Abstract
The EH domain is found in proteins associated with endocytosis and vesicle trafficking. EH domains bind to their target proteins through an asparagine-proline-phenylalanine (NPF) motif. We have measured the interaction energetics of the EH domain from EHD1 with peptides derived from two of its binding partners: Rabenosyn-5 (Ac-GPSLNPFDEED-NH2) and Rab11-Fip2 (Ac-YESTNPFTAK-NH2). HSQC spectroscopy shows that both peptides bind in the canonical binding pocket of EHD1 EH and induce identical structural changes, yet the affinity of the negatively-charged Ac-GPSLNPFDEED-NH2 (Ka=8*105 M−1) is tighter by two orders of magnitude. The thermodynamic profiles (ΔG, ΔH, ΔS) were measured for both peptides as a function of temperature. The enthalpies of binding are essentially identical and the difference in affinity is a consequence of the difference in entropic cost. Ac-GPSLNPFDEED-NH2 binding is salt-dependent, demonstrating an electrostatic component to the interaction, whereas Ac-YESTNPFTAK-NH2 binding is independent of salt. Successive replacement of acidic residues in Ac-GPSLNPFDEED-NH2 with neutral residues showed that all are important. Lysine sidechains in EHD1 EH create a region of strong positive surface potential near the NPF binding pocket. Contributions by lysine ε-amino groups to complex formation with Ac-GPSLNPFDEED-NH2 was shown using direct observe 15N NMR spectroscopy. These experiments have enabled us to define a new extended interaction motif for EHD proteins, N-P-F-[DE]-[DE]-[DE], which we have used to predict new interaction partners and hence broaden the range of cellular activities involving the EHD proteins.
The EH1 (Eps15 homology) domain is a protein interaction module found in proteins that participate in endocytosis and vesicle trafficking. It is widespread in nature, occurring in animals, plants, fungi, and unicellular organisms and it is probably a feature of all eukaryotic cells. In mammals there are 11 EH domain containing proteins which can be organized into 5 distinct families: (i) Eps15, (ii) Intersectin, (iii) Reps, (iv) γ-synergin and (v) EHD proteins (reviewed in (1)). Eps15, which contains 3 consecutive EH domains at its N-terminus, was the first family member to be discovered (2, 3) and is considered to be the prototype. Eps15 assists in the formation of clathrin-coated pits at the plasma membrane (4, 5), a process that may also involve Reps (6, 7). EHD proteins are associated with internal membranes and feature prominently in vesicle recycling pathways (8, 9).
Phage display has shown that EH domains bind to the sequence asparagine-proline-phenylalanine (NPF) in target proteins (4, 10) and this has been elaborated upon in numerous direct binding and mutagenesis experiments (10, 11). It is thought that NPF-containing proteins recruit EH domain-containing proteins to multicomponent complexes on a target membrane. Confirmed NPF-containing interaction targets in mammals include Epsin (12), stonin2 (13), Numb (4, 14), SNAP-29 (15), Disabled homolog, (16) PACSIN/syndapin (17), CALM (18), synaptojanin (19), Hrb (4, 20), SH3 domain binding protein 4 (4), SCAMP (21), EHBP1 (22), myoferlin (23), Rab11-Fip2 (7, 24) and Rabenosyn-5 (25). The great majority of these proteins contain more than one NPF motif. The EH domain is a Ca2+-binding protein module of about 100 residues. Several NMR structures have been solved (11, 26–29); all EH domains consist of a single pair of back-to-back EF hands (2 helix-loop-helix motifs) connected by a short region of β-sheet and followed by a proline-rich C-terminal tail. Typically, only the second loop possesses the canonical arrangement of Ca2+ binding ligands and EH domains usually bind only a single Ca2+ ion. NMR experiments using peptide bound to the second EH domain of Eps15 have shown that the NPF sequence is embedded within a hydrophobic pocket formed by helices 2 and 3 of the protein. The backbone adopts a type I β-turn which includes the residue following the phenylalanine; this residue was also found to contribute to specificity (30, 31).
The binding of NPF-containing peptides to EH domains is typically weak, with literature values ranging between 103 and 104 M−1 (10, 26, 28, 32). Although temporal and spatial location are likely to be important, it is still not clear how EH domains select among the large number of NPF-containing potential binding partners, or how the process is regulated. One interesting solution to the problem was found by Groemping and coworkers (33) who showed that the second EH domain of Eps15 exploits the multiple NPF motifs of stonin2 to produce a high affinity complex (Ka = 6 *106 M−1). Adjacent NPF motifs from a stonin-derived peptide bound both the primary (canonical) binding site between helices 2 and 3 and to a secondary binding pocket formed by helices 1 and 4 on the opposite side of the protein. Multiple contacts between peptide and protein were responsible for the high affinity. We are interested in the factors that lead to the selectivity and specificity of a different class of EH domain-containing proteins, the EHD proteins.
EHD proteins are of special interest because they are members of the dynamin superfamily and, like dynamin, have a capacity to remodel membranes (34–36). They contain an N-terminal G-domain, which binds ATP in preference to GTP (34) and the EH domain is at the C-terminus. Mammalian cells contain four EHD proteins (EHD1-4) whereas invertebrates such as C. elegans contain only one (known as Rme-1). The molecular weight of the monomer is about 50kD. Within the group, EHD1 and EHD3 are clearly most closely related (87% identical), but all four proteins are highly homologous and, in addition, share greater than 50% identity with the C. elegans ortholog. When overexpressed, the proteins are frequently found on membranous tubules within the cell (37) and in vitro they have been shown to bind liposomes and induce tubulation (35, 36).
The structure of EHD2 has been solved by X-ray crystallography (36) and its features are expected to be shared by all EHD proteins (38). The protein is a compact dimer with the G-domain forming the bulk of the interface. The EH domain sits on top and interacts with G-domain, specifically preventing it from binding guanine nucleotides. A framework of helices both supports the G-domain and provides the lipid-binding surface at its base. Surprisingly, the NPF binding pocket was found to be occupied by a GPF motif from the linker region that connects the EH domain to the rest of the protein. An N-terminally extended EHD1 EH domain that includes the linker region and GPF motif will dimerize in solution, implying that domain switching occurs between the subunits of the intact protein (31).
Proteins that have been specifically associated with EHD proteins in mammals are SNAP-29 (15), EHBP1 (22), Rabenosyn-5 (25), Rab11-Fip2 (24), PACSIN (syndapin) (17) and myoferlin (23). It has been noted that one or more of the NPF motifs in many of these proteins are followed by aspartic and glutamic acid residues (39) and that these residues have the potential to interact with a patch of strong positive potential on the surface of EHD1 EH (29). Furthermore, in C. elegans, negative charges following an NPF motif in amphiphysin were shown to enhance the binding of Rme1 in a GST pulldown experiment (40).
In order to understand the factors contributing to the affinity and selectivity of EH-NPF interactions, we focused on two of the most well characterized EHD binding proteins: Rabenosyn-5 and Rab11-Fip2. Both of these targets also bind Rab proteins and are active in vesicle sorting pathways. Rabenosyn-5 is an essential component of the early endosomal fusion machinery and binds Rab5 and Rab4 (41, 42). The polypeptide chain contains 6 NPF motifs (Figure 1A–C). Rab11-Fip2, as its name suggests, binds Rab11, and is found on the recycling endosome (43). It is a dimer (44, 45) and contains 3 NPF motifs on each polypeptide chain. Rab11-Fip2 is also thought to interact with a second EH domain containing protein, Reps1 (7).
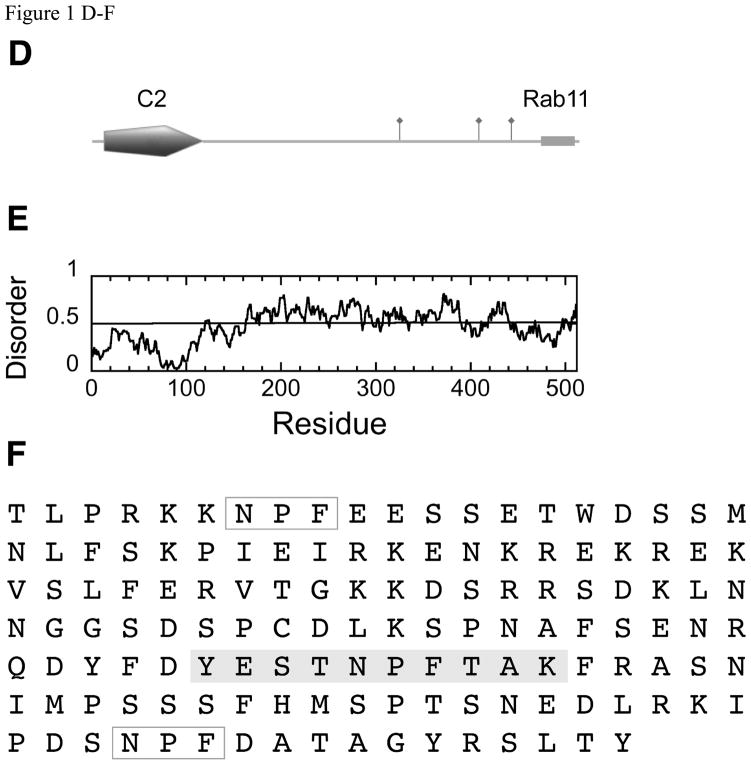
Figure 1.
