Abstract
Type I invariant NKT cells (iNKT) are a subset of αβ T cells characterized by the expression of an invariant Vα14-Jα18 TCRα chain. iNKT cells derive from CD4+CD8+ double positive thymocytes (DP), and their generation requires a long DP thymocyte half-life, to allow for Vα14-Jα18 rearrangements, expression of glycolipid-loaded CD1d on DP thymocytes, and signaling through the SLAM/SAP pathway. Here we show that c-Myb plays a central role in priming DP thymocytes to enter the iNKT lineage by simultaneously regulating CD1d expression, DP half-life, and expression of SLAMF1, SLAMF6 and SAP.
Invariant NKT (iNKT) cells are a subset of αβ T cells characterized by the expression of an invariant Vα14-Jα18 TCR (Vα24-Jα18 in humans) in combination with certain TCRβ chains (Vβ8.2, Vβ7 or Vβ2 in mice, or Vβ11 in humans). iNKT cells in mice can be CD4+ or DN (CD4−CD8−), generally have a memory or activated phenotype (CD69+CD62L−CD44hiIL-2Rhi) and express markers characteristic of NK cells, including NK1.1, NKG2D and Ly49. They are found mainly in the liver and bone marrow, but also in the thymus, spleen and blood. iNKT cells develop in the thymus from the same precursors as conventional CD4+ and CD8+ αβ T cells, the CD4+CD8+ double positive (DP) thymocytes1. In contrast to conventional αβT cells, which are selected by major histocompatibility complex (MHC)-peptide complexes presented by thymic epithelial cells, iNKT are selected by lipid antigens presented by the non-polymorphic, MHC I-like molecule CD1d, present on the surface of other DP thymocytes (reviewed in 2–5). An absolute prerequisite for the generation of iNKT cells is the stochastic rearrangement of the invariant Vα14-Jα18 TCRα chain. Given the orderly sequence of rearrangements in the TCRα locus (proximal to distal), and the distal position of Jα18 in the Jα region, Vα14-Jα18 rearrangements are always secondary6. Therefore an extended DP lifespan is necessary for the development of iNKT cells, as evidenced by the defects in iNKT development observed in RORγT and Bcl-xL deficient mice7–9. A third prerequisite for iNKT selection is the cooperative engagement of the TCR and two members of the signaling lymphocytic?ctivation molecule (SLAM) family (SLAMF1 and SLAMF6)10. SLAM engagement recruits the adaptor SLAM-associated protein (SAP) and the Src kinase Fyn, both of which are essential for the selection of the iNKT cell lineage11–15.
Once they are positively selected, iNKT cells undergo a process of differentiation and expansion that results in the acquisition of their activated NK-like phenotype. iNKT cell differentiation in the thymus occurs through stages characterized by certain phenotypic markers and sensitivity to different mutations (reviewed in2,5,16). Once a DP thymocyte expresses the invariant TCR and receives signals through the SLAM/SAP and TCR (Control Point 1) it starts downregulating HSA (CD24) and upregulating first CD44, and later DX5. These cells are small, NK1.1− and not cycling. Some of these cells can exit the thymus and mature into NK1.1+ iNKT cells in the periphery, while others continue to differentiate in the thymus. The transition from NK1.1− to NK1.1+ iNKT is accompanied by a proliferative burst (Control Point 2). Molecules important for iNKT differentiation include costimulatory molecules -CD2817, signal transduction components -Itk, Rlk, NFκB, PKCθ18, cytokines -IL-1519 and transcription factors -PLZF20,21, T-bet22, GATA-323, c-myc24.
c-Myb is a transcription factor expressed in hematopoietic stem cells (HSC) and progenitors of all hematopoietic lineages and is required for definitive hematopoiesis25. Progression through hematopoiesis requires distinct c-Myb expression thresholds26,27. In the thymus, c-Myb is expressed at highest levels in the CD4−CD8− double negative (DN) and DP thymocyte subsets, and expression declines after positive selection28,29. c-Myb−/−/Rag1−/− chimeric mice have a block at the CD44loCD25− DN stage of development, proving that c-Myb is required for early T cell development 30. A dominant interfering c-Myb blocks β-selection31 while conditional deletion of c-Myb at different stages of T cell development suggested that c-Myb influences DN to DP transition, survival of DP thymocytes and CD4 single positive (SP) thymocytes differentiation28. Along with previous evidence32,33, these experiments suggested that c-Myb may be involved in TCRα TCRβ and TCRδ recombination. In addition, c-Myb is important for GATA-3 upregulation during CD4 lineage commitment29. Interestingly, GATA-3 seems to play a role in the control of survival and functional maturation of iNKT cells23.
Given the described role of c-Myb in the regulation of TCR β, δ and α recombination28,32,33, and the role of GATA-3 in iNKT cell functional maturation and survival23, we hypothesized that c-Myb could play a role in the development and/or function of iNKT cells. Our results show that c-Myb plays a central role in priming DP thymocytes to enter the iNKT lineage by simultaneously regulating CD1d expression, DP half-life, and expression of SLAMF1, SLAMF6 and SAP.
