hours prior to irradiation. (D) Western blot analysis showing ionizing radiation-induced RPA phosphorylation in Brca1Δ11/Δ11 53BP1-/- MEFs after CtIP shRNA. Protein lysates were prepared from B cells infected with either vector (-) or CtIP shRNA (CTIP), with and without 30 Gy ionizing radiation. (E) Quantification of radial chromosome structures in metaphases from Brca1Δ11/Δ11 53BP1-/- cells treated with ATM inhibitor (5 μM) and / or PARP inhibitor (1 μM). ATMi was added at the start of the B cell culture, PARPi at 24 hours, and metaphases were made at 48 hours (n ≥ 240 metaphases; mean +/- s.d. shown from two experiments). (F) CFSE dilution analysis showing proliferation of B cells cultured with ATM inhibitor and/or PARP inhibitor for 96 hours. (G) Quantification of Rad51 foci in Brca1Δ11/Δ11 53BP1-/- cells. Rad51 foci were induced with 5Gy of ionizing radiation and cells fixed for Rad51 staining 2 hours later. Cells were either untreated, or pre-treated for 2hrs with 5μM ATM inhibitor. See also Fig. S3.
PARPi is predicted to increase the frequency of S phase-associated chromosomal aberrations (Bryant et al., 2005; Farmer et al., 2005; Jackson and Bartek, 2009). To determine whether loss of 53BP1 affects S phase-specific genomic instability, we monitored the level of chromosome breaks and radial structures in Brca1 deficient cells treated with and without PARPi. Whereas WT and 53BP1-/- cells showed very low levels of genomic instability in the presence or absence of PARPi, exposure of Brca1Δ11/Δ11 p53-/- to PARPi caused a substantial increase in the level of genomic instability (Fig. 2A). For example, treatment of Brca1Δ11/Δ11 p53-/- cells with 1 μM PARPi increased the frequency of radial chromosomes from 0.1 per cell to an average of 3.2 per cell, and increased the number of double-strand breaks (chromatid and chromosome) from 0.1 to 1.8 per cell. Thus, PARPi treatment of Brca1Δ11/Δ11 p53-/- cells leads to a greater than 20-fold increase in aberrations (Fig. 2A). In contrast, the total frequency of chromosomal aberrations in PARPi-treated Brca1Δ11/Δ11 53BP1-/- cells was just 0.5 per cell, which is 10-fold lower than PARPi-treated Brca1Δ11/Δ11 p53-/- cells. Nevertheless, PARPi-induced genomic instability in Brca1/53BP1 double mutant mice was somewhat elevated relative to WT.
Our findings were further substantiated by the observation that PARPi induced a significant amount of DNA damage signaling in Brca1Δ11/Δ11 p53-/- cells. Brca1Δ11/Δ11 p53-/- cells pre-incubated with PARPi for 24 hours showed induction of Kap1 phosphorylation (Fig. 2B), while phosphorylated Kap-1 was undetectable in PARPi -treated WT and Brca1Δ11/Δ11 53BP1-/- cells (Fig. 2B). This was not due to an intrinsic defect in ATM signaling, because similar levels of Kap1 phosphorylation were detected in cells from all genotypes treated with IR (Fig. 2B).
To determine the effects other chemotherapeutic agents that induce replication damage, we challenged WT, Brca1, and Brca1/53BP1 deficient B cells with camptothecin (CPT). CPT and its derivatives are topoisomerase I poisons that induce DSBs during replication, and are widely used as anti-cancer drugs (Jackson and Bartek, 2009). Brca1 deficient cells are hypersensitive to CPT (Nakamura et al.). Consistent with this, following treatment of Brca1Δ11/Δ11 p53+/- B cells with 4nM CPT, radial chromosomes formed at a frequency of 0.54 per cell whereas these aberrations were undetectable in CPT treated Brca1Δ11/Δ11 53BP1-/- B cells (Fig. 2C). Similarly, loss of 53BP1 suppressed CPT-induced chromosome and chromatid breaks in Brca1-deficient cells (Fig. 2C). Thus, loss of 53BP1 greatly alleviates replication-associated aberrations and toxicity that clinically relevant chemotherapeutic agents confer on Brca1-deficient cells.
Loss of 53BP1 increases HR in Brca1-mutant cells
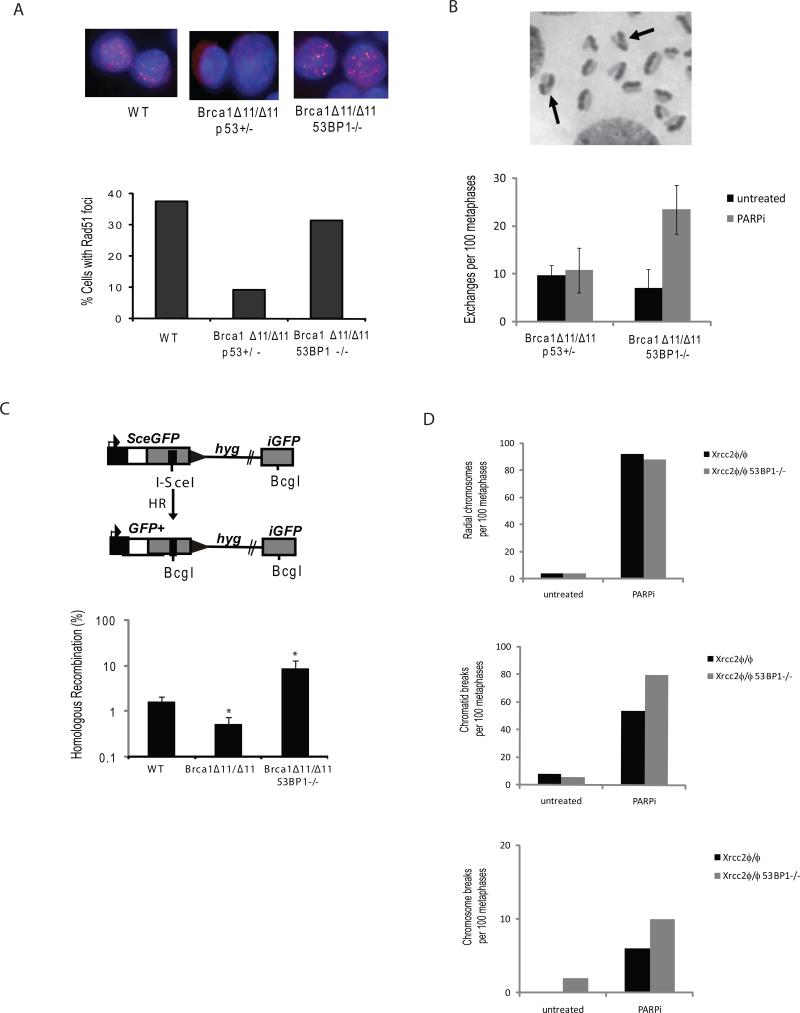
Replication-associated breaks are primarily repaired by HR. We therefore hypothesized that loss of 53BP1 might restore HR, a DNA repair pathway that is significantly compromised in the absence of Brca1 (Moynahan et al., 1999). To test this, we first examined Rad51 foci formation, a marker of HR, which is impaired in Brca1 mutant cells (Bhattacharyya et al., 2000; Scully et al., 1997). WT, Brca1Δ11/Δ11 p53+/- and Brca1Δ11/Δ11 53BP1-/- B cells were treated with IR, fixed, and stained with a Rad51 antibody (Fig. 3A). Whereas Rad51 foci formed in just 6.8% of irradiated Brca1Δ11/Δ11p53+/- cells, more than 30% of Brca1Δ11/Δ11 53BP1-/- cells exhibited Rad51 foci, similar to that observed in WT cells (Fig. 3A). Differences in the frequency of Rad51 foci formation was not due to alterations in cell cycle distribution, as the percentage of cycling cells was equivalent in all genotypes tested (Fig. S2).