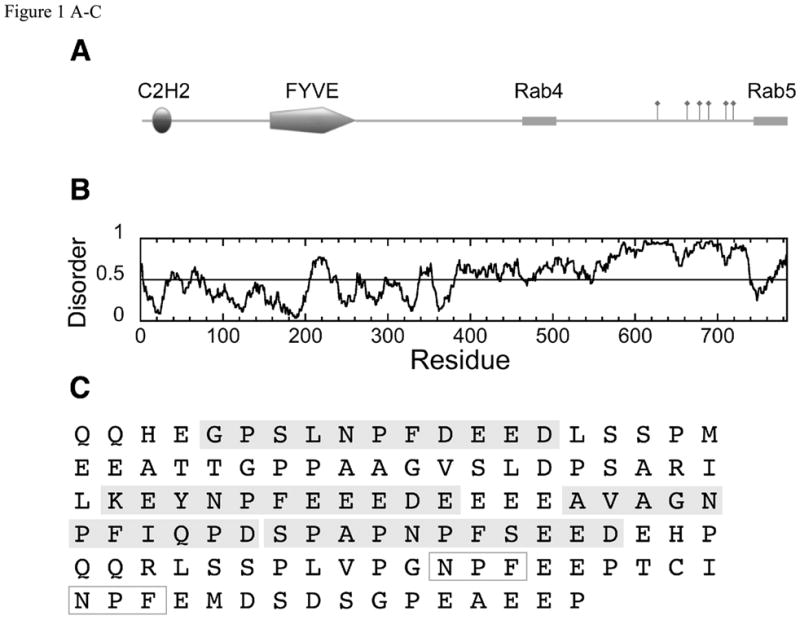
(A) Domain structure of Rabenosyn-5, showing an N-terminal C2H2 Zn-finger and a phosphatidylinositol-3-phosphate binding FYVE finger domain. The Rab4 and Rab5 binding helices (42) are marked with boxes and the 6 NPF motifs are indicated by flags. (B) Order/disorder prediction using IUPRED (http://iupred.enzim.hu/). Regions of potential disorder score >0.5. (C) Expanded sequence of the NPF-containing region (residues 618–733) showing the sequences of the peptides used in these experiments (Rsn5-NPF1, Rsn5-NPF2 etc) shaded in gray. Rsn5-NPF1 is the first sequence, GPSLNPFDEED. NPF motifs that were not used in these experiments are in open boxes. (D) Domain structure of Rab11-Fip2. The Rab11 binding region at the C-terminus is shown as a gray cylinder and the 3 NPF motifs indicated by flags. Rab11-Fip2 has a phospholipids-binding C2 domain (E) Order/disorder prediction as above. (F) Expanded sequence of the NPF region (residues 317–453) showing the sequences of the peptide Rfp2-NPF2 (YESTNPFTAK, gray box) used in these experiments. NPF motifs that were not used in these experiments are in open boxes. Images produced using MyDomains Image Creator (http://ca.expasy.org/prosite/).
We have used isothermal titration calorimetry (ITC) and NMR spectroscopy to investigate the thermodynamics and the specificity of interactions between NPF-containing peptides from Rabenosyn-5 and Rab11-Fip2. Particular attention was paid to the contribution of the negatively-charged residues following the NPF motif in peptide sequences derived from Rabenosyn-5. Based on our understanding of these interactions, we have suggested several novel interaction partners for the EHD proteins.
Methods
Constructs and Protein Preparation
A plasmid containing the coding sequence of the EH domain of EHD1 (mouse) was a generous gift from Dr. Barth Grant. Protein (residues V437 to E534) was expressed as a GST fusion and purified on glutathione-Sepharose (GE Life Sciences) using standard techniques. This construct does not contain the GPF motif (G420-F422). The EH domain was cleaved from the beads in situ using thrombin (28). Protein concentration was determined by absorbance at 280 nm using a molar absorbance coefficient of 13,980 M−1cm−1 (46).
Peptides
Peptides were synthesized and purified by the Tufts University Core Facility using standard procedures and their masses confirmed using mass spectrometry. Peptide concentration was determined by absorbance using a molar absorbance coefficient of 200 M−1cm−1 at 258 nm for phenylalanine-containing peptides and 1280 M−1cm−1 at 274 nm for tyrosine containing peptides.
Isothermal Titration Calorimetry
Protein and peptide were dialyzed exhaustively against buffer containing 25 mM MOPS pH 7.0, 1 mM CaCl2 and variable concentrations of KCl (where appropriate) at 4°C. ITC measurements were carried out at 20°C unless otherwise stated using a VP-ITC calorimeter (Microcal, Northampton, MA). The protein concentration in the calorimeter cell varied between 50 μM and 200 μM and the peptide titrant was 10-fold higher. The results were analyzed using Origin 7. Single site binding was assumed and the data fitted for the association constant (Ka), stoichiometry (n) and the molar enthalpy (ΔH).
NMR spectroscopy
HSQC spectra and directly detected 15N spectra were recorded at 25° in 10 mM imidazole-d4 pH 6.8, 10 mM NaCl, 1 mM CaCl2, 0.02% NaN3 and 10% D2O on a Bruker DPX-300 operating at 300 MHz for 1H and 30 MHz for 15N. Directly detected 15N experiments used 2.5 mM protein contained in a 5 mm tube. Peptide was dissolved in the same buffer and the pH adjusted to 6.8 before addition to the protein. 15N spectra (20,000 scans) were collected with a 2.7s acquisition time and 0.1s recycle delay with WALTZ decoupling of the protons applied continuously. The spectral width was 200 ppm. 15N chemical shifts are reported with respect to liquid ammonia at 0 ppm. HSQC assignments of EHD1 EH are those reported by (29) and assignments in the presence of saturating amounts of peptide were obtained by careful titration of the protein.
Results
EH domains bind to peptide sequences containing an asparagine-proline-phenylalanine (NPF) motif. We chose to focus our study on two NPF-containing proteins, Rabenosyn-5 and Rab11-Fip2, that are known to be targets of EHD1 and EHD3 (24, 25). They have 6 and 3 NPF sequences respectively (Figure 1). The NPF motifs of Rabenosyn-5 are frequently followed by a sequence of acidic residues. This is true of other EHD binding proteins such as PACSIN (syndapin), SNAP-29 and EHBP1 and it has been suggested that the negative charges are important for selectivity (39). Mutational analysis using intact proteins has shown that the first and second NPF sequences of Rabenosyn-5 and the second NPF sequence of Rab11-Fip2 are most important for binding to EHD proteins (24, 25). We therefore synthesized peptides corresponding to NPF1 of Rabenosyn-5 (Ac-GPSLNPFDEED-NH2, referred to as Rsn5-NPF1) and NPF2 of Rab11-Fip2 (Ac-YESTNPFTAK-NH2, referred to as Rfp2-NPF2). This pair of peptides is ideal for investigating the structural and thermodynamic contributions made by the charged residues.
Rsn5-NPF1 and Rfp2-NPF2 bind to the same site on EHD1 EH and induce similar backbone conformational changes
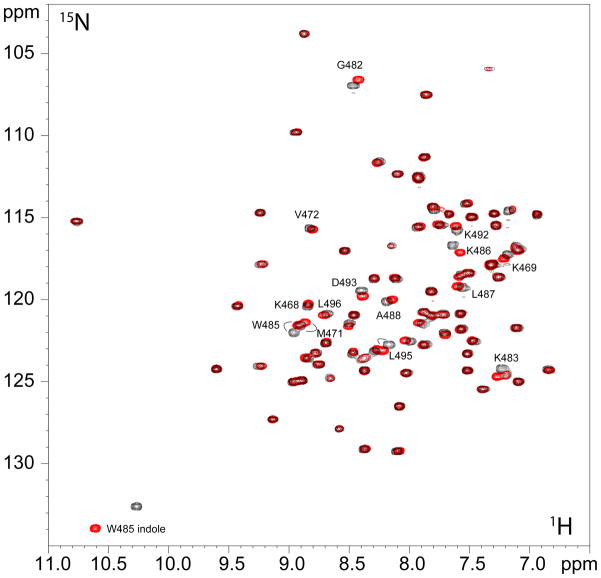
The effect of saturating amounts of Rsn5-NPF1 and Rfp2-NPF2 on the HSQC spectrum of EHD1 EH are shown in Figure 2. Both peptides induced large chemical shift changes (Figure S1) as expected from earlier experiments using EH domains from Reps1 (28) and the EH domains from Eps15 (11). These shift changes are virtually identical, with the only significant difference between them being the effect on the sidechain indole of W485. A recent NMR structure has confirmed that a peptide similar to Rfp2-NPF2 binds as expected as a β-turn in the pocket formed between helices 2 and 3 (31), thus the HSQC data show that the structure of the Rsn5-NPF1 complex is likely to be very similar. In this light, it is all the more striking to note that the peptide affinities differ greatly, with significantly more of the Rab11-Fip2 peptide (Rfp2-NPF2) than the Rabenosyn-5 peptide (Rsn5-NPF1) required to reach saturation. In order to obtain accurate Ka values and to understand the forces contributing to the difference in affinity between the two peptides, we measured the thermodynamic profile for each using isothermal titration calorimetry (ITC).
Figure 2.
1H/15N HSQC spectrum of 15N-labeled EHD1 EH in the presence of saturating amounts of Rsn5-NPF1 (black) or Rfp2-NPF2 (red) showing the almost identical chemical shift changes for the two peptides. The protein was 2.5 mM and the peptides were 3 mM and 7.5 mM respectively. Selected residues that shift upon addition of peptide are labeled.
