Abstract
Objectives
A number of common non-synonymous single nucleotide polymorphisms (SNPs) in DNA repair genes have been reported to modify bladder cancer risk. These include: APE1-Asn148Gln, XRCC1-Arg399Gln and XRCC1-Arg194Trp in the BER pathway, XPD-Gln751Lys in the NER pathway and XRCC3-Thr241Met in the DSB repair pathway.
Methods
To examine the independent and interacting effects of these SNPs in a large study group, we analyzed these genotypes in 1,029 cases and 1,281 controls enrolled in two case-control studies of incident bladder cancer, one conducted in New Hampshire, USA and the other in Turin, Italy.
Results
The odds ratio among current smokers with the variant XRCC3-241 (TT) genotype was 1.7 (95% CI 1.0–2.7) compared to wild-type. We evaluated gene-environment and gene-gene interactions using four analytic approaches: logistic regression, Multifactor Dimensionality Reduction (MDR), hierarchical interaction graphs, classification and regression trees (CART), and logic regression analyses. All five methods supported a gene-gene interaction between XRCC1-399/XRCC3-241 (p = 0.001) (adjusted OR for XRCC1-399 GG, XRCC3-241 TT vs. wild-type 2.0 (95% CI 1.4–3.0)). Three methods predicted an interaction between XRCC1-399/XPD-751 (p = 0.008) (adjusted OR for XRCC1-399 GA or AA, XRCC3-241 AA vs. wild-type 1.4 (95% CI 1.1–2.0)).
Conclusions
These results support the hypothesis that common polymorphisms in DNA repair genes modify bladder cancer risk and highlight the value of using multiple complementary analytic approaches to identify multi-factor interactions.
Key Words: DNA repair, Bladder cancer, Polymorphism, Interaction
Introduction
Bladder cancer is the fourth most common malignancy in men in Western countries and the eighth most common in women [1]. The majority of this disease is attributed to cigarette smoking; bladder cancer risk is up to four-fold higher among cigarette smokers compared with non-smokers [1]. Case-control studies provide evidence of a familial predisposition to bladder cancer [2,3,4] indicating that some susceptibility factors may be heritable. DNA repair polymorphisms are heritable factors that increase susceptibility to DNA damage resulting from exposure to bladder carcinogens [reviewed in [5]].
While many epidemiological studies have detected differences in bladder cancer susceptibility in relation to DNA repair gene polymorphisms, results are often conflicting [6,7,8,9,10,11,12,13,14]. We recently observed an increased risk of bladder cancer with the XRCC3-241 polymorphism in an Italian population (e.g. in current smokers (TT vs. CC: OR, 2.65; 95% CI, 1.21–5.80) [15]. Also some studies raise the possibility of gene-gene interactions between polymorphisms, i.e., between XRCC1-194 and XRCC3-241, XRCC1-194 and XPD-751 for bladder cancer [13, 16] and lung cancers [17, 18]. Differential findings could be related to population admixture or to the presence of gene-gene and gene-environment interactions that are not well understood due to small sample sizes and the challenges of testing for multiple genetic and environmental risk factors using traditional analytic tools.
Traditionally sized case-control studies and analytic approaches are designed to provide adequate statistical power to detect simple associations. However, it is becoming increasingly evident that many common human diseases, including sporadic forms of cancer, cannot be attributed to a single gene or exposure factor [19]. In contrast, these diseases have complex etiologies with non-additive interactions [20]. In recent years, molecular epidemiologists have been frustrated by the inconsistency of many reported gene-disease associations within and between populations [21]. For the current study, we have formed a large pool of cases and controls to evaluate bladder cancer susceptibility. We utilized both traditional analytic approaches and newer computational algorithms that specifically evaluate gene-gene and gene-environment interactions.
The current study comprises two epidemiologic studies of bladder cancer resulting in one of the largest studies of DNA repair polymorphisms and bladder cancer risk to date, with a sample size of 1,053 cases and 1,281 controls. In this study, we chose to examine DNA repair genes with polymorphisms that have previously been examined in relation to bladder cancer (XRCC1, XRCC3, XPD, APE1). Utilizing the power of both novel and traditional analytic approaches we were able to confirm previously observed associations between DNA repair gene polymorphisms and bladder cancer. Furthermore, this combination of techniques allowed us to evaluate and identify effect modification by gene-gene and gene-environment interactions.
Materials and Methods
Study Groups
New Hampshire Study
We identified all cases of bladder cancer diagnosed among New Hampshire residents, ages 25 to 74 years, from July 1, 1994 to June 30, 1998 from the State Cancer Registry. Detailed methods have been described previously [22]. Briefly, we interviewed a total of n = 857 bladder cancer cases, which was 85% of the cases confirmed to be eligible for the study. Controls less than 65 years of age were selected using population lists obtained from the New Hampshire Department of Transportation. Controls 65 year of age and older were chosen from data files provided by the Centers for Medicare & Medicaid Services (CMS) of New Hampshire. The method of control selection used in our study has been successfully employed in other case-control studies conducted in the region (e.g. Karagas et al.). For efficiency, we shared a control group with a study of non-melanoma skin cancer covering an overlapping diagnostic period of July 1, 1993 to June 30, 1995 [22]. We selected additional controls for bladder cancer cases diagnosed from July 1, 1995 to June 30, 1997 that were frequency matched to these cases on age (25–34, 35–44, 45–54, 55–64, 65–69, 70–74 years) and gender. Most (>95%) of the subjects in this study are of Caucasian origin; and thus our analyses were not appreciably altered by restricting to Caucasians. We interviewed a total n = 1,191 controls (the total shared control group and additional controls), which was 70% of the controls confirmed to be eligible for the study.
Italian Study
We did a hospital-based case-control investigation at two urology departments of S. Giovanni Battista hospital in Turin. The case group comprised unrelated Caucasian men ages 34 to 76 years, residents of the Turin metropolitan area with newly diagnosed, histologically confirmed bladder cancer treated from 1994 to 2003. Controls of a comparable age were recruited daily in random fashion (a) from patients treated at the same urology department, and (b) from patients treated at the medical and surgical departments. In Italy, we interviewed a total of 412 cases and 393 controls which was 90% of the cases and 85% of the controls that were eligible for the study.
Personal Interview
Informed consent was obtained from each participant and all procedures and study materials were approved by the institution's committee for the protection of human subjects. Both studies involved a personal interview covering sociodemographic information (including level of education), lifestyle factors such as use of tobacco (including frequency, duration and intensity of smoking) and medical history. Smoking habits were defined as current (plus ex-smokers since <1 year), former (who ceased smoking since at least 1 year), and never smokers. Both studies collected a blood or cheek cell sample.
Genotyping
We used a variety of genotyping techniques, choosing the most efficient approaches (i.e., reliable and cost-effective) for any given SNP and applying newer technologies during the course of the study. Genotyping for non-synonymous SNPs XRCC3-241 C/T, APE1-148 T/G, XPD-312 G/A and XPD-751 A/C, XRCC1-194 C/T was performed by Qiagen Genomics using their SNP mass-tagging system. For XRCC1-399 G/A, XPD-751, XRCC3-241 some genotyping was performed by PCR-RFLP as described previously [23]. Primer Extension/Denaturing High-Performance Liquid Chromatography was used for genotyping XRCC1-194 in the first 288 subjects. The 5′ Nuclease Assay (TaqMan) assay with fluorogenic minor groove binder probes was used to genotype seven polymorphisms (XPD-751, XRCC1-194, XRCC1-399, and XRCC3-241) in the recent phases of the studies. For quality control purposes, laboratory personnel were blinded to case-control status. These assays achieved greater than 95% accuracy as assessed using negative and positive quality controls (including every 10th sample as a masked duplicate). In Italy, methodologic validation included a comparison between PCR-RFLP, denaturing high performance liquid chromatography and TaqMan assay on a subset. Concordance was in the range between 99 and 100% for all the comparisons; discordant genotypes were excluded from the analysis. Ultimately, data were available for the two studies on APE1-148 (n = 1,165 controls, n = 911 cases), XRCC1-399 (n = 1,253 controls, n = 990 cases), XRCC1-194 (n = 1,203 controls, n = 978 cases), XRCC3-241 (n = 1,275 controls, n = 1,046 cases), XPD-751 (n = 1,215 controls, n = 1,009 cases). Thus, genotype data were complete on over 89% of the subjects. Hardy-Weinberg Equilibrium was evaluated among controls using a chi-square test. The patterns of missing data did not differ significantly by age, gender or smoking status (data not shown).
