Abstract
The NIAID-funded Seattle Structural Genomics Center for Infectious Disease (SSGCID) is a consortium established to apply structural genomics approaches to potential drug targets from NIAID priority organisms for biodefense and emerging and re-emerging diseases. The mission of the SSGCID is to determine ~400 protein structures over five years ending in 2012. In order to maximize biomedical impact, ligand-based drug-lead discovery campaigns will be pursued for a small number of high-impact targets. Here we review the center’s target selection processes, which include pro-active engagement of the infectious disease research and drug therapy communities to identify drug targets, essential enzymes, virulence factors and vaccine candidates of biomedical relevance to combat infectious diseases. This is followed by a brief overview of the SSGCID structure determination pipeline and ligand screening methodology. Finally, specifics of our resources available to the scientific community are presented. Physical materials and data produced by SSGCID will be made available to the scientific community, with the aim that they will provide essential groundwork benefiting future research and drug discovery.
Keywords: structure-based drug development, structural genomics, biodefense, ligand screening, SSGCID
INTRODUCTION
Over the past five to ten years, high throughput methodologies for protein expression and structure determination have been developed and implemented, leading to the discipline commonly known as “Structural Genomics”. In the academic setting, this work has been led by the National Institutes of General Medical Studies (NIGMS)-sponsored Protein Structure Initiative (PSI, http://www.nigms.nih.gov/Initiatives/PSI/), which is aimed at dramatically reducing the costs and lessening the time required to determine a three-dimensional protein structure. The ultimate goal of the PSI is to make the three-dimensional, atomic-level structures of most proteins easily obtainable from knowledge of their corresponding DNA sequences.
Recently, the National Institute of Allergy and Infectious Diseases (NIAID), Division of Microbiology and Infectious Diseases (DMID), launched a five-year initiative to establish two large-scale NIAID-funded Structural Genomics Centers for Infectious Diseases that would apply state-of-the-art high-throughput (HTP) structural biology technologies to experimentally characterize the three-dimensional atomic structure of targeted proteins from pathogens in the NIAID Category A-C priority lists and organisms causing emerging and re-emerging infectious diseases. The goal of this initiative (http://www3.niaid.nih.gov/research/resources/sg/) is to create a collection of high quality, experimentally-determined, three-dimensional (3-D) structures that are widely available to the scientific community, where they could serve as blueprints for development of structure-based drugs, vaccines and diagnostics for infectious diseases. In late 2007, the MIDWEST-based Center for Structural Genomics of Infectious Diseases (CSGID) and the Seattle Structural Genomics Center for Infectious Disease (SSG-CID) were funded on a contract basis to provide ~800 3-D atomic structures of proteins that have important biological roles in the targeted pathogens and/or are potential targets for vaccine and drug development. In this review, we will describe the approaches used and progress made by the SSGCID, while an accompanying article (by Wayne Anderson) reviews the CSGID.
SSGCID VISION AND GOALS
The primary mission of the Seattle Structural Genomics Center for Infectious Disease (SSGCID) is to establish a resource for gene-to-structure research focused on the structure determination of ~400 protein targets from NIAID Category A-C pathogens and organisms causing emerging and re-emerging infectious diseases. This mission will be accomplished through pro-active engagement of the infectious disease research and drug therapy communities, in close collaboration with NIAID program officers. In this way, our target selection plan will benefit from community expertise, and we also plan to engage the community in follow-up research as the SSGCID begins to solve structures of important disease targets. Working together with the other NIAID-funded Center for Structural Genomics of Infectious Diseases (CSGID), SSGCID intends to provide a blueprint for structure-based design of new drug and vaccine therapeutics to combat infectious diseases. This goal will be facilitated by the annual selection of several high-impact targets for a fragment-based drug lead discovery campaign.
SSGCID LEADERSHIP AND INFRASTRUCTURE
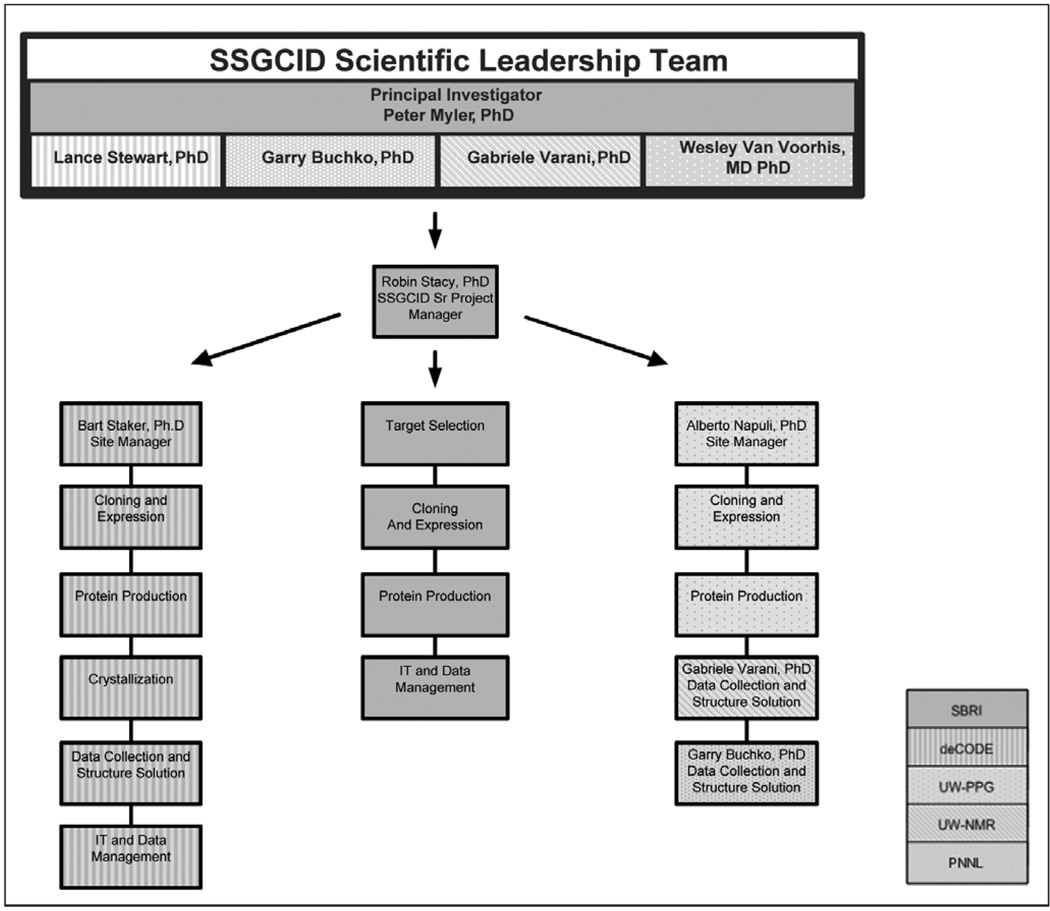
The SSGCID is divided into seven functional activities/ teams (Project Management, IT & Data Management, Target Selection, Cloning & Expression Screening, Protein Production, Crystallization, and Data Collection & Structure Solution) and is physically located at four separate institutions (Seattle Biomedical Research Institute, deCODE biostructures, the University of Washington, and the Pacific Northwest National Laboratory). Management of the SSGCID is overseen by a Scientific Leadership Team comprised of the Principal Investigators from each of the institutions, with the assistance of an overall Senior Project Manager at SBRI, and Site Managers at deCODE and UW (see Fig. 1). A Scientific Working Group (SWG) comprised of eight members from industry and academic institutions in the US meets twice annually to make recommendations on efficiently generating protein structures in a high throughput environment, and provide advice on the structural genomics needs of the scientific community.
Fig. 1. SSGCID Management and Organization.
SBRI: Seattle Biomedical Research Institute, deCODE: deCODE biostructures, UW-PPG: University of Washington Protein Production Group, UW-NMR: University of Washington NMR Group, PNNL: Pacific Northwest National Laboratory.