Figure 3. Deletion of 53BP1 restores HR in Brca1- but not Xrcc2- mutant cells.
(A) Immunofluorescence images showing Rad51 foci (red) with DAPI counterstain (blue) in cells of the indicated genotypes after treatment with ionizing radiation. Chart shows the percentage of cells with Rad51 foci (n=100 counted for each genotype). (B) B cells grown for 36 hours in BrdUTP were fixed and metaphases prepared to visualize individual sister chromatids. Sister chromatid exchanges (SCEs) in metaphase chromosomes are indicated with arrows in the image in the top panel. The chart shows the number of SCEs in B cells of the indicated genotypes treated with and without PARP inhibitor. (C) Upper panel: Structure of the HR reporter substrate DR-GFPhyg is shown, along with the HR product expressing a functional GFP+ gene. Lower panel: Frequency of HR relative to total transfected cells in WT, Brca1Δ11/Δ11 and Brca1Δ11/Δ11 53BP1-/- MEFs, as measured using the DR-GFPhyg reporter. *p<0.0001 between WT and Brca1Δ11/Δ11, *p<0.0028 between WT and Brca1Δ11/Δ11 53BP1-/-. (D) Analysis of genomic instability in metaphases from B cells treated with and without PARP inhibitor (1μM). B cells were homozygous for a conditional Xrcc2 allele that was deleted using a B cell-specific CD19-Cre transgene to produce knockout Xrcc2ϕ/ϕ, and Xrcc2ϕ/ϕ 53BP1-/- cells. Charts show the number of radial chromosomes, chromatid breaks and chromosome breaks per 100 metaphases (n=50 metaphases analyzed in each case).
See also Fig S2.
As a second measure of HR, we monitored sister chromatid exchanges (SCEs), which are dependent on Rad51 activity (Sonoda et al., 1999). While the basal level of SCEs was the same, treatment with PARPi increased the frequency of SCEs in Brca1Δ11/Δ11 53BP1-/- but not in Brca1Δ11/Δ11 p53+/- cells (Fig. 3B). PARPi treatment of Brca1Δ11/Δ11 p53+/- therefore leads to an increase in aberrant radial chromosome formation which is a marker of HR-deficiency (Fig. 2A), whereas PARPi treatment of Brca1Δ11/Δ11 53BP1-/- cells promotes sister chromatid exchange which is a marker of HR-proficiency (Fig. 3B).
As a third measure of HR, we used the DR-GFPhyg reporter system, which is designed to measure error-free HR of a site-specific break formed by the rare-cutting endonuclease I-SceI (Nakanishi et al., 2005) In this reporter, error-free HR repair of the ISceI-induced DSB leads to restoration of a GFP+ gene that uses iGFP as the template (Fig. 3C). To analyze repair of chromosomal breaks, DR-GFPhyg was integrated into WT, Brca1Δ11/Δ11, and Brca1Δ11/Δ11 53BP1-/- immortalized MEF cell lines. From these experiments, we found that HR in Brca1Δ11/Δ11 cells was reduced relative to WT (3-fold, p<0.0001), whereas HR in Brca1Δ11/Δ11 53BP1-/- cells was increased relative to WT (5-fold, p <0.0028) (Fig. 3C). Thus, in Brca1Δ11/Δ11 MEFs, loss of 53BP1 causes a 16-fold increase in HR repair of a site-specific chromosomal break. In conclusion, loss of 53BP1 increases HR in Brca1 mutant cells, as evidenced by Rad51 foci, sister chromatid exchange, and recombination reporter assays.
53BP1 does not affect the activity of XRCC2 in HR
To examine whether loss of 53BP1 could rescue the defects in cells deficient in other HR components, we examined chromosomal aberrations in cells from mice deficient for Xrcc2, a Rad51 paralog which, like Brca1, is required for Rad51 foci formation (Liu et al., 1998; Takata et al., 2001). Metaphase spreads were prepared from conditional Xrcc2ϕ/ϕ B cells, in which XRCC2 was deleted using a B cell-specific CD19-Cre transgene. As expected, Xrcc2-deficient cells stimulated to proliferate in vitro exhibited chromosome and chromatid breaks and radial chromosomes, which was increased by treatment with PARPi (Fig. 3D) . Surprisingly, whereas 53BP1 deletion promoted genome stability in Brca1-deficient cells (Fig. 2A and Fig. S1), this effect was not seen in Xrcc2 knockout cells (Fig. 3D). Rather, the frequency of breaks and asymmetric radial chromosomes was similar in Xrcc2ϕ/ϕ 53BP1-/- and Xrcc2ϕ/ϕ cells. Thus, despite the fact that both Brca1 and Xrcc2 are required for Rad51 foci formation during HR, loss of 53BP1 reverses the HR defect in Brca1- but not in Xrcc2-deficient cells. We hypothesize that Xrcc2 acts downstream of Brca1 in HR (Nagaraju et al., 2009), at a later stage that is not affected by the presence of 53BP1 (see discussion below).
