Recently, we described a β-peptide foldamer, β53–1 (Figure 1A), that assembles into a 14-helix in aqueous solution, binds the oncoprotein hDM2 with submicromolar affinity, and inhibits the interaction of hDM2 with a peptide derived from the activation domain of p53 (p53AD).1 The intact recognition epitope of β53–1, including a high degree of helical structure, is required for selective inhibition of the p53AD·hDM2 interaction. Here, we present the solution structure of β53–1 in methanol. The structure reveals details of a helix-stabilizing salt bridge on one helical face, novel “wedge into cleft” packing along another, and how distortions in the β53–1 14-helix maximize presentation of the p53AD recognition epitope. These details deepen our understanding of how β3-peptides fold and how they can be designed to form higher order structures and bind macromolecules.2,3
Figure 1.
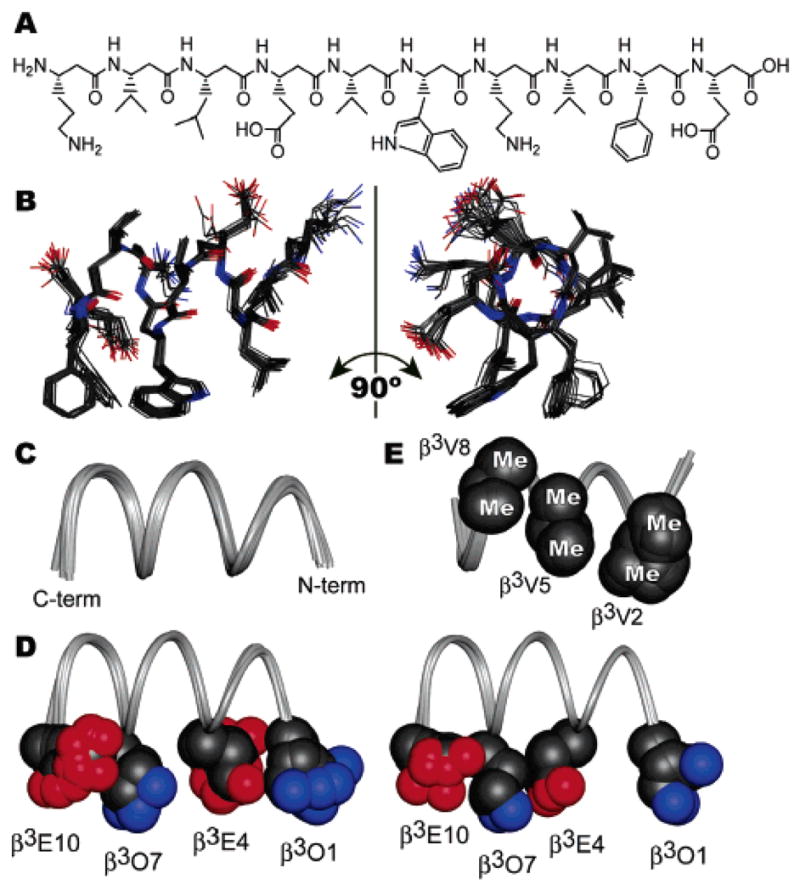
(A) Chemical structure of β53–1, shown with N-terminus at left. (B) Solution structure of β53–1 in CD3OH at 10 °C, shown as a bundle of 20 lowest-energy structures, with C-terminus at left. (C) Ribbon representation of the backbones of 20 lowest-energy structures. (D) Two subpopulations of ion pairing configurations. Superposed at left are 17 structures in which β3O1 and β3E4 are proximal; superposed at right are three structures in which β3E4 and β3O7 are proximal. (E) Conformations of β3-homovaline residues illustrating the “wedge into cleft” packing found in all 20 lowest-energy structures.