Results
c-Myb is required for the generation of iNKT cells
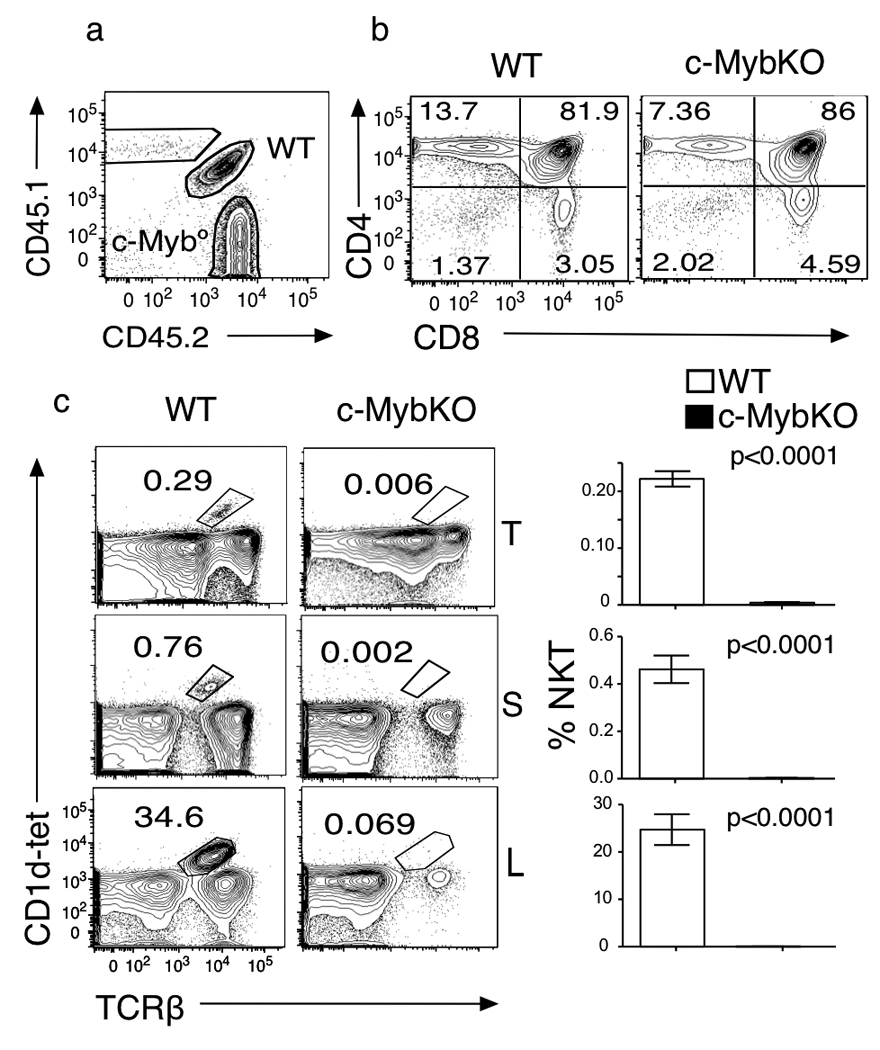
To analyze the effect of c-Myb deficiency during positive selection and avoid the confounding effects of an early deletion, we crossed c-Mybf/f mice28 with cd4-Cre mice to delete c-Myb at the DP stage of T cell development. We then compared the phenotype of c-Mybf/fcd4-Cre (c-MybKO) and GATA-3f/f cd4-Cre (GATA-3KO) mice. As previously reported for c-Mybf/f lck-Cre mice, c-Myb-deficient thymocytes have a moderate defect in the generation of mature CD4 SP cells (Fig1a). The effect is less pronounced than in GATA-3f/f cd4-Cre mice, suggesting additional contributions to GATA-3 upregulation during CD4 lineage commitment. Using a Cd1d-PBS57-tetramer34 we observed that, in comparison with GATA-3f/f cd4-Cre mice, which have normal numbers of thymic, but decreased numbers of spleen and liver iNKT cells23, c-Mybf/fcd4-Cre mice show a complete absence of iNKT cells in the thymus, spleen and liver (Fig 1a). Even after enrichment of tetramer-binding cells in the thymus35, we were unable to detect any tetramer-binding precursors in the thymus of c-Mybf/fcd4-Cre mice (Fig 1b), including HSAhi cells (stage 0), which have not yet received positive selection signals. These results indicate that c-Myb is absolutely required for the development of iNKT cells, in addition to regulating GATA-3.
Figure 1. c-Mybf/fcd4-Cre mice lack iNKT cells.
(a) Percentages of iNKT cells in the thymus, spleen and liver mononuclear cells from age-matched C57BL/6 (WT), GATA-3f/fcd4-Cre (GATA-3KO) and c-Mybf/fcd4-Cre (c-MybKO) mice stained with CD4, CD8, PBS57-loaded CD1d tetramer and TCRβ. Results representative of twelve independent c-Mybf/fcd4-Cre mice analyzed. (b) Percentages of CD1d-αGalcer tetramer+ immature (HSAhi) and mature (HSAlo) iNKT cells in the thymus of c-Mybf/wcd4-Cre (WT), or c-Mybf/fcd4-Cre (c-MybKO) thymocytes. Results shown are representative of four independent enrichment experiments.
The defect in iNKT cell development is cell intrinsic
iNKT cells are selected by DP to DP thymocyte interactions, via T cell receptor (TCR) recognition of CD1d-glycolipid complexes. We observed reduced expression of CD1d in c-Mybf/fcd4-Cre DP thymocytes (Supplementary Fig 1), so we tested whether the iNKT developmental defect in c-Mybf/fcd4Cre mice could be rescued by the presence of DP thymocytes with normal expression of CD1d. We reconstituted lethally irradiated CD45.1 wild-type hosts with a 1:1 mix of bone marrow cells from c-Mybf/fcd4-Cre mice (CD45.2) and F1 C57BL/6 (CD45.1;CD45.2) mice (Fig 2a). Chimeras were analyzed 8–12 weeks later to determine whether c-Mybf/fcd4-Cre DP thymocytes could develop into iNKT cells in the presence of normal DP thymocytes. In these conditions c-Mybf/fcd4-Cre thymocytes generated normal levels of CD8 SP cells, and, as expected, impaired numbers of CD4 SP thymocytes (Fig 2b). However, wild-type DP thymocytes could not rescue the generation of iNKT cells from c-Mybf/fcd4-Cre progenitors (Fig 2c), suggesting that the defect in CD1d expression is not sufficient to explain the iNKT phenotype observed in c-Mybf/fcd4-Cre mice.
Figure 2. The iNKT cell development defect in c-MybKO mice is cell intrinsic.
(a). Chimeric distribution of c-Mybf/fcd4-Cre (c-MybKO) and wild-type (WT) cells in the thymus of mixed bone marrow chimeras generated by injecting c-Mybf/f-cd4Cre (CD45.2) and F1(C57BL/6xB6-LY5.2/Cr) (CD45.1;CD45.2) bone marrow cells into lethally irradiated B6-LY5.2/Cr recipient mice (CD45.1) (b). Thymocyte profiles of live cells in the thymi of c-MybKO and wild-type mice. (c). Contribution of wild-type and c-MybKO bone marrow cells to iNKT populations in thymus, spleen and liver of the bone marrow chimeras described in a. Mean and SEM are shown. p-values were obtained using a two-tailed t test. Results are representative of ten chimeras analyzed in two independent experiments.