Lig4 is required for radial fusions in Brca1 mutant cells
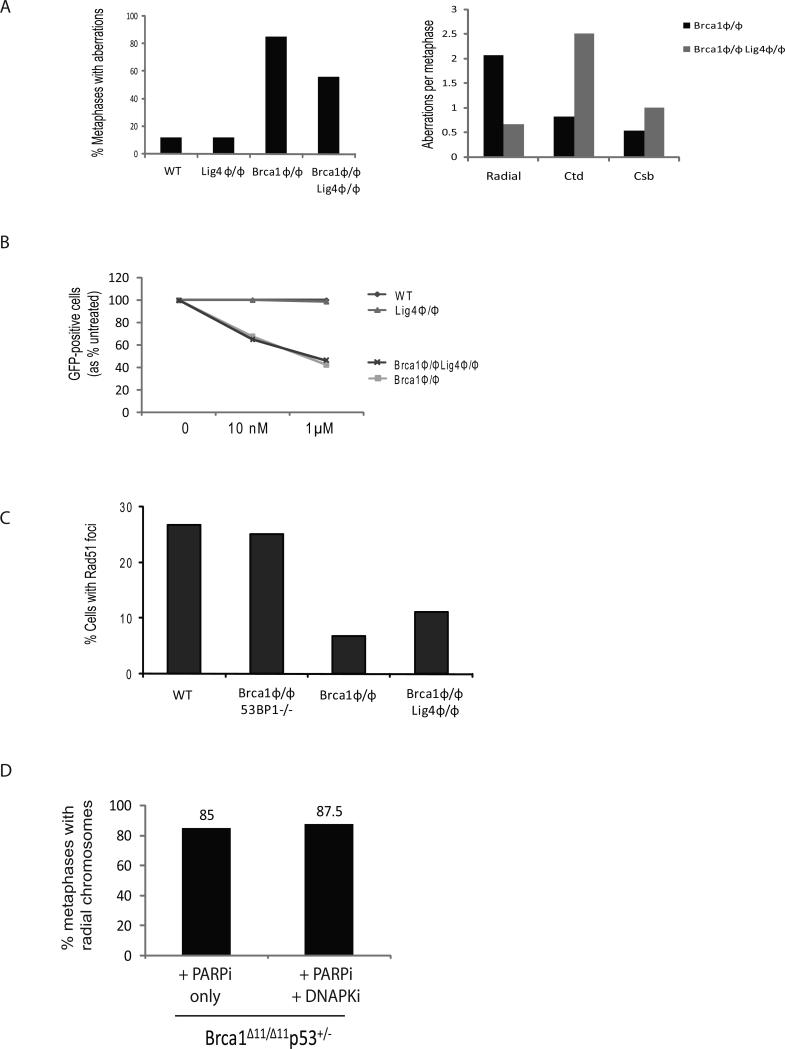
As measured by reporter substrates, the frequency of HR is enhanced in the absence of the NHEJ proteins Ku70, XRCC4, DNA-PKcs (Pierce et al., 2001) as well as by overexpression of a dominant negative 53BP1 fragment (Xie et al., 2007), suggesting that proteins of the NHEJ pathway can have suppressive effects on HR. Lig4 plays an essential role in NHEJ, so we hypothesized that Lig4 deficiency might also rescue the HR defect in Brca1 mutant cells. To test this hypothesis, we used Cre-mediated deletion to delete conditional alleles of Lig4 and Brca1 to make B cells homozygous for the knockout Lig4ϕ/ϕ and Brca1ϕ/ϕ alleles. We scored the level of chromosomal aberrations in mitotic spreads from these cells after overnight treatment with 1 μM PARPi. Whereas WT and Lig4ϕ/ϕ cells treated with PARPi did not accumulate DNA damage, 80% of Cre-infected Brca1ϕ/ϕ cells exhibited chromosomal instability (Fig. 4A). Aberrant Brca1ϕ/ϕ cells contained 0.8 chromatid breaks/cell, and most contained 2 or more radial fusions per cell, indicating that the majority of breaks are resolved into complex chromosome rearrangements in Brca1 mutant cells (Fig. 4A). 60% of PARPi-treated Brca1ϕ/ϕ Lig4ϕ/ϕ displayed genomic instability. However, only 0.66 radials/cell were present in Brca1ϕ/ϕ Lig4ϕ/ϕ cells, as opposed to Brca1ϕ/ϕ where approximately 2.1 radials/cell were observed. Instead, Brca1ϕ/ϕ Lig4ϕ/ϕ cells were characterized by high levels of unresolved chromatid breaks (average of 2.5 per cell vs. 0.8 in Brca1ϕ/ϕ cells; Fig. 4A). Thus, unlike 53BP1 deficiency, which reduces all types of chromosome aberrations in Brca1 deficient cells (Fig. 2A), Lig4 deletion only affects the frequency of radial chromosomes. Lig4 therefore appears to be specifically required for the processing of chromatid breaks into radial fusions in Brca1 mutant cells.
Figure 4. Deletion of Lig4 inhibits formation of radial chromosomes but does not rescue cell proliferation or genomic instability in Brca1Δ11/Δ11 cells.