TARGET ORGANISMS
Under the terms of the NIAID contract, efforts at both SSGCID and CSGID are focused primarily on pathogens causing emerging and re-emerging infectious diseases, including those with bioterrorism potential (see http://www3.niaid.nih.gov/topics/emerging/list.htm). These organisms include 31 different genera of bacteria, eukaryotes and viruses, which have been divided between the two centers. SSGCID will focus on the Alphaproteobacteria (Bartonella, Brucella, Ehrlichia, Anaplasma and Rickettsia), Betaproteobacteria (Burkholderia), Actinobacteria (Mycobacterium), and Spirochetes (Borrelia) among the bacteria;the Acanthamoebidae (Acanthamoeba), Aconoidasidae (Babesia), Coccidia (Cryptosporidium, Cyclospora and Toxoplasma), Diplomonadidae (Giardia), Entamoebidae (Entamoeba), Eurotiomycetes (Coccidioides) and Microsporidia (Encephalitozoon) among the eukaryotes; as well as single-stranded DNA (Erythrovirus), and negative-strand RNA (Marburg, Ebola-like, Influenza A, B & C, Arena, Hanta, Henipa, Lyssa, Nairo, Orthobunya, Phlebo and Rubula) viruses (see Table I). In general, we have chosen a single representative of each genus (usually a well-annotated strain of a pathogenic species) for initial target selection, with the option of moving to additional species/strains later in the project. Although the bacterial genomes contain between 887 and 6421 predicted protein-coding genes, and the eukaryotes contain 2030 to 10499 genes, almost all organisms (with the exception of Mycobacterium) had very few protein structures submitted to the Protein Data Bank (PDB) at the start of the project. Indeed, several genera had no published structures. A similar situation also applied to the viruses, with the exception of the Influenza viruses, despite their much smaller genomes (which contain 3–11 genes). Thus, these organisms appear to provide a fertile environment for elucidation of novel protein structures, which should prove informative to the scientific community studying their pathogenesis and control.
Table I.
SSGCID Target Organisms
| Class | Genus | Disease (species) | Genes | PDB |
|---|---|---|---|---|
| Actinobacteria | Mycobacterium | Multi-drug resistant TB (tuberculosis H37Rv) | 4049 | 623 |
| Alphaproteobacteria | Bartonella | Bacillary angiomatosis (henselae) | 1666 | 2 |
| Brucella | Brucellosis (melitensis, abortus & suis) | 3419 | 7 | |
| Ehrlichia | 1159 | 0 | ||
| Anaplasma | Human granulocytic anaplasmosis (phagocytophilia) | 1412 | 0 | |
| Rickettsia | Endemic typhus(prowazeki), Rocky Mountain Spotted fever (rickettsii) | 887 | 0 | |
| Betaproteobacteria | Burkholderia | Glanders (mallei) & melioidosis (pseudomallei) | 6421 | 52 |
| Spirochaetes | Borrelia | Lyme disease (burgdorferi) | 1702 | 34 |
| Acanthamoebidae | Acanthamoeba | Acanthamebiasis | ? | 9 |
| Aconoidasida | Babesia | Babesiosis, atypical | 3782 | 1 |
| Coccidia | Cryptosporidium | Diarrhea (parvum) | 3396 | 9 |
| Cyclospora | Diarrhea (cayetanensis) | ? | 0 | |
| Toxoplasma | Toxoplasmosis (gondii) | 8412 | 45 | |
| Diplomonadida | Giardia | Giardiasis (lamblia) | 6582 | 5 |
| Entamoebidae | Entamoeba | Dysentery & liver abcess (histolytica) | 8343 | 9 |
| Eurotiomycetes | Coccidioides | Meningitis (immitis) | 10499 | 5 |
| Microsporidia (phylum) | Encephalitozoon | (cuniculi, hellem & bieneusi) | 2030 | 7 |
| ssDNA viruses | Erythrovirus | Fifth disease (Parvovirus B19) | 5 | 1 |
| ssRNA negative-strand viruses |
(Filoviridae) Marburgvirus |
Hemorrhagic fevers (Marburg) | 7 | 0 |
| (Filoviridae) Ebola-like viruses |
Hemorrhagic fever (Ebola) | 7 | 7 | |
| (Orthomyxoviridae) Influenzavirus A |
Influenza | 11 | 83 | |
| (Orthomyxoviridae) Influenzavirus B |
Influenza | 11 | 14 | |
| (Orthomyxoviridae) Influenzavirus C |
Influenza | 9 | 1 | |
| Arenaviruses | LCM, Junin, Machupo, Guanarito, Lassa fever | 4 | 0 | |
| Hantavirus | Hanta | 3 | 2 | |
| Henipavirus | Neurological and respiratory disease (Hendra & Nipah) | 6 | 2 | |
| Lyssavirus | Rabies & Australian bat virus | 5 | 2 | |
| Nairovirus | Crimean-Congo Hemorrhagic fever | 3 | 0 | |
| Orthobunyavirus | California encephalitis, LaCrosse | 3 | 0 | |
| Phlebovirus | Rift Valley fever | 6 | 0 | |
| Rubulavrius | Mumps | 9 | 11 | |
| Total | 63848 | 931 |
The number of genes listed refers to those predicted from the genome sequence of the representative strain chosen for each genus. ‘?’ indicates that the complete genome is not yet available. The number of PDB entries refers to the total for the entire genus as of October, 2007 (SSGCID project start date).
TARGET PROTEINS
The stated purpose of the SSGCID and CSGID efforts is to provide high-quality, experimentally determined, 3-D structures that serve as blueprints for development of structure-based drugs, vaccines and diagnostics for infectious diseases. In order to provide the maximal impact on biomedical research, target selection is focused on proteins that have important biological roles, such as:
Proteins involved in pathogenesis, e.g. invasins, and adhesins.
Proteins essential to the pathogen’s life and reproductive cycle;
Proteins involved in antimicrobial/drug resistance;
Protein markers of acute or chronic infection;
Protein complexes with natural substrates, cofactors, receptors, and drug candidates;
Protein splice variants, post-translational modifications and other functionally characterized variants.
Further details of target proteins selected to date can be found below.
STRUCTURE DETERMINATION PIPELINE
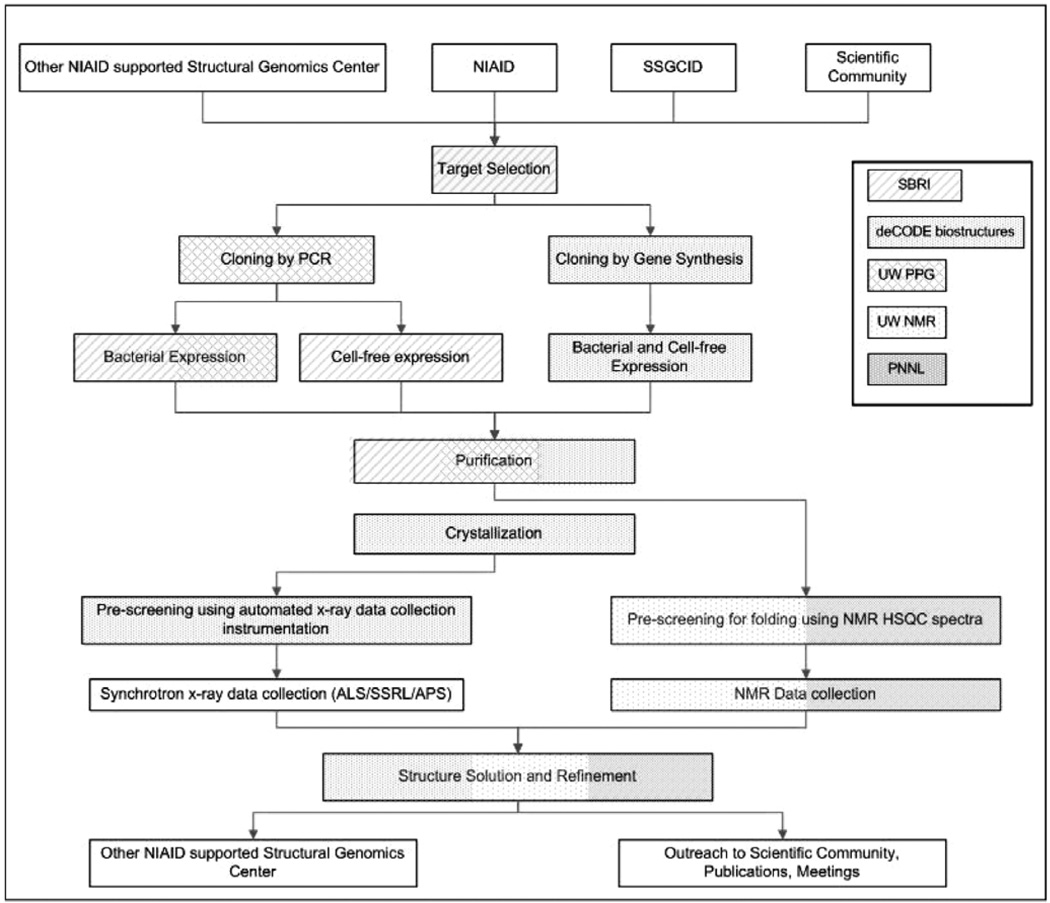
The overall SSGCID structure determination pipeline involves a number of activities distributed between the Target Selection, Cloning & Expression Screening, Protein Production, Crystallization, and Data Collection & Structure Solution teams at the five different locations (see Fig. 2). In order to maximize the likelihood of success of each target, yet minimize the cost-per-structure, we have adapted a multi-pronged serial escalation approach, whereby targets initially enter a standard high-throughput bacterial protein expression system (Tier 1), and enter more expensive “rescue pathways” (Tiers 2–9) only after failing the initial approach. In addition, there are a number of tiers dedicated to specialized activities such as NMR structure solution (Tier 10), co-crystallization (Tier 11), ligand screening (Tiers 12–15) and RNA targets (Tier 16), as well as expression of constructs supplied by requestors from the scientific community (Tier 0). A more detailed description of each activity within the SSGCID structure determination pipeline follows.