Pooled Statistical Analysis of NH and Italian Data
The goal of the statistical analysis was to assess the relationship between DNA repair gene SNPs, smoking, and bladder cancer susceptibility. To assess the independent main effects of each SNP, we conducted logistic regression analyses for individuals with one or two variant alleles in comparison to those homozygous wild type for each individual SNP. Assessment of gene-gene and gene-environment interactions was carried out using both logistic regression and multifactor dimensionality reduction (MDR).
Logistic Regression Analysis
We computed the odds ratios for the joint effects of gene pairs using individuals who are homozygous wild type at both loci as the referent group and evaluated interactions between bladder cancer risk factors (gender, smoking variables (e.g., never, former, current)), and genotype by including interaction terms in a logistic regression model. Statistical significance of the interactions as assessed using likelihood ratio tests comparing the models with and without interaction terms. Analyses were adjusted for age (less than or greater than 50), gender, smoking status (never, former, current), and study location. Additionally, we conducted an analysis restricted to men. To fully account for any study heterogeneity in other risk factors, we also conducted our logistic regression analysis with both the study-specific intercept and beta coefficients for age, gender, and smoking status (i.e., by the inclusion of interaction terms for each of these covariates with study location). These computations were done in S-Plus 6.2 (Insightful Inc., Seattle, Wash., USA).
Identification of Gene-Gene Interactions
We selected four other approaches to complement logistic regression for the analysis of gene-gene interactions: Multifactor Dimensionality Reduction (MDR), hierarchical interaction graphs, Classification and Regression Trees (CART), and Logic Regression. The genotype data were initially assessed using a hierarchical interaction graph that included both independent dominant and recessive effects for each gene. Based on this hierarchical interaction graph, a single model (either dominant or recessive) was chosen for each gene based on the combination of models that together explained the largest proportion of bladder cancer risk and was used in the analysis by all four approaches (dominant: APE1-148 entropy 0.01, XRCC1-399 entropy 0.04, XRCC1-194 entropy 0.02, XPD-751 entropy 0, recessive: XRCC3-241 entropy 0.09, interactions: XRCC1-399/XPD-751 entropy 0.15, XRCC1-399/XRCC3-241 entropy 0.26, APE1-148/XRCC3-241 entropy 0.12, XRCC1-399/XRCC1-194 entropy 0.08). Since the MDR and interaction entropy analysis tools do not permit missing values, missing values were imputed 10 independent times using PROC MI in SAS 9.1.3 and analyses were performed using each of the 10 datasets (SAS Institute, Cary, N.C., USA). The results reported were consistent across all 10 datasets and the same gene combinations were selected when the analysis was performed with the missing values deleted.
Multifactor Dimensionality Reduction. The nonparametric MDR approach is described in detail elsewhere [24,25,26,27] and reviewed by Moore [28]. MDR is a data reduction (i.e. constructive induction) approach that seeks to identify combinations of multilocus genotypes and discrete environmental factors that are associated with either high risk or low risk of disease. Thus, MDR defines a single variable that incorporates information from several loci and/or environmental factors that can be divided into high risk and low risk combinations. This new variable can be evaluated for its ability to classify and predict outcome risk status using cross-validation and permutation testing. With n-fold cross-validation, the data are divided into n equal size pieces. An MDR model is fit using (n – 1)/n of the data (i.e. the training set) and then evaluated for its generalizability on the remaining 1/n of the data (i.e. the testing set). The fitness, or value of an MDR model is assessed by estimating accuracy in the training set and the testing set. Accuracy is a function of the percentage of true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN) and is defined as (TP + TN)/(TP + TN + FP + FN). This process is repeated for all n pieces of the data and the n testing accuracies are averaged to provide an estimate of predictive ability or generalizability.
We also estimated the degree to which the same best model is discovered across n divisions of the data. This is referred to as the cross-validation consistency or CVC [24, 29]. A CVC of n in n-fold cross-validation is optimal. Here, we selected the best MDR model as the one with the lowest average prediction error. An error rate of 50% is expected under the null hypothesis. Statistical significance is determined using permutation testing. Here, the case-control labels are randomized 1,000 times and the entire MDR model fitting procedure repeated on each randomized dataset to determine the expected distribution of testing accuracies under the null hypothesis. It is the combination of cross-validation and permutationtesting that reduces the chances of making a type I error due to multiple testing [30, 31]. In this study, we used 10-fold cross-validation and 1,000-fold permutation testing. MDR results were considered statistically significant at the 0.05 level. The MDR software is open-source and freely available from http://www.epistasis.org.
Hierarchical Interaction Graphs. Hierarchical interaction graphs are another tool that helps to interpret and visualize the independent effects and interactive relationships between potential risk factors [16]. Jakulin and Bratko (2003) have provided a metric for determining the gain in information about a class variable (e.g. ability to predict case-control status) from the combination of two variables together compared with the amount of information provided by each of the variables independently [32, 33]. This measure of information gain allows us to gauge the benefit of considering two (or more) attributes as one unit. Information gain is defined in terms of Shannon entropy [[34]].
Measures of entropy are particularly useful for building interaction graphs that facilitate the interpretation of the relationship between variables. Interaction graphs are comprised of a node for each variable with pairwise connections between them. The percentage of entropy removed (i.e. information gain) by each variable is visualized for each node. The percentage of entropy removed for each pairwise Cartesian product of variables is visualized for each connection. Thus, the independent main effects of each SNP, for example, can be quickly compared to the interaction effect. Additive and non-additive interactions can be quickly assessed and used to interpret MDR models that consist of distributions of cases and controls for each genotype combination. A positive entropy (plotted in green) indicates interaction while a negative entropy (plotted in red) indicates redundancy. Interaction entropy analysis was performed using the Orange software package [35].
Classification and Regression Tree Analysis. Classification and regression tree (CART) analysis was also utilized to model gene-gene interactions. CART creates a decision tree that depicts how well each genotype or environmental factor variable predicts class (e.g. bladder cancer case-control status) [36]. Splitting rules are used to stratify data into subsets of individuals, which are represented in the CART decision tree as nodes. The CART model selects the variable used to split each branch and the split point. Each ‘child node’ is selected considering only a subset of the population within a ‘parent node’ to explain class, thus, the results are conditioned on the first splitting variable. CART was implemented using DTREG software with the Gini index to set the splitting criterion and the terminal node size at 10. It is unlikely that the CART approach will detect true epistasis – a combination of factors that have no main effect, but strong interactions in combination [37].
Logic Regression. Logic regression attempts to define the interactions between predictors to explain differences in response [see [38] for details]. Comparisons have shown that both CART and logic regression can provide complementary information in SNP analyses [39]. The algorithm constructs predictors from binary SNP data that are Boolean (logical) combinations of the original genotype data [39]. Logic expressions are depicted as trees with AND/OR operators at each branch point. White numbers on black background indicate the complement. Trees are pruned, rearranged and the optimal tree(s) are selected using a simulated annealing algorithm and permutation testing followed by cross-validation [39]. Logic regression analysis was performed in R.
