Cytotoxicity-guided fractionation of the organic extract from a Fijian Lyngbya majuscula led to the discovery of desmethoxymajusculamide C (DMMC) as the active metabolite. Spectroscopic analysis including 1D and 2D NMR, MS/MS, as well as chemical degradation and derivatization protocols, were used to assign the planar structure and stereo-configuration of this new cyclic depsipeptide. DMMC demonstrated potent and selective anti-solid tumor activity with an IC50 = 20 nM against the HCT-116 human colon carcinoma cell line via disruption of cellular microfilament networks. A linear form of DMMC was generated by base hydrolysis and the amino acid sequence was confirmed by mass spectrometry. Linearized DMMC was also evaluated in the biological assays and found to maintain potent actin depolymerization characteristics while displaying solid tumor selectivity equivalent to DMMC in the disk diffusion assay. A clonogenic assay assessing cytotoxicity to HCT-116 cells as a function of exposure duration showed that greater than 24 h of constant drug treatment was required to yield significant cell killing. Therapeutic studies with HCT-116 bearing SCID mice demonstrated efficacy at the highest dose used (%T/C = 60% at 0.62 mg/kg daily for 5 days).
Actin is a ubiquitous and highly conserved protein found in either globular (G-actin monomer) or filamentous (F-actin polymer) forms. A tightly controlled dynamic equilibrium exists between actin filaments and monomers and is essential for cell growth and division, as well as cell signaling, motility and tertiary cellular structure.1 Transformed cancer cells undergo distinct changes in actin cytoskeletal organization and protein regulation, which contribute to the abnormal growth characteristics of tumor cells. As a consequence of altered microfilament dynamics, cancer cells have enhanced capacity for tissue adhesion, tumorigenesis and an increased ability to metastasize.2,3 Metastatic tissue invasion involves a form of cellular motility usually termed ‘amoeboid motility’, a process driven by cycles of actin polymerization, cell adhesion, and acto-myosin contraction.4 These cellular processes therefore offer logical targets for the development of new anticancer drugs.
A large number of secondary metabolites have been isolated from marine invertebrates and microorganisms that show promising anticancer activities.5,6 Several of these, such as jasplakinolide, hectochlorin and doliculide, are known to stimulate actin polymerization while others such as latrunculin and various trisoxazole-containing macrolides display actin depolymerization properties.7–11 The mechanism of action has been studied in some detail for