(A) Analysis of genomic instability in metaphases from B cells treated with PARP inhibitor (1μM). Conditional Brca1 and Lig4 alleles were deleted using pMX-Cre-GFP retroviral transduction to generate B cells that were homozygous for the knockout Brca1ϕ/ϕ and Lig4ϕ/ϕ alleles. Chart (left) shows percentage of metaphases with genomic instability from cells of the indicated genotypes. Chart (right) quantifies the distribution of radial chromosomes, chromatid breaks (ctd) and chromosome breaks (csb) among the aberrant metaphases (n=50 metaphases of each genotype analyzed). (B) Cell survival after treatment with PARP inhibitor. Cultured B cells homozygous for conditional Brca1 and Lig4 alleles were infected with pMX-Cre-GFP retrovirus to produce GFP+ knockout Brca1ϕ/ϕ and Lig4ϕ/ϕ cells. The chart shows the percentage of GFP+ cells that were present in B cell cultures of the indicated genotypes five days post-infection. PARP inhibitor treatment (10nM or 1μM) led to disappearance of Brca1ϕ/ϕ and Brca1ϕ/ϕ Lig4ϕ/ϕ cells from the cultures. (C) Chart showing the percentage of cells with Rad51 foci after ionizing radiation as measured by immunofluorescence (n ≥ 160 cells). (D) Frequency of radial chromosomes in metaphase spreads from Brca1Δ11/Δ11 p53+/- B cells cultured for 24 hours with either PARP inhibitor (PARPi) or PARPi + DNA-PKcs inhibitor (DNAPKi).
To test whether the accumulation of chromatid breaks in Brca1ϕ/ϕ Lig4ϕ/ϕ cells has an effect on cell growth and survival, we compared the viability of WT, Lig4ϕ/ϕ, Brca1ϕ/ϕ and Brca1ϕ/ϕ Lig4ϕ/ϕ B cells that were treated with or without PARPi for 5 days. While Cre-infected WT and Lig4ϕ/ϕ cells were insensitive to PARPi (Fig. 4B), there was a precipitous drop in live Cre-infected (GFP+) Brca1ϕ/ϕ and Brca1ϕ/ϕ Lig4ϕ/ϕ cells in response to PARPi (Fig. 4B). Thus, Lig4 deficiency does not relieve the toxic effects of PARPi on Brca1-deficient cells.
To determine whether loss of Lig4, like 53BP1 deficiency, rescues the HR defect in Brca1 mutant cells, we examined IR induced Rad51 foci formation. Whereas more than 25% of WT and 53BP1-/- Brca1ϕ/ϕ Cre-infected cells exhibited Rad51 foci, Rad51 foci formed at a lower frequency (8-11%) in Brca1ϕ/ϕ and Brca1ϕ/ϕ Lig4ϕ/ϕ cells (Fig. 4C). Thus, despite the fact that both Lig4 or 53BP1 deficiency abrogates PARPi-induced radial fusions in Brca1 mutant cells, only 53BP1 but not Lig4 deficiency rescues PARPi induced cell death, chromosomal instability and defects in IR-induced Rad51 foci formation. Similarly, we found that inhibition of the kinase activity of DNA-dependent protein kinase (DNA-PKcs), which is considered a key component of the NHEJ repair pathway, did not reduce radial chromosome formation in PARPi-treated Brca1 deficient cells (Fig. 4D). Reversal of genomic instability in Brca1Δ11/Δ11 cells is therefore dependent on loss of 53BP1, and cannot be achieved by inactivation of any component of the non-homologous end joining pathway.
Loss of 53BP1 promotes RPA phosphorylation in a manner dependent on ATM and CtIP
Brca1 has been implicated in 5′-3′ DSB resection (Schlegel et al., 2006; Yun and Hiom, 2009a), which involves the nucleolytic processing of DSBs to produce single-stranded DNA (ssDNA) overhangs that are required for HR (Mimitou and Symington, 2009). During the process of resection, Replication Protein A (RPA) is loaded onto ssDNA, where it is then phosphorylated. To determine whether loss of 53BP1 affects resection, we irradiated WT and 53BP1-/- B cells and assessed phosphorylation of RPA and Kap-1. Kap-1 phosphorylation, which occurs independently of DSB resection, was similar in irradiated WT and 53BP1-/- cells (Fig. 5A); however, the level of RPA phosphorylation was consistently higher in 53BP1-/- cells (Fig. 5A). We conclude that 53BP1 suppresses RPA phosphorylation.
To test whether 53BP1 or Lig4 inhibits RPA phosphorylation in Brca1 mutant cells, we compared the levels of phosphorylation in WT, Brca1ϕ/ϕ, Brca1ϕ/ϕ 53BP1-/- and Brca1ϕ/ϕ Lig4ϕ/ϕ cells. Whereas Brca1ϕ/ϕ and Brca1ϕ/ϕ Lig4ϕ/ϕ B cells had similarly decreased levels of phosphorylated RPA compared to WT (Fig. 5B), importantly Brca1ϕ/ϕ 53BP1-/- B cells displayed a rescue of RPA phosphorylation to near WT levels (Fig. 5B). Thus, loss of 53BP1 but not Lig4 is able to promote ssDNA formation competent for RPA phosphorylation. This may explain why combined deficiencies in Brca1/53BP1 but not Brca1/Lig4 reverse the HR defect in Brca1 mutant cells.
To provide physical evidence for resection of DSBs in 53BP1-/- cells, we measured the degree of DNA processing at chromosomal translocation junctions. B cells containing I-SceI sites integrated into the c-myc (MycI allele) and IgH locus (IgHI allele) were infected with a retrovirus encoding I-SceI to induce translocations between c-myc and IgH (Fig. S3A). In this system, translocation breakpoints are found at various distances from the site of I-SceI induced DSBs, dependent on the degree of nucleolytic processing at the break site (Robbiani et al., 2008). We used PCR to map translocation breakpoints induced by the I-SceI system in AID-/- and AID-/- 53BP1-/- cells (Fig.S3B; use of AID-/- cells restricts translocations to breaks induced by I-SceI). All of the sequenced junctions showed processed DNA ends (Fig. S3C). However, the average total distance of the breakpoint position from the I-SceI sites on c-myc and IgH was approximately 2 fold greater in AID-/- 53BP1-/- cells compared with AID-/- (average resection, AID-/-: 448.2 nucleotides vs. AID-/- 53BP1-/-: 742.3 nucleotides, p=0.032) (Fig. S3C). Thus, loss of 53BP1 enhances nucleolytic processing of DNA ends. These findings are consistent with enhanced degradation of coding ends during V(D)J recombination in 53BP1-/- mice (Difilippantonio et al., 2008), and inhibition of DNA end resection by the yeast 53BP1 homolog, Rad9 (Lazzaro et al., 2008).
