Abstract
Breast cancer, like any other human cancer, results from the accumulation of mutations that deregulate critical cellular processes, such as cell proliferation and death. Activation of oncogenes and inactivation of tumor suppressor genes are common events during cancer initiation and progression and often determine treatment responsiveness. Thus, recapitulating these events in mouse cancer models is critical for unraveling the molecular mechanisms involved in tumorigenesis and for interrogating their possible impact on response to anticancer drugs. We have developed a novel mouse mammary epithelial cell model, which replicates the steps of epithelial tumor progression and takes advantage of the power of mouse genetics and the ability to assess three-dimensional morphogenesis in the presence of extracellular matrix to model human breast cancer.
1. Introduction
Mouse models have played an essential role in the study of human breast cancer. The first generation of such models involved studies on inbred mice and resulted in the discovery that MMTV-induced mammary tumorigenesis resulted from insertional inactivation of specific genes in the wnt and fgf families (Callahan and Smith, 2000). In addition, use of chemical carcinogens and hormones on inbred mice revealed the contribution of these factors to mammary tumorigenesis and provided model systems for prevention studies (Medina, 2006). The second generation of mouse breast cancer models involved constitutive overexpression of oncogenes, such as c-myc (Stewart et al., 1984), polyoma mt (Guy et al., 1992a), neu (Guy et al., 1992b), mutant p53 (Li et al., 1997), and cyclin D1 (Wang et al., 1994) targeted to the mammary gland. These models have provided valuable information on the role of single genes in mammary tumorigenesis but are limited by the leakiness and multi-tissue expression of the MMTV promoter and the pregnancy-type hormonal dependence of the whey acidic protein (WAP) and β-lactoglobulin promoters. There is also difficulty in controlling the level of oncogene overexpression, which is often activated constitutively throughout development, not likely representing the physiologic situation in human breast cancer. The third, and most recent, generation of mouse breast cancer models involves mammary gland–specific gene deletion and activation by use of technology such as the Cre-lox system and tetracycline-responsive transactivation to induce tissue-specific gene mutations in adult somatic tissue and, as such, provides the opportunity to more authentically model human cancer (Furth, 1997; Liu et al., 2007; Moody et al., 2002; Wijnhoven et al., 2005).
Mouse models have provided valuable information on the genetic events contributing to mammary tumorigenesis but are not easily amenable to the investigation of the biochemical and cell biological pathways involved in tumor formation. As an alternative to the use of epithelial cells cultured as monolayers on tissue culture plastic for such studies, three-dimensional (3D) culture systems have been developed and have proven very useful for interrogating the effects of oncogenes on glandular architecture and the role of epithelial-stroma interactions in mammary tumorigenesis (Bissell, 2007; Debnath and Brugge, 2005). A system extensively used in 3D-morphogenesis assays involves culture of the immortalized, nontransformed human mammary epithelial cell line MCF-10A on a reconstituted basement membrane (Debnath et al., 2003). In 3D-culture, MCF-10A cells form polarized acinar structures that resemble glandular epithelium in vivo and have been successfully used to investigate mechanisms involved in tumor initiation and progression ((Debnath and Brugge, 2005) and to perform functional screens for proteins implicated in breast cancer (Witt et al., 2006). However, MCF-10A cells are negative for estrogen receptor-alpha (ER-α), are cytogenetically abnormal with myc amplification and deletion of the locus containing p16 and p14ARF (Debnath et al., 2003), have been immortalized by events not well defined, and may not be adequately representative of the spectrum of premalignant human breast disease.
2. A Novel Mouse Mammary Epithelial Cell Model
2.1. Generation and characterization of the model
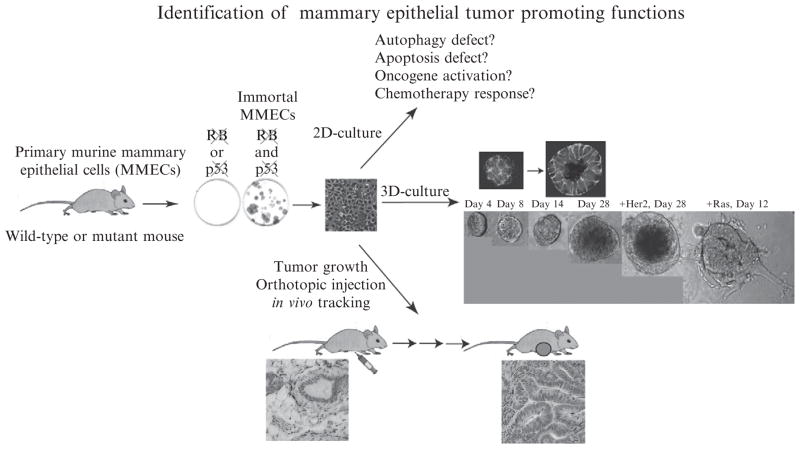
We have developed a novel mouse mammary epithelial cell model that complements and expands the applications of the mouse and human breast cancer models described earlier (Karantza-Wadsworth et al., 2007). Our model involves isolation and immortalization of primary mouse mammary epithelial cells by inactivation of the retinoblastoma and p53 pathways (Fig. 4.1), as previously described for mouse kidney epithelial cells (Degenhardt and White, 2006; Degenhardt et al., 2002a,b; Mathew et al., 2008).