Rsn5-NPF1 binds the EH domain of EHD1 with 100-fold higher affinity
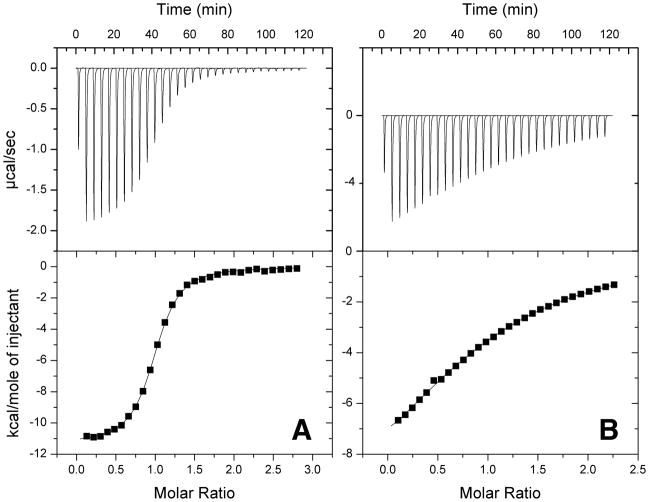
Comparable ITC datasets for the two peptides are shown in Figure 3 and the thermodynamic parameters are listed in Table 1. Binding of both peptides is enthalpically driven. Rsn5-NPF1, however, with a Ka of 8*105 M−1 at 20°C, binds more than 2 orders of magnitude more tightly than Rfp2-NPF2. This is consistently observed over a range of temperatures (Figure 4) with tighter binding found in both cases as the temperature is lowered. Most importantly, the affinity of Rsn5-NPF1 is significantly greater than that of any single-motif EH domain-NPF peptide interaction measured to date and comes within an order of magnitude of the stonin2-Eps15 EH2 complex (33) which engages 2 NPF motifs in separate pockets. The high affinity of Rsn5-NPF1 suggested that residues additional to the core NPF motif may contribute to the interaction.
Figure 3.
(A) Typical ITC dataset for Rsn5-NPF1 binding to EHD1 EH. The protein in the calorimeter cell was 50 μM and the peptide titrant was 0.5 mM. (B) Typical ITC dataset for Rfp2-NPF2 binding to EHD1 EH. The protein was 200 μM and the peptide 2 mM. Both experiments were carried out in 25 mM MOPS, 1 mM CaCl2 at 20°C.
Table 1.
Thermodynamic parameters for Rsn5-NPF1 and Rfp2-NPF2 binding to EHD1 EH.
| Rsn5-NPF1 titration as a function of temperature | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Temperature (°C) | n | Ka (M−1) | ΔG (cal mol−1) | ΔS (cal mol−1 K−1) | ΔH (cal mol−1) | TΔS (cal mol−1) | [protein] (mM) | [peptide] (mM) | c (Ka*[protein]) |
| 5.0 | 0.980 | 2.03e+06 | −8020 | −3.43 | −8986 | −954.2 | 0.0465 | 0.565 | 94 |
| 10.0 | 0.980 | 1.71e+06 | −8068 | −5.67 | −9684 | −1606 | 0.0465 | 0.565 | 80 |
| 15.0 | 0.990 | 1.10e+06 | −7958 | −8.56 | −10440 | −2467 | 0.0465 | 0.565 | 51 |
| 20.0 | 0.980 | 8.14e+05 | −7921 | −11.6 | −11330 | −3401 | 0.0465 | 0.565 | 38 |
| 25.0 | 0.980 | 6.06e+05 | −7881 | −13.6 | −11940 | −4056 | 0.0465 | 0.565 | 28 |
| Rfp2-NPF2 titration as a function of temperature | |||||||||
| 5.0 | 0.970 | 17800 | −5469 | −13.9 | −9279 | −3867 | 0.188 | 2.03 | 3.3 |
| 10.0 | 1.02 | 12800 | −5316 | −16.7 | −10040 | −4729 | 0.210 | 2.00 | 2.7 |
| 15.0 | 1.03 | 9390 | −5233 | −19.3 | −10790 | −5562 | 0.209 | 2.01 | 2.0 |
| 20.0 | 1.03 | 6640 | −5122 | −23.2 | −11920 | −6802 | 0.204 | 2.00 | 1.4 |
| 25.0 | 0.990 | 4700 | −5005 | −27.9 | −13320 | −8320 | 0.202 | 1.97 | 1.0 |
| Rsn5-NPF1 titration as a function of KCl concentration | |||||||||
| KCl (mM) | n | Ka (M−1) | ΔG (cal mol−1) | ΔS (cal mol−1 K−1) | ΔH (cal mol−1) | TΔS (cal mol−1) | [protein] (mM) | [peptide] (mM) | c |
| 0 | 1.02 | 8.61e+05 | −7954 | −11.4 | −11310 | −3343 | 0.0520 | 0.455 | 45 |
| 0 | 0.98 | 8.14e+05 | −7921 | −11.6 | −11330 | −3401 | 0.0465 | 0.565 | 38 |
| 25.0 | 0.97 | 3.02e+05 | −7444 | −10.5 | −10420 | −3079 | 0.0460 | 0.500 | 14 |
| 50.0 | 0.97 | 1.68e+05 | −7002 | −11.6 | −10420 | −3401 | 0.049 | 0.540 | 8.2 |
| 75.0 | 0.95 | 1.15e+05 | −6782 | −12.1 | −10340 | −3548 | 0.0732 | 0.750 | 8.4 |
| 100.0 | 0.99 | 86500 | −6616 | −14.5 | −10880 | −4251 | 0.102 | 1.01 | 8.8 |
| 125.0 | 1.05 | 65000 | −6450 | −13.8 | −10500 | −4046 | 0.122 | 1.22 | 7.9 |
| 150.0 | 1.09 | 53000 | −6331 | −14.2 | −10500 | −4163 | 0.1830 | 2.00 | 10 |
| 300.0 | 1.04 | 24800 | −5889 | −14.9 | −10260 | −4369 | 0.196 | 2.17 | 4.9 |
| 450.0 | 1.04 | 17800 | −5696 | −16.1 | −10430 | −4721 | 0.193 | 2.25 | 3.4 |
| Rfp2-NPF2 titration as a function of KCl concentration | |||||||||
| 0 | 1.01 | 7030 | −5155 | −22.7 | −11820 | −6656 | 0.200 | 1.96 | 1.5 |
| 75.0 | 0.960 | 6460 | −5106 | −23.0 | −11840 | −6744 | 0.227 | 1.95 | 1.5 |
| 150.0 | 1.02 | 6910 | −5145 | −23.2 | −11960 | −6802 | 0.199 | 1.96 | 1.4 |
| 300.0 | 0.990 | 7490 | −5192 | −22.4 | −11770 | −6568 | 0.197 | 2.02 | 1.5 |
| 450.0 | 1.04 | 7810 | −5217 | −20.7 | −11290 | −6069 | 0.198 | 2.03 | 1.5 |
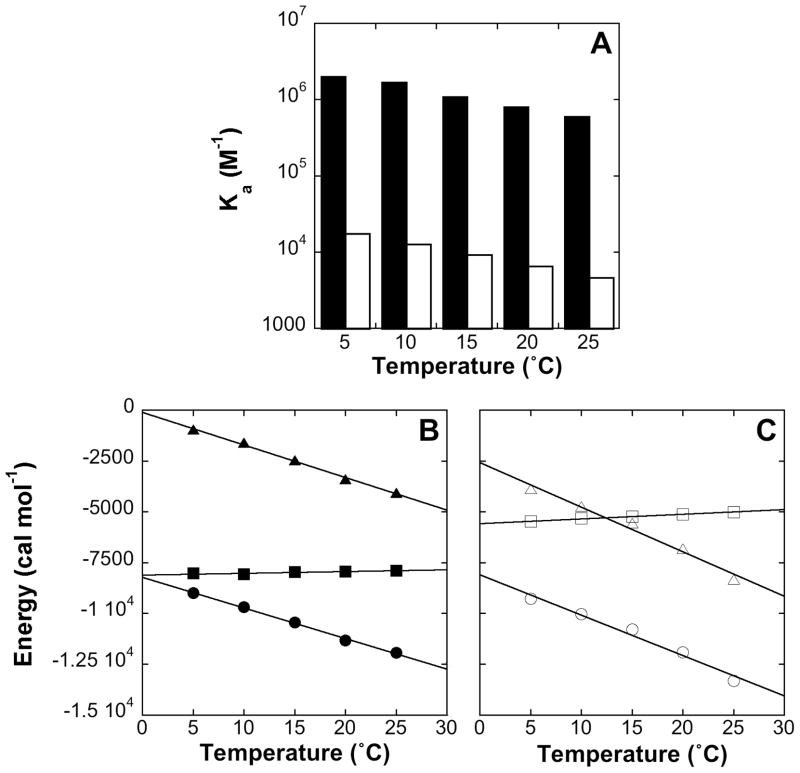
Figure 4.
(A) Association constants of Rsn5-NPF1 (filled columns) and Rfp2-NPF2 (open columns) measured by ITC as a function of temperature. (B) Thermodynamic profile of Rsn5-NPF1 binding to EHD1 EH. ΔH is in solid circles, TΔS in solid triangles and ΔG in solid squares. (C) Thermodynamic profile of Rfp2-NPF2 binding to EHD1 EH. ΔH is in open circles, TΔS is in open triangles and ΔG is in open squares.