Fig. 2. SSGCID Structure Determination Pipeline.
Inset box shows code for sites performing Target Selection, Cloning & Expression Screening, Protein Production, Crystallization, and Data Collection & Structure Solution.
Target Selection
A series of bioinformatic and manual filters are used by the SSGCID Target Selection Team (at SBRI) to select proteins predicted from a single representative genome sequence for each of the 31 bacterial, eukaryotic and viral genera indicated above. Positive selection criteria include sequence similarity to known drug targets, documented or potential roles in cell growth, pathogenesis, or drug resistance, as well as markers of infection and vaccine candidates. Negative filters include physical properties (such as size, amino acid composition, presence of transmembrane domains and low complexity sequences) predictive of difficulty in soluble expression and/or crystallization and close similarity to sequences already present in PDB or already targeted by other Structural Genomics projects. In order to achieve our annual goal of determining one or more structures from each of ~50 different proteins; we anticipate selecting at least 500 targets each year. In addition, we expect a smaller number (50–100 annually) of targets from community requests (see below).
The initial round of target selection (Batch01) involved identification of potential drug targets in three bacterial species (Burkholderia pseudomallei, Brucella melitenesis, and Rickettsia prowazekii), by virtue of their sequence similarity (>50% over >75% of their length) with protein sequences in the DrugBank database [1]. A series of physical screens were subsequently used to eliminate proteins longer than 500 amino acids, containing more than eight cysteine residues and/or containing any transmembrane spanning domains (except for N-terminal signal sequences) predicted using TmPred (http://www.ch.embnet.org/software/TMPRED_form.html) and/or TmHmm/Phobius [2]. The remaining candidate proteins were screened for near-identity (>95% similarity over >80% of their length) to proteins with known structure or those selected by other structural genomics centers by BlastP searching against TargetDB. This ultimately resulted in 196 targets from these three species, which were supplemented with 13 ftsZ orthologues selected from several different species within the Burkholderia, Brucella, and Rickettsia genera. FtsZ was chosen because of its particular interest as a bacterial drug target.
For Batch02, a list of 42 bacterial drug targets being actively pursued by pharmaceutical and academic researchers was compiled by literature survey, and their orthologues were identified within representative B. pseudomallei, B. melitenesis, R. prowazekii, Mycobacterium tuberculosis, Bartonella henselae, and Borrelia burgdorferi genomes. These were then screened through similar filters (except that the size limit was raised to 750 amino acids) as described for Batch01, resulting in 143 additional targets.
Both of these approaches were combined in Batch03 to identify an additional 1477 targets from five bacterial (M. tuberculosis, B. henselae, B. burgdorferi, Anaplasma phagocytophilum, and Ehrlichia chaffeensis) and seven eukaryotic (Babesia bovis, Coccidioides immitis, Cryptosporidium parvum, Encephalitozoon cuniculi, Entamoeba histolytica, Giardia lamblia, and Toxoplasma gondii) species.
Batch04 marked a significant departure from most structural genomics efforts by selection of five RNA riboswitch elements from three bacterial species. Riboswitches are non-coding RNA elements that bind small-molecule metabolites with high affinity and specificity and regulate the expression of associated genes [3]. These targets include a thiamine pyrophosphate-sensing (thi-box) riboswitch from M. tuberculosis, an S-adenosyl methionine (SAM-II) riboswitch from B. melitensis, two pre-queuosine-1 or 7-aminomethyl-7-deazaguanine (preQ1) riboswitches from Bacillus anthracis and one (preQ1) riboswitch from Listeria monocytogenes.
To date, the four batches described above have resulted in 1834 targets being approved for entry into the SSGCID structure determination pipeline, as well as an additional 67 targets from Community Requests (see below). Target selection Batch05 is currently in progress and will include orthologues from an additional 45 hand-selected potential drug targets in all bacterial and eukaryotic species above. We also anticipate selecting a number of viral targets (Batch06) in the coming months, as well as two different approaches (Batch07 and Batch08) to identify target proteins with potential roles in pathogenesis.
Cloning and Expression Screening
Targeted genes are initially PCR amplified from genomic DNA (bacteria) or cDNA (eukaryotes and viruses) and cloned into bacterial expression vectors (Tier 1) using a ligation-independent cloning (LIC) methodology [4]. Both vectors (BG1861 and AVA0421) are derivatives of pET14b, are regulated by the T7 promoter, and contain the amp gene encoding ampicillin resistance. BG1861 yields protein constructs with a minimal N-terminal His6-Tag: MAHHHH-HHM-ORF, while AVA0421 yields protein constructs with an N-terminal His6-Tag and a 3C protease cleavage site: MAHHHHHHMGTLEAQTQGPGSM-ORF [5]. Cleavage of the His6-Tag by 3C protease yields proteins with an N-terminal sequence: GPGSM-ORF. Cloning steps are relatively high throughput, being carried out in 96-well plates and trays. The resultant plasmids are transformed into the BL21(D3) bacterial host, grown in auto-induction medium [5], the cells lysed, the supernatant passed over Ni2+ beads, and soluble protein quantified by SDS-PAGE. Once again all steps are carried out in 96-well format, so the screening proceeds relatively rapidly. Glycerol stocks of all clones are made at this stage and DNA prepared for sequencing to confirm that the correct target has been cloned and does not contain frame-shifts or premature stop codons.
Targets that fail to express sufficient soluble protein in Tier 1 are prepared for cell-free expression (Tier 2) by PCR amplification using a common primer set and vectors (pEU-E01-LIC1 and -LIC2) re-engineered to facilitate LIC. DNA from the resultant clones is transcribed in vitro using SP6 RNA polymerase to produce sufficient RNA for small-scale expression testing using the ENDEXT® Wheat Germ cell-free protein synthesis system [6–8]. Unfortunately, attempts to use linear template obtained by PCR for small-scale expression testing generally showed low expression levels and were of limited utility in predicting the success (or failure) of large-scale purification with a plasmid template. Thus, Tier 2 screening is currently not carried out in high throughput mode. While we have thus far screened only a small number (<50) of targets in Tier 2, our results indicate that the majority produce protein, with about half of them being soluble. This is in agreement with the experience of other laboratories [7–10], and suggests that cell-free protein synthesis may provide a valuable technique for “rescuing” targets that fail to produce soluble protein in E. coli.
