Summary
Deconvoluting the natural pathway of BAT development has defined key molecular events, which enables researchers to manipulate the amount or activity of brown fat. We review recent advances on the transcriptional regulation of BAT development, and discuss the emerging questions.
Keywords: obesity, thermogenesis, brown fat, UCP1, PRDM16, PGC-1α
Introduction
In contrast to white adipose tissue (WAT), which is specialized for the storage of excess energy, brown adipose tissue (BAT) dissipates chemical energy to produce heat as a defense against cold. Interest in the development and regulation of BAT has exploded in the last few years because of a confluence of discoveries in the biology and physiology of the brown adipocyte. Clearly, much of the interest in this cell type is due to its role in the defense against hypothermia and obesity. Observations originally made in the oncology clinic with 18Fluoro-labeled 2-deoxy-glucose positron emission tomography (18FDG-PET) scanning have led to an appreciation that most, if not all, adult humans have distinct brown fat deposits, and the activity of BAT varies depending on age, adiposity, temperature and gender (Cypess et al., 2009; Saito et al., 2009; van Marken Lichtenbelt et al., 2009; Virtanen et al., 2009). Furthermore, a major transcriptional regulator of brown fat cell identity, PRDM16, was recently discovered. This is the first cell-autonomous transcriptional component that is both necessary and sufficient to stimulate the development of brown fat cells. This offers a new opportunity to investigate the developmental origins of brown fat, and provides a new pathway for the manipulation of BAT in vivo. In this article, we review current understanding of the transcriptional regulation and cellular origin of brown fat development.
Transcriptional Regulators of Brown Fat Cell Development and Differentiation
Despite the differences in the developmental origins and physiological functions of brown and white adipocytes, both cell types share a very similar transcriptional cascade that controls the process of fat differentiation. Detailed studies of white fat differentiation had previously identified PPARγ (peroxisome proliferator-activated receptor-γ) and the C/EBPs (CCAAT/enhancer-binding proteins) as key transcription factors driving fat cell differentiation (reviewed in Farmer, 2006). Indeed, PPARγ is absolutely necessary for both white fat and brown fat cell development. C/EBPs function cooperatively with PPARγ, and promote a transcriptional cascade that promotes and maintains the stable differentiated state of adipocytes. While C/EBPα is essential for the normal insulin sensitivity of mature fat cells, it is required only for the formation of white fat, but not brown fat, suggesting a possible role for other C/EBP family members in brown fat development. C/EBP-β and δ, as well as other transcription factors, also participate in the transcriptional cascade of adipogenesis by regulating PPARγ gene expression. Brown fat cell differentiation requires PPARγ but, importantly, this factor alone is not sufficient to drive mesenchymal cells into a brown fat program. This led us and other investigators to hunt for transcriptional components that would specifically promote a brown fat genetic program, including the expression of UCP1. As described below, several transcriptional factors and cofactors that affect the expression of UCP1 and that of other key brown fat-selective genes have been identified.
FoxC2 (Forkhead box C2)
FoxC2 is a member of the forkhead/winged helix transcription factor family whose gene expression is enriched in the adipose tissues of human and mouse. Enerback and colleagues reported that transgenic expression of FoxC2 in the adipose tissue induced the emergence of brown fat-like cells in WAT, with increased mitochondria and elevated expression of thermogenic genes including UCP1 and PGC-1α (Cederberg et al., 2001). Notably, transgenic expression of FoxC2 counteracts many obesity-associated pathologies, including insulin resistance and hypertriglyceridemia (Cederberg et al., 2001). This “browning” effect stimulated by FoxC2 seems mainly a result of inducing the expression of the RIα subunit of cAMP-dependent Protein Kinase (PKA). This sensitizes cells to cAMP signaling through the β-adrenergic pathway.
PGC-1α (PPARγ-co-activator-1α) and its transcriptional regulators
PGC-1α was originally identified from brown fat cells as a cold-inducible co-activator of PPARγ (Puigserver et al., 1998). Accumulating evidence indicates that PGC-1α is a master regulator of mitochondrial biogenesis and oxidative metabolism in most cell types, including brown fat and skeletal muscle. Indeed, genetic ablation of PGC-1α results in reduced capacity for cold-induced thermogenesis in vivo, and in a blunted response to cAMP signaling in cultured brown fat cells. Similarly, ectopic expression of PGC-1α in white fat cells induces a number of mitochondrial genes and thermogenic genes such as UCP1(Puigserver et al., 1998).