Circular dichroism spectroscopy showed that both peptides are probably extended in free solution (Figure S2); however, as a further test of the influence of conformational bias in the peptide structure, we used ITC to measure the affinities of both peptides for the EH domain of Reps1. Reps1 is also thought to be a target for Rab11-Fip2 (7). Like EHD1, Reps1 favors the second NPF motif of Rab11-Fip2 in vivo (7) but unlike the EHD proteins, it is not thought to be a native binding partner of Rabenosyn-5. Furthermore, Reps1 EH does not have a region of strong positive surface potential near the NPF binding pocket (28) and so it provides an excellent model for comparison. Using ITC, Reps1 EH was found to bind Rsn5-NPF1 and Rfp2-NPF2 with similar weak affinity, yielding Ka values of 2*103 M−1 for both peptides (Table S1). We can thus conclude that it is a property of the EHD1 EH domain itself that enables discrimination between Rsn5-NPF1 and Rfp2-NPF2.
Accuracy in measuring thermodynamic parameters by ITC is highly dependent on following the binding process to completion. The usual quality control measure is the ‘c’-value or product of protein concentration and Ka (assuming n = 1). The affinity of Rsn5-NPF1 with EHD1 EH is in the ideal range for ITC experiments and thermodynamic parameters are expected to be accurate (c-values > 10). Rfp2-NPF2, on the other hand, binds 100-fold more weakly and the protein concentrations that would be required to run the reaction to completion are not practical. Although our measurements are reproducible, we recognize that the errors in the Rfp2-NPF2 datasets are likely to be larger, especially for ΔH and ΔS (47). A Ka value of 4.1*103 M−1 (25°C), measured in an NMR experiment, was reported recently for a peptide similar to Rfp2-NPF2 (31). This is virtually identical to our measurement, although the limitations regarding weak complex formation apply to NMR as well as ITC.
Binding is enthalpically favorable and entropically unfavorable
In an ITC experiment, the heat of reaction is measured directly and the data fitted to determine Ka. ΔG is obtained directly from the relationship ΔG = −RTlnKa, and the entropy of binding is calculated indirectly from ΔG = ΔH - TΔS. The change in heat capacity, ΔCp, can also be obtained from variation in ΔH with temperature (ΔCp= ∂ΔH/∂T). In order to understand the forces that drive binding of the NPF-containing peptides to EHD1 EH, we recorded a thermodynamic profile for each.
Figure 4B shows the thermodynamics of binding of Rsn5-NPF1 as a function of temperature. The reaction is strongly exothermic and drives the association whereas the entropic term, TΔS, is unfavorable. At 20°C, for example, a favorable ΔH of −11.3 kcal mol−1 is offset by an unfavorable TΔS term of −3.4 kcal mol−1. ΔG, the difference between these terms, is nevertheless still large and negative and complex formation is favorable. As the temperature is increased from 5 to 25°, the ΔH and TΔS terms largely compensate and lead to a ΔG value that is only mildly sensitive to temperature. Enthalpy-entropy compensation is very commonly observed in protein-ligand binding reactions (48, 49). In a control experiment, ΔH was shown to be independent of buffer ionization enthalpy (Figure S3) so we can conclude that uptake or release of H+ does not influence the binding affinity.
The difference in affinity between Rsn5-NPF1 and Rfp2-NPF2 is entropic
A comparable graph for Rfp2-NPF2 is shown in Figure 4C. It can be seen at once that ΔH values for the two peptides are very similar whereas values of TΔS are quite different; in this case at 20° the ΔH value of −11.9 kcal mol−1 is offset by a much more unfavorable TΔS term of −6.8 kcal mol−1, leading to the weaker binding of Rfp2-NPF2. This applies across the temperature range of the experiment, with a similar entropy-enthalpy compensation as was seen for Rsn5-NPF1. Thus although binding is entropically unfavorable in both cases, it is substantially less so for Rsn5-NPF1 and the affinity is correspondingly higher.
The enthalpic and entropic contributions to ΔG reflect the molecular forces that drive the association. Enthalpy is dominated by contributions from bond formation (e.g. H-bonds and van der Waals interactions). Entropic effects include the rearrangement of water and ions in the complex and restriction in the degrees of freedom of the backbone and sidechain conformations in both protein and peptide. As the difference in the energetics of the two complexes is almost entirely entropic, it seems likely that the difference in affinity is the result of their differing effects on the layers of partially ordered ions that form in response to changes in the surface potentials of protein and peptide. Similar effects have been noted for positively charged peptides and proteins binding to DNA (50).
These experiments also yield ΔCp, the change in heat capacity upon binding, which is obtained from the response of ΔH to temperature. ΔCp is potentially very informative as it reports on the burial (extent of hydration) of polar and non-polar surfaces, which have opposite signs. ΔCp for both peptides is negative and consistent with a net increase in buried apolar surface. This presumably corresponds to the phenylalanine and proline in the NPF motif and the numerous hydrophobic residues that form the NPF binding pocket on EHD1 EH. Direct comparison of the two peptides is more problematic because although ΔH data for Rsn5-NPF1 is a linear function of temperature over the range of these experiments, yielding a ΔCp value of −150 cal mol−1 K−1, the data for Rfp2-NPF2 is curved. This may just reflect systematic inaccuracies inherent in measuring data at low c value (47). If only the lower temperature data points for Rfp2-NPF2 are taken into consideration, the ΔCp values for both peptides are about the same.
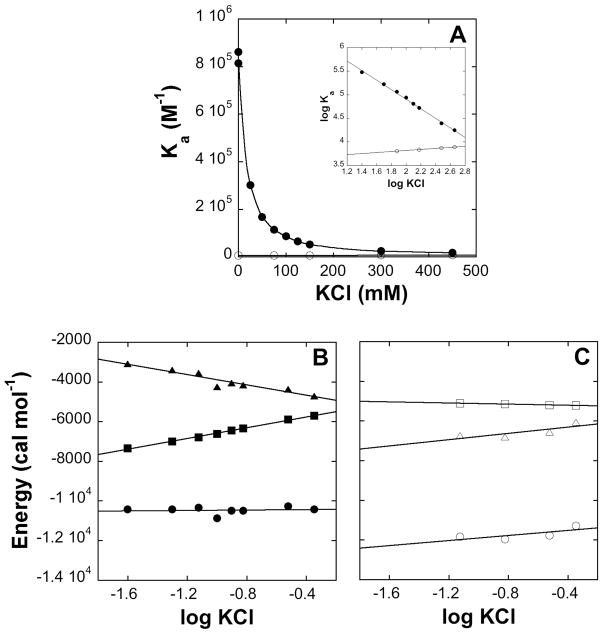
Increasing the salt concentration abolishes the difference in affinity
Electrostatic interactions are damped in solution due to screening of the charges and disruption of the long-range effects of the surface potential. We compared the effects of KCl on the thermodynamic profiles of both peptides (Figure 5, Table 1). Increasing the KCl concentration was found to have only a small effect on the binding of Rfp2-NPF2 (Figure 5A) with Ka showing a very slight increase. Rsn5-NPF1 on the other hand, showed a dramatic reduction in affinity as the ionic strength increased and the Ka value approached that of Rfp2-NPF2 at high salt (450 mM KCl). As is characteristic of electrostatic interactions (51), a plot of log Ka vs log KCl is linear (see inset). The thermodynamic profile of Rsn5-NPF1 (Figure 5B) shows that the enthalpy of binding is independent of salt concentration and that the reduction in affinity is entirely the result of a less favorable entropy of binding. This, in turn, makes ΔG less negative, which translates into a smaller association constant. The thermodynamic parameters of Rfp2-NPF2 binding (Figure 5C) are less affected by salt. These experiments taken together provide good evidence that interaction between one or more of the charged residues following the NPF motif with the positively-charged surface near the NPF binding pocket is responsible for the greater affinity of Rsn5-NPF1.
Figure 5.
(A) Dependence of the affinity of Rsn5-NPF1 (solid circles) and Rfp2-NPF2 (open circles) on KCl concentration. Inset: log-log plot showing that both sets of data are essentially linear. (B) Thermodynamic profile of Rsn5-NPF1 binding to EHD1 EH as a function of KCl concentration. ΔH is in solid circles, TΔS in solid triangles and ΔG in solid squares. (C) Thermodynamic profile of Rfp2-NPF2 binding to EHD1 EH as a function of KCl concentration. ΔH is in open circles, TΔS is in open triangles and ΔG is in open squares.
In the case of DNA binding proteins, polyelectrolyte theory has been used to analyze the salt dependence of the association constant in order to separate electrostatic from non-electrostatic contributions to the free energy and to calculate the number of ion pairs formed. The association constant Ka is given:
| (1) |
where (Ka)ne is the non-electrostatic component and n is the number of ion pairs formed upon complexation (50–52). At a KCl value of 1 M, the second term of equation (1) vanishes and Ka =(Ka)ne. Our data yield a (Ka)ne of 7.8 *103 M−1 which is very close to the binding constant of Rfp2-NPF2 in the absence of salt (6.6 *103 M−1; Table 1). At 20°C, this translates into a non-electrostatic ΔG value of −5200 cal mol−1, so we can say that about two thirds of the free energy of binding of Rsn5-NPF1 is contributed by the NPF motif and the remaining third by electrostatic interactions involving the charged residues. Because electrostatic complex formation is essentially an exchange of counterions in solution for charges on the ligand (see Discussion), the slope of the line, n, translates into the number of ion pairs formed. In this case, n=1.0 which is considerably smaller than the number of acidic residues at the terminus of Rsn5-NPF1 with the potential for ion pair formation. Thus one residue of the peptide could be significantly more important than the others, or all residues may contribute partially in a dynamic complex.