Figure 4.1.
Identification of mammary epithelial tumor–promoting functions. A novel mouse mammary epithelial cell model for investigating the impact of oncogene activation and tumor suppressor inactivation on mammary tumorigenesis is summarized here. Primary mouse mammary epithelial cells (MMECs) are isolated from wild-type or mutant mice and immortalized (iMMECs) by concurrent inactivation of the Rb and p53 pathways. iMMECs can be grown and studied in standard two-dimensional (2D) in vitro culture. The functional properties of iMMECs can also be interrogated in a 3D morphogenesis assay, where iMMECs from wild-type mice initially form a solid acinus, as seen by β-catenin staining (green) to outline the cellperiphery and Dapi counterstaining (blue) for DNA. With time (14 to 21days), cell death in the acinar center creates a hollow lumen, closely mimicking duct formation in mammary epithelium in vivo. Oncogenic events perturb 3D morphogenesis, as illustrated in the phase-contrast images of the structures generated by Her2/neu-expressing iMMECs (larger, but hollow, acini) and by activated Ras-expressing iMMECs (nonacinar structures invading the basement membrane). iMMECs from wild-type mice are poorly, clonally tumorigenic on orthotopic transplantation into the mammary fat pad. Tumors formed by wild-type iMMECs are distinctly adenocarcinomas (right) compared with normal mouse mammary tissue (left), as indicated by histologic analysis of H&E–stained sections.
Immortalized mouse mammary epithelial cells (iMMECs) from wild-type mice exhibit typical cuboidal epithelial morphology in two-dimensional (2D) culture, form tight junctions, and express ER-α and luminal epithelial cell markers (Fig. 4.1) (Karantza-Wadsworth et al., 2007). In 3D-morphogenesis assays, iMMECs form polarized acini that generate lumens through apoptosis (Fig. 4.1), similarly to MCF-10A cells (Debnath et al., 2002; 2003), and secrete β-casein into the acinar lumen upon lactogenic stimulation (Karantza-Wadsworth et al., 2007). iMMECs are not tumorigenic, forming clonal adenocarcinomas with long latency, only after acquisition of secondary genetic or epigenetic changes (Fig. 4.1) (Karantza-Wadsworth et al., 2007). Expression of different oncogenes in iMMECs affects 3D morphogenesis (Fig. 4.1) and in vivo tumorigenicity in an oncogene-dependent manner (Karantza-Wadsworth et al., 2007). Therefore, wild-type iMMECs represent an early stage in mammary tumor-igenesis and provide a facile platform for studying the role of oncogenes (expressed individually or in combination) in breast cancer progression and treatment. iMMECs can also be used for investigating the role of tumor suppressors by knocking down gene expression by RNAi technology. Alternatively, the molecular mechanisms by which oncogenes and tumor suppressor genes contribute to mammary tumorigenesis and impact response to anticancer drugs can be studied by generating iMMECs from mutant mice of interest and further genetically manipulating them in vitro as needed. Furthermore, iMMECs can be used in coculture with wild-type or mutant mouse embryonic fibroblasts (MEFs) to investigate the role of epithelial–stroma interactions in breast cancer.