Two-dimensional NMR spectroscopy was performed using 5 mM β53–1 in CD3OH at 10 °C. Previous circular dichroism and analytical ultracentrifugation experiments1 and the NMR line widths observed herein are consistent with a monomeric, 14-helical structure for β53–1 under these conditions. The proton resonances of β53–1 were assigned unambiguously using TOCSY and natural abundance 1H–13C HSQC spectra.4 ROESY experiments were then performed using mixing times of 200, 350, and 500 ms.5 The observed series of NH–CαH ROEs confirmed the sequential assignment by providing a backbone “ROE walk”. Three classes of medium-range ROEs characterize a 14-helical conformation: those between HN(i) and Hβ(i+2), HN(i) and Hβ(i+3), and Hα(i) and Hβ(i+3).6,7 All 20 potential medium-range interactions of this type were observed in the ROESY spectra of β53–1; in addition, 27 additional medium-range ROEs between side chains three positions apart were also observed.4 The large number of medium-range ROEs observed by NMR provides clear evidence for a high level of 14-helix structure in β53–1; 449 ROEs quantified using a 350 ms mixing time were subsequently assigned and integrated using SPARKY.8 Peak volumes were converted to 151 upper-limit distance constraints4 and used to perform simulated annealing torsional dynamics on 100 random starting configurations of β53–1 using DYANA.4,9 No constraint violations were reported among the resulting 20 lowest-energy structures, which are shown in Figure 1B.
The ensemble of calculated structures of β53–1 (Figure 1B) shows a 14-helix with an average backbone atom RMSD from the mean structure of 0.17 ± 0.07 Å. The backbone torsions of individual structures deviate little from the mean, even at the termini (Figure 1C), illustrating the robustness of the β53–1 14-helix in methanol. The helix is characterized by approximately 1.61 Å rise per residue and 3.0 residues per turn for residues 1–6, with a slight unwinding to approximately 1.49 Å rise per residue and 3.3 residues per turn for residues 7–10. This unwinding appears to be unique to β53–1, as it was not observed in NMR structures of unrelated β3-peptides with and without side chain ion pairing.6,7,10 Side chains are also well-defined among the lowest-energy structures, with an overall average heavy atom RMSD from the mean of 0.60 ± 0.10 Å.
β53–1 contains four charged side chains arranged to favor formation of helix-stabilizing salt bridges on one 14-helix face.11 In all 20 low-energy structures, the terminal nitrogen of β3O7 and the nearest terminal oxygen of β3E10 are characterized by a consistent separation of 5.5 ± 0.6 Å. The relative positions of the remaining two ion pairs fall into two subpopulations (Figure 2D). In 17 structures, the terminal nitrogen of β3O1 and the nearest terminal oxygen of β3E4 are closer (5.4 ± 0.9 Å) than the equivalent atoms of β3E4 and β3O7 (6.8 ± 0.9 Å). By contrast, in the remaining three structures, the terminal nitrogen of β3O7 and the nearest terminal oxygen of β3E4 are closer (3.6 ± 0.4 Å) than the equivalent atoms of β3O1 and β3E4 (7.7 ± 1.3 Å). This interplay among potential ion pairs suggests that the central salt bridge is weaker than those near the termini and supports the hypothesis that multiple interconnected ion pairs play a key helix-stabilizing role.11,12
Figure 2.
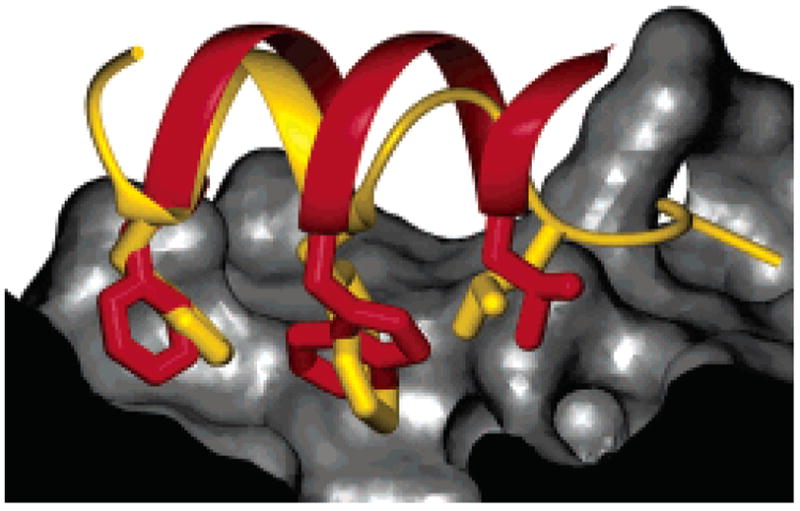
Overlay of the methanol solution structure of β53–1 (red ribbon and side chains) with the crystal structure of a p53AD-derived peptide (gold ribbon and side chains) bound to hDM2 (gray surface).23 Side chains of β53–1 not implicated in recognition have been omitted, and part of the hDM2 surface has been cut away for clarity.