A weakly binding NPF-containing sequence is converted to a strongly binding NPF-containing sequence by progressive substitution with acidic amino acids
Rabenosyn-5 contains 6 NPF motifs (Figure 1A,C) that are well conserved among vertebrate species. Several of these sequences are followed by multiple negative charges in the manner of NPF1 (Figure 1C) and all of the NPF motifs in Rabenosyn-5 have at least one negative charge in the four residues following the phenylalanine. We synthesized a further series of 11 residue peptides corresponding to motifs 2, 3 and 4 (designated Rsn5-NPF2 etc.) as a representative set of the remaining Rabenosyn-5 NPF motifs (Figure 1C) and measured their affinity for EHD1 EH. All of the Rabenosyn-5 peptides bound more tightly than Rfp2-NPF2, and furthermore, the greater the number of negative charges following the NPF motif, the more tightly the peptide bound (Figure 6A). The weakest binding peptide, Rsn5-NPF3, is the least negative. Rsn5-NPF3 has neutral residues in the +1, +2 and +3 positions following the NPF, and an aspartate residue in the +4 position. It binds with a Ka of only 1.40*104 M−1.
Figure 6.
(A) Affinity of the first 4 NPF motifs of Rabenosyn-5 for EHD1 EH in 25 mM MOPS pH 7.0, 1 mM CaCl2, 20°C measured by ITC. (B) Systematic conversion of Rsn5-NPF3 into a high affinity peptide by systematic replacement of neutral residues with acidic residues in the positions following the NPF sequence. Error bars represent the standard deviation between two measurements.
Rsn5-NPF3 was next used to test the positional contribution of the negative charges following the NPF motif. A series of peptides was synthesized in which neutral residues in the +3, +2 and +1 positions following the phenylalanine were successively replaced with aspartate or glutamate such that the C-terminus of Rsn5-NPF3 was ultimately made identical to that of Rsn5-NPF1. It was found (Figure 6B) that the affinity for EHD1 EH was enhanced 3 to 4-fold for each negative charge following the NPF motif and that the final peptide in the series, which has 4 negative charges, binds almost as well as Rsn1-NPF1. Thus all four positions following the NPF sequence are important, and it is possible that a longer run of acidic residues, as is found for NPF2 in intact Rabenosyn-5, will result in even tighter binding. Furthermore, within the constraints of this experiment, the tight binding does not appear to depend on the residues N-terminal to the NPF sequence.
Rsn5-NPF1 does not interact with Eps15 EH2
Both EHD1 EH (pI = 6.5) and Eps15 EH2 (pI = 4.8) have a net negative charge at pH 7. Whereas EHD1 EH has a distinctive region of positive potential near the NPF binding pocket, Eps15 EH2 is overwhelmingly negative (26, 31) which immediately suggests a mechanism for selectivity on the basis of charge (39). Indeed, even when the protein concentration in the ITC cell was increased to 0.5 mM, we were unable to detect any interaction between Rsn5-NPF1 and Eps15 EH2. The surface potentials of the EH domains are thus an important aspect of their structure and contribute to selectivity.
Lysine sidechain chemical shifts are differentially influenced by Rsn5-NPF1 and Rfp2-NPF2
Binding of an Rfp2-NPF2-like peptide has been shown to have very little effect on the conformation of the backbone amides of EHD1 EH (31). Our NMR experiments (Figure 2) suggest that the Rsn5-NPF1 and Rfp2-NPF2 complexes are structurally similar and our thermodynamic experiments have shown that the difference in affinity between the two peptides is an entropic effect resulting from charge-charge interactions between Rsn5-NPF1 and a region of positive potential at the protein surface. In order to see if we could detect this interaction directly, we used direct-observe 15N NMR spectroscopy to look at the nitrogen-containing sidechains of the EH domains in more detail.
15N is an insensitive nucleus due to its low gyromagnetic ratio. NMR methods that improve the sensitivity by transfer of magnetization to an attached proton (e.g. INEPT and HSQC) are unsuitable for primary amino groups due to rapid proton exchange with the solvent water. Nevertheless, aided by a large negative NOE, we found that a sample containing 15 mg protein was sufficient to obtain a good spectrum in an overnight run (Figure 7).
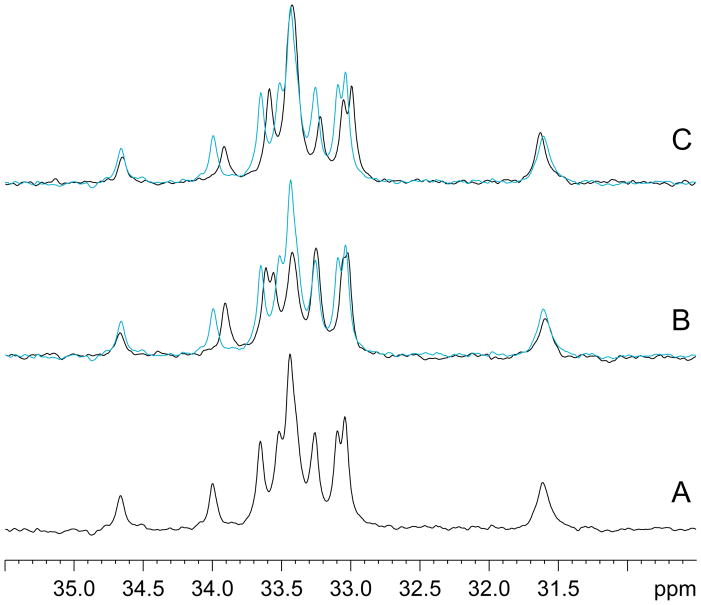
Figure 7.
30MHz proton decoupled 15N NMR spectra of the lysine region of EHD1 EH. (A) 2.5 mM protein with no added peptide; (B) complex with Rsn5-NPF1 (final concentration of 3 mM); (C) complex with Rfp2-NPF2 (final concentration of 5 mM). In (B) and (C) the uncomplexed protein is shown in cyan. The lysine ε-amino resonances are negative with respect to the amide proton envelope and have been inverted for ease of viewing.
EHD1 EH contains 13 lysine, 4 histidine, and 2 arginine residues, all of which have sidechains that are clearly visible in the 15N spectrum (Figure S4). Lysine residues K468, K469, K483 and K486 are primarily responsible for the region of positive potential near the NPF binding pocket (Figure 8). The ε-amino region of the proton-decoupled 15N spectrum of isotopically labeled EHD1 EH in the absence of peptide is shown in Figure 7A. There is significant chemical shift dispersion among the resonances, showing that they are subject to different environmental influences. As far as can be ascertained, given the degree of resonance overlap, all peaks experience a large negative NOE. This is typical and demonstrates the high mobility (≪ τc) of the ε-amino group. Although a wealth of information is potentially available from a detailed study of these 15N spectra, given that the resonances are unassigned and that the signal in the absence of the NOE is not strong enough for spectral deconvolution, we have limited ourselves to a qualitative description of the spectral changes induced by saturating amounts of the two peptides. Complete saturation was confirmed from the appearance of the HSQC spectra.
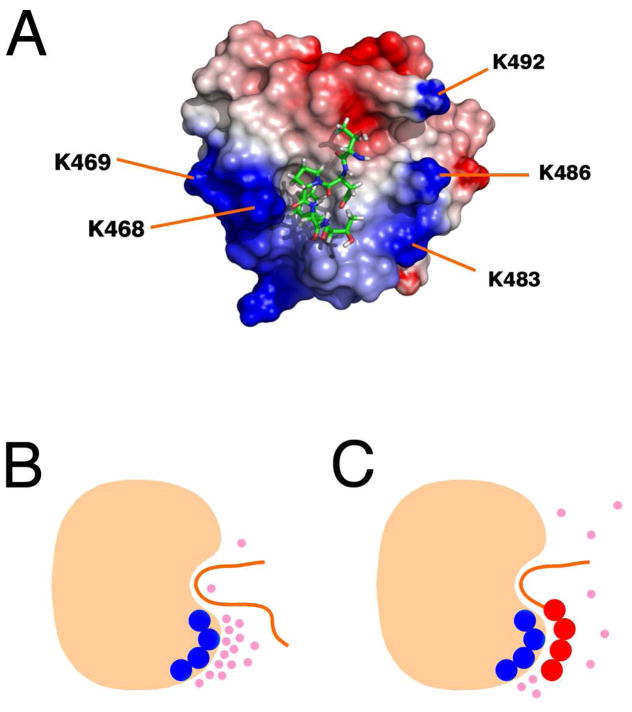
Figure 8.
(A) Structure of EHD1 EH bound to the pentapeptide TNPFT using coordinates from PDB entry 2KFF (31). The N-terminal threonine residue is at the top near the Ca2+-binding loop, and the C-terminal threonine is at the bottom. K483 and K486 are positioned very favorably for interaction with the C-terminal end of the peptide. The surface potential of the EH domain was calculated using DELPHI (62) and rendered using Pymol (www.pymol.org). (B and C) Schematic showing how displacement of ordered anions by Rsn5-NPF1 (C) but not Rfp2-NPF2 (B) can lead to a more favorable entropic component to the free energy of binding in the case of an acidic ligand. The peptide (orange) is shown in the binding pocket of the protein (beige). Ions are indicated by pink spheres, negative charges on the peptide by red circles and positive charges on the protein by dark blue circles.
Figure 7B shows that addition of the neutral peptide, Rfp2-NPF2, has relatively little effect on the overall shape of the spectral envelope, although some residues undergo small downfield shifts. In general, the intensities of the resonances remain much the same. Rsn5-NPF1, by contrast, shows a significant reduction in the intensity of several of the central peaks, indicating that the NOE becomes less negative for a subset of the lysine residues. This is presumably the result of motional restriction and shows that the aspartate and glutamate residues of the peptide exert a significant influence. It has been noted previously (30, 31) that a lysine residue (K468 in the case of EHD1 EH) has the potential to hydrogen bond with the carbonyl of the proline in the NPF motif. This interaction would be expected to be similar in both our peptides.