Several transcriptional regulators have been shown to control brown fat development and function, at least in part, through regulating the transcriptional activity or gene expression of PGC-1α. RIP140, a co-repressor of many nuclear receptors, has been shown to suppress the transcriptional activity of PGC-1α through a physical interaction (Hallberg et al., 2008). Genetic ablation of RIP140 causes the emergence of brown fat-like cells in WAT (Leonardsson et al., 2004). Similarly, SRC2/TIF2/GRIP1, a member of the steroid receptor co-activator (SRC) family, represses PGC-1α transcriptional activity. Loss of SRC2 function leads to an increase in adaptive thermogenesis and energy expenditure in vivo (Picard et al., 2002). Rb (retinoblastoma) protein and p107, another member of Rb pocket protein family, also negatively regulate PGC-1α gene expression. Adipocytes derived from pRb-deficient fibroblasts or embryonic stem cells exhibit a brown-fat phenotype with high mitochondrial content, and elevated expression of UCP1, PGC-1α and mitochondrial genes (Hansen et al., 2004). p107-deficient mice display a striking accumulation of brown fat-like cells in WAT, with multilocular lipid droplets, abundant mitochondria, and high levels of UCP1 and PGC-1α expression (Scime et al., 2005). The biological effect of p107 on brown fat development appears to be mediated through the repression of pRb, because pRb levels are significantly reduced in Sca-1+ CD31- Lin- adipogenic precursors isolated from p107 deficient mice. Lastly, twist-1, a helix-loop-helix- containing transcriptional regulator, has recently been reported as a negative regulator of PGC-1α function in brown fat. Genetic ablation or depletion of twist-1 induces the expression of brown fat-selective genes, while overexpression of twist-1 represses them in a PGC-1α-dependent fashion (Pan et al., 2009).
Taken together, these data suggest a dominant role for PGC-1α in brown fat development and its thermogenic function. However, the mass of BAT and the expression of many brown fat-selective genes are not affected by the depletion of PGC-1α. Thus, while PGC-1α is a crucial regulator of adaptive thermogenesis and mitochondrial biogenesis, it does not determine the cellular specification of brown fat.
PRDM16 (PRD1-BF-1-RIZ1 homologous domain containing protein-16)
PRDM16 is a 140kDa zinc-finger protein that was originally identified at a chromosomal breakpoint of t(1;3)(p36;q21)-positive human acute myeloid leukemia cells (Mochizuki et al., 2000). Our previous study showed that PRDM16 expression was highly enriched in BAT, compared with WAT (Seale et al., 2007). When ectopically expressed in white fat preadipocytes or myoblasts, PRDM16 induces a nearly complete brown fat genetic program. This includes mitochondrial biogenesis, increased cellular respiration and expression of brown fat-selective genes, both the cAMP-inducible thermogenic genes (UCP1, PGC-1a and deiodinase-d2) and those BAT-selective genes that are not sensitive to cAMP (such as cidea and elovl3). Furthermore, transgenic expression of PRDM16 in adipose tissue increases the formation of pockets of multilocular brown-like adipocytes in WAT depots under stimulation with a β–adrenergic agonist.