Another feature incorporated into the design of β53–1 was the inclusion of β3-homovaline (β3V) residues at positions 2, 5, and 8. It was long surmised11,13–15 and recently proven12 that β3-amino acids branched at the first side chain carbon stabilize 14-helices, in stark contrast to the effects of such side chains on α-helices.16 The β53–1 structure provides a clear rationale for these observations. All 20 low-energy structures contain a unique arrangement of β3-homovaline side chains in which one methyl group of a β3V side chain nestles into a cleft formed by the two methyl groups of another β3V side chain (Figure 2E). These interactions are especially noticeable between the side chains of β3V5 and β3V8, which are in VDW contact17 in 19 of 20 structures. Overall, interactions among the three β3V side chains bury 155 ± 13 Å2 of hydrophobic surface area from water (24% of the surfaces of these side chains). These packing interactions may explain why these and other branched residues stabilize 14-helices12,18,19 and suggest new avenues for the design of 14-helix bundles.20,21
The remaining 14-helix face consists of residues that comprise the hDM2-binding epitope, namely, β3-homoleucine (β3L3), β3-homotryptophan (β3W6), and β3-homophenylalanine (β3F9). We originally hypothesized that the side chains of these residues would form an extended hydrophobic surface that might mimic that of p53AD.1 Interestingly, the β3F9 side chain can access two specific conformations within the constraints used; the fact that this variability has been observed in another 14-helix structure7 implies that the side chain may indeed preferentially populate these rotamers within a 14-helix. The side chains of β3W6 and β3L3 are in VDW contact in all 20 structures, while the side chains of β3W6 and β3-F9 are in VDW contact in the context of only one of β3F9’s two preferred conformations (present in 6 of 20 low-energy structures). Overall, on average, the side chains of β3L3, β3W6, and β3F9 comprise a continuous, solvent-exposed hydrophobic surface area of 520 Å2. This value is comparable to the contact areas measured at the interfaces of transient homo- and heterodimeric protein complexes.22
As a consequence of the unexpected unwinding near the C-terminus of β53–1, the β3F9 side chain is not aligned perfectly with the side chains of β3L3 and β3W6 along the helix axis (see Figure 1B). This subtle distortion may avoid steric repulsions between the large side chains of β3F9 and β3W6. In fact, it is unclear whether the unwinding near the C-terminus, which is unique to β53–1, is due to more favorable ion pairing, more favorable β3V nesting interactions, or the need to avoid steric clashes on the recognition face containing large hydrophobic residues. As structures of other short, stable 14-helices are determined, it will be interesting to note what factors lead to similar distortions in the “ideal” 14-helix geometry.
Importantly, this subtle distortion allows the side chains comprising the β53–1 recognition face to better mimic those on the p53AD α-helix. Overlays between β53–1 in an idealized 14-helical conformation and p53AD bound to hDM223 revealed an imperfect alignment between the two ligands; while the β3L3, β3W6, and β3F9 side chains of β53–1 could superimpose with their counterparts on p53AD, the 14-helix backbone could not completely fit within hDM2’s binding groove.1 The comparable overlay with the solution structure of β53–1 (Figure 2) shows no such conflict. In its solution conformation, β53–1 can access all three of hDM2’s hydrophobic pockets while occupying the same binding groove as p53AD with no steric clashes. This fit demands subtle unwinding near the β53–1 C-terminus that staggers the side chains, producing a β3-peptide that is uniquely suited for α-helix mimicry. Protein–protein interactions are notoriously difficult to inhibit with most ligand classes;24,25 the solution structure of β53–1 suggests that β-peptide oligomers can present an extended, highly variable surface that could be used as a general platform to target these critical interfaces.