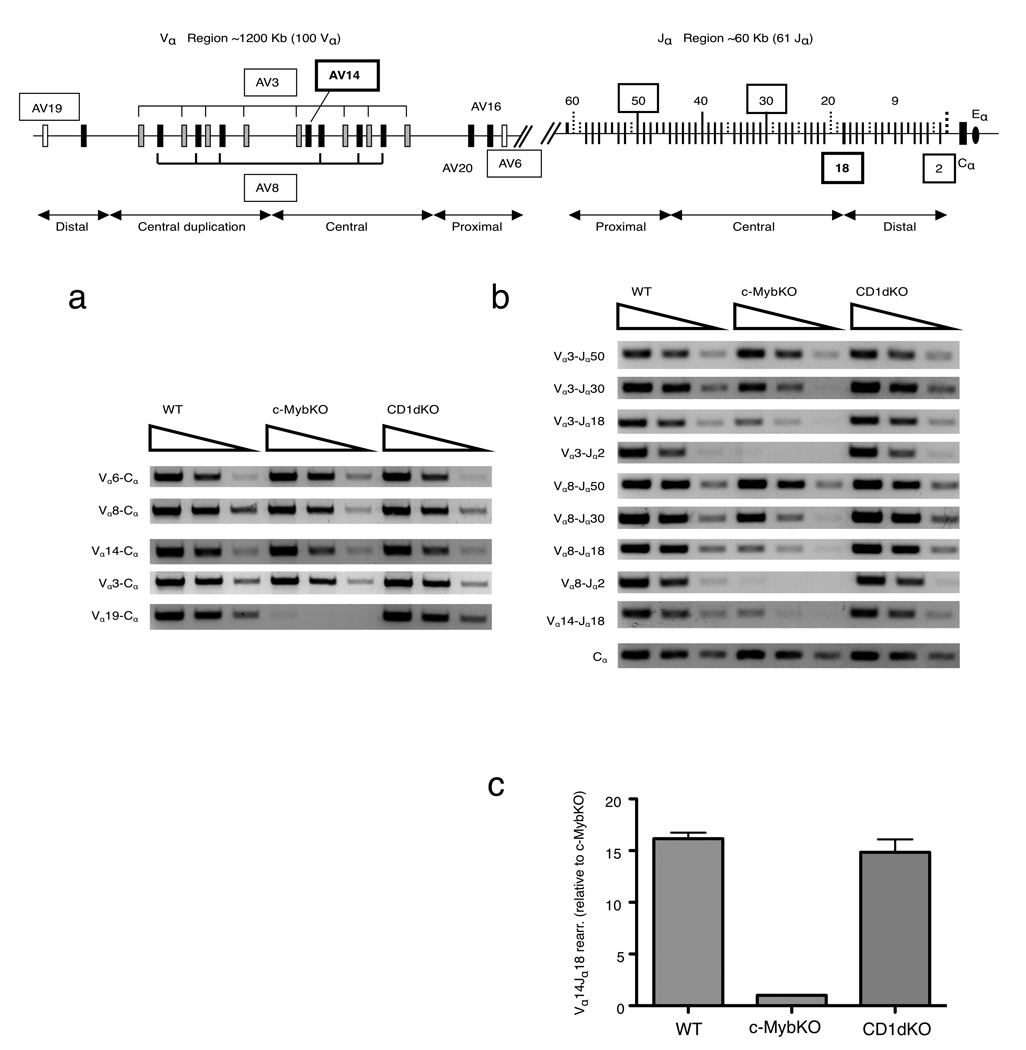
Defective distal Vα and Jα rearrangements in c-MybKO DP thymocytes
Because c-Myb is implicated in the regulation of recombination at the TCRα/δ locus, we used a semi-quantitative PCR-based assay9 for mature Vα transcripts to determine TCRα locus rearrangements in c-Mybf/f cd4-Cre mice, compared to thymocytes from wild-type C57BL/6 and CD1d-deficient mice. To assess Vα rearrangements before positive selection, the assay was performed on sorted DP thymocytes (CD4+CD8+TCRmidCD69−). Rearrangement in the Vα locus proceeds in an orderly fashion, with proximal Vα and Jα recombining first. Therefore, we tested the usage of proximal (AV6), central (AV3, AV8 and AV14) and distal (AV19) Vα segments in c-Mybf/fcd4-Cre, wild-type and CD1d-deficient thymocytes using family based AV3 and AV8 primers. There was a clear decrease in the amount of distal Vα transcripts in the c-Mybf/fcd4-Cre samples (AV19), while the usage of proximal (AV6) and central (AV8, AV14 and AV3) Vα transcripts was similar in DP thymocytes from C57BL/6, CD1d−/− and c-Mybf/fcd4-Cre mice (Fig 3a). These results suggested that c-Mybf/fcd4-Cre thymocytes have a defect in the usage of distal Vα segments.
Figure 3. c-MybKO thymocytes have defects in secondary TCRα rearragements.
(a,b) Semiquantitative RT-PCR of indicated Vα to Cα rearrangements (a) and Vα to Jα rearrangements (b) in wild-type (WT), c-Mybf/fcd4-Cre (c-MybKO) and CD1d−/− (CD1dKO) sorted DP thymocytes. Cα was used for template normalization.( c) Quantitative real-time PCR analysis of Vα14-Jα18 rearrangements in sorted CD69−TCRloCD4+CD8+ thymocytes from WT and c-Mybf/fcd4-Cre (c-MybKO) mice. Data were analyzed using the comparative CT method and normalized based on β-actin expression. Data are representative of at least three independent experiments.
To characterize Jα usage, we used family-based primers for the AV3 and AV8 families and primers to Jα segments in the proximal (Jα50), central (Jα30) and distal (Jα18 and Jα2) regions of the Jα locus7. We also analyzed the Vα14-Jα18 rearrangement characteristic for the iNKT TCR. DP thymocytes from c-Mybf/fcd4-Cre mice had normal levels of transcripts from every Vα segment to the proximal Jα50 (Vα3Jα50 and Vα8Jα50) (Fig 3b). Rearrangements to Jα30 (Vα3Jα30 and Vα8Jα30) were slightly reduced, while rearrangements to the most distal Jα2 segment (Vα3Jα2 and VαJα2) were not detected. Vα3-Jα18, Vα8-Jα18 and Vα14-Jα18 rearrangements were decreased in c-Mybf/fcd4-Cre DP thymocytes (Fig 3b, c). This shows that c-Mybf/fcd4-Cre thymocytes have a defect in distal Jα segments rearrangements, and in consequence they can not efficiently generate the Vα14Jα18 rearrangements characteristic for the TCR required for iNKT cell generation.
Bcl-xL corrects DP survival defect in c-MybKO mice
The observed defect in secondary TCRα rearrangements could be due to a direct effect of c-Myb deficiency in Vα rearrangement, or, an indirect effect on DP thymocyte survival. A similar phenotype was reported in RORγT−/− and Bcl-xL−/− mice, due to the decreased half-life of DP thymocytes in these mice7–9. Because DP thymocytes from c-Mybf/f lck-Cre mice have decreased survival28, we analyzed whether defects in survival and/or RORγT and Bcl-xL expression caused the defect in our c-Mybf/f cd4-Cre model.
We used quantitative RT-PCR to analyzed the expression of survival factors (Bcl-xL, RORγT, Bcl-2 and Mcl-1) in sorted DP thymocytes from c-Mybf/fcd4-Cre and c-Mybf/f mice. With the exception of Bcl-xL, which showed a 50% reduction, expression of these survival factors was not significantly altered in c-Mybf/fcd4-Cre DP thymocytes, (Fig 4a).
Figure 4. c-MybKO DP thymocytes have a defect in survival.