False Positive Report Probability (FPRP)
Evidence of associations between polymorphisms and complex diseases are greatly affected by the risk to be false positives. To estimate the false positive report probability (FPRP) of positive results we used a Bayesian method proposed by Wacholder et al. [40]. To compute the FPRP, we used the odds ratios from MDR models in which we considered the given classification of high risk and low risk genotype combinations with the estimated statistical power to detect an OR of 1.2, 2 and 3 and an α level equal to the observed p value. Considering the lack of information on the interactions between genes and environmental variables, in our study we have considered a wide range of prior probabilities, but lower than those previously published, i.e., from 0.00001 to 0.10 [15,40,41,42]. Given the many comparisons, we preferred to be conservative by using a cut-point of 0.2 for FPRP.
Results
The overall case group had a higher percentage of men than the control group, and a larger fraction were current or former smokers (table 1). The frequency of the minor allele in the controls by study (NH/Italy) was APE1-148 0.47/0.42, XRCC1-3990.36/0.37, XRCC1-194 0.07/0.08, XRCC3-241 0.38/0.38, XPD-751 0.36/0.42 (table 2). While we observed slight differences in frequency for the XPD-751 and APE1-148 polymorphisms between the two studies, the frequency of the other polymorphisms was similar. Hardy-Weinberg Equilibrium using a chi-square test among controls had resultant p values: APE1-148 p = 0.69, XRCC1-399 p = 0.01, XRCC1-194 p = 0.09, XRCC3-241 p = 0.34, XPD-751 p = 0.08. Deviations from the expected genotype frequency distribution for XRCC1 have been observed previously in other study populations [43, 44].
Table 1.
Selected characteristics of bladder cancer cases and controls by study
| New Hampshire, n (%) |
Italy, n (%) |
Overall, n (%) |
|||||
|---|---|---|---|---|---|---|---|
| controls n = 899 | cases n = 700 | controls n = 382 | cases n = 353 | controls n = 1281 | cases n = 1053 | adjustedd OR (95%CI) | |
| Sex | |||||||
| Female | 329 (36.6) | 168 (24.0) | – | – | 329 (25.7) | 168 (16.0) | Refa |
| Male | 570 (63.4) | 532 (76.0) | 382 (100) | 353 (100) | 952 (74.3) | 885 (84.1) | 1.6 (1.2–2.0) |
| Reference age | |||||||
| <55 | 194 (21.6) | 134 (19.1) | 151 (40.1) | 50 (14.5) | 345 (27.0) | 184 (17.6) | Ref b |
| 55–70 | 482 (53.6) | 379 (54.1) | 190 (50.4) | 214 (62.2) | 672 (52.7) | 593 (56.8) | 1.7 (1.4–2.2) |
| >70 | 223 (24.8) | 187 (26.7) | 36 (9.6) | 80 (23.3) | 259 (20.3) | 267 (25.6) | 2.2 (1.7–2.9) |
| Smoking status | |||||||
| Never | 297 (33.0) | 127 (18.2) | 116 (30.4) | 32 (9.1) | 413 (32.2) | 159 (15.1) | Refc |
| Former | 456 (50.7) | 348 (49.8) | 132 (34.6) | 115 (32.6) | 588 (45.9) | 463 (44.0) | 1.8 (1.4–2.2) |
| Current | 146 (16.2) | 224 (32.0) | 134 (35.1) | 206 (58.4) | 280 (21.9) | 430 (40.9) | 4.0 (3.1–5.1) |
| <1 Pack/day | 44 (4.9) | 42 (6.0) | 58 (15.2) | 110 (31.2) | 102 (8.0) | 152 (14.4) | 4.0 (2.9–5.6) |
| ≥1 Pack/day | 97 (10.8) | 179 (25.6) | 67 17.5) | 85 (24.1) | 164 (12.8) | 264 (25.1) | 4.3 (3.2–5.6) |
| Missing | 5 (0.6) | 3 (0.4) | 9 (2.4) | 11 (3.1) | 14 (1.1) | 14 (1.3) | – |
Data are missing on reference age (5 controls, 9 cases), smoking status (1 case).
Adjusted for age, smoking status (never, former, current).
Adjusted for smoking status, gender.
Adjusted for age, gender.
Adjusted for study.
Table 2.
Odds ratios (95%CI) for bladder cancer in relation to DNA repair gene polymorphisms overall and by smoking status
| Minor allele freq. in controls NH/Italy | Overall controls n (%) | Overall cases n (%) | Overall adjusteda OR (95% CI) | X2 for heterogeneity (p value) | Never smokerb | Former smokerb | Current smokerb | |
|---|---|---|---|---|---|---|---|---|
| BER Pathway | ||||||||
| APE1-148 | 0.47/0.42 | |||||||
| TT | 333 (28.5) | 259 (28.4) | Ref | Ref | Ref | Ref | ||
| TG | 586 (50.3) | 461 (50.6) | 1.0 (0.8–1.2) | 1.2 (0.8–2.0) | 1.1 (0.8–1.4) | 0.7 (0.5–1.1) | ||
| GG | 246 (21.2) | 191 (21.0) | 0.9 (0.7–1.2) | 0.03 (0.9) | 1.1 (0.6–2.0) | 1.0 (0.7–1.4) | 0.8 (0.5–1.3) | |
| XRCC1-399 | 0.36/0.37 | |||||||
| GG | 533 (42.5) | 412 (41.6) | Ref | Ref | Ref | Ref | ||
| GA | 536 (42.8) | 456 (46.1) | 1.1 (0.9–1.4) | 1.1 (0.8–1.7) | 1.1 (0.8–1.4) | 1.2 (0.8–1.6) | ||
| AA | 184 (14.7) | 122 (12.3) | 0.9 (0.7–1.1) | 0.03 (0.9) | 1.0 (0.6–1.8) | 0.9 (0.6–1.3) | 0.7 (0.4–1.2) | |
| XRCC1-194 | 0.07/0.08 | |||||||
| CC | 1,041 (86.5) | 857 (87.6) | Ref | Ref | Ref | Ref | ||
| CT | 152 (12.6) | 115 (11.8) | 0.9 (0.7–1.2) | 1.2 (0.7–2.2) | 0.8 (0.6–1.2) | 0.8 (0.5–1.3) | ||
| TT | 10 (0.8) | 6 (0.6) | 0.8 (0.3–2.4) | 0.05 (0.8) | 0.8 (0.1–7.0) | 0.7 (0.1–3.9) | 1.1 (0.2–6.7) | |
| DSB Pathway | ||||||||
| XRCC3-241 | 0.38/0.38 | |||||||
| CC | 482 (37.8) | 397 (38.0) | Ref | Ref | Ref | Ref | ||
| CT | 617 (48.4) | 477 (45.6) | 1.0 (0.8–1.2) | 0.9 (0.6–1.4) | 0.8 (0.6–1.1) | 1.2 (0.9–1.7) | ||
| TT | 176 (13.8) | 172 (16.4) | 1.2 (0.9–1.5) | 2.0 (0.2) | 1.2 (0.7–2.2) | 0.9 (0.6–1.3) | 1.7 (1.0–2.7) | |
| NER Pathway | ||||||||
| XPD-751 | 0.36/0.42 | |||||||
| AA | 450 (37.0) | 371 (36.8) | Ref | Ref | Ref | Ref | ||
| AC | 602 (49.6) | 483 (47.9) | 1.0 (0.8–1.2) | 0.9 (0.6–1.4) | 1.1 (0.8–1.4) | 0.8 (0.6–1.2) | ||
| CC | 163 (13.4) | 155 (15.4) | 1.1 (0.9–1.5) | 0.17 (0.7) | 1.2 (0.6–2.1) | 1.1 (0.8–1.7) | 1.1 (0.7–1.8) | |
X2 for study location heterogeneity, p value.
Data are missing for APE1-148 (n = 258); XRCC1-194 (n = 153); XRCC3-241 (n = 13); XPD-751 (n = 110); XRCC-399 (n = 91).
Adjusted for study, age, gender, smoking status (never, former, current).
Adjusted for age, gender, study.
Results of our logistic regression analysis of the single genotype effects for the pooled dataset are shown in table 2, overall and then stratified by smoking status. There was no significant heterogeneity between studies (table 2). Importantly, none of the coefficients of logistic regression with study-specific slopes differed more than 10% from the model with age, gender, smoking status and study location. Therefore, our final analysis was based on the more parsimonious models.