jasplakinolide and doliculide, which both cause cell cycle arrest at the G2/M phase by inducing actin hyperpolymerization and aggregation of the resultant F-actin. Additionally, both compounds competitively displace a fluorescent phalliodin derivative from the actin polymer.7 Interestingly, hectochlorin displayed many of the same effects on actin polymerization but is unable to displace fluorescent phalliodin from the polymerized protein.8 The cyclic depsipeptides majusculamide C and dolastatins 11 and 12 also show promising cytotoxic activity by arresting cells at cytokinesis by stimulating microfilament hyperpolymerization in a dose- and time-dependent manner.12,13 Conversely, latrunculin and the trisoxazole macrolides inhibit polymerization and nucleotide (ATP) exchange, respectively, ultimately causing microfilament depolymerization and cell death.9,10
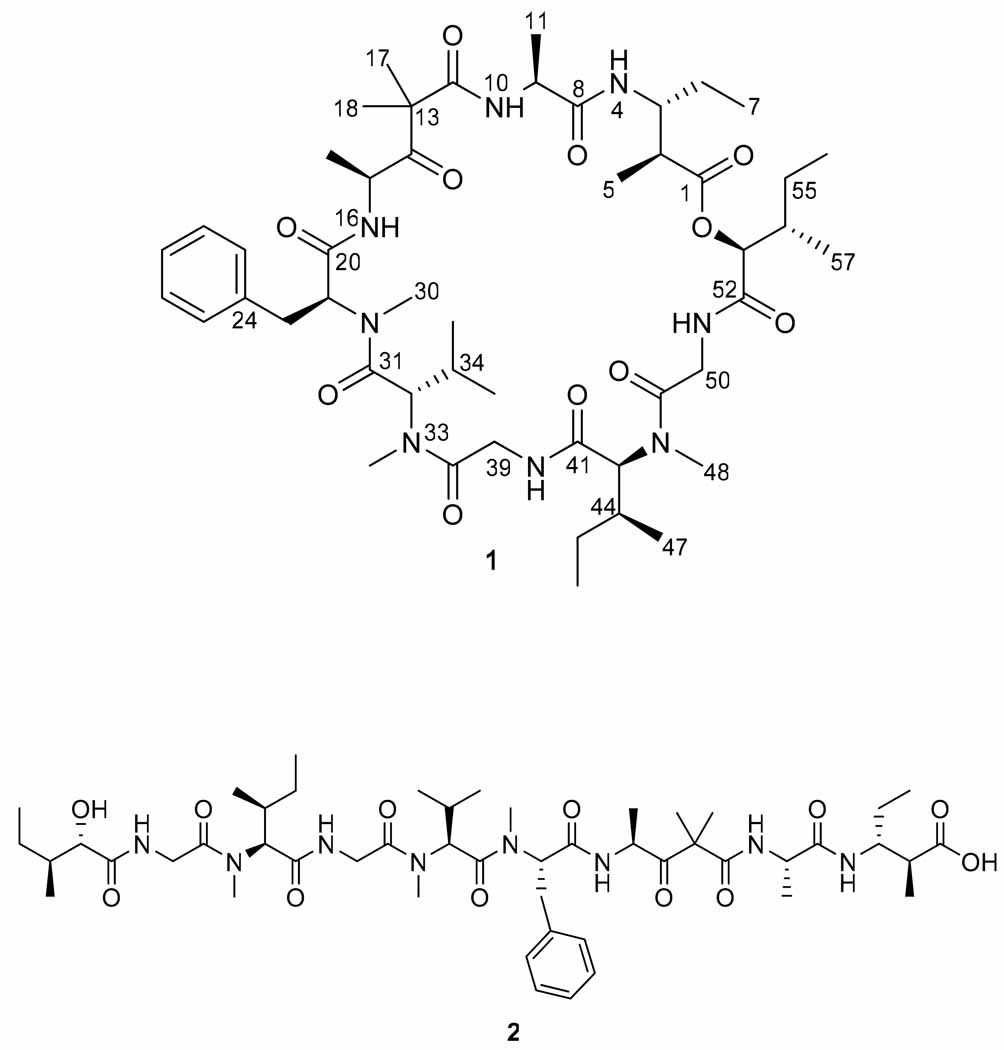
Here we report the discovery of desmethoxymajusculamide C (DMMC, 1), a new cyclic depsipeptide with potent actin depolymerization properties in both the native cyclic (1) and ring-opened forms (2). Strong cytotoxicity was observed in initial screening of the organic extract of a Fijian collection of the cyanobacterium Lyngbya majuscula. Bioassay-guided isolation led to DMMC (1) as the active constituent. Structure elucidation by extensive chemical and spectroscopic approaches revealed 1 to be an analog in a well known class of bioactive depsipeptides (Figure 3). DMMC, like its structural relatives, displays potent activity against a variety of cancer cell lines by the disruption of cytoskeletal actin microfilament networks. Moreover, we found that the ring-opened form of DMMC (2) maintains equivalent efficacy and solid tumor selectivity as the cyclic molecule (1). These findings raise significant and intriguing questions regarding the active form of the many biologically active cyclic (depsi)peptides reported in the literature to date.
Figure 3.
Representative cyclic depsipeptides related to DMMC (1) obtained from Lyngbya majuscula or the predatory sea hare Dolabella auricularia.
Results and Discussion
Isolation and Structure Determination of DMMC (1)
The marine cyanobacterium Lyngbya majuscula was collected from Yanuca Island, Fiji and repetitively extracted with 2:1 CH2Cl2/CH3OH to yield 5.5 g of crude extract. A portion of the extract (5.3 g) was sequentially fractionated by vacuum liquid chromatography (VLC) over silica gel, C18 SPE (7:3 CH3OH/H2O), and RP HPLC to yield 27.5 mg of pure 1, isolated as a light yellow glassy oil. Cytotoxic assay of fractions at each stage of the chromatography identified 1 to be the most active compound in the extract.
