Abstract
Autoxidation of linoleic acid (LA) enhanced by Fe(II)/ascorbate generates unsaturated hydroperoxides which undergo further oxidative evolution resulting in a mixture of electrophiles, including epoxyketooctadecenoic acid and dienones with intact C-18 chains as well as oxidative cleavage products such as 4-hydroxy-2(E)-nonenal (HNE), 4-oxo-2(E)-nonenal (ONE), 2(E)-octenal, 9-hydroxy-12-oxo-10(E)-dodecenoic acid, 9,12-dioxo-10(E)-dodecenoic acid and 11-oxoundec-9(E)-enoic acid. Mass spectrometric (MALDI-TOF-MS and LC-ESI-MS/MS) studies have been performed following incubation of the model protein β-lactoglobulin with LA, Fe(II), and ascorbate, which identified adducts of these electrophiles with three different protein nucleophiles. Deuterium labeled linoleic acid 17,17,18,18,18-d5-(9Z,12Z)-octadeca-9,12-dienoic acid (d5-LA) was synthesized to facilitate the detection and characterization of the protein modifications by mass spectrometry. Reduction by NaBH4 served to trap reversible adducts and to quantify the number of reducible functional groups in each adduct. This study, which mimics the distribution of reactive lipid peroxidation products generated by a continuous low level flux of reactive oxygen species present in vivo under conditions of oxidative stress, confirms that many irreversibly formed adducts previously identified following exposure of model proteins to pure electrophilic modifiers such as HNE and ONE are also generated during in situ oxidation of LA. These adducts include HNE–His Michael adducts (MA), ONE–Lys 4-ketoamide, ONE–Lys pyrrolinone, and a Cys/His–ONE–Lys pyrrole cross-link. However reversibly formed adducts, such as the HNE–Lys Schiff base, are not present at detectable levels. The isotopic labeling allowed less commonly identified “mirror-image” adducts derived from the carboxy terminus of LA to be identified. A novel 2-octenoic acid–His MA was discovered.
Introduction
Oxidative stress is implicated in the pathogenesis of various aging and neurodegenerative diseases (1-4). Reactive oxygen species such as peroxides and free radicals produced during oxidative stress can attack biomacromolecules in the cell, including DNA, proteins, lipids etc. Polyunsaturated fatty acids (PUFAs) are among the most susceptible targets. Oxidative degradation of PUFAs generates a variety of electrophilic aldehydes and ketones, called lipoxidation (LPO) products, which subsequently can modify DNA and proteins.
Protein modification by the pure LPO products, especially α,β-unsaturated aldehydes, has been extensively studied by many research groups. 4-Hydroxy-2(E)-nonenal (HNE), perhaps the most intensively studied, has been reported widely using mass spectrometry detection to modify proteins covalently (5-10). 4-Oxo-2(E)-nonenal (ONE), the 2e- oxidized cousin of HNE, which was found to be both more reactive and toxic than HNE (6, 10, 11), has received more attention recently. Many products of protein modification by ONE were elucidated by mass spectrometry (6, 7, 12-14). Other protein modifications by LPO products have been reported sporadically. 2(E)-Octenal (octenal) was found to react with histidine in bovine serum albumin through Michael addition (15). Our group recently discovered that various epoxyketooctadecenoic acid (EKODE) isomers were abundant products of non-enzymatic oxidation of linoleic acid (LA) (16), and that they can modify proteins through His Michael addition ((16), D. Lin, L. M. Sayre, unpublished data). Nε-Hexanoyllysine was also identified in protein exposed to lipid hydroperoxides (17). These modifications can potentially cause functional damage to proteins. Many enzymes have been reported to be inhibited or inactivated following covalent modification by LPO products (18-21), suggesting that these modifications may play an important role in disease progression.
To our knowledge, all previous studies have been based on either protein modification by a single pure modifier or by identification of a single specific modification of the protein following exposure to LPO products and have not attempted a comprehensive systematic study of the modifications resulting from peroxidation of a PUFA. The Tannenbaum group once investigated the covalent adducts of cytochrome c during the decomposition products of 13-hydroperoxy-octadecadienoic acid (13-HPODE) (22). However, they just monitored the change of the intact molecular weight of cytochrome c and did not characterize the reactive residue(s) by digestion and MS/MS. Liebler and coworkers recently reported the modification of apolipoprotein A1 by LPO products (23). They used a biotin-modified phospholipid as the PUFA carrier and human plasma as the matrix. This complex system made it difficult to find and characterize the protein modifications. Our goal is to study protein modification by the LPO products generated by in situ oxidation of PUFA using a model protein and a standard low flux model of oxidative stress.
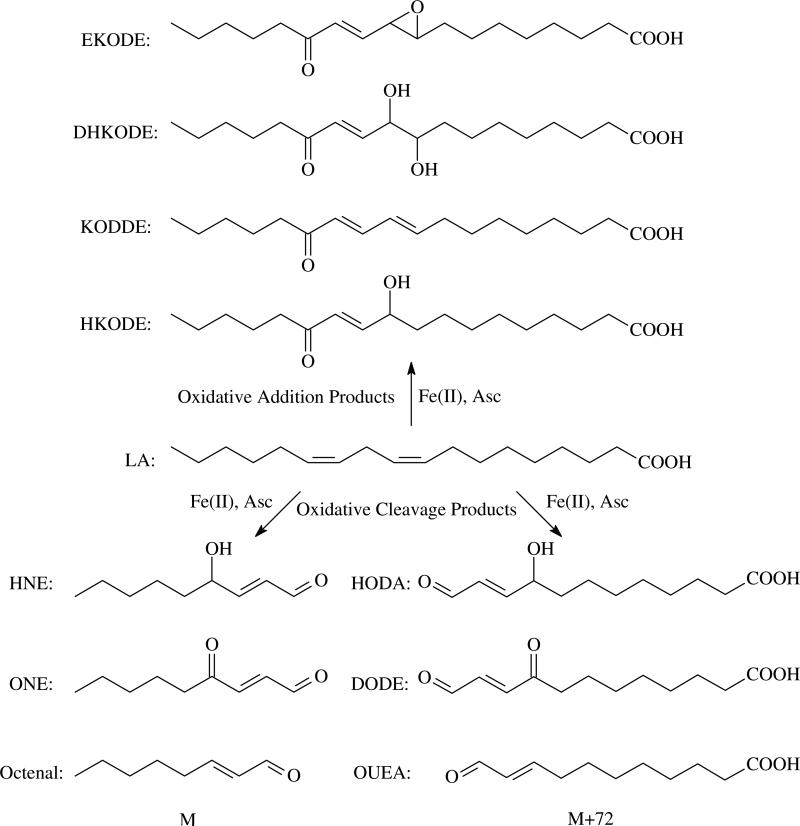
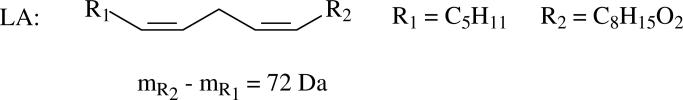
LA is the most abundant PUFA in mammalian tissue (24). Therefore, most degradation products of PUFAs are derived from LA (25, 26). Since oxidative stress and lipid peroxidation are related to many aging and neurodegenerative diseases (25), it is important to determine the protein modifications by the reactive intermediates generated from oxidized PUFA, and from LA in specific. When LA is subjected to non-enzymatic oxidation, two classes of aldehydes or ketones can be generated: those that derive from oxidative cleavage and those that result simply from oxidation. Scheme 1 shows some examples of the products from LA peroxidation. Oxidative cleavage products (OCPs) derived from the cleavage of the LA carbon chain include the well-known α,β-unsaturated aldehydes HNE, ONE, octenal etc. These products, derived from the ω–end of LA are ωOCPs. Due to the pseudo-symmetry of the homoconjugated double bond of LA, there is a corresponding set of carboxy oxidative cleavage products (cOCPs) with a MW 72 Da greater than their paired ωOCP; 9-hydroxy-12-oxo-10(E)-dodecenoic acid (HODA), 9,12-dioxo-10(E)-dodecenoic acid (DODE or KODA (27)), 11-oxoundec-9(E)-enoic acid (OUEA) are examples. Oxidative addition products (OAPs) result from the oxygenation or polyoxygenation of LA without concomitant carbon chain cleavage. These products include EKODE, dihydroxyketooctadecenoic acid (DHKODE), ketooctadecadienoic acid (KODDE), hydroxyketooctadecenoic acid (HKODE) etc.
Scheme 1.
Products Derived from Non-Enzymatic Oxidation of Linoleic Acid
β-Lactoglobulin (β-LG) is a lipid binding and transporting protein (28), which can bind long chain free fatty acids such as palmitic acid, stearic acid and oleic acid (29). When β-LG is bound with a PUFA, it is a potential target of the LPO products that may be generated in situ as observed for the protein constituents of lipoproteins (30). In addition, β-LG contains all of the common nucleophilic amino acids that are known to react with α,β-unsaturated adehydes, specifically 2 histidines, 15 lysines, 1 free cysteine and 3 arginines. These attributes make β-LG a favorable model protein for this study.
Protein modification by incubation with physiological levels of pure α,β-unsaturated aldehydes routinely results in minimal modification of any specific residue, even though these are the among the most reactive LPO products (7, 10). Therefore, detection of protein modification by LPO products requires careful analysis with mass spectrometry. In order to facilitate the detection and characterization of protein modifications by LC-ESI-MS/MS, d5-labeled linoleic acid (d5-LA) was synthesized. Any modified peptide incorporating an ωOCP or OAP with d0- or d5-labeling from oxidation of the LA (d0:d5 = 1:1) will be observed as a doublet (m:m+5) in the mass spectrum with almost identical retention times by LC-MS. The presence of this isotopic pair confirms the identification of a peak as containing the ω-carbons of LA, presumably as a result of an adduct being formed between the peptide and an LPO product.
Materials and Methods
General
All chemicals were purchased in AR or ACS grade from Aldrich, Acros Organic or Fisher Scientific unless stated specifically. 4-Bromo-butanol was purchased from TCI America (Portland, OR). 9-Bromononanoic acid was purchased from Karl Industries (Aurora, OH). Bovine β-LG and α-chymotrypsin were purchased from Sigma (St. Louis, MO). Sequencing grade modified trypsin was purchased from Promega (Madison, WI). PD-10 columns (Sephadex G-25 M) were purchased from GE Healthcare (Uppsala, Sweden). ZipTips were purchased from Millipore (Bedford, MA). Concentration of protein solutions was achieved by centrifugal evaporation under reduced pressure with a Savant Speed Vac SC110 system (Forma Scientific, Marietta, OH). HNE (31) and ONE (32) were prepared according to our previous studies. Compound 6 was prepared according to the published procedure (33). Carbonyl incorporation assay (2,4-dinitro-phenylhydrazine assay or DNPH assay) was performed according to the Levine-Stadtman method (34). SDS–PAGE was performed by the same method as the previous publication (35). All NMR spectra were acquired with either Varian Inova 400 or Gemini 200 NMR spectrometers.