(a) Quantitative real-time PCR analysis of Bcl-2, Bcl-xL, Mcl, and RORγT expression in sorted CD69−TCRloCD4+CD8+ thymocytes from c-Mybf/wcd4-Cre (WT) and c-Mybf/fcd4-Cre (c-MybKO) mice. Data were analyzed using the comparative CT method and normalized based on β-actin expression. Data shown here are averages and standard deviations of three independent experiments. (b) Contribution of c-Mybf/f-cd4-Cre and wild-type cells to the DN and DP compartments in the thymus of 1:1 mixed chimeras, expressed as a ratio between the two cell types. Two independent chimera experiments (BMQ-1 and BMQ-2) are shown. Dots represents independent mice (n=5 in each experiment). Mean and SEM are shown. p-values were obtained using a Mann-Whitney test..
The Bcl-xL expression defect is consistent with a competitive disadvantage of c-MybKO cells in the mixed bone marrow chimera described above. In the wild-type:c-Mybf/fcd4-Cre mixed chimeras, no significant differences were observed between wild-type and c-Mybf/fcd4-Cre donors at the DN level (before c-Myb has been lost), while Mybf/fcd4-Cre DP thymocytes were clearly underrepresented (Fig 4b), consistent with the previously reported role of c-Myb in regulating DP survival28. This competitive disadvantage was not observed in control experiments comparing wild-type and c-Mybf/wcd4-Cre cells (Fig 5d), excluding a Cre contribution to DP survival.
Figure 5. Bcl-xL overexpression in c-MybKO thymocytes rescues survival and rearrangement but not iNKT development.

(a) Donor contributions in bone marrow chimeras generated by injecting MiG-Bcl-xL transduced c-Mybf/fcd4-Cre (c-MybKO) (CD45.2) and wild-type (CD45.1;CD45.2) cells into lethally irradiated CD45.1 recipients, analyzed at 8–12 weeks (left), and a representative histogram of GFP expression in MiG-Bcl-xL transduced MybKO thymocytes (right). (b). Bcl-xL expression in GFP− and GFP+ c-MybKO thymocytes transduced MiG-Bcl-xL. (c) Thymic profiles (upper panel) and iNKT cells frequency (lower panels) in wild-type, c-MybKO and c-MybKO-Bcl-xL donor cells in the thymus (T) and Liver (L) of chimeras described in a. Result are representative of eight chimeric mice in two independent experiments. (d) DP thymocyte fitness across different competitive bone marrow chimeras. We compared the contribution of the test (B) versus control (A) populations in the DN and DP compartments, according to the following equation: Fitness Index (FI)=(%PopADN/%PopBDN)/(%PopADP/%PopBDP). FI=1, if the contribution of both populations to both compartments is unchanged., FI >1, if test DP thymocytes cannot compete with wild-type DP cells. Each dot represents an independent mouse. The last three columns summarize results from two independent experiments. Significance was determined using the Mann-Whitney test (E) Semiquantitative RT-PCR of indicated Vα to Cα, and Vα to Jα rearrangements in DP thymocytes from C57BL/6 (WT), c-Mybf/fcd4-Cre (GFP−) and c-Mybf/fcd4-Cre/BclxL (GFP+). Cα was used for template normalization. Data are representative of three independent experiments. (f) Quantitative real-time PCR analysis of Vα14-Jα18 rearrangements in sorted CD69−TCRloCD4+CD8+ thymocytes from WT, c-Mybf/fcd4-Cre GFP−(MybKO) and c-Mybf/fcd4-Cre GFP+ (c-MybKO;Bcl-xL) from three independent chimeras. Data were analyzed using the comparative CT method and normalized based on β-actin expression.
To directly test whether the reduction in Bcl-xL expression was responsible for the lack of iNKT cells in c-Mybf/fcd4-Cre mice, we set up competitive bone marrow chimeras with c-Mybf/fcd4-Cre cells in which Bcl-xL expression was enforced using retroviral infection. Lineage− bone marrow cells from c-Mybf/fcd4-Cre mice were transduced with a retrovirus encoding Bcl-xL and GFP (Mig-Bcl-xL). 24 h later, a 1:1 mixture of Bcl-xL transduced c-Mybf/fcd4-Cre (CD45.2) and F1 C57BL/6(CD45.1;CD45.2) Lineage− bone marrow cells was injected into lethally irradiated wild-type (CD45.1) recipients. The development of iNKT cells in the thymus, spleen and liver of the chimeras was analyzed 8–10 weeks later, comparing CD45.1+CD45.2+ cells (wild-type) to CD45.2+ GFP− (c-MybKO) and CD45.2+ GFP+ (c-MybKO-Bcl-xL) (Fig 5a). Overexpression of Bcl-xL could not rescue development of iNKT cells in c-Mybf/f cd4-Cre DP thymocytes (Fig 5b), although the virus induced high expression of Bcl-xL in c-Mybf/fcd4-Cre DP thymocytes (Fig 5c). The Bcl-xL transgene was functional, because its expression improved the ability of c-Mybf/fcd4-Cre cells to compete with wild-type thymocytes in the DP compartment (Fig 5d), and increased the percentage of CD8 SP thymocytes, as previously described36. In similar experiments, overexpression of RORγT, an upstream regulator of Bcl-xL also failed to rescue iNKT cell development in c-Mybf/fcd4-Cre thymocytes (Supplementary Fig 2). These data demonstrate that correcting the survival defect of c-Mybf/fcd4-Cre thymocytes is not sufficient to rescue iNKT cell development.
To further address if the defect in secondary TCRα rearrangements in c-Mybf/fcd4-Cre thymocytes was due exclusively to survival or to a direct effect of c-Myb on rearrangement, we sorted wild-type, c-Mybf/fcd4-Cre-GFP+ and c-Mybf/fcd4-Cre-GFP− DP thymocytes from the bone marrow chimeras, and analyzed rearrangements across the TCRα locus, as described above. c-Mybf/fcd4-Cre DP thymocytes expressing Bcl-xL had detectable levels of distal TCRα rearrangements, including Vα14-Jα8 (Fig 5e, f), suggesting that survival rescue is sufficient to restore normal secondary rearrangements.
These experiments demonstrate that c-Myb regulates the survival of DP thymocytes and that rescue of DP thymocyte survival by the forced expression of Bcl-xL restores secondary TCRα rearrangements. However, this is not sufficient to rescue iNKT cell development, suggesting that c-Myb controls additional pathways required in this process.
Partial rescue by a rearranged TCRα chain
To directly test whether lack of Vα14Jα18 rearrangements was the only defect in c-Mybf/fcd4-Cre thymocytes, we bred c-Mybf/fcd-4Cre mice with transgenic mice expressing a rearranged Vα14Jα18 TCRα chain under the control of the CD4 promoter10. Compared with wild-type mice, Vα14-Jα18+ mice have higher percentages of tetramer-reactive cells in the thymus, and these iNKT cells have a higher percentage of HSAhi and NK1.1− cells, both CD4+ and DN, probably because many more iNKT cells are undergoing positive selection (Supplementary Fig 3).