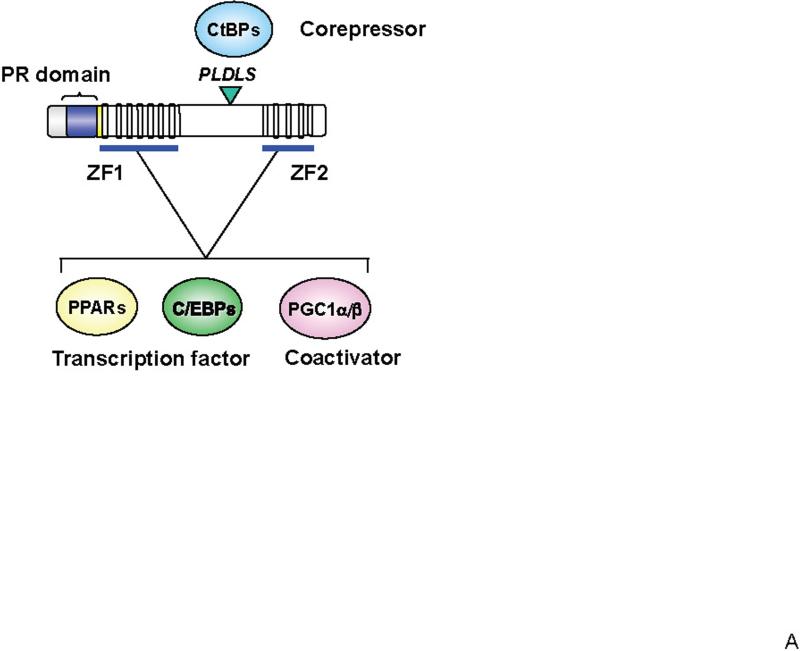
PRDM16 had previously been shown to bind directly to a specific DNA sequence via two sets of zinc-fingers (ZF1 and ZF2 domains) in vitro. Interestingly, abrogation of DNA binding using a point mutation did not substantially alter the ability of PRDM16 to induce the brown fat phenotype, when compared to the wild type protein. This suggested that PRDM16 was not working as a classical DNA-binding transcription factor. Further study demonstrated that, in addition to inducing the gene expression of PGC-1α, PRDM16 directly binds to both PGC-1α and PGC-1β to increase their transcriptional activities (Seale et al., 2007). These results directed us to search for other binding partners that further specified the actions of PRDM16. In fact, PRDM16 interacts with a variety of canonical DNA-binding transcription factors, such as PPARα, PPARγ, p53, and several members of the C/EBP family (Kajimura et al., 2009; Seale et al., 2008). In each case, binding is via one or more of PRDM16's zinc finger domains, resulting in powerful co-activation of their transcriptional activities (Figure 1A). On the other hand, PRDM16 expression in white fat precursors or in myoblasts robustly represses gene expression of selective markers for white fat cells or skeletal muscle, respectively. The repressive effect of PRDM16 on white fat cell-selective genes is mediated through its regulated association with C-terminal binding proteins (CtBP-1 and -2), well-known corepressor proteins (Kajimura et al., 2008). These results indicate that PRDM16 is a co-regulatory protein that can function as a bi-directional switch in brown fat development through multiple protein-protein interactions.
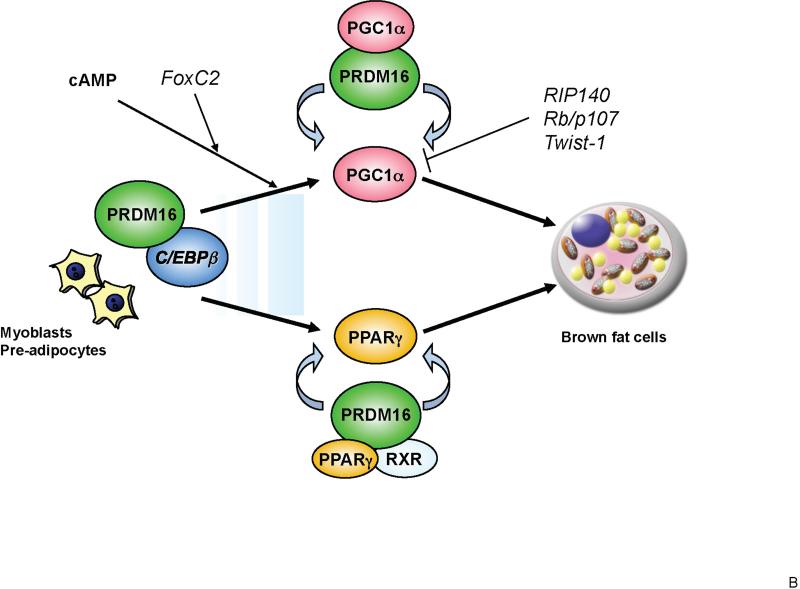
Figure 1. Transcriptional control of brown fat development through PRDM16.
(A) Structure of PRDM16 and key domains of its function. PRDM16 directly interacts with canonical transcription factors such as PPARα, PPARγ and C/EBP family members and transcriptional co-activators PGC-1α and PGC-1β through the two sets of zinc finger domains (ZF1 and ZF2). PRDM16 is also associated with the co-repressors CtBP1 and 2 through its PLDLS motif. This interaction mediates the repressive action of PRDM16 on the expression of white fat cell-specific genes. (B) PRDM16-C/EBP-β transcriptional complex acts in myf5-positive myoblastic precursors or pre-adipocytes to induce the expression of PPARγ and PGC-1α. PRDM16 co-activates PPARγ and PGC-1α, which then drives a brown fat differentiation program. The cAMP-dependent thermogenic gene program is potentiated by FoxC2 and PRDM16. RIP140, Rb/p107, and Twist-1 antagonize the expression or transcriptional activity of PGC-1α and repress brown fat genetic program.