LC-ESI-MS/MS
ESI-MS was performed with a Thermo LCQ DecaXP MAX or Advantage instrument in the positive ion mode using nitrogen as the sheath and auxiliary gas. The capillary temperature was 300 °C, the capillary voltage was 35.00 V, and the source voltage was 4.50 kV. Typically two scan events were used: (1) m/z 300–2000 full scan MS; (2) data dependent scan MS/MS on the most intense ion from event 1 or from the predefined parent mass list. The spectra were recorded using dynamic exclusion of previously analyzed ions for 0.5 min with three repeats and a repeat duration of 0.5 min. The MS/MS normalized collision energy was set to 35%. Reversed-phase HPLC was performed with a Surveyor LC system equipped with a 5 μm 2.1 × 250 mm Grace Vydac C18 column with a gradient elution program at a flow rate of 200 μL/min. Eluent A was a mixture of 95% H2O, 5% MeOH and 0.1% formic acid. Eluent B was a mixture of 95% MeOH, 5% H2O and 0.1% formic acid. The gradient program was from 80% A to 20% A over 70 min, 20% A to 80% A over 5 min, 80% A for 5 min.
Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF-MS)
5 μL of protein digest was mixed with 6 μL of 0.1% trifluoroacetic acid (TFA). A ZipTip was wet by 50% aqueous acetonitrile and then equilibrated with 0.1% TFA solution. The protein digest solution was extracted with the ZipTip and washed with 0.1% TFA. Finally, the sample was eluted with 2 μL of α-cyano-4-hydroxycinnamic acid (20 mg/mL in 70% aqueous acetonitrile and 0.1% TFA) onto the MALDI stainless steel target. The spectra were acquired with a Bruker BiFlex III MALDI-TOF mass spectrometer equipped with a pulsed nitrogen laser (3 ns pulse at 337 nm) after the samples were dried at 25 °C. All spectra were collected in the positive ion and reflectron mode with an average of 500 laser shots. All data were processed with XMass (Bruker Daltonics) or m over z (Proteometrics, LLC).
Incubation of β-LG and LA in the Presence of Fe(II) and l-Ascorbate (Asc)
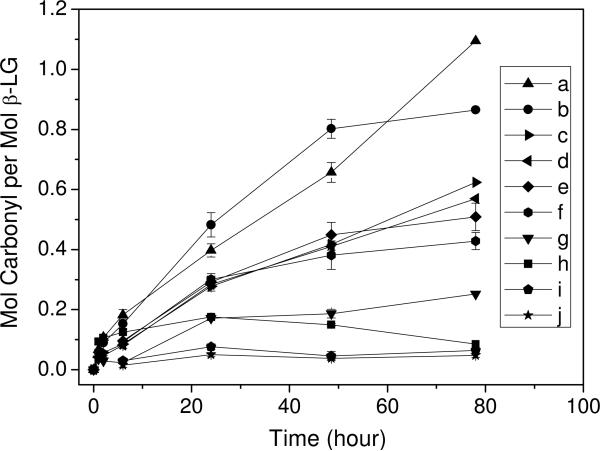
A solution of β-LG (1.0 mM, 1.0 mL) added to pH 7.4 HEPES buffer (100 mM, 8.0 mL) was incubated with a solution of LA in EtOH (200 mM), FeSO4•(NH4)2SO4 (50 mM), Asc (100 mM) and EtOH at various volumes at 37 °C. The total volume is 10.0 mL with 10% EtOH. The final concentrations of LA, Fe(II) and Asc are indicated in the legend of Figure 1. After 1, 2, 6, 24, 48 and 78 h, aliquots were assayed for carbonyl formation.
Figure 1.
β-LG (0.1 mM) carbonyl incorporation against time at various conditions in HEPES (pH 7.4, 100 mM) buffer at 37 °C. (a). 10 mM LA, 500 μM Fe(II), 1 mM Asc; (b). 5 mM LA, 500 μM Fe(II), 1 mM Asc; (c). 10 mM LA, 50 μM Fe(II), 1 mM Asc; (d). 5 mM LA, 50 μM Fe(II), 1 mM Asc; (e). 5 mM LA, 500 μM Fe(II), 15 mM Asc; (f). 5 mM LA, 1 mM Fe(II), 15 mM Asc; (g). 1 mM LA, 50 μM Fe(II), 1 mM Asc; (h). 1 mM LA, 500 μM Fe(II), 1 mM Asc; (i). 500 μM Fe(II), 15 mM Asc; (j). 500 μM Fe(II), 1 mM Asc.
Preparation of Modified β-LG by α,β-Unsaturated Aldehydes with or without NaBH4 Quenching
A solution of β-LG (1.0 mM, 200 μL) added to pH 7.4 sodium phosphate buffer (100 mM, 1.6 mL) was incubated with a solution of α,β-unsaturatedaldehyde in EtOH (20 mM HNE, ONE or octenal, 200 μL) at 37 °C. After 24 h, 1.0 mL of the solution was diluted to 2.5 mL with water and eluted through a PD-10 gel filtration column to remove unbound aldehyde and buffer salts. The eluted (3.5 mL) protein was concentrated to dryness prior to proteolytic digestion. Another l.0 mL of aliquot was treated with NaBH4 (2 M in 0.2 M NaOH solution, 100 μL) at 25 °C overnight prior to the PD-10 column elution.
Preparation of Modified β-LG by the Oxidative Products of d0- or d5-LA with or without NaBH4 Quenching
A solution of β-LG (1.0 mM, 100 μL) added to pH 7.4 HEPES buffer (100 mM, 780 μL) was incubated with a solution of d0-LA or d5-LA in EtOH (200 mM, 25 μL), Asc (100 mM, 10 μL), FeSO4•(NH4)2SO4 (50 mM, 10 μL) and 75 μL of EtOH at 37 °C. After 3 d, equal volumes of these two solutions (d0-LA and d5-LA added) were mixed together. Then 1.0 mL of the combined mixture was diluted with 3 mL of EtOH-EtOAc (1:1) to precipitate the protein. After centrifugation, the pellet was washed three times with 3 mL of EtOH-EtOAc (1:1). The remaining pellet was redissolved into 1.0 mL of water and then concentrated to dryness prior to proteolytic digestion. The other l.0 mL of combined aliquot was treated with NaBH4 (2 M in 0.2 M NaOH solution, 100 μL) at 25 °C overnight prior to the precipitation step.
Proteolytic Digestion
Modified β-LG (1.0 mg) was dissolved with 100 μL of pH 8.0 buffer containing 6 M guanidine hydrochloride and 50 mM Tris with the help of sonication. Dithiothreitol (200 mM, 2.5 μL) was added, vortexed, and incubated at 37 °C for 1 h. Then iodoacetamide (400 mM, 5.0 μL) was added, vortexed, and allowed to stand at 25 °C in a dark place for 1 h. Dithiothreitol (200 mM, 7.5 μL) was added again, vortexed, and allowed to consume the unreacted iodoacetamide at 25 °C for another 1 h. To 25 μL of this denatured sample was added 50 μL of trypsin (0.1 μg/μL) in suspension buffer or 10 μL of α-chymotrypsin (2.0 μg/μL) solution, then diluted to 200 μL with pH 7.8 NH4HCO3 solution (50 mM), and incubated at 37 °C for 24 h. The solutions were stored at –20 °C prior to MS analysis.
4-Bromo-O-(t-butyldimethyl)silylbutanol (1)
To a stirred solution of 4-bromo-butanol (2.20 g, 14.4 mmol) and imidazole (3.75 g, 53.7 mmol) in 5 mL of anhydrous DMF was added a solution of t-butyldimethylsilyl chloride (TBDMSCl) (4.57 g, 30.3 mmol) in 5 mL of anhydrous DMF at 0 °C. The mixture was gradually warmed to 25 °C. After 2 h, 50 mL of ice-water was added. The mixture was extracted with ethyl ether (4×100 mL). After the combined ethyl ether solution was washed with 20 mL of brine and dried by Na2SO4, it was concentrated by rotary evaporation and purified by flash column chromatography, eluting with ethyl acetate-hexanes (3:100). This afforded the titled compound as a colorless oil (3.33 g, 86.5%). 1H NMR (400 MHz, CDCl3) δ 3.59 (t, 2H, J = 6.4 Hz), δ 3.40 (t, 2H, J = 7.2 Hz), δ 1.89 (m, 2H), δ 1.61 (m, 2H), δ 0.84 (s, 9H), δ 0.00 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 67.49, 39.26, 36.62, 34.83, 31.27, 23.64, 0.00.
5,5,6,6,6-d5-O-(t-Butyldimethyl)silylhexanol (2)
To a suspended stirred solution of lithium granule (0.5% sodium) pieces (0.46 g, 65.71 mmol) in 20 mL of anhydrous ethyl ether was dropwise added a solution of CD3CD2Br (2.50 g, 21.93 mmol) in 25 mL of anhydrous ethyl ether in a 45 °C oil bath under the argon during a period of 3.5 h. After being refluxed for another 1 h, the ethereal solution was allowed to cool down to 25 °C. CuCN (1.03 g, 11.5 mmol) was put into a dry flask, heated in vacuo with a heat gun and 4 mL of anhydrous THF added under argon. The CuCN suspension in THF was stirred in a –70 °C dry ice-acetone bath and the above CD3CD2Li ether solution was slowly added over 15 min. After being stirred for another 7 min at –70 °C, the reaction mixture was put into a –20 °C dry ice-acetone bath for 5 min then returned to the –70 °C bath. After 10 min, it was slowly added a solution of 1 (2.12 g, 7.90 mmol) in 8 mL of anhydrous THF. The reaction mixture was warmed to –50 °C and stirred for 2.5 h, then warmed to 25 °C and stirred overnight. The reaction was quenched with a mixture of saturated NH4Cl-NH4·OH (10 mL, 9:1) and extracted with ethyl ether (3×100 mL). The combined ether extracts were washed with 10 mL of brine, dried with Na2SO4, concentrated by rotary evaporation and purified by flash column chromatography, eluting with ethyl acetate-hexanes (3:100), which afforded the titled compound as a colorless oil (760 mg, 44.4%). 1H NMR (400 MHz, CDCl3) δ 3.55 (t, 2H, J = 6.8 Hz), δ 1.46 (m, 2H), δ 1.18–1.25 (4H), δ 0.85 (s, 9H), δ 0.00 (s, 6H); 13C NMR (100 MHz, CDCl3) δ 63.59, 33.10, 31.60, 26.22, 25.68, 18.62, –5.03.