Failure to make soluble protein in Tier 2 results in entry into Tier 3 for synthetic gene construction and cloning into a different bacterial expression vector (pET28-HisSMT). Targets that fail cloning in Tier 1 and those from organisms with difficult to obtain DNA (generally community requests) are moved directly to Tier 3. At least four different constructs (terminal/internal deletions, point mutations, and codon-optimization) are designed for each synthetic gene and clones are screened for soluble expression in much the same way as Tier 1, except that eluate from the magnetic nickel bead purification is screened for protein content using a high-throughput Caliper LC90 capillary electrophoresis system. The synthetic genes are designed using Gene Composer™ software [11] to harmonize the codon usage of the gene to the E. coli expression host. A comparison of the native and codon-optimized constructs showed little difference in the success rate [12], although only bacterial targets have been tested to date.
Additional “rescue” Tiers are planned, but have net yet been implemented. Targets that produce only small amounts of soluble protein in Tiers 1–3 will be screened for improved protein production in the presence of different additives (Tier 4), and we will carry out refolding (Tier 5) to attempt rescue of clones producing insoluble protein. Expression of orthologues in bacterial (Tier 6) or by cell-free (Tier 7) systems will also be attempted, especially for targets that express soluble protein, but fail to crystallize. A subset of targets that fail to express soluble protein in Tiers 1–7 will be selected for baculovirus expression (Tier 8). This will be particularly important for eukaryotic and viral targets likely to contain post-translational modifications (e.g. viral capsid proteins). A limited number of high-value eukaryotic targets will also be tried in mammalian cell expression systems (Tier 9) before being abandoned.
Protein Purification
Cloned targets that produce soluble protein are scaled-up and protein purified in milligram quantities at the different protein production facilities. Most scaled-up growth of Tier 1 targets is carried out at the UW-PPG using a LEX 48 Bioreactor, a novel air-based system specifically designed to support the typical needs of HTP protein production labs. The LEX uses compressed air to mix and oxygenate bacterial media and regulate culture temperature, allowing simultaneous upscale of up to 48 different targets in individual 1 L bottles or 24 targets in 2 L bottles. Tier 1 protein purification is carried out at both the UW-PPG and SBRI, and involves lysis, clarification, Ni2+ affinity chromatography, size exclusion chromatography (SEC), protein characterization, and final concentration before packing and shipping. Together, these sites have the capacity to purify 10–15 proteins per week, with yields typically ranging from 10–150 mg of protein at >95% purity.
Tier 2 protein production occurs at SBRI using automated protein production and purification on the Protemist® DTII. Yields obtained in the large-scale reaction have typically been rather modest, with only low- to submilligram quantities of purified proteins (at ~0.6–0.8 mg/ml), produced from each Protemist® run. We are currently exploring whether these yields can be improved using an automated repeat-batch desk-top machine currently in beta-testing at CellFree Sciences.
Crystallization
After purification, all protein samples are delivered to deCODE for crystallization screening. High-throughput crystallization is performed using sophisticated liquid handling devices including Matrix Maker™, Drop Maker™, and the new nanovolume Microcapillary Protein Crystallization System (MPCS™). The MPCS™ technology, which was developed with funding from the Accelerated Technologies Center for Gene to 3D Structure (a PSI-2 Specialized Center) is capable of producing diffraction-ready crystals in the plastic MPCS CrystalCard, and has already been used to solve the structure of one SSGCID target [13].
Data Collection and Structure Solution
Diffraction screening of crystals is routinely conducted on deCODE’s in-house XRD (X-ray diffraction) systems, with data collection and structure solution attempted on all crystals that diffract to better than 3.0 Å resolution. Initially, most crystals required a final, high resolution data set to be collected at a synchrotron radiation source, but the recent addition of a new automated Rigaku “Ultimate Homelab” system has allowed collection of many final data-sets in-house, especially for ligand-bound structures in Tier 11 (see below). So far, all SSGCID structures have been solved by molecular replacement techniques using homologous PDB structures, pointing to one of the advantages inherent to our Target Selection strategy. Only three targets have failed structure solution by molecular replacement and these have been scheduled for selenomethionine (Se-met) protein production and MAD (multiwavelength anomalous diffraction) phasing.
Not all proteins are amenable to crystallization, and therefore, proteins less than 150 amino acids in length that fail to crystallize are 15N-labeled and two-dimensional NMR data collected for Heteronuclear Single Quantum Coherence (HSQC) screening. Those proteins that show good 1H-15N HSQC spectra are then 13C- and 15N-labeled to allow collection of the full suite of three-dimensional NMR experiments needed to assign the backbone and determine the solution structure (Tier 10). Ample spectrometer time is available to collect all the NMR data at two sites: UW-NMR and PNNL. However, while data collection is not a bottleneck, chemical shift assignment and final NMR-based structure determination is laborious and time-consuming. Because SSGCID resources are limited for this activity, we expect only 6–8 structures to be determined by NMR each year. To date the structure of two targets have been solved by this method, although several others are nearing completion.
LIGAND SCREENING
A number of protein targets from Tiers 1–3 crystallized with bound endogenous co-factors, which can often provide inspiration for structure-based drug design. Rather than rely solely on such adventitious ligand-bound structures, SSGCID has undertaken a concerted effort to elucidate the structure of a number of selected targets with both natural and synthetic ligands. Several additional Tiers (11–15) of our structure determination pipeline are devoted to this effort. Literature and database searches are conducted for every target whose structure is solved in Tiers 0–10 to determine if commercially available substrates, cofactors, or inhibitors are likely to bind the target protein. If good candidates are identified, targets are entered into Tier 11 for co-crystallization and eventual structure determination with these ligands. In addition, for a small number of high-value targets, ligands are identified experimentally by small molecule library screening using the Fragments of Life™ (FOL) co-crystallization, NMR, Surface Plasmon Resonance (SPR) and/or Fluorescence-based Thermal Stability (FTS) methods outlined below. To our knowledge, no large-scale, comprehensive, studies have been carried out to compare the results obtained from the several different assays that can be used to measure protein-ligand interaction. We intend to compare the results of screening the 384-fragment May-bridge subset of the FOL against several SSGCID proteins using FOL/co-crystallography (Tier 12), NMR (Tier 13), SPR (Tier 14) and FTS (Tier 15). The results from all screens will be made publicly available for use by the scientific community. Ligands identified by these screening methods will be co-crystallized with target proteins, or soaked with target crystals, and the ligand-bound structure solved by X-ray crystallography.
Ligand Co-crystallization or Soaking (Tier 11)
Thirteen SSGCID targets have entered Tier 11 at deCODE in a total of 80 different experiments (72 co-crystallizations and 8 soaks), of which nine different targets produced crystals. Data collection from 57 crystals revealed 13 to have ligands bound, resulting in 11 structures from four different targets.
Fragments of Life™ Screening (Tier 12)
Fragment crystallography has become a powerful and widely used method for rapid generation of inhibitor leads [14,15]. A typical fragment crystallography experiment involves co-crystallization or soaking of target protein crystals with pools or cocktails of small molecules [16]. deCODE has developed a proprietary library of small (<300 Da) metabolites and metabolite-like molecules called Fragments of Life™ (FOL), which SSGCID has employed for lead discovery against high impact targets. The current FOL library is composed of 1329 compounds and complete screening of the FOL library requires the co-crystallization or soaking of crystals into 180 pools of fragment compounds. Fragment hits are identified by examination of electron density maps from solved crystal structures. A complete X-ray data set must be collected with a resolution of at least 3.2 Å in order to properly identify potential fragment-binding hits. Two targets (BupsA.00023.a and BupsA.00027.a) have been selected for complete FOL screening, and the latter campaign is almost complete. We have demonstrated the success of this approach by obtaining three fragment-bound structures from screening 167 FOL pools. Since complete data-sets from only 34 crystals (of the 125 obtained) have been examined to date, the success rate for obtaining ligand-bound structures is ~9% per fragment pool.