Surface potential effects may be more important than specific ion pair formation
We wished to determine which lysine sidechains are close enough to a bound Rsn5-NPF1 peptide to be important in the interaction. The complex in Figure 8A shows EHD1 EH with a TNPFT peptide in the NPF binding pocket (31). This is the central portion of Rfp2NPF2 (Ac-YESTNPFTAK-NH2). The peptide is positioned with the N-terminal threonine at the top nearest to the Ca2+ binding loop and the C-terminal threonine, which corresponds to D8 in Rsn5-NPF1, at the bottom. Residues K483 and K486 of helix 3 line a channel that seems well positioned to accommodate the terminal acidic residues of Rsn5-NPF1. Alternatively, K468 and K469 form a positive patch that could lead the polypeptide chain towards helix 2 (to the left in Figure 8). Although it is tempting to pair up specific lysine residues on the protein with aspartic or glutamic acid residues on the peptide, it is probably preferable to think in terms of interaction between potential surfaces. This enables us to accommodate the thermodynamic measurements, which are explainable only in terms of the entropy of the complete system, and allows us to incorporate the potential for dynamics within the complex.
This view is illustrated schematically in Figures 8B and 8C. Layers of ordered ions (pink spheres) that are attracted to the protein surface in the absence of peptide are forced to redistribute when negatively-charged peptide is present owing to attenuation of the local field. This generates the experimentally observed positive (favorable) component to the entropy of binding for Rsn5-NPF1 (Figure 8C) that is not available to Rfp2-NPF2 (Figure 8B). Because the surface between the charged regions is never dehydrated, the enthalpic contribution remains small. The association between the acidic ‘tail’ of Rsn5-NPF1 and the EH domain may thus be quite dynamic and the contribution of the numerous lysine residues may well be additive. In addition to explaining the thermodynamics of the interaction, this model is consistent with the NMR experiments in which relatively modest changes in the 15N chemical shifts of the lysine ε-amino groups were observed when Rsn5-NPF1 bound. Furthermore, we showed with successive replacement of the acidic residues at the C-terminus, that they make approximately equal contributions to the affinity.
A high affinity EHD binding site is predicted in three new proteins
Negative charges following one or more NPF motifs have been noted in other established EHD binding proteins such as PACSIN (syndapin), SNAP29 and EHBP1 (39) and the experiments described herein provide a firm biophysical demonstration of their importance. Preliminary experiments in our laboratory (J. V. Dineen & J. D. Baleja, unpublished data) have also shown that EHDs 2–4 have similar binding preferences to those described here for the EH domain of EHD1. We can thus feel reasonably confident in defining an extended NPF-containing motif that is capable of preferentially selecting the EHD proteins over other EH domains and in using this motif to search for novel EHD binding partners.
Our criteria for selecting a viable candidate were (i) that the protein should possess the sequence N-P-F-[DE]-[DE]-[DE]; (ii) that the sequence should be conserved among vertebrate species and (iii) that the protein should be connected with endocytosis, exocytosis or vesicle trafficking. The program PATTINPROT was used to search the Uniprot protein knowledgebase and uncovered 11 matches in 9 proteins (Table 2). Four of these (Rabenosyn-5, PACSINs-1 and -2 and SNAP-29) were expected and a further two (Pygopus homolog-2 and makorin-4) have no obvious connection to endocytosis. The remaining 3 proteins, exocyst component 8 (EXOC8/Exo84), AP2-associated protein kinase 1 (AAK1) and MICAL-L1 are all associated with vesicle trafficking pathways and appear to be excellent candidates. MICAL-L1 is a Rab binding protein and was identified as a bona fide EHD1 target during the preparation of this manuscript (53).
Table 2.
Human proteins found by PATTINPROT that contain the sequence N-P-F-[DE]-[DE]-[DE]
| Protein | Sequence | Residue numbers | Accession number | Total NPF | Reference |
|---|---|---|---|---|---|
| Rabenosyn-5 | mpqqhegpsl_NPFDEE_dlsspmeeat | 626–631 | Q9H1K0 | 6 | (25) |
| dpsarilkey_NPFEEE_deeeeavagn | 662–667 | ||||
| PACSIN1 (syndapin) | fggsetngga_NPFEDD_skgvrvraly | 379–384 | Q9BY11 | 2 | (17) |
| PACSIN2 | aqsaqsqssy_NPFEDE_ddtgstvsek | 362–367 | Q9UNF0 | 3 | |
| fsstdangds_NPFDDD_atsgtevrvr | 417–422 | ||||
| SNAP29 | msaypksy_NPFDDD_gedegarpap | 9–14 | O95721 | 1 | (15) |
| MICAL-L1 | ptasleskpy_NPFEEE_eedkeeeapa | 425–430 | Q8N3F8 | 2 | (53) |
| AP2-associated protein kinase 1 | vynpsegstw_NPFDDD_nfskltaeel | 696–701 | Q2M2I8 | 1 | |
| Exocyst complex component 8 | gppqvtskat_NPFEDD_eeeepavpev | 310–315 | Q8IYI6 | 1 | |
| Pygopus homolog 2 | tpmvdhlvas_NPFEDD_fgapkvgvaa | 76–81 | Q9BRQ0 | 1 | |
| Makorin-4 | weffeegans_NPFDDE_eeavtfelge | 465–470 | Q13434 | 1 | |
Discussion
Target selection in vesicle trafficking and cell signaling cascades is frequently mediated through recognition of a short linear sequence by a globular protein domain. Well-known examples include binding of phosphotyrosine-containing motifs by SH2 domains, polyproline (PxxP) sequences by SH3 domains and YxxΦ and other motifs by the AP2 clathrin adapter (54, 55). Many of these interactions have been studied in detail, from structural, thermodynamic and kinetic points of view and the contributions made by each amino acid evaluated on a quantitative basis (56). The EH domain-NPF motif interaction, by contrast, has received relatively little quantitative treatment, possibly because EH-NPF interactions are traditionally thought of as weak and therefore difficult to measure. In this series of experiments, we showed (i) that EHD1 EH can bind tightly to NPF-containing sequences; (ii) that negatively charged residues following the NPF sequence are necessary for tight binding; (iii) that this extended interaction motif enables EHD binding proteins such as Rabenosyn-5 to discriminate between different EH domains such as those of Eps15 and Reps1. A region of positive potential near the NPF binding pocket on EHD1 EH is presumed to be critical in the interaction.
By comparing sequences derived from two EHD1-interacting proteins, Rabenosyn-5 and Rab11-Fip2, we were able to obtain a great deal of insight into the mechanism that underlies tight binding. Although the structures of the two complexes appear to be similar (on the basis of almost identical HSQC spectra), the energetics of interaction are different. Isothermal titration calorimetry showed that the negatively-charged peptide derived from Rabenosyn-5 (Rsn5-NPF1) bound with close to micromolar affinity and two orders of magnitude more tightly than a neutral peptide derived from Rab11-Fip2. Thermodynamic profiling showed that all of this difference is accounted for in the entropy of binding; although the entropy is unfavorable in both cases, it is substantially less unfavorable for Rsn5-NPF1 than it is for Rfp2-NPF2. High salt concentration abolishes the difference in affinity between the peptides and demonstrates the importance of electrostatics in Rsn5-NPF1 binding. Once again, the process is entropic. The ΔCp value is dominated by the common NPF motif and the difference between the peptides is small. We conclude that whereas Rfp2-NPF2 contacts the EH domain simply via the three residues of its NPF motif (31), Rsn5-NPF1 in addition uses the negative charges of the aspartic and glutamic acid residues that follow the NPF to contact a region of positive surface potential near the NPF binding pocket. This latter interaction is purely electrostatic in nature and contributes to both the affinity and specificity of binding.
There are strong parallels in the thermodynamic signatures of the electrostatic component of the interaction of Rsn5-NPF1 with EHD1 EH and that of certain positively-charged peptides and proteins binding to DNA (50, 52, 57, 58). These interactions are understood very well due to their importance in the recognition, specificity and affinity of transcription factors and other DNA-binding proteins for DNA. Interactions that are (i) purely entropic; (ii) show strong salt dependence which is also entropic; and (iii) have small values of ΔCp, are the characteristics of DNA binding by cations such as Mg2+, polyamines and the unstructured positively-charged ‘tails’ of proteins such as Cro and the lac repressor (50, 57, 59). The uniform charge density of DNA renders these charge-charge interactions non-specific, but clearly if the surface charge can vary, as it does with proteins, electrostatic interaction can contribute both to specificity and affinity.
These experiments are dependent on the use of short peptides as models for an intact protein, and it is reasonable to ask if this approach is valid. Typically, NPF motifs occur in regions with strong predicted propensity for disorder and Rabenosyn-5 fits comfortably into this category (Figure 1A). This is a reasonable basis for confidence; however direct evidence is obtained from the near perfect correlation of our binding constant data (Figure 6A) with the results of a series of in vivo mutagenesis experiments on whole Rabenosyn-5 by Caplan and coworkers (25). This study showed that NPF motifs 1 and 2 are most important for binding the EH domain, NPF4 to be of moderate influence and NPF3 to have no effect at all, precisely in accordance with the peptide affinities. The NPF region of Rab11-Fip2 has a smaller propensity for disorder (Figure 1C) and furthermore, it is a dimer (44, 45). Nevertheless, peptide affinities are still in agreement with mutagenesis experiments that have used intact proteins (7, 24).