5,5,6,6,6-d5-Hexyltriphenylphosphonium Bromide (3)
To a stirred solution of triphenylphosphine (0.92 g, 3.51 mmol) in 10 mL of CH2Cl2 was dropwise added a solution of bromine (0.56 g, 3.50 mmol) in 1 mL of CH2Cl2 at 0 °C. The mixture was warmed to 25 °C and stirred for 30 min. Then a solution of 2 (580 mg, 2.68 mmol) in 1 mL of CH2Cl2 was slowly added over 10 min. After 40 min, 20 mL of ethyl ether was added to the reaction mixture, and a precipitate appeared. The resulting mixture was filtered through a short silica gel column, eluting with 30 mL of ethyl ether and 10 mL of CH2Cl2 sequentially. The combined eluates were concentrated with rotary evaporation at 0 °C. The residue was dissolved in 4.5 mL of acetonitrile containing triphenylphosphine (771.8 mg, 2.94 mmol), transferred to a sealed Reacti-vial and refluxed at 110 °C for 49 h. The reaction mixture was then concentrated by rotary evaporation, and the titled compound precipitated as a white solid (0.85 g, 73.6%) by triturating with ethyl ether. 1H NMR (200 MHz, CDCl3) δ 7.63–7.94 (15H), δ 3.85 (m, 2H), δ 1.55–1.70 (4H), δ 1.23 (m, 2H). HRMS (FAB) calcd for C24H23D5P (M+) 352.2237, found, 352.2236.
Ethyl 9-Bromononanoate (4)
To a stirred solution of 9-bromononanoic acid (3.90 g, 16.4 mmol) in 15 mL of anhydrous ethanol was added dropwise redistilled thionyl chloride (3.89 g, 32.7 mmol). The reaction was allowed to stand at 25 °C overnight and then concentrated by rotary evaporation. The resulting oil was dissolved into a minimal volume of ethanol and concentrated again. This cycle was repeated for three times, yielding the titled compound as a yellow oil (4.42 g, 100%), which was used in the next step without further purification. 1H NMR (400 MHz, CDCl3) δ 4.13 (q, 2H, J = 7.1 Hz), δ 3.41 (t, 2H, J = 6.8 Hz), δ 2.29 (t, 2H, J = 7.6 Hz), δ 1.85 (m, 2H), δ 1.62 (m, 2H), δ 1.42 (m, 2H), δ 1.29–1.36 (8H), δ 1.26 (t, 3H, J = 7.2 Hz); 13C NMR (100 MHz, CDCl3) δ 174.07, 60.40, 34.54, 34.20, 32.99, 29.27, 29.22, 28.77, 28.30, 25.12, 14.48.
Ethyl 9-Oxononanoate (5)
To a solution of 4 (4.39 g, 16.6 mmol) in 125 mL of dimethyl sulfoxide was added KI (2.76 g, 16.6 mmol) and Na2CO3 (1.76 g, 16.6 mmol). After the reaction mixture was stirred at 85 °C for 11 h, 100 mL of ice-cold brine was added. The mixture was extracted with ethyl ether (3×200 mL). The combined ethyl ether solution was washed with 100 mL of water, 100 mL of brine, 100 mL of NaHCO3 and 100 mL of brine sequentially and dried by Na2SO4 overnight. After concentrating the solution by rotary evaporation, the product was purified by flash column chromatography eluting with ethyl acetate-hexanes (1:10). This afforded the titled compound as a colorless oil (1.96 g, 59.1%). 1H NMR (200 MHz, CDCl3) δ 9.76 (t, 1H, J = 1.8 Hz), δ 4.13 (q, 2H, J = 7.1 Hz), δ 2.42(dt, 2H, J1 = 1.8 Hz, J2 = 7.2 Hz), δ 2.29 (t, 2H, J = 7.6 Hz), δ 1.53–1.70 (4H), δ 1.23–1.39 (6H), δ 1.25 (t, 3H, J = 7.0).
(Z)-Ethyl 12,12-Diisopropoxydodec-9-enoate (7)
To a stirred solution of 6 (3.51 g, 7.00 mmol) in 40 mL of anhydrous THF under the argon at –40°C was added 3.0 mL of 2.0 M sodium hexamethyldisilazane (NaHMDS, 6.0 mmol) THF solution over 5 min. The reaction mixture was stirred at 0 °C for 1 h and then warmed to 25 °C for 2 h. It was then placed in a –90 °C MeOH-liquid N2 bath and a solution of 5 (330 mg, 1.65 mmol), which had been dried by azeotropic distillation of a benzene solution for three times, in 3 mL of anhydrous THF was added. After 1 h at this temperature, the reaction mixture was stirred at –78 °C (dry ice-acetone bath) for another 3.5 h. Then it was warmed to 25 °C and quenched with 4 mL of saturated NH4Cl solution and 4 mL of water. The solution was extracted with ethyl ether (3×50 mL). The combined ether extracts were washed with 10 mL of brine, concentrated under reduced pressure, and the product was purified by flash column chromatography with ethyl acetate-hexanes (1:30), affording the titled compound as a colorless oil (290 mg, 51.3%). 1H NMR (400 MHz, CDCl3) δ 5.35–5.50 (2H), δ 4.54 (t, 1H, J = 5.6 Hz), δ 4.13 (q, 2H, J = 7.2 Hz), δ 3.82–3.92 (2H), δ 2.34 (dd, 2H, J1 = J2 = 6.0 Hz), δ 2.28 (t, 2H, J = 7.6 Hz), δ 2.03 (dt, 2H, J1 = J2 = 6.8 Hz), δ 1.61 (m, 2H), δ 1.26–1.38 (8H), δ 1.26 (t, 3H, J = 7.2 Hz), δ 1.20 (d, 6H, J = 6.4 Hz), δ 1.14 (d, 6H, J = 6.0 Hz); 13C NMR (100 MHz, CDCl3) δ 174.09, 132.20, 124.53, 100.22, 67.97, 60.36, 34.57, 33.94, 29.70, 29.38, 29.32, 27.68, 25.17, 23.58, 22.76, 14.47.
17,17,18,18,18-d5-(9Z,12Z)-Ethyl Octadeca-9,12-dienoate (9)
To a solution of 7 (170 mg, 0.496 mmol) in 9.5 mL of anhydrous THF was added 252 μL of 0.1 M p-toluenesulfonic acid (0.0252 mmol) and refluxed for 20 min and then cooled in a 0 °C bath. The reaction mixture was diluted with 30 mL of hexanes, washed with 2 mL of water and 2×2 mL of brine. The aqueous phase was extracted with hexanes (2×50 mL). Then the combined hexanes solution was concentrated by rotary evaporation to give 137.7 mg (Z)-ethyl 12-oxododec-9-enoate (8), which was used for the next step without further purification. 1H NMR (400 MHz, CDCl3) δ 9.67 (t, 1H, J = 2.0 Hz), δ 5.70 (m, 1H), δ 5.54 (m, 1H), δ 4.13 (q, 2H, J = 7.1 Hz), δ 3.19 (m, 2H), δ 2.29 (t, 2H, J = 7.4 Hz), δ 2.03 (dt, 2H, J1 = J2 = 7.2 Hz), δ 1.62 (m, 2H), δ 1.26–1.40 (8H), δ 1.26 (t, 3H, J = 7.2 Hz); 13C NMR (100 MHz, CDCl3) δ 200.02, 174.11, 135.63, 118.25, 60.41, 42.80, 34.56, 29.44, 29.32, 29.27, 29.24, 27.81, 25.15, 14.49. 3 (320 mg, 0.74 mmol) was heated in vacuo at 110 °C for 1 h, and then was added 13 mL of anhydrous THF at –40 °C. After 5 min, 357 μL of 2.0 M NaHMDS (0.71 mmol) THF solution was added and stirred for 15 min. The reaction mixture was warmed to 0 °C and stirred for 2 h. Then a solution of 8 (137.7 mg), which had been dried by azeotropic distillation with benzene (3×6 mL), in 2 mL of anhydrous THF was added at –90 °C (MeOH-liquid N2 bath) over 10 min. After 1.5 h at –90 °C, the reaction mixture was stirred at –78 °C (dry ice-acetone bath) for 2 h and then warmed to 0 °C gradually. 1 N HCl was added dropwise to acidify the reaction mixture to pH 1.0. Then 2 mL of saturated NH4Cl and 2 mL of water were added. The solution was extracted with hexanes (3×20 mL). The combined hexanes solution was washed with 2 mL of brine, concentrated under reduced pressure, and purified by flash column chromatography with ethyl acetate-hexanes (1:40). This afforded the titled compound as a colorless oil (65.5 mg, 42.1%). 1H NMR (400 MHz, CDCl3) δ 5.29–5.43 (4H), δ 4.12 (q, 2H, J = 7.1 Hz), δ 2.77 (dd, 2H, J1 = J2 = 6.6 Hz), δ 2.29 (t, 2H, J = 7.4 Hz), δ 2.02–2.07 (4H), δ 1.62 (m, 2H), δ 1.24–1.39 (12H), δ 1.25 (t, 3H, J = 7.2 Hz); 13C NMR (100 MHz, CDCl3) δ 174.12, 130.46, 130.28, 128.26, 128.13, 60.39, 34.61, 31.47, 29.82, 29.55, 29.40, 29.34, 27.44, 27.42, 25.85, 25.20, 14.49. HRMS (FAB) calcd for C20H32D5O2 (M+1) 314.3102, found, 314.3095.
17,17,18,18,18-d5-(9Z,12Z)-Octadeca-9,12-dienoic Acid (d5-LA)
A solution of 9 (61.5 mg, 0.196 mmol) in 15 mL of THF and 5 mL of 0.2 M LiOH was stirred at 25 °C for 40 h. It was then rotary evaporated to remove the THF and acidified with 1 N HCl to reduce the pH to 1.0. Solid NaCl was added to saturate the aqueous solution, and the mixture was extracted with ethyl acetate-hexanes (1:1, 3×30 mL). The combined organic extracts were washed with brine (2×5 mL) and concentrated to obtain the titled compound as a colorless oil with a trace of conjugated acid isomers present. To remove the conjugated acid isomers, the product was added to a solution of maleic anhydride (37.7 mg, 385 μmol) in 1.5 mL of p-xylene and refluxed at 155 °C for 2.5 h. After the addition of 2 mL of ethyl ether and 1 mL of water, the reaction mixture was adjusted to pH 3.0 with solid LiOH and 1 N HCl and extracted with ethyl ether (3×15 mL). The combined ether solution was washed with brine (2×5 mL) and concentrated under reduced pressure. The resulting residue was purified by flash column chromatography with ethyl acetate-hexanes (1:2) to give the titled compound as a colorless oil (43.7 mg, 78.0%). 1H NMR (400 MHz, CDCl3) δ 5.28–5.42 (4H), δ 2.77 (dd, 2H, J1 = J2 = 6.6 Hz), δ 2.35 (t, 2H, J = 7.6 Hz), δ 2.02–2.07 (4H), δ 1.63 (m, 2H), δ 1.23–1.39 (12H); 13C NMR (100 MHz, CDCl3) δ 180.24, 130.46, 130.25, 128.29, 128.11, 34.25, 31.47, 29.81, 29.54, 29.37, 29.30, 29.25, 27.44, 27.41, 25.85, 24.88. HRMS (EI) calcd for C18H27D5O2 (M+) 285.2711, found, 285.2722.