Depletion of PRDM16 in primary brown fat cell precursors not only causes a near-total loss of brown fat characteristics, but also unexpectedly causes the emergence of distinct morphological and genetic characteristic of skeletal myotubes in culture. These include the formation of syncytia and ectopic activation of skeletal muscle-specific genes. Consistent with this, BAT from PRDM16-deficient mice exhibits an abnormal morphology, with reduced expression of brown fat-selective marker genes and elevated expression of skeletal muscle-specific genes (Seale et al., 2008). Conversely, expression of PRDM16 in myogenic precursors drives a robust and functional program of brown adipogenesis. These results suggest that PRDM16 is a critical determinant of the brown fat lineage from myoblast progenitors during the embryonic development.
How does PRDM16 control the conversion of myoblastic precursors to brown fat? Recent studies from our group demonstrated that PRDM16 forms a transcriptional complex with the active form of C/EBP-β (LAP) that is abundantly expressed both in brown fat and myoblasts (Kajimura et al., 2009). It has also been reported that C/EBP-β is a dominant transcription factor that controls cAMP-induced gene expression in brown fat cells (Karamitri et al., 2009). Indeed, depletion of C/EBP-β significantly blunts the ability of PRDM16 to induce the brown fat differentiation and specific fat gene program in myoblasts. Consistent with this finding, BAT from C/EBP-β deficient mice displays a similar molecular signature to BAT from PRDM16-deficient mice, with reduced expression of BAT-selective genes and elevated expression of skeletal muscle-selective genes. These studies indicate that a PRDM16-C/EBP-β complex controls the initiating events of the conversion from myoblastic precursors to brown fat cells (Figure 1B).
Surprisingly, the combination of these two factors (PRDM16 and C/EBP-β) are sufficient to induce a fully functional brown fat program in non-adipogenic cells, such as embryonic fibroblasts and skin fibroblasts, from mouse and man. Cells expressing these two factors differentiate into mature adipocytes with extremely high oxygen consumption and high expression of brown fat-selective genes including UCP1 and PGC-1α. Furthermore, when transplanted into immunocompromised mice, fibroblasts expressing these two factors give rise to an ectopic fat pad that displays a brown fat-like phenotype: multilocular adipocytes expressing UCP1. Importantly, similar to endogenous BAT, this engineered brown fat tissue can function as a sink for active glucose disposal, as determined by FDG-PET scanning (Kajimura et al., 2009).
Developmental Origin of Brown Fat
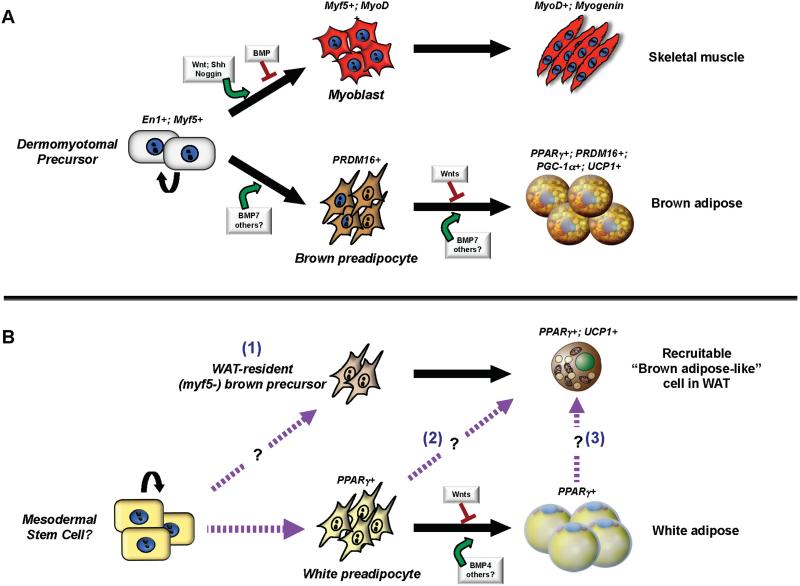
Because WAT and BAT share many common features, including a conserved PPARγ-driven transcriptional program of adipogenesis, these tissues have been assumed to share a direct common progenitor. However, recent studies indicate that brown adipocytes are developmentally closer to skeletal muscle than to white adipose cells (Figure 2). In particular, genetic fate-mapping experiments indicate that BAT in the interscapular region and skeletal muscle but not white adipose cells arise from cells that express Myf5, a gene previously assumed to be present almost exclusively in committed skeletal muscle precursors (Seale et al., 2008). Similarly, Engrailed-1 (En1) expressing cells in the central dermomyotome form BAT, skeletal muscle and dermis (Atit et al., 2006). Whether En1-expressing cells give rise to any white adipose lineages was not examined. In addition, global gene expression analyses by the Cannon and Nedergaard group show that brown but not white adipocyte precursors express a gene profile related to that of skeletal muscle cells (Timmons et al., 2007). Most recently, the mitochondrial proteomic signature of BAT was shown to be highly related to that of skeletal muscle but not to that of WAT (Forner et al., 2009).