2.2. Studying the role of autophagy in mammary tumorigenesis
We recently reported the application of our model in the study of the role of autophagy in mammary tumorigenesis (Karantza-Wadsworth et al., 2007). iMMECs were generated from beclin1+/+ and beclin1+/− mice and studied in 2D culture, 3D morphogenesis, and in vivo tumorigenicity assays. We found that allelic loss of beclin1 compromises the autophagy potential of iMMECs and results in increased susceptibility of iMMECs to metabolic stress and accelerated lumen formation in mammary acini. Defective autophagy also activates the DNA damage response in iMMECs and mammary acini in vitro and in mammary tumors in vivo. Furthermore, we showed that monoallelic deletion of beclin1 promotes gene amplification in vitro and accelerates mammary tumorigenesis, supporting the hypothesis that autophagy limits metabolic stress to protect the genome, whereas autophagy defects increase DNA damage and genomic instability that may ultimately facilitate breast cancer progression (Karantza-Wadsworth and White, 2007; Karantza-Wadsworth et al., 2007). Similar results were obtained with beclin1+/− and atg5−/− kidney epithelial cells (Degenhardt et al., 2006; Mathew et al., 2007b), indicating that defective autophagy can promote genomic instability and epithelial tumor progression independent of cell type or means of autophagy inactivation. Thus, use of our novel mouse mammary epithelial cell model has provided valuable insight into the role of autophagy as a tumor suppressive mechanism in mammary tumor-igenesis and, together with parallel studies on kidney epithelial cells (Degenhardt et al., 2006; Mathew et al., 2007b), has set the foundation for understanding the complex interplay between autophagy, metabolic stress management, and cancer progression and for identifying ways to manipulate autophagy for maximum therapeutic benefit (Mathew et al., 2007a).
3. Protocols
This chapter provides detailed protocols for iMMEC generation and culture, 3D-morphogenesis assays, and in vivo tumorigenicity studies and will hopefully serve as a valuable addition to the technical repertoire used in breast cancer research.
3.1. Mouse mammary epithelial cell isolation and immortalization
3.1.1. Mouse mammary gland harvesting
Six- to 8-week-old female mice (wild-type or mutant, virgin or pregnant) are euthanized as required by the local Institutional Animal Care and Use Committee. Each mouse is then placed on its back on a corkboard, pinned in place through the feet, and swabbed with 70% ethanol. A ventral midline incision through the skin is made to expose the five pairs of subcutaneous mammary glands (Rasmussen et al., 2000). Additional incisions from the midline down each rear leg and below each ear facilitate the removal of the No. 4 and No. 1 fat pads, respectively. The skin flaps, with mammary glands attached, are carefully separated from the peritoneum with a blunt-edged instrument. The free edge of each skin flap is pinned to the corkboard, thereby exposing the adherent mammary glands, which can then be excised from the skin flap. Removal of the thoracic glands Nos. 2 and 3 should be performed with additional care to leave lymph nodes and muscle behind. The No. 1 fat pad may not be removed, if one is unable to distinguish it from the salivary gland, which is darker in color than the mammary tissue. Expected mammary tissue yield is 0.3 to 0.5 g for a virgin mouse and 1.0 to 1.2 g for a pregnant mouse. The mammary tissue is collected in phosphate-buffered saline (PBS) with antibiotics (pen/strep) on ice.
3.1.2. Mammary gland digestion and epithelial cell recovery
The mammary tissue pieces that float are transferred to a 10-cm sterile petri dish, whereas any tissue piece that precipitates is discarded. Mammary tissue is mechanically minced with two scalpels inside the petri dish under sterile conditions until uniform and oily in appearance (0.5- to 1-mm pieces), and then transferred to a small flask containing PBS (10 ml/g tissue) and antibiotics. Appropriate volume of 1% collagenase A stock Table 4.1 is added to achieve a final collagenase concentration of 0.05 to 0.1%, and tissue is stirred at 37° for 30 to 90 min, depending on collagenase activity. Stirring should be fast enough to cause thorough mixing without splashing of medium and tissue onto flask walls. The completeness of digestion can be checked starting at 30 min by aseptically removing small aliquots for low-power microscopic examination (40× total magnification) with an inverted stage, phase microscope. The desired endpoint is an epithelial preparation with no visible tissue pieces remaining and more than 80% of epithelial organoids free of adhering stromal tissue. If cells have lysed and DNA has been released producing a “stringy” clumping of organoids, DNAse stock solution Table 4.1 can be added until all clumping is cleared. On completion of digestion, 20 ml of F12 plus 5% fetal bovine serum (FBS) are added to the tissue digest and clumps are allowed to settle for 2 min. Epithelial organoids (mouse mammary epithelial cells, MMECs) are collected from the supernatant by centrifugation at 1500 rpm for 5 min and are then washed three times with F12 plus 5% FBS. Percoll gradient purification of epithelial cells is not routinely performed, because a highly enriched epithelial cell population is usually obtained.
Table 4.1.