Results and Discussion
Formation of Protein-Bound Carbonyls by the Products of LA Peroxidation
Quantitation of protein-bound carbonyls is a commonly used marker of oxidative protein damage. The presence of carbonyl protein adducts is taken to reflect the extent of oxidative protein modification in specific and oxidative stress in general. While the initial presumption was that protein carbonyls reflected direct oxidation of protein side chains, proteins modified by many LPO products will result in the incorporation of carbonyls. The extent of carbonyl incorporation is expected to reflect the degree of overall modification and that protein with more extensive carbonyl incorporation will also yield a greater variety of detectable modifications both in the number of identified sites and the chemical nature of the products. Therefore, we optimized our experimental conditions of protein modification, to produce significant carbonyl incorporation.
Protein based carbonyls are readily quantified by DNP-hydrazone formation in the presence of DNPH (34). With this assay, we determined time courses of carbonyl generation in β-LG in the presence of LA, Fe(II) and Asc under various conditions (Figure 1). The time courses revealed that carbonyl formation was both time and concentration dependent. The optimal oxidative conditions obtained in this study, incubation with 500 μM Fe(II), 1 mM Asc and 5 or 10 mM LA for 3 d, i.e. conditions a or b (Figure 1) were employed to chemically characterize the protein modifications. SDS–PAGE (Figure S1, Supporting Information) indicated that β-LG was still present after 3 d of oxidation (lane 3) with a significant formation of both dimers and trimers, suggesting that β-LG was cross-linked during this time course but not extensively oxidatively fragmented.
Strategy to Explore Protein Modification with d0-LA and d5-LA
Protein modification by LPO products is widely studied by mass spectrometry. Many studies have incubated test proteins with a single LPO product (5-9, 12-15). In these studies, even if the incorporation into the protein is stoichiometric (1:1) the modification may be distributed over a variety of residue types, viz His, Cys and Lys, as well as different residues of the same type. The complexity is increased when different products are formed by reaction with the same residue, e.g. HNE can form a Michael adduct (MA), a Schiff base or a pyrrole following reaction with any Lys residue. Thus, the low level of each specific modification is always an obstacle for the identification of modified peptides. This complexity is amplified further in this study where the LPO products are generated in situ by oxidation of PUFA, where multiple reactive electrophiles are generated over time, instead of a bolus addition of a single pure modifier.
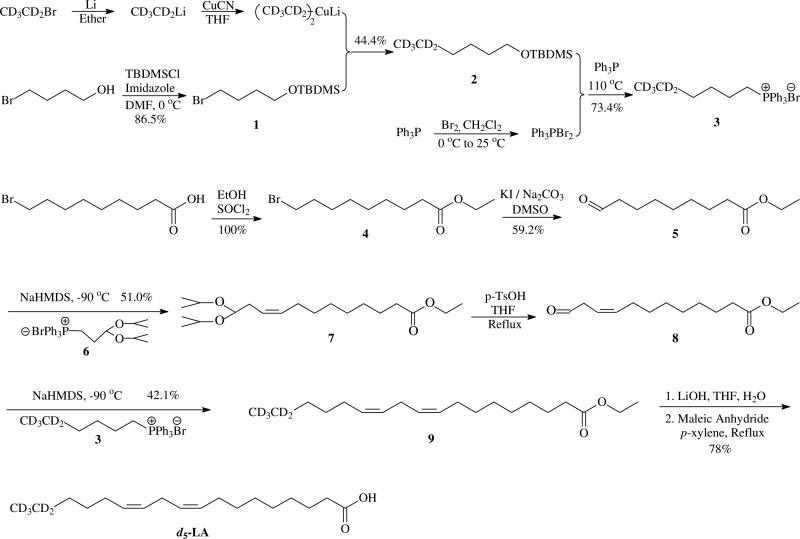
In order to increase the sensitivity and rule out false positives in the mass spectra, a mixture of light and heavy (1:1) stable isotope labeling can be introduced (10, 36). We synthesized d5-labeled linoleic acid (d5-LA) (Scheme 2) for this purpose. Five deuterium atoms, which are stable and nonexchangeable, are introduced into the ω-ethyl terminus of the LA. As the ω-terminus is not involved in any oxidation process (37), the aldehydes generated from the ω-terminus of d5-LA are always 5 Da heavier than those derived from d0-LA. The peptides which are modified by either d0- or d5-labeled aldehyde will appear as a 1:1 doublet (m:m+5) in the mass spectra, which are readily indentified according to common intensities, retention times and tandem mass spectra of each parent ion.
Scheme 2.
Preparation of d5-LA
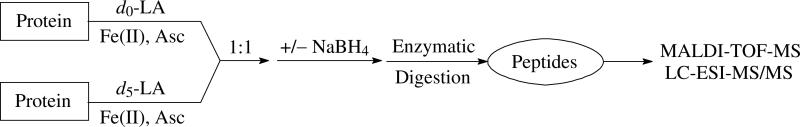
Therefore, our strategy (Scheme 3) is to incubate the protein with either d0-LA or d5-LA at the same concentration in the presence of Fe(II) and Asc. The protein will be modified by either light or heavy isotope coding aldehydes from both forms of LA. These two solutions are mixed together in a ratio of 1:1 and half of the mixture is reduced by NaBH4. Ketone, aldehyde and Schiff base adducts will be reduced with a mass increase of 2 Da. This reduction helps to confirm the structure of the products by allowing the susceptible double bonds to be counted. Additionally, the reduction stabilizes the reversibly-formed products such as Lys Schiff bases and some MAs (10). Both unreduced and reduced modified proteins are then subjected to enzymatic digestion with trypsin or chymotrypsin and analyzed with MALDI-TOF-MS and LC-ESI-MS/MS.
Scheme 3.
Strategy in this Study
Preliminary Study on Protein Modification by MALDI-TOF-MS
MALDI-TOF-MS is one of the most straightforward and simple ways to detect the peptides from proteolytic digests. It has the special virtue of facile visual detection of the mass doublets introduced by the isotopic labeling (Figure 2). Therefore, we first tried to use MALDI-TOF-MS to seek modified peptides from the β-LG incubated with d0-LA and d5-LA (1:1) in the presence of Fe(II) and Asc for 3 d. If the peptide is modified by ωOCPs such as HNE and ONE (Scheme 1) which contain the ω-terminus, labeled with either C2H5- or C2D5-, it will appear as a 1:1 doublet peak (m:m+5) in the mass spectrum. As the OAPs of LA, such as EKODEs (Scheme 1), also retain the ω-terminus, any peptide modified by OAPs will also be observed as a doublet peak (m:m+5). However, if the peptide is modified by a cOCP, such as HODA and DODE which instead contain the carboxy terminus of LA, it will appear as a singlet peak (m') in the spectrum. Figure 3 shows that LA is pseudosymmetric since R1, which contains the ω-terminus, and R2, which contains the carboxy terminus, are different. The pseudosymmetry of R1 and R2 relative to the double allylically-activated CH2 group results in related OCPs being formed. Many oxidations of LA are initiated by the abstraction of a hydrogen from this activated CH2 group. The resulting mesomeric radical attacks oxygen molecules at either end of this system with approximately equal probability (37). The consequential decomposition reactions generate related products which contain either R1 such as HNE and ONE or R2 such as HODA and DODE, respectively. As the mass difference of R2 and R1 is 72 Da, the related ωOCP and cOCP containing R1 and R2 will exhibit a 72 Da difference. Therefore, a peptide (m') modified by the cOCPs of LA is predicted to be observed at 72 Da greater mass than the low mass peak of a d0:d5 doublet (m:m+5, m' = m+72) of a peptide modified by ωOCP. This m+72 satellite, however, will not be observed for the OAPs such as EKODE since these products contain both ω and carboxy termini. This difference distinguishes the peptides modified by OCPs from those modified by OAPs.
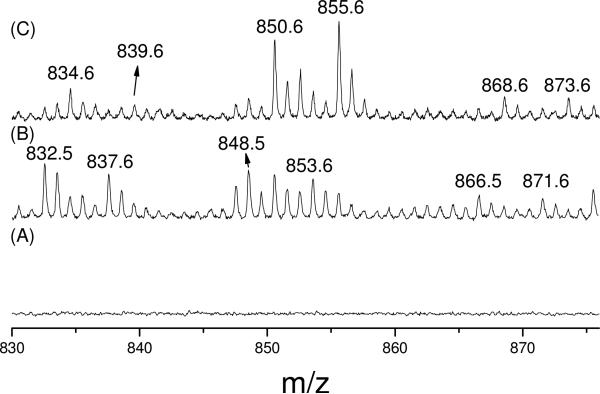
Figure 2.
MALDI-TOF mass spectra of the chymotryptic digest of β-LG. (A). control; (B). β-LG incubated with LA (d0:d5 = 1:1), Fe(II) and Asc; (C). β-LG incubated with LA (d0:d5 = 1:1), Fe(II) and Asc with NaBH4 reduction. 832.5/837.6, 848.5/853.6 and 866.5/871.6 are modified HIRL at H146 by d0-/d5-KODDE, d0-/d5-EKODE and d0-/d5-DHKODE, respectively, through Michael addition. 834.6/839.6, 850.6/855.6 and 868.6/873.6 are reduced forms of above products, respectively.
Figure 3.
Asymmetric structure of LA.
When the tryptic and chymotryptic digests were subjected to MALDI-TOF-MS, several doublet peaks were observed. Figure 2 shows an example of three doublets (See their assignments in Table 1) detected in contrast to the control spectrum in the range shown. In the MALDI-TOF-MS of the parallel sample reduced by NaBH4, three doublets (See their assignments in Table 2) were observed with 2 Da increments (Figure 2C), indicating that they each contained a single reducible carbonyl or Schiff base. All observed modified peptides and their assignments are listed on Table 1. Both His and Lys adducts were detected. For the His modification, HNE–His MAs have been widely observed by mass spectrometry when various proteins were treated with pure HNE (6, 7, 22, 38). Liebler and coworkers first found this product on ApoA1 from biotin-modified linoleoylglycerylphosphatidycholine supplemented oxidized plasma (23). Our result corroborates the existence of this product in the protein incubated with a PUFA subjected to non-enzymatic oxidative conditions. The anticipated HODA–His MA, an m' singlet peak 72 Da higher than HNE–His MA, was also observed. An EKODE–His MA and its hydrated product DHKODE–His MA (Figures 4, S2 and S3, Supporting Information), which were discovered recently (D. Lin, L. M. Sayre, unpublished data), were also detected. In addition, the mass shift of 294 Da matches that predicted for a KODDE–His MA (Figure 4). KODDE was recently found to be an oxidative product of LA (37, 39) where the presence of an α,β-unsaturation predicts the formation of MAs. An unknown modification of His with a mass shift of 346 Da was also observed. This modification is predicted to be derived from an OAP since the isotopic m:m+5 doublet was not accompanied by a peak at m+72. The modifier is presumed to be a highly oxidized species derived from LA, it would be consistent with a hydroxylated form of DHKODE.