NMR-based screening (Tier 13)
In the last 10–15 years, ligand screening by Saturation Transfer Difference Nuclear Magnetic Resonance (STD-NMR) and Transfer Nuclear Overhauser Enhancement (TR-NOE) has been shown to be a very efficient avenue for the development of clinical candidates, both in the pharmaceutical and biotechnology industry [17]. Unlike other high-throughput screens, NMR ligand screening has the advantage of identifying low affinity binders. In addition, since direct information is obtained for every compound, false-negatives can be avoided. The UW-NMR group has developed a fragment library of 520 compounds divided into 64 mixtures of six to eight compounds with favorably resolved 1D NMR spectra, which can be screened relatively quickly (five to fourteen days per target). The two targets described above (BupsA.00023.a and BupsA.00027.a) have been screened using a combination of STD-NMR and TR-NOE spectroscopy, and we have identified 99 hits for the former and 61 for the latter. Several of the stronger hits for BupsA.00023.a have been validated by inter-ligand NOEs, indicating that they likely bind to similar regions of the target protein.
Surface Plasmon Resonance-based Screening (Tier 14)
Surface Plasmon Resonance (SPR) provides a rapid, high-throughput, and quantitative method for screening small molecule binding to proteins [18]. In this approach, the target protein of interest is immobilized on a chip surface in a microfluidic chamber, and the ligand/fragment solutions passed over the surface, where their binding is detected by a change in refractive index of the surface. We have screened sub-sets of deCODE’s FOL library for binding to BupsA.00023.a using a Fujifilm AP-3000 and GE Biacore T-100 and found that 112/384 and 44/96, respectively, showed measurable interaction. The first screen contained 68 fragments identified as binders by NMR, of which 32 were detected as binding by SPR. In the second screen, 13 of the 25 fragments identified as hits by NMR gave measurable responses by SPR. Interestingly, 9 of these 13 hits were also identified as hits in the first screen, including one fragment that appeared to be a super-stoichiometric binder. These results indicate a much stronger correlation in data between the two SPR instruments than between SPR and NMR, presumably reflecting the different ranges of binding affinity detected using the two approaches.
Fluorescence-based Thermal Shift screening (Tier 15)
Fluorescence-based Thermal Shift (FTS) assays, also know as Differential Scanning Fluorimetry, ThermoFluor™ or Thermal melting, provide a rapid and inexpensive method to identify ligands and fragments that bind to, and stabilize, purified proteins [19]. The protein-ligand mix is heated in a Real-Time PCR machine and the temperature at which the protein “melts” is determined by measuring the increase in fluorescence of a dye with affinity for the hydrophobic parts of the proteins that are exposed as the protein unfolds. Ligands that stably interact with the protein will cause a “thermal shift” in the denaturation curve towards a higher temperature. We have just begun to evaluate the utility of this method for ligand/fragment screening with SSGCID targets.
COMMUNITY INTERACTIONS
SSGCID, along with CSGID, offers gene-to-structure service to the infectious disease scientific community without cost. All materials and information generated from these services will become publicly available through structure deposition with the PDB, materials deposition with the Biodefense and Emerging Infections Research Resource Repository (BEIRRR), and other database resources such as the PSI TargetDB and PepcDB. SSGCID currently has 67 community request targets entered into the structure determination pipeline, with one structure already in the PDB.
Individual or collaborative groups of investigators interested in proposing a target for structure determination at the SSGCID or CSGID are requested to submit a Target Selection Proposal to the appropriate center http://www.ssgcid.org and http://www.csgid.org. Following submission to SSGCID, members of the Target Selection team contact the submitter directly to confirm submission acceptance and clarify details of the request. Sequence analyses are performed to ensure that the target is suitable to attempt structure determination, including identification of potential domain boundaries for large targets. SSGCID personnel then work with the requestor to clarify the precise details of any materials (template DNA, expression constructs and/or protein) available and an appropriate entry point (generally Tier 0, Tier 1, or Tier 3) into the structure determination pipeline. The proposal is then submitted to NIAID for approval. Community requests are given priority status at SSGCID.
SSGCID PROGRESS TO DATE
Summary
As of March 2009, the SSGCID consortium has selected 1901 targets for entry into the structure determination pipeline, including 67 from the scientific community (see Table II). A total of 332 soluble proteins have been purified from 305 different targets and we have submitted 55 structures (from 39 targets) to the PDB, with an additional 28 proteins (from 16 targets) in the final stages of structure solution or awaiting deposition. While the majority of structures have been solved by X-ray crystallography, we have completed NMR assignment for five targets, with two having been submitted to the BMRB as well as PDB. The 55 solved structures are listed in Table III. For more up-to-date statistics and the current status of all targets in our pipeline, please visit http://www.ssgcid.org/home/Target_Status.asp.
Table II.
Cumulative Status of SSGCID Targets
| Status | Targets | Proteins |
|---|---|---|
| Target Approved | 1901 | -- |
| Selected | 1104 | -- |
| Cloned | 984 | -- |
| Expressed | 746 | -- |
| Soluble | 542 | -- |
| Purified | 305 | 332 |
| Crystallized | 97 | 152 |
| Diffraction | 80 | 93 |
| Crystal structure | 55 | 83 |
| HSQC | 10 | 10 |
| NMR assigned | 6 | 6 |
| NMR structure | 2 | 2 |
| In PDB | 39 | 55 |
Table III.
SSGCID Targets Submitted to the PDB
| Organism | Target_ID | PDB | Description | Ligand |
|---|---|---|---|---|
| Bartonella henselae | BaheA.00657.a | 3GIR | Glycine cleavage system protein t | |
| BaheA.00113.b | 3E60 | 3-oxoacyl-(acyl carrier protein) synthase II | ||
| Brucella melitenesis | BrabA.00002.a | 3EG4 | Tetrahydropyridine 2-carboxylate N-succinyl transferase | |
| BrabA.00006.a | 3E7D | Precorrin-8X methylmutase CbiC/CobH | ||
| BrabA.00010.b | 3EMK | Glucose/ribitol dehydrogenase | ||
| BrabA.00010.b | 3ENN | Glucose/ribitol dehydrogenase | ||