Rescue of Brca1 mutant cells by 53BP1 deletion is dependent on ATM
ATM kinase activity is required for nucleolytic processing of DSBs to generate RPA-coated ssDNA (Jazayeri et al., 2006). The CtIP protein is phosphorylated by ATM after DNA damage induced by IR (Li et al., 2000), and forms a complex with Mre11/Rad50/Nbs1 and Brca1, which is required for DSB resection (Chen et al., 2008; Sartori et al., 2007; Yun and Hiom, 2009a). Prompted by these findings, we tested whether ATM activity and CtIP mediates the resection observed in Brca1Δ11/Δ11 53BP1-/- cells. We found that Brca1Δ11/Δ11 53BP1-/- B cells pretreated with ATM inhibitor (ATMi, KU55993) (Hickson et al., 2004) showed a significantly lower level of phosphorylated RPA after irradiation (Fig. 5C). Similarly, Brca1Δ11/Δ11 53BP1-/- MEFs infected with CtIP shRNA showed impairment of RPA phosphorylation (Fig. 5D). Thus, the increase in DSB resection promoted by the absence of 53BP1 is ATM- and CtIP-dependent.
Since increased RPA phosphorylation in Brca1/53BP1 double-deficient cells correlates with rescued genomic stability, we tested whether inhibition of the ATM-dependent pathway leading to RPA phosphorylation would sensitize Brca1Δ11/Δ11 53BP1-/- cells to the effects of PARPi. Treatment of Brca1Δ11/Δ11 53BP1-/- cells with ATMi induced a small increase in the frequency of radial chromosomes, and combination of ATMi and PARPi led to a 15-fold enhancement in radials relative to ATMi alone (Fig. 5E). Moreover, ATMi re-sensitized Brca1Δ11/Δ11 53BP1-/- cells to PARPi, evidenced by its ability to inhibit the growth of B cells (Fig. 5F). Finally, ATMi reduced Rad51 foci formation in Brca1Δ11/Δ11 53BP1-/- cells (Fig. 5G). Taken together, these results suggest that loss of 53BP1 promotes HR in Brca1 deficient cells in a manner dependent on ATM.
DISCUSSION
Interplay between NHEJ and HR proteins in S phase
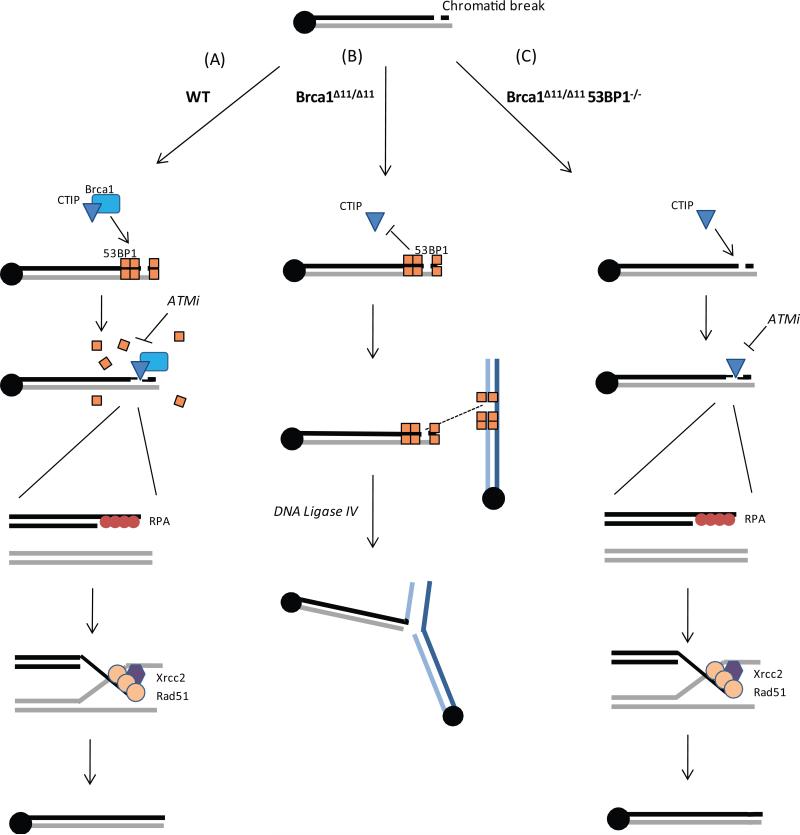
DNA double-strand breaks can be repaired by one of two major pathways: homology-based repair using the intact chromatid as a template (HR) or direct joining across the break site (NHEJ). Our data are consistent with the idea that these two DSB repair pathways compete for repair of replication-associated chromatid breaks (Saberi et al., 2007; Shrivastav et al., 2008; Sonoda et al., 2006). In WT cells, HR-mediated Xrcc2/Rad51-dependent sister chromatid recombination is the predominant repair pathway, and inter-chromosomal radial fusions are only rarely produced (Fig. 6A). In contrast, Brca1 deficiency prevents normal HR, so that chromatid breaks, instead of being faithfully repaired, are resolved into aberrant chromatid fusions between heterologous chromosomes. These fusions are dependent on Lig4, a component of the classical NHEJ pathway (Figure 6B). This shows that in the absence of normal Brca1-mediated HR, a toxic NHEJ pathway mediates rejoining of chromatid breaks.
Figure 6. Model for reversal of genomic instability in Brca1Δ11/Δ11 cells by 53BP1 deletion.
Chromatid breaks accumulate in S-phase cells as a consequence of errors in DNA replication or through failure of single-strand break repair (e.g. after PARP inhibition). (A) In WT cells, Brca1 displaces 53BP1 from DSBs, enabling resection at the break site by factors such as CtIP, which promotes RPA loading onto single-stranded regions of DNA. RPA is displaced by Rad51, which enables strand invasion at the homologous region of the sister chromatid. Rad51 acts in complex with several effectors of HR, including Xrcc2, leading to error-free, template-directed repair of the double-strand break. In Brca1Δ11/Δ11 cells, 53BP1 is not displaced, and inhibits resection. In the absence of resection, the break persists and if more than one chromatid break is present, these breaks can be joined by a Lig4-dependent NHEJ pathway. Joining of the chromatid breaks produces radial chromosome structures. (C) In Brca1Δ11/Δ11 53BP1-/- cells, 53BP1 is not present at the double-strand break site, enabling resection even in Brca1-deficient cells. RPA and downstream effectors of homologous recombination are able to load normally at the break site, permitting error-free DNA repair. See also Fig. S4.