Table 1.
Modified Peptides Detected by MALDI-TOF-MS from Enzymatic Digests
| Peptide sequence$ | Position | Mass of modified peptide | Assignment (Mass Shift) | Enzyme used |
|---|---|---|---|---|
| VRTPEVDDEALEKF | 123–136 | 1802.0/1807.0 | ONE–Lys 4-Ketoamide (154/159 Da) | Chymotrpsin |
| VRTPEVDDEALEKF | 123–136 | 1874.0 | DODE–Lys 4-Ketoamide (226 Da) | Chymotrpsin |
| VRTPEVDDEALEKF | 123–136 | 1784.0/1789.0 | ONE–Lys Pyrrolinone (136/141 Da) | Chymotrpsin |
| VRTPEVDDEALEKF | 123–136 | 1856.0 | DODE–Lys Pyrrolinone (208 Da) | Chymotrpsin |
| HIRL | 146–149 | 694.5/699.5 | HNE–His MA (156/161 Da) | Chymotrpsin |
| HIRL | 146–149 | 766.5 | HODA–His MA (228 Da) | Chymotrpsin |
| HIRL | 146–149 | 832.5/837.6 | KODDE–His MA (294/299 Da) | Chymotrpsin |
| HIRL | 146–149 | 848.5/853.6 | EKODE–His MA (310/315 Da) | Chymotrpsin |
| HIRL | 146–149 | 866.5/871.6 | DHKODE–His MA (328/333 Da) | Chymotrpsin |
| HIRL | 146–149 | 884.5/889.6 | His Unknown (346/351 Da) | Chymotrpsin |
| ALPMHIR | 142–148 | 1147.8/1152.8 | EKODE–His MA (310/315 Da) | Trypsin |
| ALPMoHIR | 142–148 | 1163.7/1168.7 | EKODE–His MA (310/315 Da) | Trypsin |
| ALPMHIR | 142–148 | 1165.7/1170.8 | DHKODE–His MA (328/333 Da) | Trypsin |
bold and underlined residue was modified
Table 2.
Modified Peptides Detected by MALDI-TOF-MS from Enzymatic Digests after Reduction
| Peptide sequence$ | Position | Mass of modified peptide | Assignment (Mass Shift) | Enzyme used |
|---|---|---|---|---|
| HIRL | 146–149 | 696.5/701.4 | HNE–His MA (158/163 Da) | Chymotrpsin |
| HIRL | 146–149 | 768.5 | HODA–His MA (230 Da) | Chymotrpsin |
| HIRL | 146–149 | 834.6/839.6 | KODDE–His MA (296/301 Da) | Chymotrpsin |
| HIRL | 146–149 | 850.6/855.6 | EKODE–His MA (312/317 Da) | Chymotrpsin |
| HIRL | 146–149 | 868.6/873.6 | DHKODE–His MA (330/335 Da) | Chymotrpsin |
| HIRL | 146–149 | 886.5/891.5 | His Unknown (348/353 Da) | Chymotrpsin |
| ALPMHIR | 142–148 | 1149.7/1154.7 | EKODE–His MA (312/317 Da) | Trypsin |
| ALPMoHIR | 142–148 | 1165.6/1170.6 | EKODE–His MA (312/317 Da) | Trypsin |
bold and underlined residue was modified
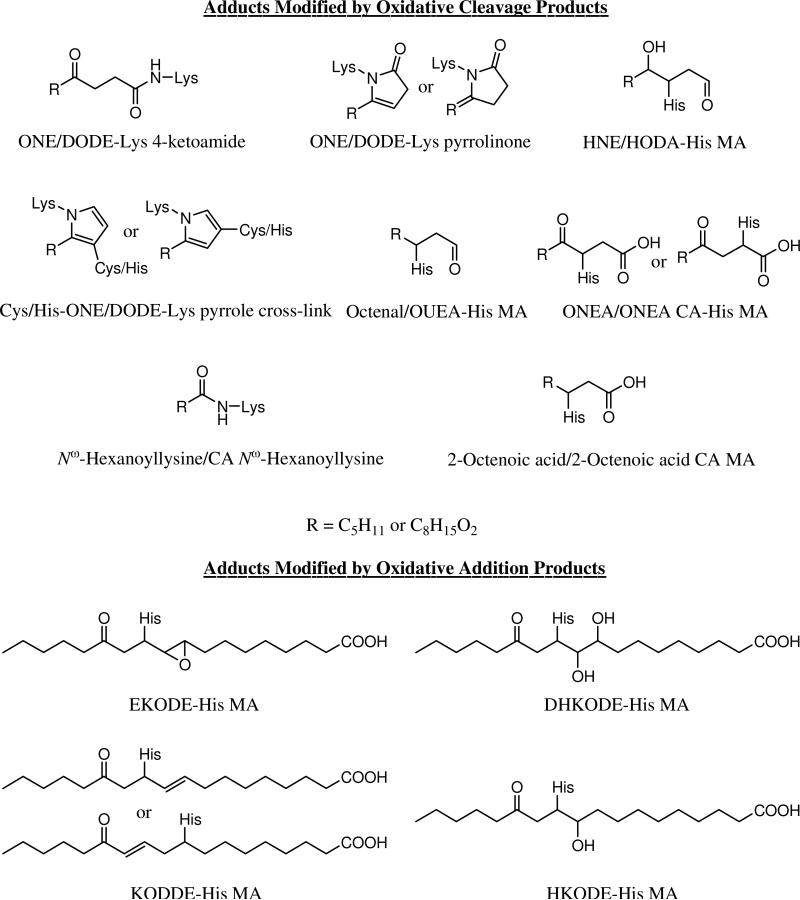
Figure 4.
Protein adducts modified by LA peroxidation products.
Four different Lys modifications by OCPs were detected: ONE or DODE–Lys 4-ketoamide and ONE or DODE–Lys pyrrolinone adducts (Figure 4). The structure of the ONE/DODE–Lys 4-ketoamide adduct was clarified recently by our group and identified as a major non-cross-linking Lys product (14). The Tannenbaum group also showed that DODE 4-ketoamide was the major product of cytochrome c exposed to 13-HPODE, an initial product of LA peroxidation, in the presence of Asc (22). The ONE–Lys pyrrolinone product was first detected by mass spectrometry in our previous study when apomyoglobin was treated with pure ONE (7). These four modifications were also found to be the major products of Lys modified by the products of LA peroxidation in a preliminary model study (X. Zhu, V. E. Anderson, L. M. Sayre, submitted).
In addition to the modification by the LPO products, protein side chains can also be oxidized by metal-catalyzed oxidation systems (40). We found that the Met was predominantly oxidized to methionine sulphoxide (Mo) with a mass shift of 16 Da after the standard 3-day incubation. Both EKODE–His146 MA of ALPMHIR and ALPMoHIR doublet signals were detected.
Sodium borohydride reduction will trap two common reversible modifications, Lys Schiff base formation and MA formation with α,β-unsaturated aldehydes. The reduction additionally assists in the structure assignment of the product by comparing the observed mass shifts in the mass spectra of the reduced and unreduced samples. Table 2 shows the modified peptides and their assignments after borohydride reduction of the protein. No new products were found compared to the results without reduction. These data indicate that all HNE, HODA, KODDE, EKODE and DHKODE–His MAs were reduced and displayed the attendant mass increment of 2 Da, as predicted by the single reducible carbonyl in each product. The unknown product m+346 presumably contains a single carbonyl since it exhibited a mass shift of 2 Da (m+348) after NaBH4 reduction.
Characterization of β-LG Modification by the Products of LA Peroxidation with LC-ESI-MS/MS
After the preliminary study of the proteolytic digests by MALDI-TOF-MS, several modified peptides were found and assigned based on the predicted mass shift, isotopic incorporation and potential to be reduced by NaBH4. However, as these assignments are based on the m/z of predicted cleavage peptides following digestion, the sequence of the peptide is unconfirmed, and the modified residue cannot be unambiguously identified. Therefore, the proteolytic digests were also subjected to LCESI-MS/MS. Tandem mass spectrometry can provide confirmation of the peptide sequence, localize the modification, and consequently assist in confirming the identification of the modification.
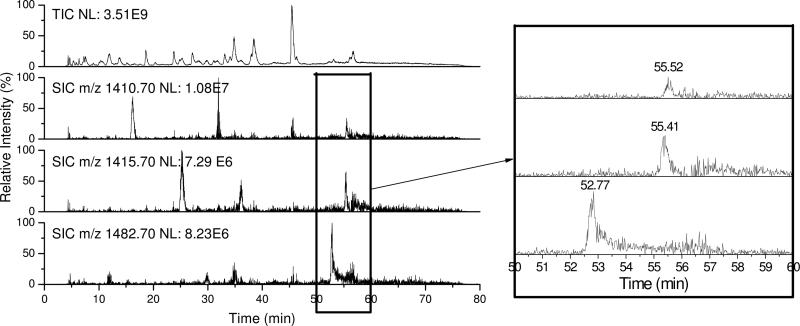
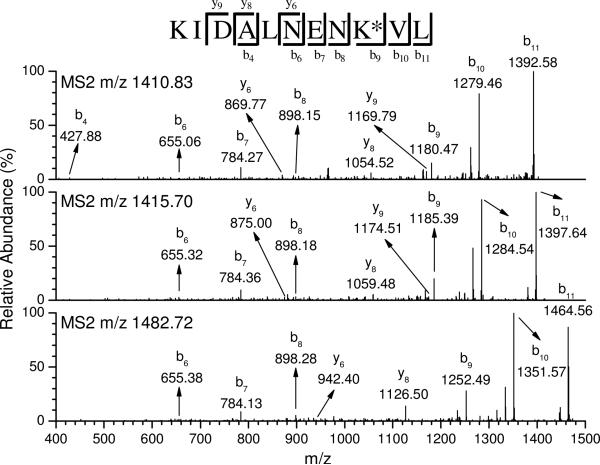
d5-LA was extremely helpful for probing the modifications identified in this study, because peptides modified by d0- and d5-LA LPO products behaved similarly both chromatographically and by mass spectrometry. The modified peptides (m+5) by d5-labeled LPO products had only minor variations in their HPLC retention times from that (m) of the unlabeled counterpart, with the labeled compound eluting slightly earlier than the unlabeled as expected for D-labeling (41). Figure 5 shows an example of the LC chromatograms of modified peptide KIDALNENKVL (P83–93) at K91 by ONE/DODE 4-ketoamide formation. The top trace is the total ion chromatogram (TIC) of the chymotryptic digest of β-LG incubated with LA (d0:d5 = 1:1) in the presence of Fe(II) and Asc. The second trace is the selected ion chromatogram (SIC) of d0-ONE-modified P83–93 (m/z 1410.70) which elutes at 55.52 min. The third trace is the SIC of d5-ONE-modified P83–93 (m/z 1415.70) which elutes at 55.41 min. The retention times exhibit minimal difference so that two peaks effectively overlap. As previously noted, for the modified peptide (m:m+5) by the ω-OCP, there is an accompanying identical in sequence peptide (m+72) modified by the cOCP, which always eluted several minutes earlier, because the alkyl carboxylate is more polar than the alkyl chain of the ωOCP. The fourth trace shows that the SIC of DODE-modified P83–93 (m/z 1482.70) eluting at 52.77 min, around 3 min earlier than ONE-modified P83–93. This pattern lends confidence to the structural assignment of the m:m+5 doublets and reinforces the conclusion that they are modified by ωOCPs.