Supplementary Material
Acknowledgments
This work was supported by the NIH (GM 59843 to A.S. and AI 01806 to M.H.), the National Foundation for Cancer Research, and in part by a grant to Yale University, in support of A.S., from the Howard Hughes Medical Institute. J.A.K. is grateful to the NSF for a Predoctoral Fellowship.
Footnotes
Supporting Information Available: Assignment tables, ROE-derived upper-distance limits. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Kritzer JA, Lear JD, Hodsdon ME, Schepartz A. J Am Chem Soc. 2004;126:9468. doi: 10.1021/ja031625a. [DOI] [PubMed] [Google Scholar]
- 2.Seebach D, Beck AK, Bierbaum DJ. Chem Biodiversity. 2004;1:1111. doi: 10.1002/cbdv.200490087. [DOI] [PubMed] [Google Scholar]
- 3.Cheng RP, Gellman SH, DeGrado WF. Chem Rev. 2001;101:3219. doi: 10.1021/cr000045i. [DOI] [PubMed] [Google Scholar]
- 4.Please see Supporting Information for details.
- 5.ROE intensities were linear in this range.
- 6.Arvidsson PI, Rueping M, Seebach D. Chem Commun. 2001:649. [Google Scholar]
- 7.Etezady-Esfarjani T, Hilty C, Wuthrich K, Rueping M, Schreiber J, Seebach D. Helv Chim Acta. 2002;85:1197. [Google Scholar]
- 8.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco, CA: 2004. [Google Scholar]
- 9.Guntert P, Mumenthaler C, Wuthrich K. J Mol Biol. 1997;273:283. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 10.Rueping M, Mahajan YR, Jaun B, Seebach D. Chem–Eur J. 2004;10:1607. doi: 10.1002/chem.200305571. [DOI] [PubMed] [Google Scholar]
- 11.Hart SA, Bahadoor ABF, Matthews EE, Qiu XYJ, Schepartz A. J Am Chem Soc. 2003;125:4022. doi: 10.1021/ja029868a. [DOI] [PubMed] [Google Scholar]
- 12.Kritzer JA, Tirado-Rives J, Hart SA, Lear JD, Jorgensen WL, Schepartz A. J Am Chem Soc. 2005;127:167. doi: 10.1021/ja0459375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raguse TL, Lai JR, Gellman SH. Helv Chim Acta. 2002;85:4154. [Google Scholar]
- 14.Hamuro Y, Schneider JP, DeGrado WF. J Am Chem Soc. 1999;121:12200. [Google Scholar]
- 15.Gung BW, Zou D, Stalcup AM, Cottrell CE. J Org Chem. 1999;64:2176. [Google Scholar]
- 16.Chakrabartty A, Baldwin RL. Adv Protein Chem. 1995;46:141. [PubMed] [Google Scholar]
- 17.Creighton TE. Proteins. W. H. Freeman and Co; New York: 1993. [Google Scholar]
- 18.Martinek TA, Fulop F. Eur J Biochem. 2003;270:3657. doi: 10.1046/j.1432-1033.2003.03756.x. [DOI] [PubMed] [Google Scholar]
- 19.Glattli A, Seebach D, van Gunsteren WF. Helv Chim Acta. 2004;87:2487. [Google Scholar]
- 20.Raguse TL, Lai JR, LePlae PR, Gellman SH. Org Lett. 2001;3:3963. doi: 10.1021/ol016868r. [DOI] [PubMed] [Google Scholar]
- 21.Cheng RP, DeGrado WF. J Am Chem Soc. 2002;124:11564. doi: 10.1021/ja020728a. [DOI] [PubMed] [Google Scholar]
- 22.Nooren IMA, Thornton JM. J Mol Biol. 2003;325:991. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 23.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Science. 1996;274:948. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 24.Schepartz A, Kim PS. Curr Opin Chem Biol. 1998;2:9. doi: 10.1016/s1367-5931(98)80029-x. [DOI] [PubMed] [Google Scholar]
- 25.Sakurai K, Chung HS, Kahne D. J Am Chem Soc. 2004;126:16288. doi: 10.1021/ja044883w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