The base excision repair (BER) pathway polymorphism APE1-148 was un-related to bladder cancer risk overall (table 2) or in either study (NH 1.0 (95% CI 0.7–1.3), Italy 0.9 (95% CI 0.5–1.7)). The odds ratio for XRCC1-399 variants was slightly below one (table 2, NH 0.9 (95% CI 0.6–1.2), Italy 0.8 (95% CI 0.5–1.3)). XRCC1-194 variants were rare, with an overall OR slightly below 1 (table 2, strata were too small to compute risks for NH and Italy separately). In the double strand break (DSB) repair pathway, individuals variant for XRCC3-241 had a slightly elevated risk of bladder cancer overall (table 2), (NH 1.1 (95% CI 0.8–1.5), Italy 1.7 (95% CI 1.0–2.7)) that was highest among XRCC3-241 variant homozygous current smokers (table 2). Likewise, we did not observe a clear association with the nucleotide excision repair (NER) pathway polymorphism XPD-751 (table 2, NH 1.2 (95% CI 0.9–1.7), Italy 1.1 (95% CI 0.7–1.9)). We did not detect any statistically significant interactions between smoking and any of the genotypes. Further, odds ratios did not differ markedly by gender, and the odds ratios for analyses restricted to males were similar to those performed on the entire cohort (data not shown).
To evaluate the large number of possible combinations of genotypes, we used MDR, hierarchical interaction graphs, CART and logic regression approaches. We then used traditional logistic regression to evaluate the interactions between genotypes that were predicted by at least three of these four methods (MDR, hierarchical interaction graphs, CART, logic regression) (table 4). The interaction predicted by all four methods was reaffirmed by logistic regression (increased risk with XRCC1-399 GG/XRCC3 TT vs. XRCC1-399 GG/XRCC3CC, adjusted OR 1.9 (95% CI 1.3–2.9) p = 0.001).
Table 4.
Interactions between genotypes by logistic regression by smoking status
| Controls | Cases | Overall OR (95%CI)a | Never smokerb | Former smokerb | Current smokerb | ||
|---|---|---|---|---|---|---|---|
| Interactions predicted by all methods (MDR, hierarchical interaction graph, CART, logic regression) | |||||||
| XRCC-399 | XRCC3-241 | ||||||
| GG | CC | 201 | 153 | Ref | Ref | Ref | Ref |
| GA | TT | 84 | 73 | 1.2 (0.8–1.7) | 1.2 (0.5–2.9) | 1.0 (0.6–1.8) | 1.4 (0.7–2.7) |
| GG | TT | 56 | 75 | 1.9 (1.3–2.9) | 1.5 (0.6–3.7) | 1.3 (0.7–2.4) | 4.8 (1.9–12.1) |
| GA | CT | 250 | 207 | 1.2 (0.9–1.6) | 1.0 (0.5–1.9) | 0.9 (0.6–1.3) | 2.0 (1.2–3.4) |
| GG | CC or CT | 476 | 336 | Ref | Ref | Ref | Ref |
| GA or AA | CC or CT | 598 | 484 | 1.2 (1.0–1.5) | 1.9 (0.8–4.3) | 1.1 (0.9–1.5) | 1.3 (0.9–1.9) |
| GG | TT | 56 | 75 | 2.0 (1.4–3.0) | 1.2 (0.8–1.8) | 1.5 (0.9–2.6) | 4.3 (1.8–10) |
| GA or AA | TT | 119 | 91 | 1.1 (0.8–1.5) | 1.3 (0.7–2.7) | 0.9 (0.5–1.4) | 1.3 (0.8–2.3) |
| p valuec | 0.001 | 0.4 | 0.06 | 0.006 | |||
| Interactions predicted by 3 methods (MDR, hierarchical interaction graph, logic regression) | |||||||
| XRCC1-399 | XPD-751 | ||||||
| GG | AA | 200 | 133 | Ref | Ref | Ref | Ref |
| GG | AC | 232 | 204 | 1.4 (1.0–1.9) | 1.7 (0.8–3.4) | 1.6 (1.0–2.4) | 1.1 (0.6–1.8) |
| GA | AA | 177 | 171 | 1.5 (1.1–2.1) | 1.9 (0.9–4.1) | 1.5 (1.0–2.4) | 1.4 (0.8–2.6) |
| GA | AC | 274 | 198 | 1.1 (0.8–1.5) | 1.3 (0.6–2.6) | 1.1 (0.7–1.6) | 1.1 (0.7–1.9) |
| GA | CC | 61 | 64 | 1.6 (1.1–2.5) | 1.3 (0.5–3.8) | 1.9 (1.0–3.5) | 1.6 (0.7–3.4) |
| GG | AA | 200 | 133 | Ref | Ref | Ref | Ref |
| GA or AA | AA | 243 | 218 | 1.4 (1.1–2.0) | 1.9 (1.0–3.9) | 1.4 (0.9–2.1) | 1.4 (0.8–2.4) |
| GG | AC or CC | 304 | 266 | 1.3 (1.0–1.8) | 1.7 (0.9–3.3) | 1.4 (0.9–2.1) | 1.1 (0.7–1.8) |
| GA or AA | AC or CC | 445 | 333 | 1.2 (0.9–1.5) | 0.9 (0.5–1.7) | 0.9 (0.6–1.4) | 1.8 (1.9–3.4) |
| p valuec | 0.008 | 0.5 | 0.6 | 0.4 | |||
Adjusted for age, gender, smoking status (never, former, current) and study.
Adjusted for age, gender and study.
p value for gene-gene interaction using dominant/recessive model grouping.