Although radial chromosomes are reduced in Brca1-deficient cells that also lack Lig4, other types of DNA damage (notably chromatid breaks) persist, and there is no rescue of HR. By contrast, deletion of 53BP1 rescues Rad51-dependent HR almost to WT levels in Brca1-deficient cells. 53BP1 promotes formation of radial fusions in Brca1-mutant cells by a different mechanism than Lig4. According to our model, 53BP1-binding to chromatid breaks in Brca1 deficient cells interferes with HR, not by promoting end-joining per se, but rather by partially blocking ATM-dependent resection at the break site, which is necessary for HR-mediated repair (Fig. 6B and C). Thus, the previous puzzling observation that loss of 53BP1 suppresses both Brca1Δ11/Δ11 lethality and tumorigenesis (Cao et al., 2009) can now be explained by our finding that 53BP1 deficiency rescues HR by a mechanism dependent on ATM-mediated resection.
Our results show that DSB repair proteins have very different abilities to affect the choice of HR versus NHEJ. 53BP1 plays a major role in pathway choice in Brca1-deficient cells, whereas Lig4 does not affect the initial choice of HR versus NHEJ. Defects in HR in mammalian cells therefore might not be rescued simply by inhibiting the activity of any protein associated with NHEJ. Consistent with this, treatment of Brca1Δ11/Δ11 cells with an inhibitor of the NHEJ factor DNA-dependent protein kinase (DNA-PKcs) does not decrease the frequency of radial chromosome formation (Fig. 4D).
Loss of 53BP1 does not have the general effect of rescuing any defect in HR; for example, it does not normalize the genomic instability in Xrcc2-deficient cells (Fig. 3D). According to our model, 53BP1 affects the initial stage of HR, that is, exonuclease-mediated resection of the DSB. Xrcc2 might be active at a downstream stage of HR, as a cofactor of Rad51 during strand exchange (Fig 6A and C). In this case, deficiency in Xrcc2 would not be rescued by increased resection. Consistent with this idea, Xrcc2 has been shown to act late during gene conversion (Nagaraju et al., 2009).
53BP1 and Brca1 regulate the choice between NHEJ and HR
The precise role of 53BP1 in NHEJ is a matter of ongoing investigation. Previous work has shown that 53BP1 is required for long-range intra-chromosomal end-joining during V(D)J recombination and class switch recombination (Difilippantonio et al., 2008; Manis et al., 2004; Reina-San-Martin et al., 2007) as well as fusions between dysfunctional telomeres (Dimitrova et al., 2008). Other NHEJ-mediated events, such as the repair of DNA damage induced by ionizing radiation, are less profoundly affected by the absence of 53BP1 (Ward et al., 2004). This has led to the hypothesis that 53BP1 acts as a ‘synapsis factor’ required specifically for ‘long-range’ joining of distant DNA breaks by NHEJ (Difilippantonio et al., 2008; Reina-San-Martin et al., 2007). We found that radial chromosomes are present in Xrcc2-/- 53BP1-/- cells (Fig. 3D) as well as in Brca1Δ11/Δ11 53BP1-/- cells treated with ATMi (Fig. 5E), demonstrating that 53BP1 is not absolutely required for long-range interchromosomal joining of DNA DSBs. Consistent with this notion, 53BP1 is dispensable for AID-dependent (Ramiro et al., 2006) and I-SceI-induced (Fig. S3) translocations between c-myc and IgH. However, by inhibiting DSB resection, 53BP1 may increase the probability that blunt DNA ends can be ligated by Lig4-mediated classical NHEJ. According to this hypothesis, a key mechanism by which 53BP1 regulates the choice of HR versus NHEJ and promotes long range end joining could be via inhibition of DSB resection.
Brca1 has been implicated in several processes including DSB repair, chromatin remodeling, cell cycle progression, and transcription (Yun and Hiom, 2009b). Nevertheless, very little is known about how Brca1 contributes to these processes. Brca1 possesses ubiquitin-ligase activity, but this function does not appear to be essential for maintaining genomic stability (Reid et al., 2008). Based on our results, we propose that one critical function for Brca1, and perhaps other factors that commit cells to HR, might be to remove end-joining proteins such as 53BP1 from replication-associated breaks (Fig. 6A). Interestingly, the retention of Ku (Postow et al., 2008) and 53BP1 (Watanabe et al., 2009) is dependent on DSB-induced ubiquitylations of these factors. While active removal of end joining proteins by protein ubiquitylation may promote resection (Lee et al., 1998; Zhang et al., 2007), conversely, end resection itself could promote dissociation of end joining proteins (Fig. 6A). For example, Mre11/CtIP dependent DSB resection (Lengsfeld et al., 2007; Sartori et al., 2007) and/or Mre11-dependent nucleosome displacement (Tsukuda et al., 2005) might facilitate the generation of nucleosome-free ssDNA regions that would be incompatible with continued 53BP1 chromatin binding but accessible for recruitment of HR proteins. Since Brca1 forms a complex with the Mre11 and CtIP (Chen et al., 2008; Sartori et al., 2007; Yu et al., 1998), Brca1 might play a similar role in the dynamic removal of NHEJ proteins from DSBs that would promote faithful sister chromatid repair and avoid inappropriate end-ligation.
Implications for cancer therapy
Germline mutations in Brca1 confer susceptibility of developing breast and ovarian cancer with high penetrance (Antoniou et al., 2003). Cancers that develop in heterozygous carriers typically lose the second functional Brca1 allele during tumor progression. It is possible that loss of one allele confers a defect in HR (Cousineau and Belmaaza, 2007), and reduction in gene dosage (haploinsufficiency) fuels sufficient genomic instability to accelerate cancer promoting mutations, including loss of the remaining Brca1 WT allele. Heterozygous Brca1 mutant mice do not develop mammary tumors; however, mammary tumors develop with a long latency in mutant mice in which Brca1 is deleted in mammary epithelium. Notably, loss of 53BP1 significantly reduces mammary tumorigenesis in Brca1 deficient mice (Fig. 1A). Our study suggests that one therapeutic strategy to treat Brca1-mutation carriers might be the systemic use of compounds that inhibit 53BP1, which we predict would render Brca1 heterozygous carriers more HR competent. One potential side-effect of this approach could be defects in B cell immunoglobulin isotype switching, which is strictly dependent on 53BP1 (Fig. S4).