Figure 2. Hierarchical developmental relationships in adipose.
BAT (A) and WAT (B) have separate developmental origins in the embryo. (A) BAT and skeletal muscle originate from precursors in the dermomyotome that express Engrailed-1 (En1) and Myf5. Brown adipose fate in the somite may be controlled by members of the transforming growth factor (TGF)-β superfamily of secreted factors such as BMP-7. Canonical Wnt signaling represses the differentiation of brown preadipocytes into mature brown adipose cells. PRDM16, PPARγ, PGC-1α and UCP1 are functional markers of brown adipose cells in the developmental, homogenous deposits of BAT. MyoD expression and skeletal muscle commitment is positively regulated by Wnts, Sonic Hedgehog (Shh) and Noggin. BMPs suppress myogenic commitment. (B) The embryonic stem cells of the white adipose lineage remain to be well defined. White preadipocytes that express PPARγ differentiate into mature white adipose cells in a process that also appears to be stimulated by certain BMPs. The adaptive UCP1-expressing brown-like adipose cells that develop in WAT in response to cold or β-adrenergic stimulation are not descendent from Myf5-expressing cells. These cells may be derived from (1) a specialized compartment of Myf5-negative but committed brown precursors; (2) directed differentiation from white preadipocytes and/or (3) transdifferentiation from mature white adipocytes (as indicated). The broken purple arrows depict hypothetical precursor-product relationships.
As described above, PRDM16 appears to function as a major determinant of a brown adipose /skeletal muscle fate switch from a common cellular precursor compartment. Interestingly, genetic ablation of PRDM16 in mouse caused a significant but relatively modest reduction in the molecular and morphological characteristics of brown fat (Seale et al., 2008). This suggests a parallel or independent pathway(s) that directs a subset of myoblasts into brown adipocyte lineage in vivo. There are 17 members of PRDM16 family in mammals, and it is possible that one of more of these may compensate the chronic loss of PRDM16 during embryogenesis. Notably, myogenin-deficient mice that completely lack differentiated skeletal muscle have an expanded BAT depot in the interscapular region. Together, these findings are consistent with the hypothesis that BAT shares a direct common upstream precursor with skeletal muscle cells. It will now be important to examine whether Myf5-expressing cells can clonally give rise to both cell lineages.
The signaling molecules that control the timing and specificity of PRDM16 expression and commitment to the brown adipose lineage are unknown. Bone morphogenic proteins (BMP), members of the TGF-β superfamily of secreted factors, have been suggested to facilitate adipogenic differentiation. Of particular interest is BMP7, which has been described as a selective and potent inducer of brown but not white adipogenesis in preadipocyte and multipotent fibroblast cultures (Tseng et al., 2008). BMP7 treatment of fibroblast cultures is associated with induction of key brown adipogenic regulators such as PRDM16 and PGC-1α, although the mechanisms that mediate this effect are not known. Importantly, BMP7-deficient embryos possess significantly reduced amounts of BAT that lack UCP1 expression. The cellular specificity and timing of BMP7 signaling in brown adipose tissue is an important open question for future experiments. Interestingly, certain BMPs also negatively regulate skeletal myogenesis, suggesting that BMP7 or a related family member may act on early somitic precursor cells to direct brown adipocyte versus skeletal muscle cell determination. Another member of the TGF-β superfamily, GDF-3, has been shown to negatively regulate a thermogenic gene program in white fat depots and hence, this factor could also play a role in BAT development (Shen et al., 2009)
Activation of the canonical Wingless (Wnt) signaling pathway is also associated with opposing effects on adipose and skeletal muscle lineages. Specifically, Wnt activation represses both brown and white adipogenesis by suppressing the induction of PPARγ and C/EBPα in precursor cells (Ross et al., 2000). By contrast, Wnt ligands are crucial inductive cues for embryonic muscle formation through direct activation of muscle determination factors. Fibroblast Growth Factors (FGFs) -16, -19 (FGF15 in mouse), and 21 have also been implicated in the regulation of brown adipogenesis and thermogenic activity of brown fat (Konishi et al., 2000).