Vα14-Jα18+ c-Mybf/fcd4-Cre (Vα14Tg c-MybKO) mice had decreased percentages and absolute numbers of iNKT cells in the thymus, spleen and liver when compared with Vα14-Jα18+ c-Mybf/wcd4-Cre (Vα14Tg) mice (Fig. 6a,b). Compared with Vα14Tg, Vα14Tg c-MybKO mice had lower percentages and numbers of the most mature thymic iNKT cell populations, CD44+NK1.1+ and HSAlo iNKT cells (Fig. 6c), suggesting that positive selection and intrathymic maturation of DP thymocytes bearing a PBS57-CD1d tet-reactive TCR is impaired in the absence of c-Myb.
Figure 6. Partial rescue of iNKT cell defect in c-MybKO mice by a rearranged Vα14Jα18 TCR transgene.
(a) Thymic profile of Vα14 Tg and Vα14 Tg c-MybKO mice between seven to nine weeks of age. (b) Percentages and absolute numbers of iNKT cells in the thymus (T), spleen (S) and liver (L) of Vα14 Tg and Vα14 Tg c-MybKO. Mean and SEM are shown. p-values were obtained using a two-tailed t test. (c) Percentages and absolute numbers of iNKT cell subpopulations in gated TCR-βhiPBS57-CD1dtet+ thymocytes from Vα14Tg and Vα14 Tg c-MybKO mice. p values were corrected for multiple hypothesis testing using the Bonferroni's Multiple Comparison Test. (d) IL-4 and IFNg secretion from iNKT isolated from wild-type, Vα14 Tg and Vα14 Tg c-MybKO mice and left unstimulated (U) or stimulated for 4h in vitro with PDBU and ionomycin (P+I). Cells were first stained for PBS57-CD1d tetramer and TCR-β and then intracellularly stained for IL-4 and IFNγ. Results shown are representative of three experiments. (a–d) n=6 for both Vα14 Tg and Vα14 Tg c-MybKO mice
To test whether the PBS57-CD1d-Tet+ cells generated in Vα14-Jα18+ c-Mybf/fcd4-Cre mice behave as iNKT cells, we stimulated thymocytes in vitro for 4 h with PMA and Ionomycin in the presence of Monensin, and determined interleukin-4 (IL-4) and interferon γ (IFN©) cytokine production by intracellular staining. PBS57-CD1d-Tet+ thymocytes from Vα14-Jα18+ c-Mybf/fcd4-Cre and Vα14-Jα18+/c-Mybf/wcd4-Cre mice behaved similarly to wild-type iNKT cells (Fig. 6d), with a high percentage of cells able to produce IL-4 and IFN-γ simultaneously after a short stimulation. This suggests that c-MybKO cells that can go through positive selection develop into apparently normal iNKT cells.
These results suggest that c-Myb plays additional roles in iNKT cell development, since not even expression of a CD1d-reactive TCR in many DP thymocytes is able to rescue normal development of iNKT cells in the absence of c-Myb. Furthermore, the phenotype of the iNKT cells generated, with an accumulation of more immature phenotypes, including HSAhi and NK1.1− suggests that the defect may be in early stages of positive selection of the iNKT lineage.
Expression of SAP, SLAMF1 and SLAMF6 in c-MybKO DP thymocytes
Besides the signals mediated by the TCR-CD1d interaction, development of iNKT cells requires signals initiated by homophilic interactions between members of the SLAM family -primarily SLAMF1 and SLAMF610, and mediated by the adaptor protein SAP11–15. Given the partial blockade in positive selection in DP thymocytes bearing a Tet-reactive TCR in c-Mybf/fcd4-Cre mice we analyzed the expression of these proteins in c-Mybf/fcd4-Cre DP thymocytes. Quantitative RT-PCR on sorted DP thymocytes from c-Mybf/fcd4-Cre and c-Mybf/wcd4-Cre mice showed decreased expression of SLAMF1, SLAMF6 and SAP mRNA in DP thymocytes from c-Mybf/fcd4-Cre (Fig 7a). As previously noted, RORγT expression was not altered (not shown). The surface expression of SLAMF1, and SLAMF6 is decreased in DP thymocytes from c-Mybf/fcd4-Cre mice when compared with c-Mybf/wcd4-Cre thymocytes (Fig 7b). This is only evident at the DP stage, and expression of other SLAMF family members, such as SLAMF3 and SLAMF5, is not affected. In addition, expression of SAP, as assessed by intracellular staining, was decreased in DP thymocytes from c-Mybf/fcd4-Cre mice. These results suggest that SLAM/SAP expression is impaired in c-Mybf/fcd4-Cre DP thymocytes and this could explain the positive selection defect observed in the Vα14-Jα18+/c-Mybf/fcd4-Cre mice.
Figure 7. Slamf1 and Slamf6 expression is decreased in c-MybKO mice.
(a) Quantitative real-time PCR of RORγT, Slamf1, Slamf6 and SAP expression in DP thymocytes sorted from c-Mybf/wcd4-Cre (WT) and c-Mybf/fcd4-Cre (c-MybKO) mice. Data were analyzed using the comparative CT method and normalized based on β-actin expression. Data represent averages of three independent experiments. (b) Expression of Slamf1, Slamf3, Slamf6, Slamf5 and SAP in DN and DP thymocytes from c-Mybf/wcd4-Cre(WT) and c-Mybf/fcd4-Cre (c-MybKO) mice as assessed by flow cytometry. Representative histograms and the mean and SEM of MFI in DP and DN populations, normalized to the average MFI in wild-type mice, are shown. n=7, compiled from three different experiments. p values were corrected for multiple hypothesis testing using the Bonferroni's Multiple Comparison Test.
SLAM/SAP signals are defective in c-MybKO DP thymocytes
To test whether signaling through the SLAM/SAP axis is impaired in c-Mybf/fcd4-Cre DP thymocytes we set up mixed bone marrow chimeras in which DP thymocytes from c-Mybf/fcd4-Cre or Jα18−/− mice37 were used as "helpers" for iNKT differentiation of CD1d−/− DP thymocytes. (Supplementary Fig. 4). Lethally irradiated wild-type hosts (CD45.1) were reconstituted with a mix of bone marrow cells from CD1d−/− mice (CD45.2;CD1d−) and c-Mybf/fcd4-Cre or Jα18−/− mice (CD45.2;CD1d+)(Fig. 8a). Given the competitive defect of c-Mybf/fcd4-Cre DP thymocytes in this experimental set-up (Fig. 4b), we used an excess of c-Mybf/fcd4-Cre bone marrow (3:1 CD1d−/−:Jα18−/− versus 1:1 CD1d−/−:c-Mybf/fcd4-Cre) to obtain similar ratios of helper:helped in these chimeras, and only included chimeras where the ratio of CD1d−/−:helper was approximately 3:1(Fig. 8b). To ensure that host DP thymocytes do not contribute to the iNKT selection process, we excluded chimeras with a significant host contribution. Jα18−/− thymocytes can support CD1d−/− iNKT cells development , which migrate to the liver, while c-Mybf/fcd4-Cre thymocytes are unable to do so (Fig. 8a, c). These results show that the defect in SLAM and CD1d expression in c-Mybf/fcd4-Cre thymocytes contributes to the block in iNKT cell generation observed in these mice.