Figure 5.
TIC (1st trace) of the chymotryptic digest of modified β-LG by LA (d0:d5 = 1:1) peroxidation products and SIC of modified KIDALNENKVL by d0-ONE (2nd trace), d5-ONE (3rd trace) or DODE (4th trace) through 4-ketoamide formation. Part of the left figure was enlarged and shown on the right.
Although the doublet SIC peaks (d0- and d5-labeled) assist in the identification of modified peptides, their sequences still need to be elucidated in consideration of the coincidental coelution of two unrelated peptides with a 5 Da mass difference. This can be realized by tandem mass (MS/MS). The tandem mass spectra of the peptides modified by d0- and d5-LA peroxidation end product and the carboxy analog (if available) have similar MS/MS spectra as shown in Figure 6. The three tandem mass spectra of m/z 1410.83, m/z 1415.70 and m/z 1482.72 correspond to the modified peptide peaks at 55.52, 55.41 and 52.77 min on the right of the Figure 5, respectively. As K91 is modified by d0-ONE, d5-ONE or DODE with a mass shift of 154, 159 or 226 Da, b9–b11 and y3–y10 ions from each spectrum should have a corresponding mass shift, while all other b and y fragment ions are identical. For example, the b10 ions for d0-ONE, d5-ONE and DODE-modified P83–93 appear at m/z 1279.46, 1284.54 and 1351.57, respectively, which are 154, 159 or 226 Da higher than the native b10 ion (m/z 1125.54, spectrum not shown). However, the b7 ions are the same in all three spectra. These tandem mass spectra identify the modified peptides, whose spectra have similar pattern, confirm the peptide sequence, locate the modified residue, and exclude the potential for false positives resulting from coelution of different peptides, whose tandem spectra would have a different pattern.
Figure 6.
Tandem mass spectra of the modified KIDALNENKVL at K91 by d0-ONE (top), d5-ONE (middle) or DODE (bottom) through 4-ketoamide formation.
Our characterization of β-LG modification by the LA peroxidation products using MALDI-TOF-MS identified several protein adducts (Tables 1 and 2). From preliminary model studies with highly sensitive pseudo dipeptides containing His and Lys (X. Zhu, V. E. Anderson, L. M. Sayre, submitted), we found additional modifications generated by in situ oxidation of LA. We anticipated that these modifications should also appear in proteins similarly treated. Therefore, we calculated the predicted masses of modified peptides from both tryptic and chymotryptic digests according to the mass shift observed in model studies and added these masses to the target mass list for data-dependent LC-ESI-MS/MS. The resulting tandem mass spectra were first searched by Bioworks TurboSEQUEST using the criteria of Delta Cn greater than 0.08 and XCorr greater than 1.8, 2.5, and 3.5 for singly, doubly, and triply charged ions, respectively, and then investigated manually. Numerous protein adducts, including all of the modifications found by MALDI-TOF-MS, were identified in consideration of doublet SIC peaks and the tandem mass spectra.
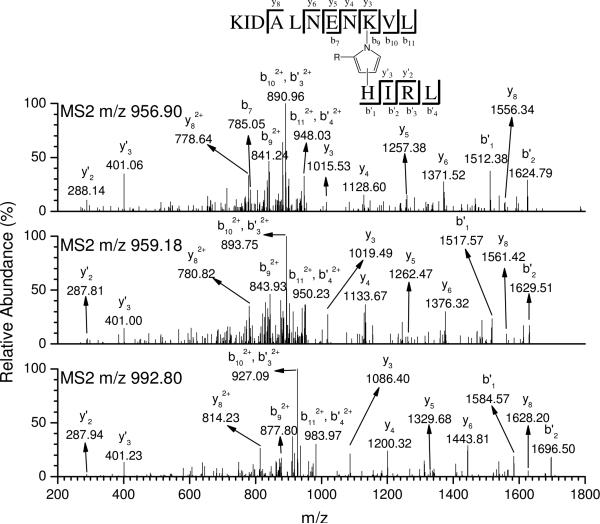
Tables 3 and 4 list modifications (The sequence and mass of the modified peptides are listed in Tables S1 and S2, Supporting Information) detected by LC-ESI-MS/MS from chymotryptic and tryptic digests, respectively. All modified peptides in the tables were confirmed manually by the following criteria. If the peptide was identified as modified by ωOCP or OAP, it had to meet two requirements. First, there had to be a doublet (m:m+5), which had similar retention times and intensities. Second, the tandem mass spectra of those two peaks had to exhibit identical major fragments except for the differences which were reasonably explained by the isotopic modification. If the peptide was identified as modified by cOCP, it also had to meet two requirements. First, it appeared as a singlet (m') and was accompanied by a doublet (m'–72:m' – 67) if the doublet can be detected. Second, the tandem mass spectrum of the singlet was reasonably explained and had identical fragment ions that could be interpreted from the peptide sequence and did not contain the putative modified residue. Most products found were reported previously when the protein was incubated with the pure modifier, including the HNE–His MA, ONE–Lys 4-ketoamide adduct, Cys/His–ONE–Lys pyrrole cross-link, ONE–Lys pyrrolinone and octenal–His MA (Figure 4). Both His146 and His161 were found to be modified by HNE/HODA through Michael addition. ONE, an oxidative product of ω-6 PUFAs, has been reported to be more reactive than HNE (6, 11). This enhanced reactivity is reflected in β-LG being widely modified by ONE and its cOCP analog DODE (Tables 3 and 4). ONE and DODE are the only LA peroxidation end products we found which modified three different amino acids: Lys, His and Cys. All of these ONE adducts are long-lived products (12) and may serve as markers of intracellular oxidative stress. Moreover, the Cys/His–ONE–Lys pyrrole cross-link (7, 10, 12, 32, 42) was very abundant and the only cross-linked product detected. The MS/MS spectrum of this inter-peptide cross-link is complicated since it contains fragment ions from two independent peptide sequences. Figure 7 shows the MS/MS spectrum of doubly charged peptide P83–93 cross-linked to HIRL by d0-ONE, d5-ONE and DODE. y3–y8 and b7 ions clearly show that K91 is modified but not K83. b'1–b'2 and y'2–y'3 ions indicate that H146 is modified but not R148. Therefore, K91 and H146 are cross-linked by ONE/DODE with the formation of pyrrole. A His–HNE–Lys Michael adduct-Schiff base cross-link has been reported previously (43, 44). However, we did not find evidence for this cross-link in β-LG, even after reduction by NaBH4. This is probably either because the 3 day incubation disfavors the formation of this reversibly formed product or because the amino acids neighboring the target nucleophile potentially influence the reactivity of the nucleophile in His–HNE–Lys Michael adduct-Schiff base cross-link. Octenal was also found to modify the His146 through Michael addition. In order to confirm these products, β-LG was incubated with pure HNE, ONE and octenal separately, then subjected to the enzymatic digestion and analyzed with LC-ESI-MS/MS. It was found that all of the above products had the same retention time and tandem mass spectra as those from the pure aldehyde modified controls, which further confirmed the assignments of the structure of the adducts.
Table 3.
Modifications Detected by LC-ESI-MS/MS from Chymotryptic Digest#
| Modified Residues | Assignment | Mass Shift* (Da) |
|---|---|---|
| H146, H161 | HNE–His MA | 156/161 |
| H146, H161 | HODA–His MA | 228 |
| K47, K60, K77, K83, K91, K100, K101, K135, K138 | ONE–Lys 4-Ketoamide | 154/159 |
| K47, K60, K77, K83, K91, K100, K101, K135, K138, K141 | DODE–Lys 4-Ketoamide | 226 |
| C121 & K135 | Cys–ONE–Lys Pyrrole Cross-link | 118/123 |
| C121 & K135 | Cys–DODE–Lys Pyrrole Cross-link | 190 |
| H146 & L1, H146 & K8, H146 & K47, H146 & K77, H146 & K83, H146 & K91, H146 & K100, H146 & K101, H146 & K135, H146 & K138 | His–ONE–Lys Pyrrole Cross-link | 118/123 |
| H146 & L1, H146 & K8, H146 & K47, H146 & K77, H146 & K83, H146 & K91, H146 & K101, H146 & K135, H146 & K138 | His–DODE–Lys Pyrrole Cross-link | 190 |
| K47, K60, K77, K83, K91, K100, K101, K135 | ONE–Lys Pyrrolinone | 136/141 |
| K47, K60, K77, K83, K91, K100, K101, K135, K138 | DODE–Lys Pyrrolinone | 208 |
| K91 | Nε-Hexanoyllysine | 98/103 |
| K91 | CA Nε-Hexanoyllysine | 170 |
| H146 | Octenal–His MA | 126/131 |
| H146 | OUEA–His MA | 198 |
| H146 | 2-Octenoic acid–His MA | 142/147 |
| H146 | 2-Octenoic acid CA–His MA | 214 |
| H146 | ONEA–His MA | 170/175 |
| H146 | KODDE–His MA | 294/299 |
| H146 | EKODE–His MA | 310/315 |
| H146 | HKODE–His MA | 312/317 |
| H146 | DHKODE–His MA | 328/333 |
| H146 | His Unknown | 346/351 |
Sequence and mass of modified peptides are listed in Table S1 (Supporting Information)
mass shifts indicating both d0- and d5-adducts of ωOCP and OAP, and only d0-adduct of cOCP; CA: carboxy analog, which is the corresponding cOCP partner of ωOCP.
Table 4.
Modifications Detected by LC-ESI-MS/MS from Tryptic Digest#
| Modified Residues | Assignment | Mass Shift* (Da) |
|---|---|---|
| H146 | HNE–His MA | 156/161 |
| H146 | HODA–His MA | 228 |
| L1, K70, K77, K138 | ONE–Lys 4-Ketoamide | 154/159 |
| L1, K70, K77, K138 | DODE–Lys 4-Ketoamide | 226 |
| K70 | ONE–Lys Pyrrolinone | 136/141 |
| K70 | DODE–Lys Pyrrolinone | 208 |
| H146 | Octenal–His MA | 126/131 |
| H146 | OUEA–His MA | 198 |
| H146 | 2-Octenoic acid–His MA | 142/147 |
| H146 | 2-Octenoic acid CA–His MA | 214 |
| H146 | EKODE–His MA | 310/315 |
Sequence and mass of modified peptides are listed in Table S2 (Supporting Information)
mass shifts indicating both d0- and d5-adducts of ωOCP and OAP, and only d0-adduct of cOCP; CA: carboxy analog, which is the corresponding cOCP partner of ωOCP.