Reagents for iMMEC culture
| Reagent | Source | Stock | Storage |
|---|---|---|---|
| Collagenase A | Roche | 1% in H2O | −20° |
| DNAse | Sigma | 0.4 mg/ml in PBS | −20° |
| EGF | Sigma | 10 μg/ml in PBS | 4° |
| Fetuin | Sigma | 2 mg/ml in 20% FBS | 4° |
| F12 | Invitrogen | 4° | |
| FBS | Invitrogen | −20° | |
| Hydrocortisone | Sigma | 1 mg/ml in EtOH | −20° |
| Insulin | Sigma | 10 mg/ml in 25 mM HEPES, pH 8.2 | 4° |
| Matrigel, growth factor reduced | BD | −20° | |
| Pen/Strep | Invitrogen | 10,000 units | −20° |
| Prolactin | Sigma | 4° | |
| Prolong Antifade | Molecular Probes | −20° | |
| Trypsin, regular strength | Invitrogen | 0.05% | −20° |
| Trypsin, lower strength | Clonetics | 0.025% | −20° |
3.1.3. Primary MMEC culture
For primary culture, MMECs from three mice can be pooled during washing, pelleted, resuspended in 2× hormone plating medium (F12, 10 μg/ml insulin, 2 μg/ml hydrocortisone, 10 ng/ml EGF) Table 4.2 and plated in two 6-cm or one 10-cm plate (4 or 8 ml of resuspended cells per plate, respectively) that have been precoated with equal volume of 100 μl/cm2 fetuin-serum solution (2 mg/ml fetuin in 20% FBS) Table 4.1 for 4 to 5 h at 37° (Rijnkels and Rosen, 2001). Cells are not routinely counted before plating, because single-cell suspensions required for accurate cell counting negatively impact viability of MMECs in primary culture. MMECs are allowed to settle for 36 to 48 h in the resultant 1× plating medium with 10% FBS before switching to reduced-FBS growth medium (F12, 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 5 ng/ml EGF) Table 4.2 to minimize fibroblast growth and are kept in a humidified incubator with 8.5% CO2 at 37°. Medium changes are performed every 4 days. Primary MMECs do not tolerate trypsinization well but remain viable for approximately 10 to 14 days in culture without cell passage.
Table 4.2.
Medium recipes for iMMECs
| Component | 2× hormone plating medium | Reduced-FBS growth medium | Regular growth medium | Cloning medium | Differentiation medium |
|---|---|---|---|---|---|
| F12 | 250 ml | 500 ml | 500 ml | 50 ml | 500 ml |
| FBS | No FBS! (already in fetuin-serum solution) | 25 ml (5% final) | 50 ml (10% final) | 10 ml (20% final) | |
| EGF (10 μg/ml stock in PBS | 250 μl (10 ng/ml final) | 250 μl (5 ng/ml final) | 250 μl (5 ng/ml final) | 25 μl (5 ng/ml final) | |
| Hydrocortisone (1 mg/ml stock in EtOH) | 500 μl (2 μg/ml final) | 500 μl (1 μg/ml final) | 500 μl (1 μg/ml final) | 50 μl (1 μg/ml final) | 500 μl (1 μg/ml final) |
| Insulin (10 mg/ml stock) | 250 μl (10 μg/ml final) | 250 μl (5 μg/ml final) | 250 μl (5 μg/ml final) | 25 μl (5 μg/ml final) | 250 μl (5 μg/ml final) |
| Prolactin | 1.5 mg (3 μg/ml final) | ||||
| Pen/Strep (10,000 units) | 5 ml | 5 ml | 5 ml | 0.5 | 5 ml |
3.1.4. Primary MMEC transfection and plating
MMECs from the mammary glands of a single mouse are resuspended in 250 μl F12 plus 5% FBS and electroporated in the presence of 10 μg ScaI-linearized cytomegalovirus (CMV)-driven adenovirus type 5 E1A plasmid (pCMVE1A [White et al., 1991]), 10 μg ScaI-linearized dominant negative mouse p53 plasmid (p53DD [Shaulian et al., 1992]), and 100 μg salmon sperm carrier DNA, as previously described for baby mouse kidney (BMK) epithelial cells (Degenhardt and White, 2006; Degenhardt et al., 2002b). During electroporation, the cells are pulsed at 220 V and 950 μF. MMECs from one electroporation are resuspended in 16 ml of 2× hormone plating medium Table 4.2 and plated in four 6-cm or two 10-cm fetuin-coated plates Table 4.1, as described previously. Again, MMECs are allowed to settle for 36 to 48 h in 1× plating medium with 10% FBS before switching to reduced-FBS growth medium Table 4.2 and are kept in a humidified incubator with 8.5% CO2 at 37°. Medium changes are performed every 4 days without cell passage.