Figure 8. c-MybKO DP thymocytes are unable to support the development of CD1d deficient iNKT cells.
(a) Percentages of iNKT cells derived from CD1dKO, c-MybKO and Jα18KO donor competitors in c-MybKO-CD1dKO and Jα18KO-CD1dKO mixed bone marrow chimeras. Representative chimeras out of five each, generated in two different experiments, are shown. Top: gating strategy used to identify donor cells (CD45.2 positive) and cells derived from c-MybKO or Jα18KO donors (CD1d positive) and CD1dKO donors (CD1d negative) (b) Donor chimerism in the DP thymic compartment in c-MybKO-CD1dKO and Jα18KO-CD1dKO chimeras. (c) Percentages and absolute number of CD1dKO, c-MybKO and Jα18KO donor iNKT cells in the liver of c-MybKO-CD1dKO and Jα18KO-CD1dKO chimeras. p value was calculated using the Mann-Whitney test.
Discussion
Development of iNKT cells has at least three basic prerequisites-productive rearrangement of a Vα14-Jα18 TCR, expression of glycolipid loaded CD1d, and signaling through the SLAM/SAP pathway. Furthermore, for productive rearrangement of Vα14-Jα18, DP thymocytes need a minimum lifespan, because the α chain is always generated as a result of secondary Vα rearrangements. Here we provide evidence that c-Myb is critical for the fulfillment of each of these prerequisites. c-Mybf/fcd4-Cre DP thymocytes have decreased expression of Bcl-xL, which results in a reduced lifespan, and consequently a lack of secondary TCRα rearrangements, and reduced expression of CD1d and several critical molecules in the SLAM/SAP signaling pathway. As a result of these defects, no iNKT precursors can be generated.
Although c-Myb does not absolutely control any of the prerequisites for iNKT cell development, it positively regulates each of them, allowing DP thymocytes to live long enough to rearrange the canonical Vα14-Jα18 TCR and to express high enough levels of CD1d and molecules of the SLAM/SAP signaling pathway to generate the signals that enable iNKT positive selection.
c-Myb deficiency leads to alterations in Bcl-xL expression, however this effect is independent of RORγT and TcF/Lef expression (data not shown), two factors that have been previously described to control Bcl-xL expression38,39. This survival defect seems to be the primarily cause for the defective rearrangement of distal Vα-Jα segments in c-Mybf/fcd4-Cre thymocytes, although a more detailed analysis is required to completely discard a direct effect of c-Myb on TCRα rearrangement, especially given its described role in rearrangement of the β and δ loci 28,32,33.
Although Bcl-xL was able to rescue c-Mybf/fcd4-Cre DP thymocytes survival and TCRα rearrangement, a complete block in iNKT development was still observed. Partial rescue, in terms of numbers and maturation status of the iNKT cells also occurred after expressing a pre-rearranged TCRα. This is probably due to independent defects in expression of SLAMF1, SLAMF6 and SAP.
The effect of c-Myb on SLAMF1 and SLAMF6 expression in DP thymocytes is very selective, as other family members located in the same genomic locus (SLAMF2, SLAMF3 and SLAMF5) are not affected. These observations may have important implications, because the SLAM locus has been identified as a susceptibility locus in several murine autoimmunity models, including systemic lupus erythematosus -the Sle1b allele40 and NOD -the Nkt1 allele41. The Nkt1 allele is responsible for the decreased number of NKT cells in the NOD strain42 and, remarkably, the SLAM family members expression pattern is similarly altered in the thymi of Nkt1 and c-Mybf/f-cd4-Cre mice41. This suggests that the polymorphism(s) that determine the altered SLAM expression in Nkt1 mice may alter c-Myb function. Whether this is a direct effect due to c-Myb binding to regulatory elements that control SLAM family member expression remains to be determined.
Our work shows how a series of relatively small changes in the expression of different molecules orchestrated by c-Myb completely determines the existence of a critical lineage of immune cells. Given the important role of iNKT cells in the responses to multiple diseases, and the wide variability in iNKT cell numbers in human populations43, polymorphisms in c-Myb, or in some of its binding sites may play a significantrole in susceptibility to disease.
Methods
Mice
All mice were maintained in a specific pathogen-free facility at Oklahoma Medical Research Foundation and were handled in compliance with guidelines established by the Institutional Animal Care and Use Committees. Generation of conditional c-Mybfl/fl mice were described previously28. Although originally generated using a 129/ola ES cell line, c-Mybf/f mice have been backcrossed nineteen times to the C57BL/6 background, and carry the C57BL/6 SLAM allele, as determined by PCR with the markers DMit17 and DMit4744. Deletion of the floxed c-Myb allele was accomplished by crossing to cd4-Cre Tg mice. cd4-Vα14-Jα18 transgenic mice, Jα18−/− and Cd1d−/− have been described10,37,45. B6-LY5.2/Cr (CD45.1) congenic mice were obtained from the National Cancer Institutes Frederick animal production program. Mice were used between 8–12 weeks.
Cell preparation and flow cytometry
Single-cell suspensions were prepared from the thymus, spleen and liver. Liver suspensions were purified using a Percoll (Amersham Biosciences) gradient. Hepatic mononuclear cells were collected from the 700% interface, red cell lysed, and then stained for FACS analysis. Cells were incubated with Fc blocking antibodies before staining with specific antibodies and tetramer. Dead cells and doublets were excluded from analysis. Samples were collected on a LSRII(BD) and analyzed with Flowjo (Treestar Inc.). For Bcl-xL intracellular staining, cells first fixed with 2% paraformaldehyde at 37°C and then permeabilized with 0.1% Triton X-100. Fluorochrome labeled monoclonal antibodies (clone indicated in parentheses) against CD45.1 (A20), CD45.2 (104), TCR beta (H57-597), CD24 (M1/69), CD1d (1B1), CD4 (L3T4) (GK1.5), CD8a (ly-2) (53-6.7), CD44 (IM7) NK1.1(PK136), CD19 (bio1D3), SLAMF1 (TC15-12F12.2), SLAMF3 (Ly9ab3), SLAMF5 (mCD87.7) and SLAMF6 (13G3-19D) were from eBioscience or Biolegend. Rabbit anti-Bcl-xL (54H6) was from Cell Signaling. iNKT cells were identified using a murine APC-conjugated CD1d tetramer loaded with PBS57, an analogue of α-galactosylceramide34, provided by the National Institutes of Health Tetramer Facility.