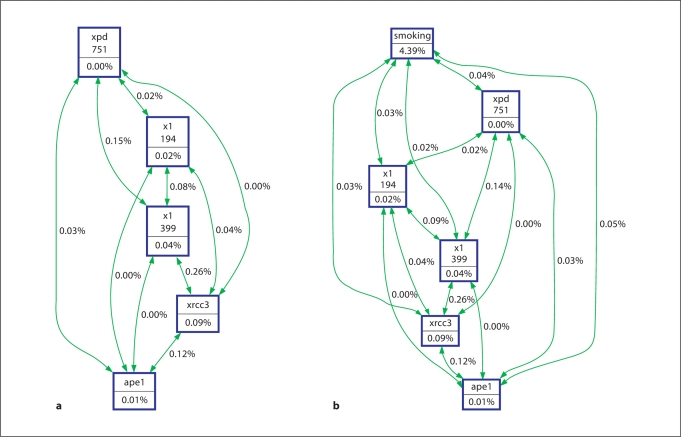
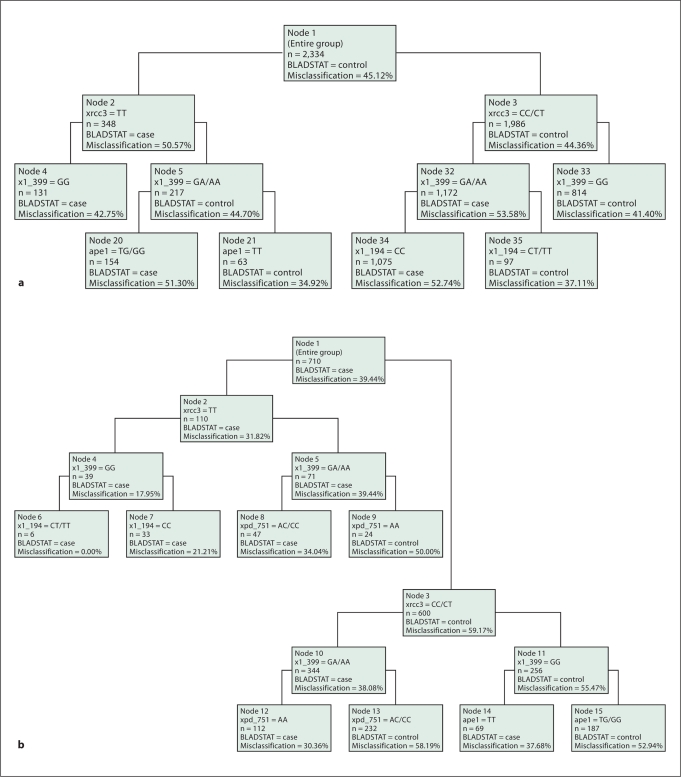
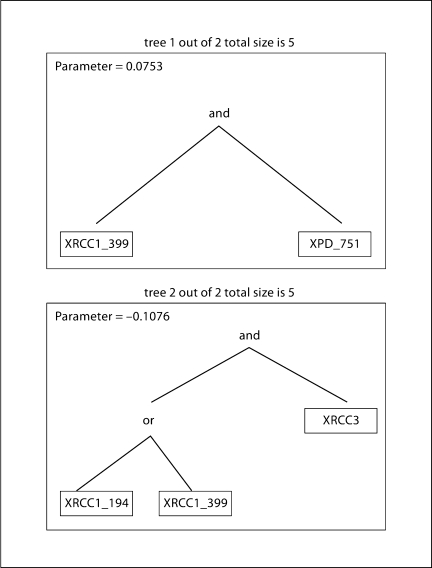
In addition to assessing the concordance between the models, we also examined the complementary information provided by each analytic method to detect gene-gene interactions. MDR interaction modeling (table 3) identified, XPD-751, XRCC1-399, XRCC3-241 as the combination of SNPs that most accurately predicts bladder cancer status (average prediction error 45%, CVC 8/10, permutation test p = 0.003). Table 3 also indicates that the single most important predictor of bladder cancer risk is smoking status (average prediction error 44%, CVC 10/10, permutation test p = 0.001). Likewise, the strongest two way interaction shown in the hierarchical interaction graph (fig. 1A, green arrows) was between XRCC3-241and XRCC1-399. This interaction remains strong when smoking is included in the model (fig. 1B). XRCC3 was the most important single gene in the models (fig. 1A, B). Likewise, the classification tree shown in figure 2 selected XRCC3 for the initial binary split (fig. 2A SNPs only, fig. 2B SNPs in current smokers). In figure 2A, within the XRCC3-CC/CT group, daughter nodes predict increased risk among individuals who are XRCC1-399 GA/AA and XRCC1-194 CC (figure 2, nodes 32,35). From the XRCC3-TT branch, XRCC1-399 GA/AA (node 4) or a combination of XRCC1-399 GG and APE1-TG/GG is associated with increased risk (nodes 5, 20). As observed previously, the initial split was on smoking status (current smokers vs. former/never smokers). Focusing on current smokers (fig. 2B), the model with the least misclassification (0%) includes XRCC3-TT, XRCC1-399 GG, and XRCC1-194 CT/TT. We also examined gene-gene interactions in this dataset using logic regression (fig. 3). The optimal model predicted two independent sets of interactions: between XRCC1 399 and XPD 751 (tree 1), and between XRCC3 and either one of the two XRCC1 SNPs − 194 or 399 (tree 2).
Table 3.
Multifactor dimensionality reduction (MDR) interaction model
| Number of factors | Model | Low risk | High risk | Cross validation consistency | Avg. prediction error (%) | p value permutation test |
|---|---|---|---|---|---|---|
| 1 | Smoking | Never smoker, Former smoker | Current smoker | 10/10 | 43.63 | 0.001 |
| 2 | APE1-148 | Never smoker except APE1-148 TG, | Current smoker, | 4/10 | 45.16 | 0.003 |
| Smoking | Former smoker | Never smoker, APE1-148 TG | ||||
| 3 | XPD-751 | Never smoker except for XPD-751 AC, XRCC1- | Current smoker except for XPD-751 | 10/10 | 46.43 | 0.05 |
| XRCC1-399 | 399 GA; XPD-751 CC, XRCC1-399 GG | AC, XRCC1-399 AA; | ||||
| Smoking | Former smoker except for XPD-751 AA, XRCC1- 399 GA; XPD-751 CC, XRCC1-399 GG/GA | |||||
| 4∗ | XPD-751 | Former smoker, XRCC1-399 AA | Current smoker except XRCC3-241 | 8/10 | 45.28 | 0.003 |
| XRCC1-399 | Never smoker, XRCC3-241 TT, XRCC1-399 AA | CT, XPD-751 AA, XRCC1-399 GG, or Former smoker XRCC3-241 TT, | ||||
| XRCC3-241 | Never smoker except XPD-751 AA, XRCC1-399 | |||||
| Smoking | GA, XRCC3-241 CT | XPD-751 AC/CC | ||||
| 5 | XPD-751 | XPD_751 AC; Former smoker, APE1-148 TG, | Current smoker; APE1-148 TG | 10/10 | 46.22 | 0.01 |
| XRCC1-399 | XRCC3-241 CT, XRCC1-399 GA, XPD_751 AC | Never smoker, XPD_751 AC, | ||||
| XRCC3-241 | Never smoker, APE1-148 TG, XRCC3-241 CC, | XRCC1-399 GA, XRCC3-241 CC, | ||||
| APE1-148 | XRCC1-399 GG, XPD_751 AC | APE1-148 TG |
Denotes the genetic model with the highest cross-validation consistency and accuracy.
Fig. 1.
Hierarchical interaction graph of genotypes. The percentage of entropy removed (i.e. information gain) by each variable is visualized for each node (box). The percentage of entropy removed for each pairwise Cartesian product of variables is visualized for each connection. A positive entropy (plotted in green) indicates interaction while a negative entropy (plotted in red) indicates redundancy. a Overall analysis of genotype. b Analysis of genotype and smoking status.
Fig. 2.
Classification and regression tree (CART) model of genotypes. Splitting rules are used to stratify data into subsets of individuals, which are represented in the CART decision tree as nodes. Each ‘child node’ is selected considering only a subset of the population within a ‘parent node’ to explain class, thus, the results are conditioned on the first splitting variable. a Overall analysis of genotype. b Analysis of genotype within current smokers.
Fig. 3.
Logic Regression model of genotype interactions. The algorithm constructs predictors from binary SNP data that are Boolean (logical) combinations of the original genotype data. Logic expressions are depicted as trees with AND/OR operators at each branch point.
Three methods (MDR, hierarchical interaction graph, logic regression) predicted an interaction between XRCC1-399 and XPD-751 (table 4). Relative to individuals with XRCC1-399 GG and XPD-751 AA genotypes, those with at least a variant allele for either XRCC1-399 or XPD-751 had increased bladder cancer risk (e.g. XRCC1-399GA, XPD-751 AA OR 1.5 (95%CI 1.1–2.1)). The interaction p value for heterozygotes/variants compared with wild-type was statistically significant (p = 0.008) from logistic regression analysis.
We further evaluated potential gene-environment interactions by stratifying our logistic regression analysis of the genotype combinations that were selected in our initial screen by smoking status (table 4). Bladder cancer risk was particularly elevated in the current smokers with XRCC1-399 GG/XRCC3 TT genotypes versus XRCC1-399 GG/XRCC3CC (adjusted OR 4.8 (95% CI 1.9–12.1)). When age, gender and smoking history were added to the initial predictive models with all genotypes, the four analytic methods consistently selected smoking status, followed by male gender and age above 50 years as most highly predictive for bladder cancer risk (data not shown). The strongest four-factor MDR model without smoking, included the polymorphisms XPD-751, XRCC1-399, XRCC3-241, and APE1-148 (average prediction error 46.54%, cross-validation consistency 10/10). The best gene-only model was the two locus with XRCC1-399 GA, and XRCC3-241 TT as the high risk genotype combination (average prediction error 47%, cross-validation consistency 9/10).