Although peptides have proven to be useful models for intact Rabenosyn-5 and Rab11-Fip2, it would be naïve to suppose that they deliver a complete picture. EHD proteins are dimers and can presumably bind two NPF motifs simultaneously. If both motifs were contributed by the same target molecule, then for entropic reasons this would substantially increase the binding affinity. In addition, the NPF motif must displace the GPF motif from its binding pocket if complex formation is to occur (31). It would obviously be desirable in future experiments to measure affinities of the intact proteins.
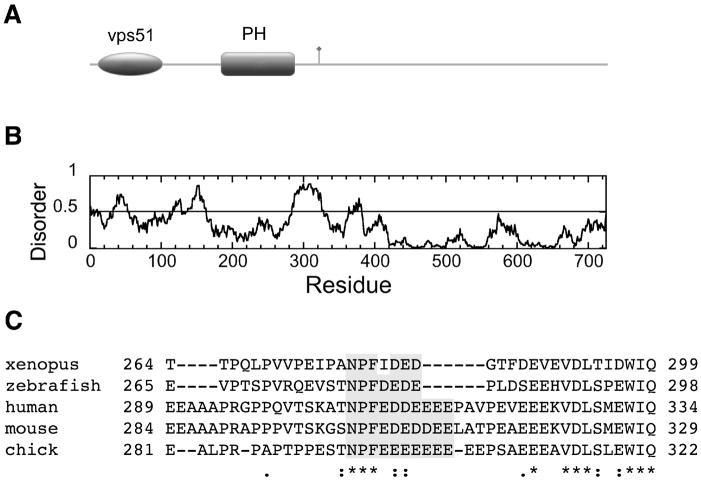
Having established an extended binding motif (N-P-F-[DE]-[DE]-[DE]) that is selective for the EH domains of the EHD proteins, we searched for novel occurrences of the sequence in proteins that play a role in vesicle trafficking pathways. Three good candidates emerged: exocyst component 8 (EXOC8), AAK1 and MICAL-L1 and this last protein was very recently verified experimentally (53). A fourth promising vesicle trafficking protein, vps53, which is part of the GARP complex, contains a slightly relaxed version of the EHD binding sequence (N-P-F-L-D-E-D). These potential interaction partners are important, because if experimentally validated, they will establish new roles and sites of action for the EHD proteins. The exocyst, for example, is important in abscission (the last step of cytokinesis), GLUT4 transport, provision of membranous material for phagocytosis and cell migration, and maintenance of polarity of polarized cell membranes (60). EXOC8 binds the small GTPase RalA through a pleckstrin homology domain (61) and contains a single well-conserved NPF motif in a region of predicted structural disorder (Figure 9) that should capable of interaction with an EH domain. If EXOC8 proves to be a genuine binding partner, then this establishes an important connection with between EHD proteins, exocytosis and the Ral signaling network.
Figure 9.
Domain structure of human EXOC8, a putative novel EHD binding partner. The N-terminal vps51 domain is found in a number of vesicle sorting proteins, notably the GARP (Golgi-associated retrograde protein) complex. The pleckstrin homology (PH) domain binds to RalA (61). The NPF motif is indicated with a flag. (B) Order/disorder prediction using IUPRED (http://iupred.enzim.hu/). Regions of potential disorder score >0.5. (C) ClustalW alignment shows that the NPF motif and following acidic residues (shaded gray) of EXOC8 are conserved from fish to humans (accession numbers: human, Q8IYI6; chick Q5ZJ43; zebrafish Q4RF58; xenopus Q5U247; mouse Q6PGF7).
We began this study by asking how EH domains discriminate between the numerous potential NPF-containing binding partners at their disposal. Doubtless, temporal and spatial restrictions in a cellular environment play a significant part, however a number of different selective strategies employed by nature are beginning to emerge. Our own experiments have shown that the interaction motif itself can be extended with concomitant increase in the binding affinity. Limited experiments with the EH domain of Reps1 and the EH2 domain of Eps15 showed that the extended motif was also highly selective for the EH domains of EHD proteins. A different method of increasing NPF-EH domain affinity was discovered in Eps15 (33). EH2 of Eps15 binds tightly to its partner stonin2 by using two NPF motifs; one bound in the canonical pocket formed between helices 2 and 3, and a second in a novel pocket on the other side of the protein. EHD1 EH, in fact, contains a similar pocket, but preliminary experiments using a Rabenosyn-5 peptide containing two NPF motifs resulted only in the binding of 2 EH domains at normal affinity (D. Corrigan and J. Baleja, unpublished work). A third way of enhancing EH-NPF affinity is to present the NPF motif as a preformed β-turn which reduces the unfavorable loss of configurational entropy upon binding. This strategy may be employed by myoferlin (23), which contains an NPF motif as part of a structured C2 domain. Lastly, it is plausible that the EH domains of EHD proteins will bind tightly to NPF motif followed by phosphorylated serine or threonine residues. The NPF motifs of Rab11-Fip2 are in a serine/threonine rich region of the protein and the peptide Rfp2-NPF2 contains an NPF motif that is followed directly by a threonine residue. This is one possible explanation why genetic and cell-based assays identify Rab11-Fip2 as a bona fide EHD target whereas the in vitro experiments described here suggest binding would be relatively weak and non-selective.
Supplementary Material
Acknowledgments
We thank Dr. Barth Grant for generously providing the EHD1 gene and Patrick Magoon for carrying out some early NMR titrations.
Footnotes
Abbreviations: CD, circular dichroism; Eps15, epidermal growth factor phosphorylation substrate 15; EH, Eps15 homology; EHD, EH domain-containing; EXOC8, exocyst component 8, also known as exo84; GST, glutathione-S-transferase; Hrb1, HIV Rev binding protein 1, also known as RAB; ITC, isothermal titration calorimetry; NMR, nuclear magnetic resonance; NPF aspargine-proline-phenylalanine; Rab11-Fip2, Rab11 family interaction protein 2;
Supported in part by NIH Grant GM067985.
Supporting Information Available
An example of an HSQC titration of EHD1 EH, verification the ITC data is independent of the buffer ionization constant, CD spectroscopy of the peptides, the complete 15N NMR spectrum of EHD1 EH, and a table summarizing EH binding constant data is provided as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Polo S, Confalonieri S, Salcini AE, Di Fiore PP. EH and UIM: endocytosis and more. Sci STKE 2003. 2003:re17. doi: 10.1126/stke.2132003re17. [DOI] [PubMed] [Google Scholar]
- 2.Fazioli F, Minichiello L, Matoskova B, Wong WT, Di Fiore PP. Eps15, a novel tyrosine kinase substrate, exhibits transforming activity. Mol Cell Biol. 1993;13:5814–5828. doi: 10.1128/mcb.13.9.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong WT, Schumacher C, Salcini AE, Romano A, Castagnino P, Pelicci PG, Di Fiore PP. A protein-binding domain, EH, identified in the receptor tyrosine kinase substrate Eps15 and conserved in evolution. Proc Natl Acad Sci U S A. 1995;92:9530–9534. doi: 10.1073/pnas.92.21.9530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salcini AE, Confalonieri S, Doria M, Santolini E, Tassi E, Minenkova O, Cesareni G, Pelicci PG, Di Fiore PP. Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11:2239–2249. doi: 10.1101/gad.11.17.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benmerah A, Bayrou M, Cerf-Bensussan N, Dautry-Varsat A. Inhibition of clathrin-coated pit assembly by an Eps15 mutant. J Cell Sci. 1999;112(Pt 9):1303–1311. doi: 10.1242/jcs.112.9.1303. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi A, Urano T, Goi T, Feig LA. An Eps homology (EH) domain protein that binds to the Ral-GTPase target, RalBP1. J Biol Chem. 1997;272:31230–31234. doi: 10.1074/jbc.272.50.31230. [DOI] [PubMed] [Google Scholar]
- 7.Cullis DN, Philip B, Baleja JD, Feig LA. Rab11-FIP2, an adaptor protein connecting cellular components involved in internalization and recycling of epidermal growth factor receptors. J Biol Chem. 2002;277:49158–49166. doi: 10.1074/jbc.M206316200. [DOI] [PubMed] [Google Scholar]
- 8.Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat Cell Biol. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- 9.Lin SX, Grant B, Hirsh D, Maxfield FR. Rme-1 regulates the distribution and function of the endocytic recycling compartment in mammalian cells. Nat Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- 10.Paoluzi S, Castagnoli L, Lauro I, Salcini AE, Coda L, Fre S, Confalonieri S, Pelicci PG, Di Fiore PP, Cesareni G. Recognition specificity of individual EH domains of mammals and yeast. EMBO J. 1998;17:6541–6550. doi: 10.1093/emboj/17.22.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enmon JL, de Beer T, Overduin M. Solution structure of Eps15’s third EH domain reveals coincident Phe-Trp and Asn-Pro-Phe binding sites. Biochemistry. 2000;39:4309–4319. doi: 10.1021/bi9927383. [DOI] [PubMed] [Google Scholar]
- 12.Chen H, Fre S, Slepnev VI, Capua MR, Takei K, Butler MH, Di Fiore PP, De Camilli P. Epsin is an EH-domain-binding protein implicated in clathrin-mediated endocytosis. Nature. 1998;394:793–797. doi: 10.1038/29555. [DOI] [PubMed] [Google Scholar]