Enrichment of CD1d-α-GalCer Tetramer positive cells
Thymocytes were enriched for tetramer positive cells with anti-biotin microbeads (Miltenyi Biotec). In brief, cells were stained with APC-conjugated PBS57 loaded CD1d tetramer after anti-FcγR block. After washing, cells were incubated with biotinylated anti-APC mAb, washed again and incubated with anti-biotin microbeads. Magnetic separation was performed according to manufacturer? instructions(Miltenyi Biotec Inc.).
Retroviral transduction of hematopoietic progenitors
Mig-Bcl-xL was obtained from Addgene and Mig-RORγT vector is a gift of X. Sun. Preparation of viral supernatant was performed essentially as described46. For progenitor enrichment, total blood marrow cells were depleted of mature cells using a mouse lineage depletion cocktail (BD Biosciences) -containing biotinylated anti-CD3ε(145-2C11), -CD45R (RA3-6B2), -TER-199 (TER-119), -CD11b (M1/70) -Ly6G and Ly6C (RB6-8C5)- and streptavidin-coated magnetic particles . The resulting cells (Lin−) were infected as described47
Generation of mixed bone marrow chimeras
Bone marrow cells were harvested from the femurs and tibiae of F1 (C57BL/6x B6-Ly5.2), Mybf/fcd4Cre (CD45.2). B6-LY5.2/Cr (CD45.1) recipient mice were lethally irradiated with 1000 rad from a cesium source. At least 4 h after irradiation, a 1:1 mixture of F1(CD45.1/CD45.2) and Mybf/fcd4Cre (total of 1 × 106 cell) bone marrow cells were retro-orbitally injected into the hosts. Animals were given antibiotic-containing water and housed in sterile microisolator cages. Mice were analyzed eight to twelve weeks after reconstitution.
A similar protocol was followed in the mixed bone marrow chimeras used to compare the "helper" ability of c-Myb? and Jα18? thymocytes.
FACS Sorting and real-time RT-PCR
Preselected double positive thymocytes from Mybf/wcd4Cre and Mybf/f cd4Cre mice were sorted on a MoFlo (Dako) and total RNA was extracted using Qiagen RNeasy mini kit according to the manufacturer? protocol. Reverse transcription (RT) was performed on 2µg of RNA using Taqman reverse transcription reagents (Applied Biosystems). 10ng of cDNA was used for each assay. SYBR Green real time PCR was performed using an ABI 7500 Real Time PCR systems. Gene specific forward (F) and reverse (R) primers were designed across exon-exon junction using Primer Express software (Applied Biosystems) with equivalent efficiency to the β-actin primers. Data were analyzed using the comparative CT method as described by ABI 7500 system SDS software v1.4.
RorγtF 5CCGCTGAGAGGGCTTCAC-3
RorγtR 5 TGCAGGAGTAGGCCACATTACA-3’
Bcl-2F 5GTGGATGACTGAGTACCT GAACC-3
Bcl-2R 5AGCCAGGAGAAATCAAACAGAG-3’
Bcl-xLF 5TGA CCACCTAGAGCCTTGGA-3
Bcl-xLR 5AGAACCACACCAGCCACAGT-3’
MclF 5GGCCAAACACTTAAAGAGCG-3
MclR 5TGGAAGAACTCCACA AACCC-3’
β-actinF 5CAACGAGCGGTTCCGATG-3’
β-actinR 5GCCACAGGATTCCATACCCA-3’
Vα14F: 5' GGATGACACTGCCACCTACA 3'
Jα18R: 5' CTGAGTCCCAGCTCCAAA A 3'
VαJα rearrangement analysis
Cell sorting, total RNA extraction and RT were performed as described above. PCR was performed by using 1:3 serially diluted cDNA template resulting from the above RT reaction and forward (F) and reverse (R) primers:
Vα3F 5CCCAGTGGTTCAAGGAGTGA-3’
Vα6F 5CTGACTCATGTCAGCCTGAGAG-3
Vα8F 5CAACAAGAGGACCGAGCACC-3
Vα14F 5-TGGGAGATACTCAGCAACTCTGG-3’
Vα19F 5 CTGCTTCTGACAGAGCTCCAG-3
Jα2R 5ACCACTTAGTCCTCCAGTATTC-3’
Jα18R 5CAGGTATGACAATCAGCTGAGTCC-3’
Jα30R 5 AGATGTGTCCCTTTTCCAAAGATG-3’
CαF 5 TTCAAAGAGACCAACGCCAC-3
CαR 5TTCAGCAGGAGGATTCGGAG-3’
PCR products were separated by agarose gel electrophoresis and visualized by ethidium bromide staining.
iNKT in-vitro activation assay
Thymocytes were incubated for 4 h at 4 × 106/ml in R10 medium (RPMI 1640 with 10% FCS, HEPES, penicillin and streptomycin, pyruvate, non-essential amino acids, L-glutamine, and 2-ME) in the presence of 50 ng/ml of PDBU, 500 ng/ml ionomycin and 3 µM monensin. Cells were extracellularly stained with anti-TCRβ and CD1d tetramer. After washing, cells were intracellularly stained for IL-4 and IFNγ using BD cytofix/cytoperm kit (BD biosciences).
Statistical Methods
Normal distribution of the data was assessed using the Kolmogorov-Smirnov test. Depending on the results, statistical significance was determined by the Student's t test (for parametric data) or by the Mann-Whitney test (for non-parametric data). In some instances, p values were corrected for multiple hypothesis testing using the Bonferroni's Multiple Comparison Test.
Supplementary Material
Acknowledgments
This work was supported by grants AI059302 (NIH/NIAID) and 0855002F (AHA) to JAI, and CA85842 (NIH/NCI) to TBP. JY was supported in part by a Robert R. Wagner Fellowship. The authors thank A. Bendelac (Howard Hughes Medical Institute), A. Veillette (Clinical Research Institute, Montreal), X-H.. Sun (Oklahoma Medical Research foundation, Oklahoma City) and M. Lang University of Oklahoma Health Science Center, Oklahoma City), and the NIH tetramer facility, for providing reagents, and S. Kovats (Oklahoma Medical Research foundation, Oklahoma City) for discussions.
Footnotes
Author Contributions
T.H. designed the study, did experiments, analyzed data, and wrote the manuscript, A.S. did experiments, J.Y and T.P.B provided experimental mice, data and discussions, J.A-I designed the study, did experiments, analyzed data and wrote the manuscript.