False Positive Report Probability (FPRP)
Table 5 reports the FPRP values calculated using the statistical power to detect an OR of 1.2, 2.0 and 3.0 with an α level equal to the observed p value. Results show a good reliability on a 3-loci gene-only model (XRCC1-399-GG + XRCC3-241-TT + APE1-148-TG/GG vs. the remaining ‘low-risk’ genotypes) in the overall population with very low prior probabilities (0.0001) for OR = 2 or 3. Among all the two-loci significant models, the comparison XRCC1-399-GG + XRCC3-241-TT vs. XRCC1-399-GG + XRCC3-241-CC/TT is still interesting at a prior probability of 0.01 (OR = 2 or 3), as well as for the 4 loci model involving XPD, XRCC1, XRCC3and APE1 genes. On the other hand, other two-loci models (XRCC1-399-GA + XRCC3-241-TT vs. ‘low-risk’ genotypes; XRCC1-399-GG + XRCC3-241-TT vs. XRCC1-399-GG + XRCC3-241-CC/TT) require higher prior probabilities (0.1 for OR = 2 or 3).
Table 5.
False positive report probabilities
| OR | p value | OR = 1.2 power | Prior probability |
OR = 2.0 power | Prior probability |
OR = 3.0 power | Prior probability |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.1 | 0.01 | 0.001 | 0.0001 | 0.00001 | 0.1 | 0.01 | 0.001 | 0.0001 | 0.00001 | 0.1 | 0.01 | 0.001 | 0.0001 | 0.00001 | ||||||
| Single gene | ||||||||||||||||||||
| XRCC3-TT vs. CC Current smokers | 1.7 (1.0–2.7) | 0.025 | 0.070 | 0.760 | 0.972 | 0.997 | 1.000 | 1.000 | 0.754 | 0.227 | 0.763 | 0.970 | 0.997 | 1.000 | 0.992 | 0.182 | 0.710 | 0.961 | 0.996 | 1.000 |
| XRCC1-GA vs. others Overall | 1.2 (1.0–1.4) | 0.020 | 0.500 | 0.269 | 0.802 | 0.976 | 0.998 | 1.000 | 1.000 | 0.155 | 0.669 | 0.953 | 0.995 | 1.000 | 1.000 | 0.155 | 0.669 | 0.953 | 0.995 | 1.000 |
| 2 Genes | ||||||||||||||||||||
| (XRCC1-399-GA+ XRCC3-241-TT) vs. others Overall | 1.2 (1.0–1.4) | 0.020 | 0.500 | 0.269 | 0.802 | 0.976 | 0.998 | 1.000 | 1.000 | 0.155 | 0.669 | 0.953 | 0.995 | 1.000 | 1.000 | 0.155 | 0.669 | 0.953 | 0.995 | 1.000 |
| (XRCC1-399-GG + XRCC3-241-TT) vs. (XRCC1-399-GG + XRCC3-241-CC/TT) Overall | 2.0 (1.4–3.0) | 0.001 | 0.007 | 0.517 | 0.922 | 0.992 | 0.999 | 1.000 | 0.500 | 0.014 | 0.138 | 0.617 | 0.942 | 0.994 | 0.975 | 0.007 | 0.076 | 0.452 | 0.892 | 0.988 |
| (XRCC1-399-GG + XRCC3-241-TT) vs. (XRCC1-399-GG + XRCC3-241-CC/TT) Current smokers | 4.3 (1.8–10.3) | 0.001 | 0.002 | 0.821 | 0.981 | 0.998 | 1.000 | 1.000 | 0.043 | 0.182 | 0.711 | 0.961 | 0.996 | 1.000 | 0.210 | 0.044 | 0.335 | 0.835 | 0.981 | 0.998 |
| (XRCC1-399-GA/AA + XPD-751-AA) vs. (XRCC1-399-GG + XPD-751-AA) Overall | 1.4 (1.1–2.0) | 0.064 | 0.198 | 0.745 | 0.970 | 0.997 | 1.000 | 1.000 | 0.975 | 0.373 | 0.867 | 0.985 | 0.998 | 1.000 | 1.000 | 0.367 | 0.865 | 0.985 | 0.998 | 1.000 |
| (XRCC1-399-GA/AA + XPD-751-AA) vs. (XRCC1-399-GG + XPD-751-AA) Never smokers | 1.9 (1.0–3.9) | 0.080 | 0.105 | 0.873 | 0.987 | 0.999 | 1.000 | 1.000 | 0.556 | 0.565 | 0.935 | 0.993 | 0.999 | 1.000 | 0.893 | 0.447 | 0.899 | 0.989 | 0.999 | 1.000 |
| (XRCC1-399-GA/AA + XPD-751-AC/CC) vs. (XRCC1-399-GG + XPD-751-AA) Overall | 1.3 (1.0–1.8) | 0.114 | 0.315 | 0.765 | 0.973 | 0.997 | 1.000 | 1.000 | 0.995 | 0.508 | 0.919 | 0.991 | 0.999 | 1.000 | 1.000 | 0.507 | 0.919 | 0.991 | 0.999 | 1.000 |
| 3 or more genes | ||||||||||||||||||||
| (XRCC1-399-GG + XRCC3-241-TT + APEX-148-TG/GG) vs. others Overall | 1.9 (1.4–2.5) | 0.000 | 0.001 | 0.074 | 0.467 | 0.898 | 0.989 | 0.999 | 0.643 | 0.000 | 0.001 | 0.007 | 0.066 | 0.415 | 0.999 | 0.000 | 0.000 | 0.005 | 0.044 | 0.313 |
| (XPD-751-wt + XRCC1-399-GG + RCC3-241-TT + APEX-148-TG/GG) vs. others or (XPD-751-het + XRCC1-399-GG + XRCC3-241-TT + APEX-148-TT) vs. others Overall | 2.7 (1.5–4.9) | 0.001 | 0.004 | 0.719 | 0.966 | 0.996 | 1.000 | 1.000 | 0.162 | 0.057 | 0.400 | 0.870 | 0.985 | 0.999 | 0.636 | 0.015 | 0.145 | 0.631 | 0.945 | 0.994 |
Discussion
Our combined analysis of two completed case-control studies improved our statistical power for investigating the risk factors for bladder cancer. This malignancy, like many others, likely has a complex, and as yet uncertain genetic architecture. In the current study, we investigated the hypothesis that individuals with prevalent SNPs in DNA repair genes modify genetic susceptibility to bladder cancer using a multifaceted analytical approach that combines traditional statistical methods with newer computational algorithms to screen for gene-gene interactions. Our study supports previous reports that the XRCC3-241 and the XRCC1-399 SNPs modify bladder cancer risk. The most consistently predicted gene-gene interactions were XRCC1-399/XRCC3-241 and XRCC1-399/XPD-751.
Using this approach, we observed a slightly reduced risk of bladder cancer among XRCC1-399 variants that is consistent with our previous finding of a 40% reduction in risk of bladder cancer among those with at least one XRCC1-399 variant allele compared with those with one or two wild-type alleles in the New Hampshire study population [23]. XRCC1 acts as a scaffolding protein throughout the BER process [45]. The codon 399 polymorphism occurs in the BRCT1 domain, a region involved in binding polyADPribose polymerase (PARP) and APE1 [46].
We also previously reported an increased bladder cancer risk associated with the XRCC3-241 polymorphism and increased DNA adduct levels in the Italian study [8, 15]. Overall we found a modest association with bladder cancer risk for the XRCC3-241polymorphism that was highest among variant current smokers (table 2). XRCC3 variant genotype was included in the best MDR model (table 3) and explained the most entropy in bladder cancer case control status by hierarchical interaction graph (fig. 1). The initial split on XRCC3 in the CART and logic regression models reaffirmed this effect (fig. 2, 3). In the U.S., Stern and colleagues found an elevated bladder cancer risk among individuals who carry at least one Met variant allele at codon 241 (233 cases, 209 controls), particularly among heavy smokers [13]. Results of other independent analyses of the XRCC3-241 polymorphism and bladder cancer risk have been inconsistent [8,9,10, 13]. XRCC3 is required for stabilization of the RAD51 complex in repair of double strand breaks and crosslinks, and for maintaining chromosome stability during cell division [47, 48]. Polymorphic variants for XRCC3-241 appear to be functionally capable of homology-directed double strand DNA repair ex vivo and were not especially sensitive to DNA damaging agents [49]. It is possible that the observed relationship between XRCC3 and bladder cancer is due to another biologic function that is modified by the codon 241 amino acid substitution, or that 241 is in linkage disequilibrium with another causal polymorphism.