3.1.5. Cloning and expansion of immortalized MMEC colonies
Over a period of 4 to 6 weeks, primary (nontransfected) or singly transfected MMECs and any contaminating fibroblasts undergo cell death, whereas colonies of doubly transfected, immortalized MMECs (iMMECs) arise and start growing. iMMEC colonies first appear at 2 to 3 weeks after initial plating (typically, 1 to 5 colonies per 10-cm plate) and are composed of tightly packed, cuboidal cells with typical epithelial morphology (Fig. 4.1) (Karantza-Wadsworth et al., 2007). When colonies reach 0.6 to 0.8 cm in diameter, the medium is removed, and the plate is washed once with PBS. Individual colonies are surrounded by glass cloning cylinders (6 × 8 mm to 10 × 10 mm, depending on colony size; Bellco Biotechnology) secured in place by autoclaved vacuum grease (VWR). iMMECs are recovered by limited trypsin digestion with lower strength trypsin (0.025%) Table 4.1 for 5 to 10 sec and are transferred to 24-well plates in coloning medium F12, 20% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 5 ng/ml EGF, Table 4.2 (0.5 ml medium per well, no need for fetuin-precoating). Colonies isolated from different plates are truly independent. However, as many colonies as possible are recovered from every plate, because colony survival after transfer to 24-well plates is approximately 50 to 70%. iMMECs are subsequently expanded in regular growth medium (F12, 10% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 5 ng/ml EGF) Table 4.2 and can be frozen in 92% FBS-8% DMSO from 10-cm plates at approximately 70% confluency for long-term storage in vapor phase nitrogen. During regular tissue culture, iMMECs require refeeding every 3 days and passage every 3 to 5 days, usually at 1:5 split.
3.1.6. iMMEC characterization by WB and IF
Once multiple iMMEC colonies have been generated from a particular mouse strain, iMMECs are examined by Western blotting (WB) for expression of E1A and p53DD (immortalizing proteins), ER-α and epithelial cell markers, such as cytokeratin (CK) 5/6, CK8, CK14, E-cadherin, β-catenin, Ep-CAM, vimentin Table 4.3. For immunofluorescence (IF), iMMECs can be grown on glass coverslips to 70% confluency and then fixed with 1:1 methanol/acetone at −20° for 10 min. Coverslips are washed with PBS three times for 5 min each time (5 min × 3), and then incubated with primary antibody (at 1:100–1:400 dilution, depending on antibody) in 5% bovine serum albumin (BSA, Sigma), PBS, 0.1% Tween-20 (PBST) for 1 h at 37°. After PBST washes (5 min × 3), coverslips are incubated with fluorescein- or rhodamine-conjugated secondary antibody (at 1:100 dilution) for 40 min at room temperature (RT). Coverslips are again washed with PBST (5 min × 3), incubated with 0.5 ng of DAPI (4′, 6′-diamidino-2-phenylindole, Sigma) for 15 min at RT, and finally washed with PBS for 5 min, before being mounted with the antifade agent Prolong (Molecular Probes).
Table 4.3.
Useful antibodies and fluorescent reagents for iMMEC analysis
| Antibody or stain | Purpose | Source |
|---|---|---|
| Activated (cleaved) caspase-3 | Apoptosis marker | Cell signaling |
| β-casein | Milk protein | Santa Cruz |
| β-catenin | Cell–cell junctions | Zymed |
| Cytokeratin 5/6 | Myoepithelial cell marker | Covance |
| Cytokeratin 8 | Luminal cell marker | Abcam |
| Cytokeratin 14 | Myoepithelial cell marker | Covance |
| DAPI | Nuclear counterstain | Sigma |
| E1A | Immortalization marker | Oncogene |
| E-cadherin | Cell–cell junctions | RDI |
| Ep-CAM | Epithelial cell marker | Santa Cruz |
| ER-α | Hormone receptor | Santa Cruz |
| Occludin | Cell–cell junctions | Zymed |
| p53 (for p53DD) | Immortalization marker | Oncogene |
| Smooth muscle actin (SMA) | Myoepithelial cell marker | Sigma |
| Vimentin | Myoepithelial cell marker | Santa Cruz |
| ZO-1 | Cell–cell junctions | Zymed |
3.2. Lactogenic stimulation
3.2.1. In 2D culture
Cells are grown on plastic culture dishes until confluent and are subsequently induced with differentiation medium (F12, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 3 μg/ml prolactin) Table 4.2 ±2% Matrigel (Streuli et al., 1995) for 6 days, with medium changes every 2 days.
3.2.2. In 3D culture
Mammary acini are grown on Matrigel for 12 days as described section 3.4.1 and then induced with differentiation medium containing 2% Matrigel for 2 additional days. Mammary acini are subsequently fixed and processed for IF, as described section 3.4.2.