| BrabA.00010.e | 3GAF | 7-alpha-hydroxysteroid dehydrogenase | ||
| BrabA.00014.a | 3E5B | Isocitrate lyase | ||
| BrabA.00014.a | 3EOL | Isocitrate lyase | ||
| BrabA.00020.a | 3EK1 | Aldehyde dehydrogenase | ||
| BrabA.00023.a | 3FQ3 | Inorganic pyrophosphatase | ||
| BrabA.00028.a | 3FVB | Bacterioferritin | ||
| BrabA.00052.a | 3DOC | TrkA glyceraldehyde-3-phosphate dehydrogenase | ||
| BrabA.00102.a | 3FS2 | 2-dehydro-3-deoxyphosphooctonate aldolase | ||
| BrabA.00136.a | 3GE4 | Ferritin:DNA-binding protein Dps | ||
| Burkholderia pseudomallei | BupsA.00001.a | 3DMO | Cytidine deaminase | |
| BupsA.00005.a | 3D5T | Malate dehydrogenase | ||
| BupsA.00008.a | 3ECD | Serine hydroxymethyltransferase | ||
| BupsA.00010.a | 3FTP | 3-ketoacyl-(acyl-carrier-protein) reductase | ||
| BupsA.00010.b | 3EK2 | Enoyl-(acyl carrier protein) reductase | ||
| BupsA.00010.e | 3EZL | Acetyacetyl-CoA reductase | ||
| BupsA.00014.b | 3EOO | Methylisocitrate lyase | ||
| BupsA.00023.a | 3D63 | Inorganic pyrophosphatase | ||
| BupsA.00023.a | 3EIY | Inorganic pyrophosphatase | Pyrophosphate | |
| BupsA.00023.a | 3EIZ | Inorganic pyrophosphatase | ||
| BupsA.00023.a | 3EJ0 | Inorganic pyrophosphatase | Frag110 | |
| BupsA.00023.a | 3EJ2 | Inorganic pyrophosphatase | Frag928 | |
| BupsA.00025.a | 3DPI | NAD+ synthase | Acetate | |
| BupsA.00027.a | 3D6B | Glutaryl-CoA dehydrogenase | Frag239 | |
| BupsA.00027.a | 3EOM | Glutaryl-CoA dehydrogenase | ||
| BupsA.00027.a | 3EON | Glutaryl-CoA dehydrogenase | Frag75 | |
| BupsA.00032.a | 3D64 | S-adenosyl-homocysteine hydrolase | NAD | |
| BupsA.00033.a | 3CEZ | Methionine-R-sulfoxide reductase | Acetate | |
| BupsA.00033.a | 3CXK | Methionine-R-sulfoxide reductase | Acetate | |
| BupsA.00035.a | 3DAH | Ribose-phosphate pyrophosphokinase | AMP, Phosphate | |
| BupsA.00072.a | 3E5Y | RNA methyltransferase, TrmH | ||
| BupsA.00076.a | 3DMP | Uracil phosphoribosyltransferase | ||
| BupsA.00085.b | 3ENK | UDP-glucose 4 epimerase | ||
| BupsA.00092.a | 3DMS | Isocitrate dehydrogenase | ||
| BupsA.00112.a | 3EZ4 | 3-methyl-2-oxobutanoate hydroxymethyltransferase | Tris | |
| BupsA.00114.a | 3EZN | Phosphoglycerate mutase | ||
| BupsA.00114.a | 3FDZ | Phosphoglycerate mutase | 3-phospho-glycerate | |
| BupsA.00122.a | 3F0D | 2C–methyl-D-erythritol 2,4-cyclodiphosphate synthase | ||
| BupsA.00122.a | 3F0E | 2C–methyl-D-erythritol 2,4-cyclodiphosphate synthase | Mg | |
| BupsA.00122.a | 3F0F | 2C–methyl-D-erythritol 2,4-cyclodiphosphate synthase | CMP | |
| BupsA.00122.a | 3F0G | 2C–methyl-D-erythritol 2,4-cyclodiphosphate synthase | CMP | |
| BupsA.00130.a | 2KE0 | Peptidyl-prolyl cis-trans isomerase | ||
| BupsA.00141.a | 3EZO | Acyl-carrier-protein S-malonyltransferase | ||
| BupsA.00141.c | 3G87 | Malonyl CoA-acyl carrier protein transacylase | ||
| Giardia lamblia | GilaA.00333.a | 3GBZ | Kinase, CMGC CDK | |
| GilaA.00333.a | 3GC0 | Kinase, CMGC CDK | AMP | |
| Plasmodium falciparum | PlfaA.01650.a | 2KDN | PFE0790c hypothetical protein, conserved | |
| Rickettsia prowazekii | RiprA.00010.b | 3F9I | 3-ketoacyl-(acyl-carrier-protein) reductase | |
| RiprA.00023.a | 3D53 | Inorganic pyrophosphatase | ||
| RiprA.00023.a | 3EMJ | Inorganic pyrophosphatase |
So far, our success rates for soluble expression and crystallization have exceeded expectation. For Tier 1, 73% (418/573) of bacterial targets and 72% (124/173) of eukaryotic targets produced soluble protein. Most (52%) targets produced soluble protein with both Tier 1 vectors (BG1861 and AVA0421), with only a small proportion (8%) showing differential solubility between vectors. Thus, we now usually upscale and purify targets cloned into AVA0421, since the success rate was somewhat higher (52% vs. 44%); and this vector offers the option of shipping targets cleaved and/or un-cleaved. Cleavage of the N-terminal His6-tag typically yielded protein preparations of slightly higher purity. Of the 332 proteins shipped to deCODE for crystallization, 152 (46%) have yielded crystals. However, evidence is emerging that eukaryotic targets have a lower success rate than bacterial targets by roughly half, although the number of trials is still small and not yet complete. The majority (>61%) of crystallized proteins have yielded usable diffraction data, but seven crystals are required, on average, to produce a dataset, and four datasets necessary to produce a final structure. Moreover, in several cases, the structure has not yet reached sufficient resolution (2.5 Å) for submission to PDB. Sixteen of the structures submitted to PDB contain bound ligands, of which eight are products of Tier 11 & 12 ligand screening/co-crystallization efforts.
Selected Structures
All structures solved by SSGCID can be viewed at our web-site (http://www.ssgcid.org/home/Structures.asp) and at the PDB. It is our intention to publish manuscripts describing some, but not all, of these structures. Below, we describe six examples that illustrate the types of insight that can be gained from these structures.
BolA-like protein
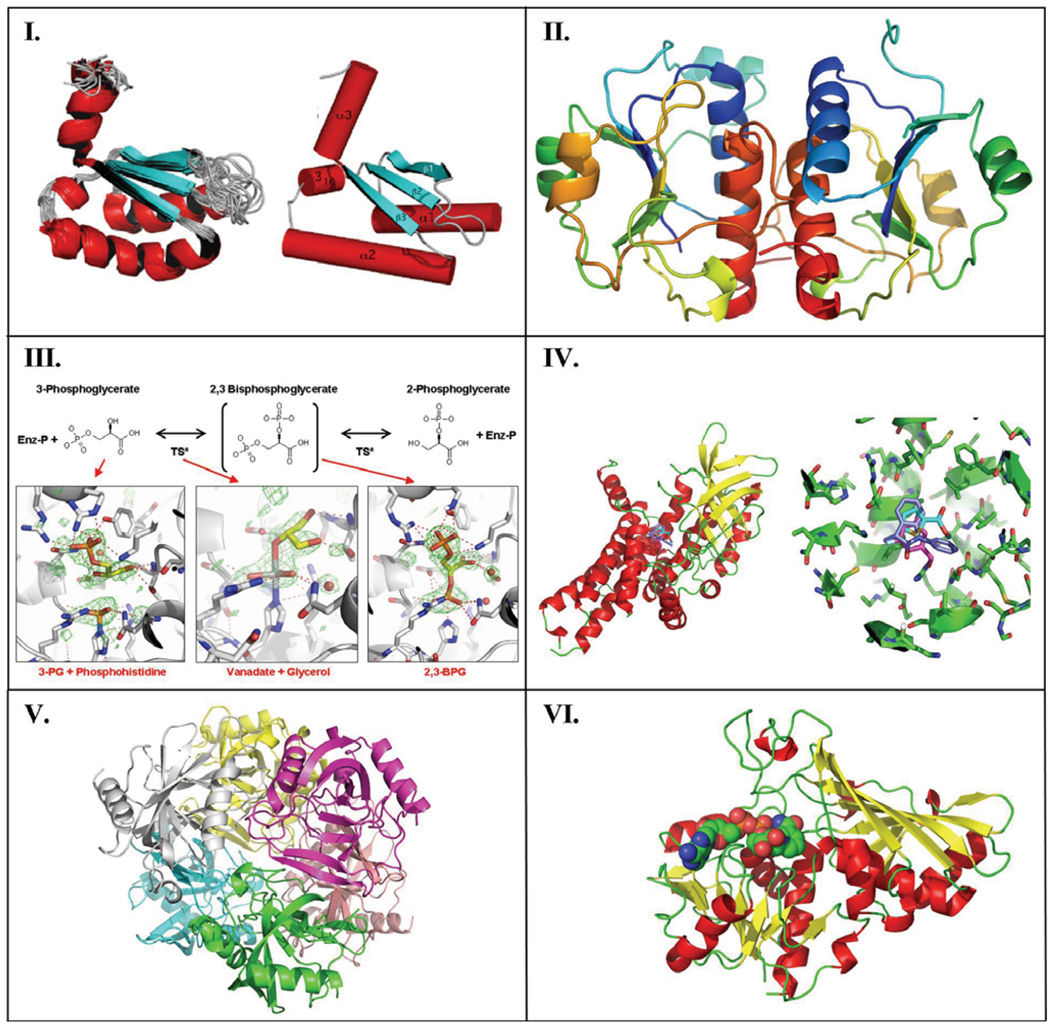
The first Community Request target and first NMR structure determined by SSGCID was for the Plasmodium falciparum protein PFE0790c. P. falciparum is the deadliest of the four species responsible for human malaria, a disease contracted by 350–500 million people annually. The target was a request from the Malaria Group led by Dr. Raymond Hui at the University of Toronto and while not on the SSGCID organism list, the request was specially approved due to its relevance as a potential drug target. PFE0790c is a member of a highly conserved family of BolA-like proteins found in both prokaryotes and eukaryotes. While the molecular function of BolA-like proteins are unknown, their expression has been associated with stress-response [20], and consequently, these proteins represent potential drug targets. Because PFE0790c failed repeated crystallization attempts made by the Malaria Group, it was placed into our Tier 10 NMR pipeline. As shown in Panel I of Fig. (3), the overall topology of the protein is αββαβα with (β2 parallel to β3 and a one-turn 310-helix between α2 and β3. While the fold is similar to the fold observed for the BolA-like protein from Mus musculus [21] and Xanthomonas campestris [22], significant differences exist especially in the relative orientations of α1 and α2. Note that the latter two structures (1V6O and 1V9J, respectively) were also obtaining using NMR-based methods, suggesting that perhaps it may be difficult to crystallize a member of the BolA-like family of proteins.