Since Brca1 is an essential gene, it remains unclear why deficiency in Brca1 is tolerated in tumor cells. One possibility is that Brca1 loss is specifically tolerated in breast and ovary. A more likely scenario is that secondary genetic alterations arise, in addition to loss of heterozygosity at Brca1, which promote the survival of Brca1 mutant cells. For example, loss of p53 seems to be an essential step in Brca1-associated tumorigenesis (Holstege et al., 2009; Xu et al., 1999). Tumors deficient in p53 and Brca1 are severely impaired in HR, and therefore sensitive to therapies which utilize PARPi to achieve synthetic lethality. However, treatment with PARPi provides a strong selective pressure for mutations that restore DSB repair capacity. Interestingly, many Brca1 deficient tumors overexpress Rad51 (Martin et al., 2007), which might partially restore DSB repair in these cells. Based on our study, loss of 53BP1 is another secondary mutation which would render Brca1-mutant cells HR competent and resistant to PARP inhibitors. Similar to 53BP1 deficiency, our model predicts that other mutations that enhance DSB resection would make Brca1 mutant cells resistant to PARP inhibition. However, our finding that loss of 53BP1 rescues Brca1-deficient HR by promoting ATM-dependent resection opens a therapeutic window for ATM inhibitors as a second line of chemotherapy for PARP inhibitor-resistant tumors (Fig. 5E and F). Thus, ATM inhibitors might be used therapeutically to re-sensitize Brca1 deficient tumors that develop resistance to PARPi.
EXPERIMENTAL PROCEDURES
Mice
Mice carrying the Brca1 exon 11 allele were obtained from the NCI mouse repository, 53BP1-/- mice were a gift from Junjie Chen, and Xrcc2 and Lig4 conditional mice were gifts from Peter McKinnon.
B cell cultures, retroviral transduction, and metaphase preparation
Resting B lymphocytes were isolated from mouse spleens and cultured with LPS (25μg/ml, Sigma) and IL-4 (5 ng/ml, Sigma) as described (Callen et al., 2007). ATM inhibitor (KU55993, KuDOS Pharmaceuticals) was used at 5 μM, DNA-PK inhibitor (NU7026, Sigma) was used at 20 μM and PARP inhibitor (KU58948, KuDOS) was used at various concentrations depending on the experiment. Irradiation was performed with a caesium-137 source. B cells were infected 24 hrs after culture with pMX-Cre-IRES-GFP as described (Robbiani et al., 2008). GFP+ B cells were sorted 4.5 days after infection using a FACSAria (Becton-Dickinson). For analysis of cell proliferation, CFSE (5 μM, Molecular Probes) labeling was at 37°C for 10 min. To quantify apoptotic cells, the PhiPhiLux G1D2 kit was used according to manufacturer's protocol (OncoImmunin, Inc.). For FISH analysis, metaphases were prepared and imaged as described (Callen et al., 2007).
Western blotting and Immunofluorescence
Primary antibodies were used at the following dilutions: anti-tubulin (1:15,000, Sigma), anti-Kap-1 pS824 (1:700, Bethyl Labs), anti-RPA pS4/S8 (1:1000 Bethyl Laboratories), and anti-CtIP (mAb 14-1, used at 1:50, gift from Richard Baer, Columbia University). MEFs were prepared for immunofluorescence by growth on 18mm x 18mm glass cover slips. Lymphocytes were dropped onto slides coated with CellTak (BD). Cells were fixed with methanol, incubated with anti-Rad51 primary antibody (H-92, Santa Cruz; used at 1:100) and detected with anti-rabbit-Alexa546 antibody (Invitrogen; used at 1:200).
MTT assay
MEFs were plated at 10,000 cells / well. After growth for four days, MTT substrate (Sigma) was added for 2 hours and solubilized in isopropanol according to the manufacturer's protocol. Plates were read using a Wallac Victor2 1420 multi-label counter.
shRNA
For CTIP depletion by shRNA, we obtained a construct containing the following hairpin sequence targeting CTIP exons 7 and 8, cloned into pMX-pie-IRES-GFP (gift of Davide Robbiani, Rockefeller University):
TGCTGTTGACAGTGAGCGAGCTCTCTATGTACAAATGAATTAGTGAAGCCAC AGATGTAATTCATTTGTACATAGAGAGCGTGCCTACTGCCTCGGA
Passage-immortalized mouse embryonic fibroblasts were infected with this construct or empty pMX-GFP vector. Infected cells were selected by growth in medium containing 2 μg/ml puromycin for one week. Cells containing the shRNA construct were maintained in 1 μg/ml puromycin.