3.3. Generation of stable cell lines
Proteins of interest can be easily expressed or down regulated in iMMECs, so that the impact of oncogene activation and tumor suppressor inactivation on mammary tumorigenesis and treatment responsiveness can be readily investigated. So far, iMMECs have been engineered to express human Bcl-2, H-RasV12, myr-Akt, wild-type human HER2/neu, and the vector control Table 4.5 by electroporation (as described insection 3.1.4) with pcDNA3.1hBcl-2, pcDNA3.1H-RasV12, pcDNA3.1Myr-Akt, pcDNA3.1wtHER2/neu, and pcDNA3.1 vector (Invitrogen), respectively, followed by selection with geneticin (Karantza-Wadsworth et al., 2007) Table 4.5. For studying the autophagy potential of iMMECs generated from beclin1+/+ and beclin1+/− mice, stable expression of EGFP-LC3 is performed by electroporation with pcDNA3.EGFP-LC3, followed by selection with geneticin for apoptosis-competent iMMECs, and by electroporation with pcDNA3.EGFP-LC3 and pcDNA3.1zeo (Invitrogen), followed by double selection with geneticin and zeocin for Bcl-2 expressing iMMECs (Karantza-Wadsworth et al., 2007) Table 4.5. Geneticin and zeocin are used at 300 and 100 μg/ml respectively Table 4.4 for 10 to 14 days. Individual drug-resistant colonies are isolated and expanded to stable cell lines, as described in section 3.1.5. The drug used for selection is routinely kept in the regular growth medium thereafter.
Table 4.5.
Mouse (C57BL/6, Wild-type or mutant) mammary epithelial cell lines, E1A and p53DD-derived (Karantza-Wadsworth et al., 2007)
| Genotype | Transgene | iMMEC cell line name |
|---|---|---|
| Wild-type | WTA (or 21), WTB, WTC, WTD, WT3, WT5 | |
| Wild-type | pcDNA3.1 vector | WTA.V, WT3.V |
| Wild-type | Bcl-2 (human) | WTA.B1, WTA.B4, WT3.B1, WT3.B2, WT3.B3, WT3.B8 |
| Wild-type | EGFP-LC3 | WTA-LC3, WT3-LC3 |
| Wild-type | Bcl-2, EGFP-LC3 | WTA.B4-LC3, WT3.B3-LC3 |
| Wild-type | HER2/neu (human, wild-type) | WTA.H2, WTA.H3 |
| Wild-type | myr-Akt | WTA.A5, WTA.A7 |
| Wild-type | H-RasV12 | WTA.R3, WTA.R5 |
| beclin1+/− | BLN2 (or 2.1), BLN4 | |
| beclin1+/− | pcDNA3.1 vector | BLN2.V |
| beclin1+/− | Bcl-2 | BLN2.B2, BLN2.B4, BLN2.B5, BLN2.B8 |
| beclin1+/− | EGFP-LC3 | BLN2-LC3 |
| beclin1+/− | Bcl-2, EGFP-LC3 | BLN2.B4-LC3.5 |
Table 4.4.
Drug selection for stable transfection of iMMECs
| Drug | Source | Final concentration |
|---|---|---|
| Geneticin | Invitrogen | 300 μg/ml |
| Zeocin | Invitrogen | 100 μg/ml |
3.4. Three-dimensional (3D) morphogenesis
Three-dimensional culture of iMMECs on a reconstituted basement membrane is performed according to a modified version of the protocol previously described for the immortalized, nontransformed human mammary epithelial cell line MCF-10A (Debnath et al., 2003).
3.4.1. iMMEC trypsinization and plating on matrigel
Eight-well RS glass slides (BD Falcon) are coated with 40 μl per well of growth factor-reduced Matrigel thawed overnight at 4°. Trypsin (1 ml) is added to a 10-cm confluent plate of iMMECs, swirled around, and immediately aspirated to leave a thin film behind, which prevents cell clumping and ensures a single-cell suspension. After 15 to 20 min in a 8.5% CO2 humidified incubator at 37°, iMMECs get dislodged and are collected in 2 ml growth medium with repeated pipetting to break up cell clumps, and are then counted. iMMECs are pelleted, resuspended in growth medium at a concentration of 25,000 cells/ml, and mixed 1:1 with growth medium containing 4% Matrigel; 400 μl of the resultant solution (12,500 cells/ml in growth medium with 2% Matrigel) is plated in each well. Growth medium containing 2% Matrigel is replaced every 4 days.