In addition to the dedicated depots of brown adipocytes that are formed before birth, “brown adipocyte-like” cells are also found interspersed in white adipose tissues of adult animals that have been acclimated to cold or chronically treated with selective β3-adrenergic agonists. These induced “brown adipocyte-like” cells express UCP1 and have a multilocular morphology similar to their counterparts in preformed depots. However, these cells have a distinct developmental origin, since they are not descendent from a Myf5-expressing progenitor (Seale et al., 2008). It thus remains to be determined whether mature white adipose cells, committed populations of preadipocytes, or undetermined stem cells are the source of these brown fat-like cells. Strain-dependent variability in the expression of UCP1 in brown adipose cells from retroperitoneal WAT but not in those from interscapular BAT depots suggests that these cell types are genetically dissimilar (Xue et al., 2007). The developmental origins and molecular characteristics of these cells will be very important to understand. While the supraclavicular depots of brown fat in humans form distinct pads in a predictable location and are thus likely analogous to the BAT arising from muscle in mice, brown fat cells interspersed in white depots are also found in humans.
Emerging questions and therapeutic opportunities
Recent advances in brown fat biology and physiology further suggest its pivotal role in controlling energy homeostasis, and its promise as a therapeutic approach to combat obesity in mice and humans. For example, a number of studies in animal models (e.g. fat-selective Ucp1 transgenic mouse) have demonstrated that increases in amount or activity of brown fat can counteract obesity through increasing whole body energy expenditure. The new findings on the regulation of brown fat development have identified key cellular and molecular events, as reviewed above, which now enable researchers to ask a variety of fundamental questions in the field. For example, how much brown fat is needed to alter whole-body energy expenditure and counteract obesity? This can be determined either by transplantation of brown fat cells or with transgenic animal models. Since skin fibroblasts or the stromal vascular fraction from adipose tissues are relatively easy to obtain, even from humans, autologous transplantation of engineered brown fat cells generated by expressing PRDM16 and C/EBP-β would be a feasible way to explore this question. It is also extremely important to clarify if there is a single, unified pathway/mechanism(s) via key transcriptional regulators, such as FoxC2, pRb, RIP140, PRDM16 and PGC-1α, acting to induce brown fat development. Better understanding of the molecular circuits that regulate brown fat specification and development will lead to identification of novel and specific pharmacological targets for anti-obesity drugs.
One of the major unsolved questions is the molecular identity and cellular origins of the cAMP-inducible “brown fat-like” cells residing in the WAT. As described above, it does seem clear that a cAMP-induced transformation of WAT to BAT, presumably from a Myf5-independent cell lineage, is associated with a protection against obesity and metabolic diseases (Cederberg et al., 2001; Leonardsson et al., 2004). Identifying specific cell surface markers and establishing methods to isolate this cell population will advance our understanding of this issue. In addition, the role of BAT developmental regulators such as PRDM16 and PGC-1α in the formation of cAMP-induced “brown fat-like” cells and metabolic benefit must now be assessed.
Since gene transfer or cellular transplantation may not be an optimal method for human therapeutics in the metabolic syndrome, we also need to explore alternative ways to induce brown fat activity and development. Screening chemical compounds or drugs to induce dominant brown fat regulators such as PRDM16, FoxC2 or PGC-1α is certainly plausible. Alternatively, characterizing the upstream inductive components, such as endogenous hormones/polypeptides that stimulate the formation of brown fat cells during development will be valuable. Likewise, new drugs or polypeptides that increase differentiation or activity of BAT that exists in adult humans may offer new treatments for obesity and diabetes.