Fig. 3. Selected protein structures from SSGCID.
Panel I. The solution structure for BolA-like protein PFE0790c from P. falciparum (PDB ID: 2KDN). The ribbon image on the left represents the ensemble of the final 20 NMR structures superimposed on the average structure, while the right cartoon represents the structure closest to the average structure with the three α-helices and three β-strands labeled. For clarity, the unstructured, N-terminal 22-residue tag has been removed from both structures. Color scheme: Helices = red, β-strands = cyan, loops and turns = grey. Panel II. Ribbon drawing of the RNA methyltransferase BupsA.00072.a from B. pseudomallei (3E5Y). The asymmetric unit contains a dimer of two molecules, which is the biological unit. On the left dimer, the thread of the knot can be seen as the orange-red section passing through the yellow-green section. These are roughly residues 80–120 of the 156 amino acid protein. Panel III. Ligand-bound structures of BupsA.00114.a, phosphoglycerate mutase from B. pseudomallei (3EZN). The reaction catalyzed by this enzyme is shown in the top panel. Close-ups of the active site of the phosphoglycerate mutase reveal the 3PG substrate and a transition-state intermediate as a covalently-bound phosphate (left panel), which can be mimicked by vanadate + glycerol (center panel). The final product, 2,3-BPG is shown in the right panel. Panel IV. Fragment-bound Structures from BupsA.00027.a, a glutaryl-CoA dehydrogenase from B. pseudomallei (3D6B). The left panel contains a ribbon diagram of BupsA.00027.a colored by secondary structure, showing α-helices (red), and β-sheets (yellow). Three different fragments (cyan, pink and purple) are superimposed/bound in the active-site. The right panel shows a close-up up of active-site binding pocket, showing the same three fragments. Panel V. Hexameric structure of inorganic pyrophosphatase, RiprA.00023.a, from Rickettsia prowazekii (3D53). Each 20 kDa monomer is colored differently (magenta, gray, green, yellow, pink, peach). Panel VI. Endogenous co-factor (NAD) bound to the glyceraldehyde-3-phosphate dehydrogenase, BrabA.00052.a, from B. melitenesis (3DOC) The NAD molecule is shown as a space-filling model within the ribbon diagram of the protein.
2’-O-methyl RNA methyltransferase
The 2’-O-methylation of ribosomal RNA is one of the most common ways bacteria can obtain antibiotic resistance [23]. The structure of the 2’-O-methyl RNA methyltransferase from B. pseudomallei (BupsA.00072.a) contains a 31 (trefoil) protein knot. According to the protein knot server (http://knots.mit.edu/), which confirmed that this structure has a knot, there are only 40 similar structures in the PDB, including several SpoU-like RNA methyltransferases. The fold of these RNA methyltransferases is quite different from the classical methyltransferase fold, although related to the other SpoU-like RNA methyltransferases. The ribbon diagram of the BupsA.00072.a structure, in Panel II of Fig. (3), shows it to be a dimer and the thread of the knot can be seen as the orange-red section passing through the yellow-green section in the left dimer. Most SpoU-like RNA methyltransferases also contain an RNA binding domain (RBD), but BupsA.00072.a does not contain this domain (nor does the homologous protein from H. influenzae), suggesting it may bind an accessory protein, or perhaps target a different substrate than the other enzymes.
Phosphoglycerate mutase
This target was selected from B. pseudomallei (BupsA.00114.a) for Tier 11 ligand co-crystallization. A BupsA.00114.a crystal was soaked with the enzyme substrate, 3-phophoglycerate (3PG) and an X-ray data set collected after 1 hour. Two protein molecules were present in the crystallographic asymmetric unit with electron density corresponding to 3PG clearly present in one molecule, as shown in Panel III of Fig. (3). However, additional electron density surrounding the active site histidine was also present. This electron density fit and refined well when interpreted as a covalently bound phosphate. No phosphohistidine electron density was apparent in the non-soaked apo-crystal structure, suggesting that the phosphate adduct represents a covalent intermediate. Investigation into the reaction mechanism revealed that phosphoglycerate mutase does indeed form a covalent intermediate with phosphate and then adds the phosphate to 3PG creating a reaction intermediate, 2,3-bisphosphoglycerate (2,3-BPG), which subsequently reforms the transition-state intermediate and the final product, 2-phosphoglycerate (2PG). Inspection of the electron density in the second molecule of the asymmetric unit revealed density that was too large to fit 3PG, but no density near the histidine was visible. It was possible to fit 3PG in two opposing conformations, neither of which could wholly fit all of the electron density. However, the electron density was aptly explained by building in 2,3-PG. Thus, one crystal structure revealed two different steps in the reaction pathway. One molecule contains the reaction intermediate (2,3-BPG), while the other contains the substrate (3PG), and a transition-state intermediate (the phospho-histidine residue). Since previous studies had suggested that vanadate may be used as a transition state mimic [24], we undertook vanadate soaks. The vanadate reacted with glycerol in the cryosolvent, producing an interesting transition state mimic between the histidine residue, vanadate and glycerol. Glycerol substitutes for 3PG and a covalent ternary complex representing the covalent transition-state intermediate can be seen in Panel III of Fig. (3).
Glutaryl-CoA dehydrogenase
This enzyme, from B. pseudomallei (BupsA.00027.a), was subjected to a full Fragments of Life™ screen (Tier 12). Crystals grew readily in the presence of at least 114 fragment pools. To date, 34 crystals have been examined, resulting in three fragment-bound structures. One such structure is shown in Panel IV of Fig. (3). Purified BupsA.00027.a has a distinct yellow color suggestive of FAD (flavin adenine dinucleotide) binding and BupsA.00027.a crystals also have a distinct coloration. However, FAD is not visible in any crystal structure. Fragment-bound crystal structures identify a fragment-binding ‘hot spot’ where all bound fragment molecules have been identified so far. This ‘hot spot’ is located in the putative acyl-CoA binding region in the heart of the catalytic active site of the protein [25].
Inorganic pyrophosphatase (PPase)
PPase is a soluble enzyme that catalyzes the hydrolysis of pyrophosphate to two phosphate ions. This essential activity is believed to drive many biosynthetic reactions by depleting the cellular pyrophosphate concentration and therefore its inhibition could provide a way to inhibit bacterial growth. We have determined structures of the PPase from several bacterial species, including that from R. prowazekii (RiprA.00023.a), which represents the first structure ever reported for this organism. The molecule crystallizes as a homohexamer, similar to the well-described PPase from Escherichia coli. The PPase from R. prowazekii forms a tightly packed spherical structure as seen in Panel V of Fig. (3), which we hypothesize to be the active and soluble hexamer. The active-site pocket of each PPase monomer is solvent exposed and open on the surface of the hexamer.
Glyceraldehyde-3-phosphate dehydrogenase
As indicated above, we found several examples where targets from Tiers 1–3 crystallized with bound endogenous co-factors without their deliberate addition during protein expression, purification, or crystallization. This finding is congruent with those of structural genomics efforts in general where about 20% of all novel protein crystal structures feature either a bound metal or an endogenous ligand (see http://smb.slac.stanford.edu/public/jcsg/cgi/jcsg_ligand_check.pl). One interesting example from our SSGCID project, as shown in Panel VI of Fig. (3), is the presence of NAD (nicotinamide-adenine dinucleotide) bound in the active site of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) from B. melitenesis (BrabA.00052.a). This enzyme carries out the sixth step of glycolysis by catalyzing the conversion of glyceraldehyde-3-phosphate to D-glycerate-1,3-bisphosphate in two steps, which are linked to the reduction of NAD+ to NADH.