3.4.2. Fixation and immunofluorescence
Mammary acini are fixed in 4% formalin for 25 min at room temperature. Fixed structures are washed with PBS-glycine (PBS, 100 mM glycine) three times for 15 min each time. The structures are then blocked with IF buffer (PBS, 0.1% BSA, 0.2% Triton X-100, 0.05% Tween-20) plus 10% goat serum for 30 min at 37°, followed by secondary block [IF buffer containing 10% goat serum and 20 μg of goat anti-mouse F(ab′)2/ml] for 10 min at 37°, and then incubation with primary antibody (in secondary block solution, usually at 1:100 dilution) for 90 min at 37 degrees. Structures are then washed three times in IF buffer for 15 min each. Anti-mouse or anti-rabbit secondary antibodies coupled with fluorescein or rhodamine are diluted in IF buffer containing 10% goat serum (usually at 1:100 dilution), followed by incubation for 40 min at RT. After three washes with IF buffer for 15 min each, structures are incubated with 0.5 ng of DAPI for 15 min. Structures are washed with PBS for 5 min before being mounted with the antifade agent Prolong (Molecular Probes). Confocal laser scanning microscopy can be done with a Zeiss LSM510-META confocal microscope system. The percentage of acini with lumen formation is the mean of two independent experiments (for each experiment, 100 acini are scored for each cell line at each time point).
3.4.3. Histology
Mammary acini are grown on Matrigel for 12 days as described previously, fixed in 10% neutral buffered formalin, scraped from the glass slide with a razor blade, pelleted, embedded in paraffin, and processed for H&E staining.
3.4.4. Electron microscopy
Mammary acini grown for 12 days on Matrigel as described previously are fixed with electron microscopy fixative (1.2% paraformaldehyde/2.5% glutaraldehyde/0.03% picric acid) in 100 mM cacodylate buffer for 1 h at RT and then overnight at 4°. Fixed acini are scraped from the coverslip with a razor blade, pelleted, and processed for EM with standard procedure.
3.5. Orthotopic tumor growth
Cells are harvested by trypsinization, washed, and resuspended in PBS (107 cells/ml). Orthotopic mammary gland implantation of iMMECs is performed with IACUC-approved protocol; 5- to 8-week-old NCR nude female mice are anesthetized with ketamine (100 mg/kg intraperitoneally, IP) and xylazine (10 mg/kg IP). A small incision is made to reveal the right second or third mammary gland, and 106 cells are injected into the mammary fat pad. The incision is closed with surgical clips that are removed 10 days later. Tumor outgrowth is monitored by weekly measurements of tumor length (L) and width (W). Tumor volume is calculated as πLW2/6. At the time of animal euthanasia and mammary tumor dissection, the left second or third mammary gland is collected as a normal control. Clonal mammary tumors generated by wild-type iMMECs after acquisition of secondary genetic or epigenetic changes appear at 3 to 4 months with 60% penetrance (in 3 of 5 mice), whereas highly tumorigenic iMMECs expressing activated H-Ras or wild-type Her2/neu form mammary tumors in 3 to 5 weeks with 100% penetrance (in 5 of 5 mice).
4. Concluding Remarks
The mouse mammary epithelial cell model presented takes advantage of the strength of mouse genetics in combination with 3D morphogenesis and orthotopic tumor growth assays for the study of oncogene and tumor suppressor functions as they pertain to mammary tumorigenesis, and by extension to human breast cancer. Generation of iMMECs from beclin1+/+ and beclin1+/− mice and assessment of their properties in 2D culture, 3D morphogenesis, and in vivo tumorigenicity (Karantza-Wadsworth et al., 2007) has provided valuable insight into the role of autophagy in mammary tumorigenesis and has set the stage for future investigations focused on the intriguing relationship between autophagy, metabolism, stress response, and cancer progression and treatment (Karantza-Wadsworth and White, 2007; Mathew et al., 2007a). The in vitro and in vivo protocols described here will hopefully be widely applicable in breast cancer research involving mouse modeling and will provide new tools for successfully investigating the molecular mechanisms implicated in breast cancer progression and treatment responsiveness.