Acknowledgements
We are grateful to Drs. R. Gupta and M. Khandekar for their critical comments on the manuscript. We apologize for the inability to cite a vast number of papers that contribute to this field due to a space limit. S.K. is supported by AHA scientist development grant (0930125N). P.S. is supported by a NIH grant (DK081605). This work is funded by a NIH grant to B.M.S (DK31405).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, Conlon RA. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol. 2006;296:164–176. doi: 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- Cederberg A, Gronning LM, Ahren B, Tasken K, Carlsson P, Enerback S. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. The New England journal of medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell metabolism. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell metabolism. 2009;10:324–335. doi: 10.1016/j.cmet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Hallberg M, Morganstein DL, Kiskinis E, Shah K, Kralli A, Dilworth SM, White R, Parker MG, Christian M. A functional interaction between RIP140 and PGC-1alpha regulates the expression of the lipid droplet protein CIDEA. Molecular and cellular biology. 2008;28:6785–6795. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JB, Jorgensen C, Petersen RK, Hallenborg P, De Matteis R, Boye HA, Petrovic N, Enerback S, Nedergaard J, Cinti S, te Riele H, Kristiansen K. Retinoblastoma protein functions as a molecular switch determining white versus brown adipocyte differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4112–4117. doi: 10.1073/pnas.0301964101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Seale P, Tomaru T, Erdjument-Bromage H, Cooper MP, Ruas JL, Chin S, Tempst P, Lazar MA, Spiegelman BM. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes & development. 2008;22:1397–1409. doi: 10.1101/gad.1666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamitri A, Shore AM, Docherty K, Speakman JR, Lomax MA. Combinatorial transcription factor regulation of the cyclic AMP-response element on the Pgc-1alpha promoter in white 3T3-L1 and brown HIB-1B preadipocytes. The Journal of biological chemistry. 2009;284:20738–20752. doi: 10.1074/jbc.M109.021766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi M, Mikami T, Yamasaki M, Miyake A, Itoh N. Fibroblast growth factor-16 is a growth factor for embryonic brown adipocytes. The Journal of biological chemistry. 2000;275:12119–12122. doi: 10.1074/jbc.275.16.12119. [DOI] [PubMed] [Google Scholar]
- Leonardsson G, Steel JH, Christian M, Pocock V, Milligan S, Bell J, So PW, Medina-Gomez G, Vidal-Puig A, White R, Parker MG. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8437–8442. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N, Shimizu S, Nagasawa T, Tanaka H, Taniwaki M, Yokota J, Morishita K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood. 2000;96:3209–3214. [PubMed] [Google Scholar]
- Pan D, Fujimoto M, Lopes A, Wang YX. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell. 2009;137:73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Gehin M, Annicotte J, Rocchi S, Champy MF, O'Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell. 2002;111:931–941. doi: 10.1016/s0092-8674(02)01169-8. [DOI] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, MacDougald OA. Inhibition of adipogenesis by Wnt signaling. Science (New York, N.Y. 2000;289:950–953. doi: 10.1126/science.289.5481.950. [DOI] [PubMed] [Google Scholar]
- Saito M, Okamatsu-Ogura Y, Matsushita M, Watanabe K, Yoneshiro T, Nio-Kobayashi J, Iwanaga T, Miyagawa M, Kameya T, Nakada K, Kawai Y, Tsujisaki M. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scime A, Grenier G, Huh MS, Gillespie MA, Bevilacqua L, Harper ME, Rudnicki MA. Rb and p107 regulate preadipocyte differentiation into white versus brown fat through repression of PGC-1alpha. Cell metabolism. 2005;2:283–295. doi: 10.1016/j.cmet.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell metabolism. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JJ, Huang L, Li L, Jorgez C, Matzuk MM, Brown CW. Deficiency of growth differentiation factor 3 protects against diet-induced obesity by selectively acting on white adipose. Molecular endocrinology (Baltimore, Md. 2009;23:113–123. doi: 10.1210/me.2007-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons JA, Wennmalm K, Larsson O, Walden TB, Lassmann T, Petrovic N, Hamilton DL, Gimeno RE, Wahlestedt C, Baar K, Nedergaard J, Cannon B. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:4401–4406. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. The New England journal of medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. The New England journal of medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- Xue B, Rim JS, Hogan JC, Coulter AA, Koza RA, Kozak LP. Genetic variability affects the development of brown adipocytes in white fat but not in interscapular brown fat. J Lipid Res. 2007;48:41–51. doi: 10.1194/jlr.M600287-JLR200. [DOI] [PubMed] [Google Scholar]