Figure 7.
Tandem mass spectra of doubly charged peptide KIDALNENKVL cross-linked to HIRL by d0-ONE (top), d5-ONE (middle) or DODE (bottom).
The other protein adducts found by MALDI-TOF-MS were also confirmed by LC-ESI-MS/MS, such as EKODE–His146 MA, DHKODE–His146 MA, KODDE–His146 MA and the unknown modification of His146 with a mass shift of 346 Da. In addition, an adduct with a mass shift of 312 Da matches the His146 MA of HKODE (Figure 4) and as predicted for an OAP, lacked an m+72 accompanying peak. Another adduct with a mass shift of 170 Da matches 4-oxo-2-nonenoic acid (ONEA)–His MA (Figure 4) which was also found in the model study (X. Zhu, V. E. Anderson, L. M. Sayre, submitted).
For the Lys modifications, except for the ONE–Lys products, Nε-hexanoyllysine (Figure 4) was the only modification found on the amine group. Nε-Hexanoyllysine was first reported to be a Lys product formed by reaction with LPO products in 1999 (17). The mechanism of its formation was elucidated recently by Uchida and our group (45). However, only Lys91 was found to be acylated, which suggests that it was not a major Lys modification under the oxidative conditions of this study.
In our model study (X. Zhu, V. E. Anderson, L. M. Sayre, submitted), we discovered a novel stable His modification, 2-octenoic acid–His MA (Figure 4). In this study, His146 was also found to be modified by a mass shift of 142 Da, consistent with formation of a 2-octenoic acid–His MA. This assignment was further corroborated by the NaBH4 treatment which left the observed MS unchanged, indicating the absence of reducible double bonds.
Met was shown to be predominantly oxidized into Mo from the MALDI results. MS/MS spectra of modified peptides containing Mo such as ALPMoHIR and LIVTQTMoK showed that base peaks [M + H – 64]+ were routinely observed (data not shown). The 64 Da neutral loss corresponding to CH3SOH is a hallmark of peptides that contain Mo (46).
Characterization of β-LG Modification by the Products of LA Peroxidation after Reduction with LC-ESI-MS/MS
Some protein modifications by LPO products are reversible such as the HNE–Lys MA and the ONE–Lys Schiff base adduct. This reversibility limits their detection by mass spectrometry after the standard denaturation and enzymatic digestion protocols. However, these products may play an important role biologically. For example, an HNE–Lys MA was reported to inactivate glucose-6-phosphate dehydrogenase (20), a cytosolic enzyme in the pentose phosphate pathway. Therefore, NaBH4 was used to stabilize the reversible protein modifications by reduction. However, even after reduction, the HNE–Lys MA and Schiff base adducts were not identified, suggesting that these modifications were only present below our limit of detection. ONE–Lys MA and Schiff base adducts were not observed either. All reversibly formed products would need some free aldehyde to maintain the equilibrium in the incubation mixture at the end of three days. Thus, it is not surprising, considering that ONE itself is not stable under the experimental reaction conditions, that it could not sustain a sufficient steady state level of free ONE to drive the formation of sufficient ONE–Lys MA or Schiff base adducts to be detectable. An alternative fate of reversibly formed protein adducts is irreversible reaction to form stable protein adducts, such as the conversion of the ONE–Lys Schiff base into the 4-ketoamide. These conversions also serve to reduce the steady concentrations of the anticipated reversible intermediate adduct forms.
Another function of NaBH4 is to corroborate the structure of detected protein modification products by detecting the mass shift in the mass spectra following reduction. If the protein adduct contains an aldehyde, ketone or Schiff base, its mass increases 2 Da after reduction. If the product contains two of these reducible functional groups, its mass increases 4 Da. Numerous products were found without borohydride reduction (Tables 3 and 4), and the proposed structures were consistent with the mass increment observed after reduction (Tables S3 and S4, Supporting Information). For example, an ONE–Lys MA and the 4-ketoamide adduct have the same mass shift of 154 Da prior to reduction, which resulted in the misidentification of the 4-ketoamide adduct as a MA (6), but the MA has two reducible carbonyls while the 4-ketoamide adduct has only one. Therefore, the MA should exhibit a mass shift of 4 Da after reduction while the mass shift predicted for the 4-ketoamide adduct is 2 Da. Tables S3 and S4 (Supporting Information) show that all of the Lys+154 adducts in Tables 3 and 4 are the ONE–Lys 4-ketoamide adducts since they all exhibit only a 2 Da shift.
Summary and Conclusions
One of the major sources of in vivo protein modification during oxidative stress is thought to be the oxidative products of PUFAs. While many previous studies have focused on identifying protein adducts derived from a single oxidation product, this study has extended this approach to characterizing the products that are formed when LPO products are generated by in situ radical promoted aerobic oxidation of LA, the most abundant PUFA in mammalian tissue (24). This study should more closely model both the distribution of LPO products present in vivo and also the consistent low level steady state flux of reactive intermediates that then have time to evolve into more stable end products. Thus, the ONE Schiff base, thought to be a prominent adduct formed (11), is not detected in this study, but its stable end product the 4-ketoamide is a prominently observed modification. Although previous mass spectrometric studies on the protein modification detected a number of products (5-9, 12-15, 17, 22, 23), this is the first study to elucidate the chemical nature of protein modification exposed to the LA peroxidation with mass spectrometry. At least 2 His, 11 Lys and 1 Cys of β-LG, which contains 2 His, 15 Lys, 1 free Cys and 3 Arg residues, were found to be modified by LA peroxidation products, suggesting that the electrophiles from the autoxidation of LA widely modify the protein without obvious selectivity. Interestingly, more modified peptides were detected from the chymotryptic digest than the tryptic digest of modified protein. If Lys is modified, its C-terminus cannot be cleaved by trypsin, but this doesn't affect the digestion efficiency of chymotrypsin which cleaves at C-terminus of amino acids containing the hydrophobic side chains. Lys residues were widely modified by LA peroxidation products in this study suggesting that the digestion efficiency of trypsin was reduced, which might partially contribute to the detection of fewer modified peptides from the tryptic digest. Many adducts found in this study contain carbonyl groups such as ONE–4-ketoamide adduct and EKODE–His MA. The results confirm that in addition to the metal-catalyzed oxidation of protein side chain, protein-bound carbonyls can also come from protein modifications by the LPO products. Tannenbaum group (22) and Liebler and coworkers (23) reported that they both found DODE–Lys 4-ketoamide adduct under similar conditions from ours and we also observed a number of ONE/DODE–Lys 4-ketoamide adducts in this study. That may indicate that the 4-ketoamide adduct is one of the major protein modifications from the LA peroxidation products. Therefore, products found in this study may provide some guidance to seek biomarkers of oxidative stress in vivo.
Supplementary Material
Acknowledgement
This work was supported by NIH grants HL 53315 and AG 15885.
1 Abbreviations
- Asc
L-ascorbic acid
- cOCP
oxidative cleavage product containing carboxy terminus
- DHKODE
dihydroxyketooctadecenoic acid
- DNPH
2,4-dinitrophenylhydrazine
- EKODE
epoxyketooctadecenoic acid
- ESI
electrospray ionization
- HKODE
hydroxyketooctadecenoic acid
- HNE
4-hydroxy-2-nonenal
- 13-HPODE
13-hydroperoxy-octadecadienoic acid
- HODA
9-hydroxy-12-oxo-10(E)-dodecenoic acid
- DODE or KODA
9,12-dioxo-10(E)-dodecenoic acid
- KODDE
ketooctadecadienoic acid
- LA
linoleic acid
- d5-LA
17,17,18,18,18-d5-(9Z,12Z)-octadeca-9,12-dienoic acid
- β-LG
bovine β-lactoglobulin
- LPO
lipoxidation
- MA
Michael adduct
- MALDI-TOF-MS
matrix-assisted laser desorption ionization time-of-flight mass spectrometry
- Mo
methionine sulphoxide
- NaHMDS
sodium hexamethyldisilazane
- OAP
oxidative addition product
- OCP
oxidative cleavage product
- ωOCP
oxidative cleavage product containing ω-terminus
- Octenal
2(E)-Octenal
- ONE
4-oxo-2-nonenal
- ONEA
4-oxo-2-nonenoic acid
- OUEA
11-oxoundec-9(E)-enoic acid
- PUFA
polyunsaturated fatty acid
- SIC
selected ion chromatogram
- TFA
trifluoroacetic acid
- TIC
total ion chromatogram
Footnotes
Supporting Information Available: Tables S1–S4 and Figures S1–S3 are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Jenner P. Oxidative stress in Parkinson's disease. Ann. Neurol. 2003;53:S26–S38. doi: 10.1002/ana.10483. [DOI] [PubMed] [Google Scholar]
- 2.Chauhan V, Chauhan A. Oxidative stress in Alzheimer's disease. Pathophysiology. 2006;13:195–208. doi: 10.1016/j.pathophys.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Perry G, Nunomura A, Hirai K, Zhu X, Prez M, Avila J, Castellani RJ, Atwood CS, Aliev G, Sayre LM, Takeda A, Smith MA. Is oxidative damage the fundamental pathogenic mechanism of Alzheimer's and other neurodegenerative diseases? Free Radical Biol. Med. 2002;33:1475–1479. doi: 10.1016/s0891-5849(02)01113-9. [DOI] [PubMed] [Google Scholar]
- 4.Berliner Judith A, Watson Andrew D. A role for oxidized phospholipids in atherosclerosis. N. Engl. J. Med. 2005;353:9–11. doi: 10.1056/NEJMp058118. [DOI] [PubMed] [Google Scholar]
- 5.Bruenner BA, Jones AD, German JB. Direct characterization of protein adducts of the lipid peroxidation product 4-hydroxy-2-nonenal using electrospray mass spectrometry. Chem. Res. Toxicol. 1995;8:552–559. doi: 10.1021/tx00046a009. [DOI] [PubMed] [Google Scholar]