- 13.Martina JA, Bonangelino CJ, Aguilar RC, Bonifacino JS. Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J Cell Biol. 2001;153:1111–1120. doi: 10.1083/jcb.153.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith CA, Dho SE, Donaldson J, Tepass U, McGlade CJ. The cell fate determinant Numb interacts with EHD/Rme-1 family proteins and has a role in endocytic recycling. Mol Biol Cell. 2004;15:3698–3708. doi: 10.1091/mbc.E04-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y, Shi H, Wei S, Wong SH, Hong W. Mutually exclusive interactions of EHD1 with GS32 and syndapin II. Mol Membr Biol. 2004;21:269–277. doi: 10.1080/09687680410001716871. [DOI] [PubMed] [Google Scholar]
- 16.Mishra SK, Keyel PA, Hawryluk MJ, Agostinelli NR, Watkins SC, Traub LM. Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J. 2002;21:4915–4926. doi: 10.1093/emboj/cdf487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun A, Pinyol R, Dahlhaus R, Koch D, Fonarev P, Grant BD, Kessels MM, Qualmann B. EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol Biol Cell. 2005;16:3642–3658. doi: 10.1091/mbc.E05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan JR, Prasad K, Jin S, Augustine GJ, Lafer EM. Eps15 homology domain-NPF motif interactions regulate clathrin coat assembly during synaptic vesicle recycling. J Biol Chem. 2003;278:33583–33592. doi: 10.1074/jbc.M304346200. [DOI] [PubMed] [Google Scholar]
- 19.Haffner C, Takei K, Chen H, Ringstad N, Hudson A, Butler MH, Salcini AE, Di Fiore PP, De Camilli P. Synaptojanin 1: localization on coated endocytic intermediates in nerve terminals and interaction of its 170 kDa isoform with Eps15. FEBS Lett. 1997;419:175–180. doi: 10.1016/s0014-5793(97)01451-8. [DOI] [PubMed] [Google Scholar]
- 20.Doria M, Salcini AE, Colombo E, Parslow TG, Pelicci PG, Di Fiore PP. The Eps15 homology (EH) domain-based interaction between Eps15 and Hrb connects the molecular machinery of endocytosis to that of nucleocytosolic transport. J Cell Biol. 1999;147:1379–1384. doi: 10.1083/jcb.147.7.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fernandez-Chacon R, Achiriloaie M, Janz R, Albanesi JP, Sudhof TC. SCAMP1 function in endocytosis. J Biol Chem. 2000;275:12752–12756. doi: 10.1074/jbc.275.17.12752. [DOI] [PubMed] [Google Scholar]
- 22.Guilherme A, Soriano NA, Furcinitti PS, Czech MP. Role of EHD1 and EHBP1 in perinuclear sorting and insulin-regulated GLUT4 recycling in 3T3-L1 adipocytes. J Biol Chem. 2004;279:40062–40075. doi: 10.1074/jbc.M401918200. [DOI] [PubMed] [Google Scholar]
- 23.Doherty KR, Demonbreun AR, Wallace GQ, Cave A, Posey AD, Heretis K, Pytel P, McNally EM. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J Biol Chem. 2008;283:20252–20260. doi: 10.1074/jbc.M802306200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Naslavsky N, Boehm M, Backlund PS, Jr, Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Beer T, Carter RE, Lobel-Rice KE, Sorkin A, Overduin M. Structure and Asn-Pro-Phe binding pocket of the Eps15 homology domain. Science. 1998;281:1357–1360. doi: 10.1126/science.281.5381.1357. [DOI] [PubMed] [Google Scholar]
- 27.Koshiba S, Kigawa T, Iwahara J, Kikuchi A, Yokoyama S. Solution structure of the Eps15 homology domain of a human POB1 (partner of RalBP1) FEBS Lett. 1999;442:138–142. doi: 10.1016/s0014-5793(98)01644-5. [DOI] [PubMed] [Google Scholar]
- 28.Kim S, Cullis DN, Feig LA, Baleja JD. Solution structure of the Reps1 EH domain and characterization of its binding to NPF target sequences. Biochemistry. 2001;40:6776–6785. doi: 10.1021/bi002700m. [DOI] [PubMed] [Google Scholar]
- 29.Kieken F, Jovic M, Naslavsky N, Caplan S, Sorgen PL. EH domain of EHD1. J Biomol NMR. 2007;39:323–329. doi: 10.1007/s10858-007-9196-0. [DOI] [PubMed] [Google Scholar]
- 30.de Beer T, Hoofnagle AN, Enmon JL, Bowers RC, Yamabhai M, Kay BK, Overduin M. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- 31.Kieken F, Jovic M, Tonelli M, Naslavsky N, Caplan S, Sorgen PL. Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1. Protein Sci. 2009;18:2471–2479. doi: 10.1002/pro.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK. Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273:31401–31407. doi: 10.1074/jbc.273.47.31401. [DOI] [PubMed] [Google Scholar]
- 33.Rumpf J, Simon B, Jung N, Maritzen T, Haucke V, Sattler M, Groemping Y. Structure of the Eps15-stonin2 complex provides a molecular explanation for EH-domain ligand specificity. EMBO J. 2008;27:558–569. doi: 10.1038/sj.emboj.7601980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee DW, Zhao X, Scarselletta S, Schweinsberg PJ, Eisenberg E, Grant BD, Greene LE. ATP binding regulates oligomerization and endosome association of RME-1 family proteins. J Biol Chem. 2005;280:17213–17220. doi: 10.1074/jbc.M412751200. [DOI] [PubMed] [Google Scholar]
- 35.Blume JJ, Halbach A, Behrendt D, Paulsson M, Plomann M. EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp Cell Res. 2007;313:219–231. doi: 10.1016/j.yexcr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Daumke O, Lundmark R, Vallis Y, Martens S, Butler PJ, McMahon HT. Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature. 2007;449:923–927. doi: 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- 37.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.George M, Ying G, Rainey MA, Solomon A, Parikh PT, Gao Q, Band V, Band H. Shared as well as distinct roles of EHD proteins revealed by biochemical and functional comparisons in mammalian cells and C. elegans. BMC Cell Biol. 2007;8:3. doi: 10.1186/1471-2121-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grant BD, Caplan S. Mechanisms of EHD/RME-1 protein function in endocytic transport. Traffic. 2008;9:2043–2052. doi: 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pant S, Sharma M, Patel K, Caplan S, Carr CM, Grant BD. AMPH-1/Amphiphysin/Bin1 functions with RME-1/EHD1 in endocytic recycling. Nat Cell Biol. 2009;11:1399–1410. doi: 10.1038/ncb1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eathiraj S, Pan X, Ritacco C, Lambright DG. Structural basis of family-wide Rab GTPase recognition by Rabenosyn-5. Nature. 2005;436:415–419. doi: 10.1038/nature03798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindsay AJ, McCaffrey MW. Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J Biol Chem. 2002;277:27193–27199. doi: 10.1074/jbc.M200757200. [DOI] [PubMed] [Google Scholar]
- 44.Jagoe WN, Lindsay AJ, Read RJ, McCoy AJ, McCaffrey MW, Khan AR. Crystal structure of Rab11 in complex with Rab11 family interacting protein 2. Structure. 2006;14:1273–1283. doi: 10.1016/j.str.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 45.Wei J, Liu Y, Bose K, Henry GD, Baleja JD. Disorder and structure in the Rab11 binding domain of Rab11 family interacting protein 2. Biochemistry. 2009;48:549–557. doi: 10.1021/bi8020197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 48.Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 49.Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 50.Crane-Robinson C, Dragan AI, Privalov PL. The extended arms of DNA-binding domains: a tale of tails. Trends Biochem Sci. 2006;31:547–552. doi: 10.1016/j.tibs.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 51.Record MT, Jr, Lohman ML, De Haseth P. Ion effects on ligand-nucleic acid interactions. J Mol Biol. 1976;107:145–158. doi: 10.1016/s0022-2836(76)80023-x. [DOI] [PubMed] [Google Scholar]
- 52.Saecker RM, Record MT., Jr Protein surface salt bridges and paths for DNA wrapping. Curr Opin Struct Biol. 2002;12:311–319. doi: 10.1016/s0959-440x(02)00326-3. [DOI] [PubMed] [Google Scholar]
- 53.Sharma M, Panapakkam Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 Links EHD1 to Tubular Recycling Endosomes and Regulates Receptor Recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Evans PR, Owen DJ. Endocytosis and vesicle trafficking. Curr Opin Struct Biol. 2002;12:814–821. doi: 10.1016/s0959-440x(02)00395-0. [DOI] [PubMed] [Google Scholar]
- 55.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 56.McNemar C, Snow ME, Windsor WT, Prongay A, Mui P, Zhang R, Durkin J, Le HV, Weber PC. Thermodynamic and structural analysis of phosphotyrosine polypeptide binding to Grb2-SH2. Biochemistry. 1997;36:10006–10014. doi: 10.1021/bi9704360. [DOI] [PubMed] [Google Scholar]
- 57.Mascotti DP, Lohman TM. Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. Proc Natl Acad Sci U S A. 1990;87:3142–3146. doi: 10.1073/pnas.87.8.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dragan AI, Read CM, Makeyeva EN, Milgotina EI, Churchill ME, Crane-Robinson C, Privalov PL. DNA binding and bending by HMG boxes: energetic determinants of specificity. J Mol Biol. 2004;343:371–393. doi: 10.1016/j.jmb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 59.Takeda Y, Ross PD, Mudd CP. Thermodynamics of Cro protein-DNA interactions. Proc Natl Acad Sci U S A. 1992;89:8180–8184. doi: 10.1073/pnas.89.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- 61.Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J. 2005;24:2064–2074. doi: 10.1038/sj.emboj.7600699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.