References
- Bissell MJ. Modelling molecular mechanisms of breast cancer and invasion: Lessons from the normal gland. Biochem Soc Trans. 2007;35:18–22. doi: 10.1042/BST0350018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan R, Smith GH. MMTV-induced mammary tumorigenesis: Gene discovery, progression to malignancy and cellular pathways. Oncogene. 2000;19:992–1001. doi: 10.1038/sj.onc.1203276. [DOI] [PubMed] [Google Scholar]
- Debnath J, Brugge JS. Modelling glandular epithelial cancers in three-dimensional cultures. Nat Rev Cancer. 2005;5:675–688. doi: 10.1038/nrc1695. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, Chen G, Lindsten T, White E. BAX and BAK mediate p53-independent suppression of tumorigenesis. Cancer Cell. 2002a;2:193–203. doi: 10.1016/s1535-6108(02)00126-5. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degenhardt K, Sundararajan R, Lindsten T, Thompson C, White E. Bax and Bak independently promote cytochrome C release from mitochondria. J Biol Chem. 2002b;277:14127–14134. doi: 10.1074/jbc.M109939200. [DOI] [PubMed] [Google Scholar]
- Degenhardt K, White E. A mouse model system to genetically dissect the molecular mechanisms regulating tumorigenesis. Clin Cancer Res. 2006;12:5298–5304. doi: 10.1158/1078-0432.CCR-06-0439. [DOI] [PubMed] [Google Scholar]
- Furth PA. Conditional control of gene expression in the mammary gland. J Mammary Gland Biol Neoplasia. 1997;2:373–383. doi: 10.1023/a:1026399329934. [DOI] [PubMed] [Google Scholar]
- Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol Cell Biol. 1992a;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992b;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–1635. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karantza-Wadsworth V, White E. Role of autophagy in breast cancer. Autophagy. 2007;3:610–613. doi: 10.4161/auto.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Rosen JM, McMenamin-Balano J, Muller WJ, Perkins AS. neu/ERBB2 cooperates with p53-172H during mammary tumorigenesis in transgenic mice. Mol Cell Biol. 1997;17:3155–3163. doi: 10.1128/mcb.17.6.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, Kerkhoven RM, van Vliet MH, Wessels LF, Peterse JL, Berns A, Jonkers J. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104:12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007a;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007b;21:1367–1381. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Karp CM, White E. A mouse epithelial cell model to study the role of apoptosis and autophagy in cancer. Methods Enzymol. 2008 doi: 10.1016/S0076-6879(08)01605-4. [DOI] [PubMed] [Google Scholar]
- Medina D. Chemical carcinogenesis of rat and mouse mammary glands. Breast Dis. 2006;28:63–68. doi: 10.3233/bd-2007-28107. [DOI] [PubMed] [Google Scholar]
- Moody SE, Sarkisian CJ, Hahn KT, Gunther EJ, Pickup S, Dugan KD, Innocent N, Cardiff RD, Schnall MD, Chodosh LA. Conditional activation of Neu in the mammary epithelium of transgenic mice results in reversible pulmonary metastasis. Cancer Cell. 2002;2:451–461. doi: 10.1016/s1535-6108(02)00212-x. [DOI] [PubMed] [Google Scholar]
- Rasmussen SB, Young LJT, Smith GH. Preparing mammary gland whole mounts from mice. In: Ip MM, Asch BB, editors. Methods in Mammary Gland Biology and Breast Cancer Research. Kluwer Academic/Plenum Publishers; New York: 2000. pp. 75–85. [Google Scholar]
- Rijnkels M, Rosen JM. Adenovirus-Cre–mediated recombination in mammary epithelial early progenitor cells. J Cell Sci. 2001;114:3147–3153. doi: 10.1242/jcs.114.17.3147. [DOI] [PubMed] [Google Scholar]
- Shaulian E, Zauberman A, Ginsberg D, Oren M. Identification of a minimal transforming domain of p53: Negative dominance through abrogation of sequence-specific DNA binding. Mol Cell Biol. 1992;12:5581–5592. doi: 10.1128/mcb.12.12.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart TA, Pattengale PK, Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TC, Cardiff RD, Zukerberg L, Lees E, Arnold A, Schmidt EV. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 1994;369:669–671. doi: 10.1038/369669a0. [DOI] [PubMed] [Google Scholar]
- White E, Cipriani R, Sabbatini P, Denton A. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991;65:2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijnhoven SW, Zwart E, Speksnijder EN, Beems RB, Olive KP, Tuveson DA, Jonkers J, Schaap MM, van den Berg J, Jacks T, van Steeg H, de Vries A. Mice expressing a mammary gland-specific R270H mutation in the p53 tumor suppressor gene mimic human breast cancer development. Cancer Res. 2005;65:8166–8173. doi: 10.1158/0008-5472.CAN-05-1650. [DOI] [PubMed] [Google Scholar]
- Witt AE, Hines LM, Collins NL, Hu Y, Gunawardane RN, Moreira D, Raphael J, Jepson D, Koundinya M, Rolfs A, Taron B, Isakoff SJ, Brugge JS, LaBaer J. Functional proteomics approach to investigate the biological activities of cDNAs implicated in breast cancer. J Proteome Res. 2006;5:599–610. doi: 10.1021/pr050395r. [DOI] [PMC free article] [PubMed] [Google Scholar]