References
- 1.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2:971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 2.Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–518. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda JL, Mallevaey T, Scott-Browne J, Gapin L. CD1d-restricted iNKT cells, the 'Swiss-Army knife' of the immune system. Curr Opin Immunol. 2008;20:358–368. doi: 10.1016/j.coi.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 5.Kronenberg M, Engel I. On the road: progress in finding the unique pathway of invariant NKT cell differentiation. Curr Opin Immunol S. 2007 doi: 10.1016/j.coi.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Krangel MS, Carabana J, Abbarategui I, Schlimgen R, Hawwari A. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor alpha/delta locus. Immunol Rev. 2004;200:224–232. doi: 10.1111/j.0105-2896.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, et al. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 8.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4+8+ stage of thymic ontogeny. Proc Natl Acad Sci U S A. 2005;102:5114–5119. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borowski C, Bendelac A. Signaling for NKT cell development: the SAP-FynT connection. J Exp Med. 2005;201:833–836. doi: 10.1084/jem.20050339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nichols KE, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nat Med. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasquier B, et al. Defective NKT cell development in mice and humans lacking the adapter SAP, the X-linked lymphoproliferative syndrome gene product. J Exp Med. 2005;201:695–701. doi: 10.1084/jem.20042432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung B, Aoukaty A, Dutz J, Terhorst C, Tan R. Signaling lymphocytic activation molecule-associated protein controls NKT cell functions. J Immunol. 2005;174:3153–3157. doi: 10.4049/jimmunol.174.6.3153. [DOI] [PubMed] [Google Scholar]
- 15.Eberl G, Lowin-Kropf B, MacDonald HR. Cutting edge: NKT cell development is selectively impaired in Fyn- deficient mice. J Immunol. 1999;163:4091–4094. [PubMed] [Google Scholar]
- 16.Gapin L. The making of NKT cells. Nat Immunol. 2008;9:1009–1011. doi: 10.1038/ni0908-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams JA, et al. Regulation of thymic NKT cell development by the B7-CD28 costimulatory pathway. J Immunol. 2008;181:907–917. doi: 10.4049/jimmunol.181.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felices M, Berg LJ. The Tec kinases Itk and Rlk regulate NKT cell maturation, cytokine production, and survival. J Immunol. 2008;180:3007–3018. doi: 10.4049/jimmunol.180.5.3007. [DOI] [PubMed] [Google Scholar]
- 19.Matsuda JL, et al. Homeostasis of V alpha 14i NKT cells. Nat Immunol. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 20.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savage AK, et al. The Transcription Factor PLZF Directs the Effector Program of the NKT Cell Lineage. Immunity. 2008 doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 23.Kim PJ, et al. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- 24.Dose M, et al. Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc Natl Acad Sci U S A. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mucenski ML, et al. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto H, et al. Proper levels of c-Myb are discretely defined at distinct steps of hematopoietic cell development. Blood. 2006;108:896–903. doi: 10.1182/blood-2005-09-3846. [DOI] [PubMed] [Google Scholar]
- 27.Greig KT, Carotta S, Nutt SL. Critical roles for c-Myb in hematopoietic progenitor cells. Semin Immunol. 2008 doi: 10.1016/j.smim.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nat Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 29.Maurice D, Hooper J, Lang G, Weston K. c-Myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J. 2007;26:3629–3640. doi: 10.1038/sj.emboj.7601801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen RDr, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pearson R, Weston K. c-Myb regulates the proliferation of immature thymocytes following beta-selection. EMBO J. 2000;19:6112–6120. doi: 10.1093/emboj/19.22.6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hernandez-Munain C, Lauzurica P, Krangel MS. Regulation of T cell receptor delta gene rearrangement by c-Myb. J Exp Med. 1996;183:289–293. doi: 10.1084/jem.183.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsiang YH, Goldman JP, Raulet DH. The role of c-Myb or a related factor in regulating the T cell receptor gamma gene enhancer. J Immunol. 1995;154:5195–5204. [PubMed] [Google Scholar]
- 34.Liu Y, et al. A modified alpha-galactosyl ceramide for staining and stimulating natural killer T cells. J Immunol Methods. 2006;312:34–39. doi: 10.1016/j.jim.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 35.Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages of thymic NKT cell development. J Exp Med. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao DT, Korsmeyer SJ. BCL-XL-regulated apoptosis in T cell development. Int Immunol. 1997;9:1375–1384. doi: 10.1093/intimm/9.9.1375. [DOI] [PubMed] [Google Scholar]
- 37.Cui J, et al. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- 38.Hossain MZ, Yu Q, Xu M, Sen JM. ICAT expression disrupts beta-catenin-TCF interactions and impairs survival of thymocytes and activated mature T cells. Int Immunol. 2008;20:925–935. doi: 10.1093/intimm/dxn051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie H, Huang Z, Sadim MS, Sun Z. Stabilized beta-catenin extends thymocyte survival by up-regulating Bcl-xL. J Immunol. 2005;175:7981–7988. doi: 10.4049/jimmunol.175.12.7981. [DOI] [PubMed] [Google Scholar]
- 40.Wandstrat AE, et al. Association of Extensive Polymorphisms in the SLAM/CD2 Gene Cluster with Murine Lupus. 2004;21:769–780. doi: 10.1016/j.immuni.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Jordan MA, Fletcher JM, Pellicci D, Baxter AG. Slamf1, the NKT cell control gene Nkt1. J Immunol. 2007;178:1618–1627. doi: 10.4049/jimmunol.178.3.1618. [DOI] [PubMed] [Google Scholar]
- 42.Esteban LM, et al. Genetic control of NKT cell numbers maps to major diabetes and lupus loci. J Immunol. 2003;171:2873–2878. doi: 10.4049/jimmunol.171.6.2873. [DOI] [PubMed] [Google Scholar]
- 43.Berzins SP, Cochrane AD, Pellicci DG, Smyth MJ, Godfrey DI. Limited correlation between human thymus and blood NKT cell content revealed by an ontogeny study of paired tissue samples. Eur J Immunol. 2005;35:1399–1407. doi: 10.1002/eji.200425958. [DOI] [PubMed] [Google Scholar]
- 44.Morel L, Yu Y, Blenman KR, Caldwell RA, Wakeland EK. Production of congenic mouse strains carrying genomic intervals containing SLE-susceptibility genes derived from the SLE-prone NZM2410 strain. Mamm Genome. 1996;7:335–339. doi: 10.1007/s003359900098. [DOI] [PubMed] [Google Scholar]
- 45.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 46.Hernandez-Hoyos G, Alberola-Ila J. Analysis of T-cell development by using short interfering RNA to knock down protein expression. Methods Enzymol. 2005;392:199–217. doi: 10.1016/S0076-6879(04)92012-5. [DOI] [PubMed] [Google Scholar]
- 47.Lauritsen JP, et al. Egr2 Is Required for Bcl-2 Induction during Positive Selection. J Immunol. 2008;181:7778–7785. doi: 10.4049/jimmunol.181.11.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.