Supplementary Material
ACKNOWLEDGEMENTS
This research was funded by NIAID under Federal Contract No. HHSN272200700057C. Special thanks to Tom Edwards and Doug Davies for contributions to BupsA. 00052.a and BupsA00114.a. The authors acknowledge support in part from NIGMS-NCRR co-sponsored PSI-2 Specialized Center Grant U54 GM074961 through the Accelerated Technologies Center for Gene to 3D Structure (www.ATCG3D.org), which funded the development of the Microcapillary Protein Crystallization System. Part of the research was performed at the W.R. Wiley Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by U.S. Department of Energy’s Office of Biological and Environmental Research (BER) program located at Pacific Northwest National Laboratory (PNNL). PNNL is operated for the U.S. Department of Energy by Battelle. We thank Dr. Sam Miller for providing us with Burkholderia pseudomallei 1710b DNA and acknowledge ATCC as source of Giardia lamblia DNA (ATCC_50803) and BEIR Repository as source of Brucella melitensis strain biovar abortus 2308 DNA (DD-156). The authors also thank the entire SSGCID team.
ABBREVIATIONS
- 2PG
2-phosphoglycerate
- 2,3-BPG
2,3-bisphosphoglycerate
- 3-D
Three-dimensional
- 3PG
3-phophoglycerate
- BEIRRR
Biodefense and Emerging Infections Research Resource Repository
- CSGID
Center for Structural Genomics of Infectious Diseases
- DMID
Division of Microbiology and Infectious Diseases
- FAD
Flavin adenine dinucleotide
- FOL
Fragments of Life™
- FTS
Fluorescence-based thermal stability
- GADPH
Glyceraldehyde-3-phosphate dehydrogenase
- HSQC
Heteronuclear single quantum coherence
- HTP
High-throughput
- LIC
Ligation-independent cloning
- MAD
Multiwavelength anomalous diffraction
- MPCS™
Microcapillary Protein Crystallization System
- NAD
Nicotinamide-adenine dinucleotide
- NIAID
National Institute of Allergy and Infectious Diseases
- NIGMS
National Institutes of General Medical Studies
- NMR
Nuclear magnetic resonance
- PCR
Polymerase chain reaction
- PDB
Protein Data Bank
- PNNL
Pacific Northwest National Laboratory
- PSI
Protein Structure Initiate
- SBRI
Seattle Biomedical Research Institute
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SE-MET
Selenomethionine
- SPR
Surface plasmon resonance
- SSGCID
Seattle Structural Genomics Center for Infectious Disease
- STD-NMR
Saturation transfer difference nuclear magnetic resonance
- SWG
Scientific Working Group
- TR-NOE
Transfer nuclear overhauser enhancement resonance
- UW-PPG
University of Washington protein production group
- UW-NMR
University of Washington NMR group
- XRD
X-ray diffraction
REFERENCES
- 1.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. Nucleic Acids Res. 2006;34(Database issue):D668–D672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krogh A, Larsson B, Von H, Sonnhammer EL. J. Mol. Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 3.Edwards TE, Ferre-D'Amare AR. Structure. 2006;14(9):1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Aslanidis C, De Jong PJ. Nucleic. Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov A, Vignali M, LaCount DJ, Quartley E, de Vries C, De Rosa D, Babulski J, Mitchell SF, Schoenfeld LW, Fields S, Hol WG, Dumont ME, Phizicky EM, Grayhack EJ. Mol. Cell. Proteomics. 2004;3(9):934–938. doi: 10.1074/mcp.T400008-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.Madin K, Sawasaki T, Ogasawara T, Endo Y. Proc. Natl.Acad. Sci. USA. 2000;97(2):559–564. doi: 10.1073/pnas.97.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinarov DA, Loushin Newman CL, Markley JL. FEBS. J. 2006;273(18):4160–4169. doi: 10.1111/j.1742-4658.2006.05434.x. [DOI] [PubMed] [Google Scholar]
- 8.Vinarov DA, Lytle BL, Peterson FC, Tyler EM, Volkman BF, Markley JL. Nat. Methods. 2004;1(2):149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- 9.Tyler RC, Aceti DJ, Bingman CA, Cornilescu CC, Fox BG, Frederick RO, Jeon WB, Lee MS, Newman CS, Peterson FC, Phillips GN, Jr, Shahan MN, Singh S, Song J, Sreenath HK, Tyler EM, Ulrich EL, Vinarov DA, Vojtik FC, Volkman BF, Wrobel RL, Zhao Q, Markley JL. Proteins. 2005;59(3):633–643. doi: 10.1002/prot.20436. [DOI] [PubMed] [Google Scholar]
- 10.Busso D, Kim R, Kim SH. J. Struct. Funct. Genomics. 2004;5(1–2):69–74. doi: 10.1023/B:JSFG.0000029197.44728.c5. [DOI] [PubMed] [Google Scholar]
- 11.Stewart L, Burgin AB. In: Frontiers in Drug Design and Discovery. Atta UR, Springer BA, Caldwell GW, editors. San Francisco: Bentham Science Publishers; 2005. pp. 297–391. [Google Scholar]
- 12.Raymond A, Lovell S, Lorimer D, Walchli J, Mixon M, Wallace E, Thompkins K, Archer K, Burgin A, Stewart L. BMC Biotechnol. 2009;9(37) doi: 10.1186/1472-6750-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerdts CJ, Elliott M, Lovell S, Mixon MB, Napuli AJ, Staker BL, Nollert P, Stewart L. Acta. Crystallogr. D. Biol. Crystallogr. 2008;64(Pt 11):1116–1122. doi: 10.1107/S0907444908028060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erlanson DA. Curr. Opin. Biotechnol. 2006;17(6):643–652. doi: 10.1016/j.copbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Erlanson DA, McDowell RS, O'Brien T. J. Med. Chem. 2004;47(14):3463–3482. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 16.Nienaber VL, Richardson PL, Klighofer V, Bouska JJ, Giranda VL, Greer J. Nat. Biotechnol. 2000;18(10):1105–1108. doi: 10.1038/80319. [DOI] [PubMed] [Google Scholar]
- 17.Hajduk PJ, Greer J. Nat. Rev. Drug Discov. 2007;6(3):211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 18.Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. Anal. Chim. Acta. 2008;620(1–2):8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Anal. Biochem. 2006;357(2):289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 20.Santos JM, Freire P, Vicente M, Arraiano CM. Mol.Microbiol. 1999;32(4):789–798. doi: 10.1046/j.1365-2958.1999.01397.x. [DOI] [PubMed] [Google Scholar]
- 21.Kasai T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, Seki E, Matsuda T, Matsuda N, Tomo Y, Shirouzu M, Terada T, Obayashi N, Hamana H, Shinya N, Tatsuguchi A, Yasuda S, Yoshida M, Hirota H, Matsuo Y, Tani K, Suzuki H, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kigawa T. Protein Sci. 2004;13(2):545–548. doi: 10.1110/ps.03401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin KH, Lin FY, Hu YC, Sze KH, Lyu PC, Chou SH. Biomol. NMR. 2005;31(2):167–172. doi: 10.1007/s10858-004-7804-9. [DOI] [PubMed] [Google Scholar]
- 23.Cundliffe E. Biochem. Soc. Symp. 1987;53:1–8. [PubMed] [Google Scholar]
- 24.Bond CS, White MF, Hunter WN. J. Mol. Biol. 2002;316(5):1071–1081. doi: 10.1006/jmbi.2002.5418. [DOI] [PubMed] [Google Scholar]
- 25.Fu Z, Wang M, Paschke R, Rao KS, Frerman FE, Kim JJ. Biochem. 2004;43(30):9674–9684. doi: 10.1021/bi049290c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.