Our analytic strategy utilized multiple interaction modeling approaches to efficiently assess potential gene-gene interactions. All methods concordantly predicted an interaction between XRCC1-399 and XRCC3-241 and a related interaction (XRCC1-194) was observed previously in another study [13]. In vitro studies indicate that XRCC3 is required for the assembly and stabilization of the Rad51 complex with heteroduplex DNA [47] and modulates progression of replication forks [50]. The BER pathway enzyme XRCC1 co-localizes with Rad51 in response to DNA damage [51]. Thus, the possibility of an interaction between these polymorphisms warrants further consideration.
Three analytic methods (MDR, hierarchical interaction graph, logic regression) predicted an interaction between XRCC1-399 and XPD-751 that was supported by our logistic regression analysis (table 4). The CART model conditioned its single tree analysis on XRCC3, therefore, it is not surprising that an independent XRCC1/XPD interaction was not included in the tree (fig. 2). Logic regression showed this interaction as a separate tree (fig. 3). MDR selects the high risk combinations of factors, which may include both additive and multiplicative effects. Hierarchical interaction graphing is useful for interpreting the exact nature of the interactions. The line between XRCC1-399 and XPD-751 is green accompanied by a positive percent of entropy indicating a greater than additive effect. XPD/ERCC2 is a member of the TFIIH complex that unwinds the damaged DNA following DNA damage recognition. This complex is also involved in normal gene transcription and controlled cellular apoptosis [52]. The highest odds ratios were observed in never and former smokers, coinciding with a previously observed association between higher DNA adduct levels and XPD-751 or XRCC1-399 genotype in never smokers [11]. A possible explanation for this finding is that the effects of DNA repair genotype on risk may be overwhelmed by the carcinogenic effects of tobacco constituents. The phenotype is more apparent among never smokers, probably reflecting impaired removal of more subtle, environmentally related, DNA damage events.
Not surprisingly, when all variables were entered into the model, current smoking was the strongest single risk factor, followed by male gender and age [53]. XPD-751, XRCC1-399 and XRCC3-241 genotypes combined with smoking predicted bladder cancer risk well. Bladder cancer risk associations with these three genes were previously observed in the individual studies (XPD-751 and XRCC1-399 in New Hampshire [16, 23] and XRCC3-241 in Italy [15]).
Associations between polymorphisms and complex diseases may simply be false positive findings. After applying a method for the estimation of the number of false positive results, many of the above reported combinations of polymorphisms remained plausibly true associations. In particular, the combination of XRCC1-399-GG + XRCC3-241-TT + APE1-148-TG/GG versus the remaining ‘low-risk’ genotypes consistently appeared at higher risk even considering the very low prior probability of 0.0001 (OR = 2 or 3). While we observe associations between DNA repair SNPs and bladder cancer risk, these findings could be due to chance, and may not be causal.
Our results highlight the utility of our comprehensive analytic approach for efficiently picking the important associations out of a large group of potentially related factors. Further investigation of these interactions in other epidemiologic studies and experimental systems will be required to support these observations and elucidate their mechanisms.
Acknowledgements
We would like to thank all members of the New Hampshire Health Study team for making this project possible. This publication was funded in part by grant numbers CA102327, CA099500, CA82354, CA57494, ES00002, 5 P42 ES05947, RR018787, and ES07373 from the National Cancer Institute, NIH and from the National Institute of Environmental Health Sciences, NIH. Additional support for Dr. Andrew was kindly provided through a fellowship from the American Society of Preventive Oncology and the Cancer Research Foundation of America. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIEHS, NIH, ASPO, CRFA. The Italian study has been granted by the Compagnia di San Paolo (Turin, Italy; P. Vineis) and by the Associazione Italiana per le Ricerche sul Cancro (G. Matullo).
References
- 1.Kirkali Z, Chan T, Manoharan M, et al. Bladder cancer: Epidemiology, staging and grading, and diagnosis. Urology. 2005;66:4–34. doi: 10.1016/j.urology.2005.07.062. [DOI] [PubMed] [Google Scholar]
- 2.Cartwright RA. Genetic association with bladder cancer. Br Med J. 1979;2:798. doi: 10.1136/bmj.2.6193.798-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sullivan JW. Epidemiologic survey of bladder cancer in greater New Orleans. J Urol. 1982;128:281–283. doi: 10.1016/s0022-5347(17)52886-4. [DOI] [PubMed] [Google Scholar]
- 4.Kantor AF, Hartge P, Hoover RN, Fraumeni JF., Jr Familial and environmental interactions in bladder cancer risk. Int J Cancer. 1985;35:703–706. doi: 10.1002/ijc.2910350602. [DOI] [PubMed] [Google Scholar]
- 5.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–1530. [PubMed] [Google Scholar]
- 6.Stern MC, Johnson LR, Bell DA, Taylor JA. XPD codon 751 polymorphism, metabolism genes, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1004–1011. [PubMed] [Google Scholar]
- 7.Schabath MB, Delclos GL, Grossman HB, et al. Polymorphisms in XPD exons 10 and 23 and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2005;14:878–884. doi: 10.1158/1055-9965.EPI-04-0235. [DOI] [PubMed] [Google Scholar]
- 8.Matullo G, Guarrera S, Carturan S, et al. DNA repair gene polymorphisms, bulky DNA adducts in white blood cells and bladder cancer in a case-control study. Int J Cancer. 2001;92:562–567. doi: 10.1002/ijc.1228. [DOI] [PubMed] [Google Scholar]
- 9.Sanyal S, Festa F, Sakano S, et al. Polymorphisms in DNA repair and metabolic genes in bladder cancer. Carcinogenesis. 2004;25:729–734. doi: 10.1093/carcin/bgh058. [DOI] [PubMed] [Google Scholar]
- 10.Shen M, Hung RJ, Brennan P, et al. Polymorphisms of the DNA repair genes XRCC1, XRCC3, XPD, interaction with environmental exposures, and bladder cancer risk in a case-control study in northern Italy. Cancer Epidemiol Biomarkers Prev. 2003;12:1234–1240. [PubMed] [Google Scholar]
- 11.Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22:1437–1445. doi: 10.1093/carcin/22.9.1437. [DOI] [PubMed] [Google Scholar]
- 12.Stern MC, Umbach DM, van Gils CH, Lunn RM, Taylor JA. DNA repair gene XRCC1 polymorphisms, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2001;10:125–131. [PubMed] [Google Scholar]
- 13.Stern MC, Umbach DM, Lunn RM, Taylor JA. DNA Repair Gene XRCC3 Codon 241 Polymorphism, Its Interaction with Smoking and XRCC1 Polymorphisms, and Bladder Cancer Risk. Cancer Epidemiol Biomarkers Prev. 2002;11:939–943. [PubMed] [Google Scholar]
- 14.Garcia-Closas M, Malats N, Real FX, et al. Genetic variation in the nucleotide excision repair pathway and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2006;15:536–542. doi: 10.1158/1055-9965.EPI-05-0749. [DOI] [PubMed] [Google Scholar]
- 15.Matullo G, Guarrera S, Sacerdote C, et al. Polymorphisms/Haplotypes in DNA repair genes and smoking: A bladder cancer case-control study. Cancer Epidemiol Biomarkers Prev. 2005;14:2569–2578. doi: 10.1158/1055-9965.EPI-05-0189. [DOI] [PubMed] [Google Scholar]