GFP HR assay
HPRT-DRGFPhyg (Nakanishi et al., 2005) was electroporated into various MEF cell lines and integrants were selected in 90-125 μg/ml Hygromycin (Roche). For each integrant, an intact copy of the DR-GFPhyg reporter was confirmed by Southern Blot with the iGFP fragment as the probe. To measure HR, 4×105 cells were plated in a 12-well tissue culture dish 24 hr prior to transfection. Cells were transfected with either an I-SceI expression vector (pCBASce) to measure HR, or a GFP expression vector (pCAGGS-NZEGFP) to measure transfection efficiency (Pierce et al., 1999). HR relative to total transfected cells was determined by dividing the %GFP+ cells from each I-SceI transfection by the %GFP+ cells from a parallel GFP transfection. Values represent the mean of at least six independent transfections for each genotype, and error bars represent the standard deviation from the mean. Statistical analysis was performed using an unpaired t-test.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Richard Baer for the CtIP antibody, Davide Robbiani for the CtIP shRNA, Junjie Chen and Peter McKinnon for mice, Susan Sharrow and Larry Granger for flow cytometry, and Jeremy Daniel, Jaqueline Barlow for comments on the manuscript. MCN is an HHMI investigator. N.F. is a fellow of the Leukemia and Lymphma society. This research was supported in part by NIH grant AI037526 to MCN, an NIH/NCI grant RO1CA120954 to J.M.S., and the Intramural Research Program of the National Institutes of Health, USA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andreassen PR, Ho GP, D'Andrea AD. DNA damage responses and their many interactions with the replication fork. Carcinogenesis. 2006;27:883–892. doi: 10.1093/carcin/bgi319. [DOI] [PubMed] [Google Scholar]
- Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, et al. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem. 2000;275:23899–23903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- Brodie SG, Xu X, Qiao W, Li WM, Cao L, Deng CX. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene. 2001;20:7514–7523. doi: 10.1038/sj.onc.1204929. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Callen E, Jankovic M, Difilippantonio S, Daniel JA, Chen HT, Celeste A, Pellegrini M, McBride K, Wangsa D, Bredemeyer AL, et al. ATM prevents the persistence and propagation of chromosome breaks in lymphocytes. Cell. 2007;130:63–75. doi: 10.1016/j.cell.2007.06.016. [DOI] [PubMed] [Google Scholar]
- Cao L, Kim S, Xiao C, Wang RH, Coumoul X, Wang X, Li WM, Xu XL, De Soto JA, Takai H, et al. ATM-Chk2-p53 activation prevents tumorigenesis at an expense of organ homeostasis upon Brca1 deficiency. Embo J. 2006;25:2167–2177. doi: 10.1038/sj.emboj.7601115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35:534–541. doi: 10.1016/j.molcel.2009.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- Cousineau I, Belmaaza A. BRCA1 haploinsufficiency, but not heterozygosity for a BRCA1-truncating mutation, deregulates homologous recombination. Cell Cycle. 2007;6:962–971. doi: 10.4161/cc.6.8.4105. [DOI] [PubMed] [Google Scholar]
- Deng CX. BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic acids research. 2006;34:1416–1426. doi: 10.1093/nar/gkl010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiTullio RA, Jr., Mochan TA, Venere M, Bartkova J, Sehested M, Bartek J, Halazonetis TD. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol. 2002;4:998–1002. doi: 10.1038/ncb892. [DOI] [PubMed] [Google Scholar]
- Edwards SL, Brough R, Lord CJ, Natrajan R, Vatcheva R, Levine DA, Boyd J, Reis-Filho JS, Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Chen HT, Celeste A, Ward I, Romanienko PJ, Morales JC, Naka K, Xia Z, Camerini-Otero RD, Motoyama N, et al. DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nat Cell Biol. 2002;4:993–997. doi: 10.1038/ncb884. [DOI] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer research. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Holstege H, Joosse SA, van Oostrom CT, Nederlof PM, de Vries A, Jonkers J. High incidence of protein-truncating TP53 mutations in BRCA1-related breast cancer. Cancer research. 2009;69:3625–3633. doi: 10.1158/0008-5472.CAN-08-3426. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. Embo J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Goodarzi AA, Jeggo PA, Paull TT. 53BP1 promotes ATM activity through direct interactions with the MRN complex. Embo J. 2009 doi: 10.1038/emboj.2009.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- Liu N, Lamerdin JE, Tebbs RS, Schild D, Tucker JD, Shen MR, Brookman KW, Siciliano MJ, Walter CA, Fan W, et al. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. doi: 10.1038/ni1067. [DOI] [PubMed] [Google Scholar]
- Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer research. 2007;67:9658–9665. doi: 10.1158/0008-5472.CAN-07-0290. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- Nagaraju G, Hartlerode A, Kwok A, Chandramouly G, Scully R. XRCC2 and XRCC3 regulate the balance between short- and long-tract gene conversions between sister chromatids. Molecular and cellular biology. 2009;29:4283–4294. doi: 10.1128/MCB.01406-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS genetics. 6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, Wang ZQ, Jasin M. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & development. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes & development. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postow L, Ghenoiu C, Woo EM, Krutchinsky AN, Chait BT, Funabiki H. Ku80 removal from DNA through double strand break-induced ubiquitylation. J Cell Biol. 2008;182:467–479. doi: 10.1083/jcb.200802146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid LJ, Shakya R, Modi AP, Lokshin M, Cheng JT, Jasin M, Baer R, Ludwig T. E3 ligase activity of BRCA1 is not essential for mammalian cell viability or homology-directed repair of double-strand DNA breaks. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20876–20881. doi: 10.1073/pnas.0811203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina-San-Martin B, Chen J, Nussenzweig A, Nussenzweig MC. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur J Immunol. 2007;37:235–239. doi: 10.1002/eji.200636789. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Molecular and cellular biology. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–1120. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegel BP, Jodelka FM, Nunez R. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer research. 2006;66:5181–5189. doi: 10.1158/0008-5472.CAN-05-3209. [DOI] [PubMed] [Google Scholar]
- Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T, Livingston DM. Association of BRCA1 with Rad51 in mitotic and meiotic cells. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. Embo J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Molecular and cellular biology. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M, Sasaki MS, Tachiiri S, Fukushima T, Sonoda E, Schild D, Thompson LH, Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Molecular and cellular biology. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkitaraman AR. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat Rev Cancer. 2004;4:266–276. doi: 10.1038/nrc1321. [DOI] [PubMed] [Google Scholar]
- Wang B, Matsuoka S, Carpenter PB, Elledge SJ. 53BP1, a mediator of the DNA damage checkpoint. Science. 2002;298:1435–1438. doi: 10.1126/science.1076182. [DOI] [PubMed] [Google Scholar]
- Ward IM, Reina-San-Martin B, Olaru A, Minn K, Tamada K, Lau JS, Cascalho M, Chen L, Nussenzweig A, Livak F, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165:459–464. doi: 10.1083/jcb.200403021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic acids research. 2009;37:2176–2193. doi: 10.1093/nar/gkp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28:1045–1057. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC, Deng CX. Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nature genetics. 2001;28:266–271. doi: 10.1038/90108. [DOI] [PubMed] [Google Scholar]
- Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A, Deng CX. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nature genetics. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009a;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. Understanding the functions of BRCA1 in the DNA-damage response. Biochem Soc Trans. 2009b;37:597–604. doi: 10.1042/BST0370597. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hefferin ML, Chen L, Shim EY, Tseng HM, Kwon Y, Sung P, Lee SE, Tomkinson AE. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat Struct Mol Biol. 2007;14:639–646. doi: 10.1038/nsmb1261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.