- 6.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem. Res. Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Minkler PE, Sayre LM. Mass spectroscopic characterization of protein modification by 4-hydroxy-2-(E)-nonenal and 4-oxo-2-(E)-nonenal. Chem. Res. Toxicol. 2003;16:901–911. doi: 10.1021/tx0300030. [DOI] [PubMed] [Google Scholar]
- 8.Isom AL, Barnes S, Wilson L, Kirk M, Coward L, Darley-Usmar V. Modification of cytochrome c by 4-hydroxy-2-nonenal: evidence for histidine, lysine, and arginine-aldehyde adducts. J. Am. Soc. Mass Spectrom. 2004;15:1136–1147. doi: 10.1016/j.jasms.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 9.Aldini G, Dalle-Donne I, Vistoli G, Facino RM, Carini M. Covalent modification of actin by 4-hydroxy-trans-2-nonenal (HNE): LC-ESI-MS/MS evidence for Cys374 Michael adduction. J. Mass Spectrom. 2005;40:946–954. doi: 10.1002/jms.872. [DOI] [PubMed] [Google Scholar]
- 10.Sayre LM, Lin D, Yuan Q, Zhu X, Tang X. Protein adducts generated from products of lipid oxidation: focus on HNE and ONE. Drug Metab. Rev. 2006;38:651–675. doi: 10.1080/03602530600959508. [DOI] [PubMed] [Google Scholar]
- 11.Lin D, Lee H.-g., Liu Q, Perry G, Smith MA, Sayre LM. 4-Oxo-2-nonenal is both more neurotoxic and more protein reactive than 4-hydroxy-2-nonenal. Chem. Res. Toxicol. 2005;18:1219–1231. doi: 10.1021/tx050080q. [DOI] [PubMed] [Google Scholar]
- 12.Zhu X, Sayre LM. Mass spectrometric evidence for long-lived protein adducts of 4-oxo-2-nonenal. Redox Rep. 2007;12:45–49. doi: 10.1179/135100007X162176. [DOI] [PubMed] [Google Scholar]
- 13.Yocum AK, Oe T, Yergey AL, Blair IA. Novel lipid hydroperoxide-derived hemoglobin histidine adducts as biomarkers of oxidative stress. J. Mass Spectrom. 2005;40:754–764. doi: 10.1002/jms.847. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X, Sayre LM. Long-lived 4-oxo-2-enal-derived apparent lysine Michael adducts are actually the isomeric 4-ketoamides. Chem. Res. Toxicol. 2007;20:165–170. doi: 10.1021/tx600295j. [DOI] [PubMed] [Google Scholar]
- 15.Alaiz M, Giron J. Modification of histidine residues in bovine serum albumin by reaction with (E)-2-octenal. J. Agric. Food Chem. 1994;42:2094–2098. [Google Scholar]
- 16.Lin D, Zhang J, Sayre LM. Synthesis of six epoxyketooctadecenoic acid (EKODE) isomers, their generation from nonenzymatic oxidation of linoleic acid, and their reactivity with imidazole nucleophiles. J. Org. Chem. 2007;72:9471–9480. doi: 10.1021/jo701373f. [DOI] [PubMed] [Google Scholar]
- 17.Kato Y, Mori Y, Makino Y, Morimitsu Y, Hiroi S, Ishikawa T, Osawa T. Formation of Nε-(hexanonyl)lysine in protein exposed to lipid hydroperoxide. A plausible marker for lipid hydroperoxide-derived protein modification. J. Biol. Chem. 1999;274:20406–20414. doi: 10.1074/jbc.274.29.20406. [DOI] [PubMed] [Google Scholar]
- 18.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Cysteine modification by lipid peroxidation products inhibits protein disulfide isomerase. Chem. Res. Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 19.Crabb JW, O'Neil J, Miyagi M, West K, Hoff HF. Hydroxynonenal inactivates cathepsin B by forming Michael adducts with active site residues. Protein Sci. 2002;11:831–840. doi: 10.1110/ps.4400102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szweda LI, Uchida K, Tsai L, Stadtman ER. Inactivation of glucose-6-phosphate dehydrogenase by 4-hydroxy-2-nonenal. Selective modification of an active-site lysine. J. Biol. Chem. 1993;268:3342–3347. [PubMed] [Google Scholar]
- 21.Ishii T, Tatsuda E, Kumazawa S, Nakayama T, Uchida K. Molecular basis of enzyme inactivation by an endogenous electrophile 4-hydroxy-2-nonenal: identification of modification sites in glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 2003;42:3474–3480. doi: 10.1021/bi027172o. [DOI] [PubMed] [Google Scholar]
- 22.Williams MV, Wishnok JS, Tannenbaum SR. Covalent adducts arising from the decomposition products of lipid hydroperoxides in the presence of cytochrome c. Chem. Res. Toxicol. 2007;20:767–775. doi: 10.1021/tx600289r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szapacs ME, Kim H-YH, Porter NA, Liebler DC. Identification of proteins adducted by lipid peroxidation products in plasma and modifications of apolipoprotein A1 with a novel biotinylated phospholipid probe. J. Proteome Res. 2008;7:4237–4246. doi: 10.1021/pr8001222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esterbauer H, Rotheneder M, Striegl G, Waeg G, Ashy A, Sattler W, Juergens G. Vitamin E and other lipophilic antioxidants protect LDL against oxidation. Fett Wiss. Technol. 1989;91:316–324. [Google Scholar]
- 25.Spiteller G. Peroxidation of linoleic acid and its relation to aging and age dependent diseases. Mech. Ageing Dev. 2001;122:617–657. doi: 10.1016/s0047-6374(01)00220-2. [DOI] [PubMed] [Google Scholar]
- 26.Spiteller D, Spiteller G. Oxidation of linoleic acid in low-density lipoprotein: an important event in atherogenesis. Angew. Chem., Int. Ed. 2000;39:585–589. doi: 10.1002/(sici)1521-3773(20000204)39:3<585::aid-anie585>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 27.Podrez EA, Poliakov E, Shen Z, Zhang R, Deng Y, Sun M, Finton PJ, Shan L, Gugiu B, Fox PL, Hoff HF, Salomon RG, Hazen SL. Identification of a novel family of oxidized phospholipids that serve as ligands for the macrophage scavenger receptor CD36. J. Biol. Chem. 2002;277:38503–38516. doi: 10.1074/jbc.M203318200. [DOI] [PubMed] [Google Scholar]
- 28.Banaszak L, Winter N, Xu Z, Bernlohr DA, Cowan S, Jones TA. Lipid-binding proteins: a family of fatty acid and retinoid transport proteins. Adv. Protein Chem. 1994;45:89–151. doi: 10.1016/s0065-3233(08)60639-7. [DOI] [PubMed] [Google Scholar]
- 29.Spector AA, Fletcher JE. Binding of long chain fatty acids to beta - lactoglobulin. Lipids. 1970;5:403–411. doi: 10.1007/BF02532106. [DOI] [PubMed] [Google Scholar]
- 30.Steinbrecher UP. Oxidation of human low density lipoprotein results in derivatization of lysine residues of apolipoprotein B by lipid peroxide decomposition products. J. Biol. Chem. 1987;262:3603–3608. [PubMed] [Google Scholar]
- 31.Nadkarni DV, Sayre LM. Structural definition of early lysine and histidine adduction chemistry of 4-hydroxynonenal. Chem. Res. Toxicol. 1995;8:284–291. doi: 10.1021/tx00044a014. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W-H, Liu J, Xu G, Yuan Q, Sayre LM. Model studies on protein side chain modification by 4-oxo-2-nonenal. Chem. Res. Toxicol. 2003;16:512–523. doi: 10.1021/tx020105a. [DOI] [PubMed] [Google Scholar]
- 33.Viala J, Santelli M. Three-carbon homologating agent. New preparation of (3,3-diisopropoxypropyl)triphenylphosphonium bromide. Synthesis. 1988:395–397. [Google Scholar]
- 34.Levine RL, Williams JA, Stadtman ER, Schacter E. Carbonyl assays for determination of oxidatively modified proteins. Meth. Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 35.Yuan Q, Zhu X, Sayre LM. Chemical nature of stochastic generation of protein-based carbonyls: metal-catalyzed oxidation versus modification by products of lipid oxidation. Chem. Res. Toxicol. 2007;20:129–139. doi: 10.1021/tx600270f. [DOI] [PubMed] [Google Scholar]
- 36.Chen X, Chen YH, Anderson VE. Isolation and Characterization of Protein Cross-links by Isotopic Derivatization and ESI-MS. Anal. Biochem. 1999;273:192–203. doi: 10.1006/abio.1999.4243. [DOI] [PubMed] [Google Scholar]
- 37.Spiteller G. Linoleic acid peroxidation - the dominant lipid peroxidation process in low density lipoprotein - and its relationship to chronic diseases. Chem. Phys. Lipids. 1998;95:105–162. doi: 10.1016/s0009-3084(98)00091-7. [DOI] [PubMed] [Google Scholar]
- 38.Carini M, Aldini G, Facino RM. Mass spectrometry for detection of 4-hydroxy-trans-2-nonenal (HNE) adducts with peptides and proteins. Mass Spectrom. Rev. 2004;23:281–305. doi: 10.1002/mas.10076. [DOI] [PubMed] [Google Scholar]
- 39.Spiteller P, Kern W, Reiner J, Spiteller G. Aldehydic lipid peroxidation products derived from linoleic acid. Biochim. Biophys. Acta, Mol. Cell Biol. Lipids. 2001;1531:188–208. doi: 10.1016/s1388-1981(01)00100-7. [DOI] [PubMed] [Google Scholar]
- 40.Amici A, Levine RL, Tsai L, Stadtman ER. Conversion of amino acid residues in proteins and amino acid homopolymers to carbonyl derivatives by metal-catalyzed oxidation reactions. J. Biol. Chem. 1989;264:3341–3346. [PubMed] [Google Scholar]
- 41.Zhou L, Thakker DR, Voyksner RD, Anbazhagan M, Boykin DW, Hall JE, Tidwell RR. Metabolites of an orally active antimicrobial prodrug, 2,5-bis(4-amidinophenyl)furan-bis-O-methylamidoxime, identified by liquid chromatography/tandem mass spectrometry. J. Mass Spectrom. 2004;39:351–360. doi: 10.1002/jms.591. [DOI] [PubMed] [Google Scholar]
- 42.Oe T, Arora JS, Lee SH, Blair IA. A novel lipid hydroperoxide-derived cyclic covalent modification to Histone H4. J. Biol. Chem. 2003;278:42098–42105. doi: 10.1074/jbc.M308167200. [DOI] [PubMed] [Google Scholar]
- 43.Cohn JA, Tsai L, Friguet B, Szweda LI. Chemical characterization of a protein-4-hydroxy-2-nonenal cross-link: immunochemical detection in mitochondria exposed to oxidative stress. Arch. Biochem. Biophys. 1996;328:158–164. doi: 10.1006/abbi.1996.0156. [DOI] [PubMed] [Google Scholar]
- 44.Stewart BJ, Doorn JA, Petersen DR. Residue-specific adduction of tubulin by 4-hydroxynonenal and 4-oxononenal causes cross-linking and inhibits polymerization. Chem. Res. Toxicol. 2007;20:1111–1119. doi: 10.1021/tx700106v. [DOI] [PubMed] [Google Scholar]
- 45.Ishino K, Shibata T, Ishii T, Liu Y-T, Toyokuni S, Zhu X, Sayre LM, Uchida K. Chem. Res. Toxicol. ACS ASAP; Protein N-Acylation: H2O2-Mediated Covalent Modification of Protein by Lipid Peroxidation-Derived Saturated Aldehydes. [DOI] [PubMed] [Google Scholar]
- 46.Lagerwerf FM, van de Weert M, Heerma W, Haverkamp J. Identification of oxidized methionine in peptides. Rapid Commun. Mass Spectrom. 1996;10:1905–1910. doi: 10.1002/(SICI)1097-0231(199612)10:15<1905::AID-RCM755>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.