- 16.Andrew AS, Nelson HH, Kelsey KT, et al. Concordance of multiple analytical approaches demonstrates a complex relationship between DNA repair gene SNPs, smoking, and bladder cancer susceptibility. Carcinogenesis. 2006;27:1030–1037. doi: 10.1093/carcin/bgi284. [DOI] [PubMed] [Google Scholar]
- 17.Zhou W, Liu G, Miller DP, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2, smoking, and lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2003;12:359–365. [PubMed] [Google Scholar]
- 18.Chen S, Tang D, Xue K, et al. DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis. 2002;23:1321–1325. doi: 10.1093/carcin/23.8.1321. [DOI] [PubMed] [Google Scholar]
- 19.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4:850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 20.Moore JH. The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Hum Hered. 2003;56:73–82. doi: 10.1159/000073735. [DOI] [PubMed] [Google Scholar]
- 21.Caporaso NE. Why have we failed to find the low penetrance genetic constituents of common cancers? Cancer Epidemiol Biomarkers Prev. 2002;11:1544–1549. [PubMed] [Google Scholar]
- 22.Karagas MR, Tosteson TD, Blum J, et al. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population. Environ Health Perspect. 1998;106(suppl 4):1047–1050. doi: 10.1289/ehp.98106s41047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelsey KT, Park S, Nelson HH, Karagas MR. A population-based case-control study of the XRCC1 Arg399Gln polymorphism and susceptibility to bladder cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:1337–1341. [PubMed] [Google Scholar]
- 24.Ritchie MD, Hahn LW, Roodi N, et al. Multifactor-dimensionality reduction reveals high-order interactions among estrogen-metabolism genes in sporadic breast cancer. Am J Hum Genet. 2001;69:138–147. doi: 10.1086/321276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ritchie MD, Hahn LW, Moore JH. Power of multifactor dimensionality reduction for detecting gene-gene interactions in the presence of genotyping error, missing data, phenocopy, and genetic heterogeneity. Genet Epidemiol. 2003;24:150–157. doi: 10.1002/gepi.10218. [DOI] [PubMed] [Google Scholar]
- 26.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene-gene and gene-environment interactions. Bioinformatics. 2003;19:376–382. doi: 10.1093/bioinformatics/btf869. [DOI] [PubMed] [Google Scholar]
- 27.Hahn LW, Moore JH. Ideal discrimination of discrete clinical endpoints using multilocus genotypes. In Silico Biol. 2004;4:183–194. [PubMed] [Google Scholar]
- 28.Moore JH. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Rev Mol Diagn. 2004;4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- 29.Moore JH. Cross-validation consistency for the assessment of genetic programming results in microarray studies. Lecture Notes in Computer Science. 2003;2611:99–106. [Google Scholar]
- 30.Coffey CS, Hebert PR, Ritchie MD, et al. An application of conditional logistic regression and multifactor dimensionality reduction for detecting gene-gene interactions on risk of myocardial infarction: The importance of model validation. BMC Bioinformatics. 2004;5:49–59. doi: 10.1186/1471-2105-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coffey CS, Hebert PR, Krumholz HM, et al. Reporting of model validation procedures in human studies of genetic interactions. Nutrition. 2004;20:69–73. doi: 10.1016/j.nut.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Jakulin A, Bratko I. Analyzing attribute dependencies. In: Lavrac N, Gamberger D, Blockeel H, Todorovski L, editors. PKDD 2003, LNAI 2838. Cavtat: Croatia: Springer-Verlag; 2003. pp. 229–240. [Google Scholar]
- 33.Jakulin A, Bratko I, Smrke D, Demsar J, Zupan B: Attribute interactions in medical data analysis. Artificial intelligence in medicine Europe. Protaras, Cyprus, 2003, pp 229–238.
- 34.Pierce JR. An introduction to information theory – Symbols, signals and noise. New York: Dover Publications; 1980. [Google Scholar]
- 35.Demsar J, Zupan B. Orange: From Experimental Machine Learning to Interactive Data Mining, White Paper. Ljublijana, Slovenia: Faculty of Computer and Information Science, University of Ljublijana; 2004. [Google Scholar]
- 36.Breiman L, Friendman JH, Olshen RA, Stone CJ. Classification and regression trees. Belmont: Wadsworth; 1984. [Google Scholar]
- 37.Cook NR, Zee RY, Ridker PM. Tree and spline based association analysis of gene-gene interaction models for ischemic stroke. Stat Med. 2004;23:1439–1453. doi: 10.1002/sim.1749. [DOI] [PubMed] [Google Scholar]
- 38.Ruczinski I, Kooperberg C, LeBlanc M. Logic regression. J Comput Graph Stat. 2003;12:475–511. [Google Scholar]
- 39.Ruczinski I, Kooperberg C, LeBlanc M. Exploring interactions in high dimensional genomic data: An overview of logic regression, with applications. J Mult Anal. 2004;90:178–195. [Google Scholar]
- 40.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: An approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung RJ, Brennan P, Canzian F, et al. Large-scale investigation of base excision repair genetic polymorphisms and lung cancer risk in a multicenter study. J Natl Cancer Inst. 2005;97:567–576. doi: 10.1093/jnci/dji101. [DOI] [PubMed] [Google Scholar]
- 42.Matullo G, Dunning AM, Guarrera S, et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27:997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- 43.Duell EJ, Holly EA, Bracci PM, Wiencke JK, Kelsey KT. A population-based study of the Arg399Gln polymorphism in X-ray repair cross-complementing group 1 (XRCC1) and risk of pancreatic adenocarcinoma. Cancer Res. 2002;62:4630–4636. [PubMed] [Google Scholar]
- 44.Ye W, Kumar R, Bacova G, et al. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis. 2006;27:1835–1841. doi: 10.1093/carcin/bgl017. [DOI] [PubMed] [Google Scholar]
- 45.Kubota Y, Nash RA, Klungland A, et al. Reconstitution of DNA base excision-repair with purified human proteins: Interaction between DNA polymerase beta and the XRCC1 protein. EMBO J. 1996;15:6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 46.Marsin S, Vidal AE, Sossou M, et al. Role of XRCC1 in the coordination and stimulation of oxidative DNA damage repair initiated by the DNA glycosylase hOGG1. J Biol Chem. 2003;278:44068–44074. doi: 10.1074/jbc.M306160200. [DOI] [PubMed] [Google Scholar]
- 47.Bishop DK, Ear U, Bhattacharyya A, et al. Xrcc3 is required for assembly of Rad51 complexes in vivo. J Biol Chem. 1998;273:21482–21488. doi: 10.1074/jbc.273.34.21482. [DOI] [PubMed] [Google Scholar]
- 48.Ronen A, Glickman BW. Human DNA repair genes. Environ Mol Mutagen. 2001;37:241–283. doi: 10.1002/em.1033. [DOI] [PubMed] [Google Scholar]
- 49.Araujo FD, Pierce AJ, Stark JM, Jasin M. Variant XRCC3 implicated in cancer is functional in homology-directed repair of double-strand breaks. Oncogene. 2002;21:4176–4180. doi: 10.1038/sj.onc.1205539. [DOI] [PubMed] [Google Scholar]
- 50.Henry-Mowatt J, Jackson D, Masson JY, et al. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol Cell. 2003;11:1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 51.Taylor RM, Moore DJ, Whitehouse J, Johnson P, Caldecott KW. A cell cycle-specific requirement for the XRCC1 BRCT II domain during mammalian DNA strand break repair. Mol Cell Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Araujo SJ, Wood RD. Protein complexes in nucleotide excision repair. Mutat Res. 1999;435:23–33. doi: 10.1016/s0921-8777(99)00042-7. [DOI] [PubMed] [Google Scholar]
- 53.Silverman DT, Morrison AS, Devesa SS. Bladder Cancer. In: Schottenfeld DFJ, editor. Cancer Epidemiology and Prevention. New York: Oxford University Press; 1996. pp. 1156–1179. [Google Scholar]