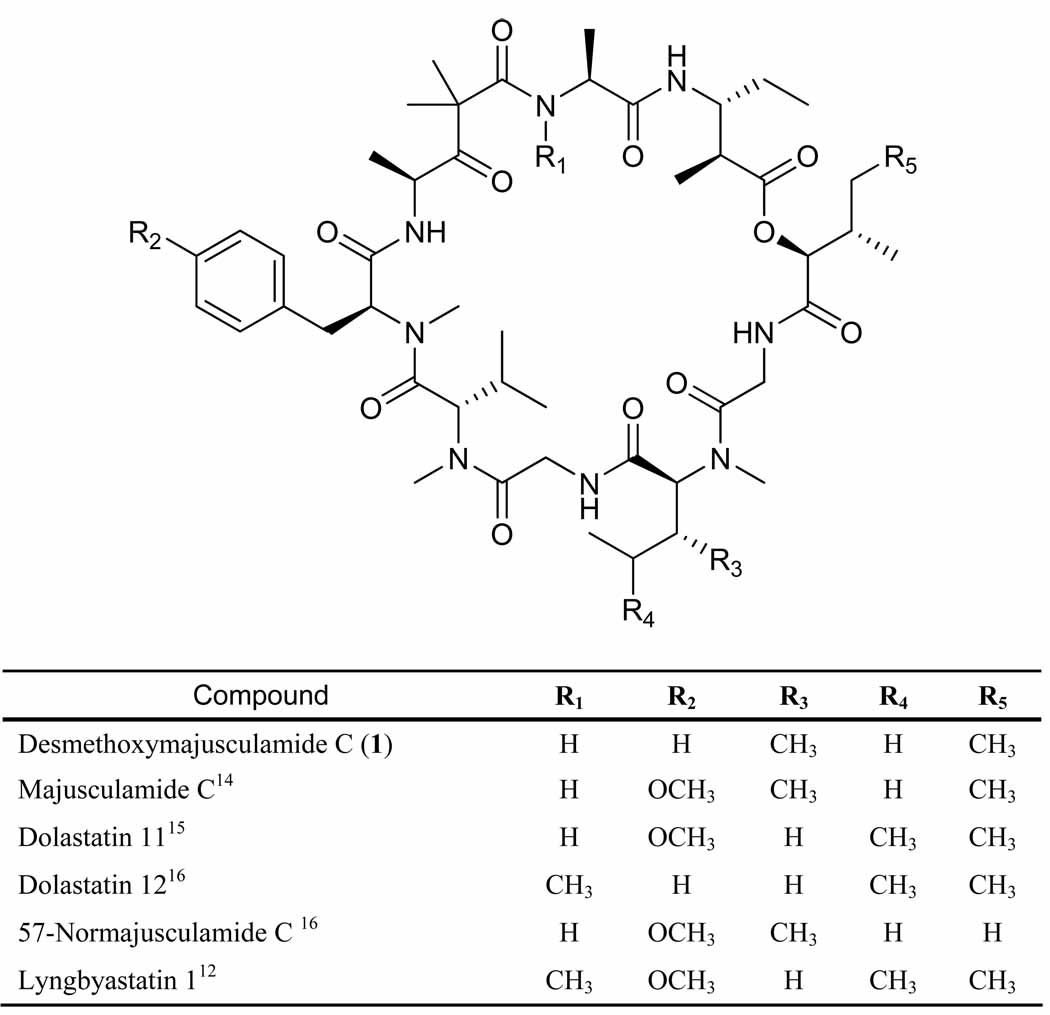
Desmethoxymajusculamide C (1) possessed a molecular formula of C49H78N8O11 as determined by HR FAB MS (obsd m/z [M + H]+ 955.5867; calcd for m/z 955.5868). Combined with IR absorption bands at 1739 and 1641 cm−1 for ester and amide functionalities, respectively, these data indicated a peptide-type natural product. The 1H and 13C NMR spectra for 1 were well dispersed in CDCl3 (Table 1 and supporting information) and allowed the construction of nine partial structures by 2D NMR (COSY, HMBC), accounting for all atoms in the molecular formula for 1. Six standard amino acids were deduced as alanine (Ala), N-methyl-valine (N-MeVal), N-methyl-phenylalanine (N-MePhe), N-methyl-isoleucine (N-MeIle) and two glycine (Gly) units. Two additional residues were similarly deduced as the α-hydroxy acid, 2-hydroxy-3-methylpentanoate (Hmpa) and the β-amino acid, 3-amino-2-methylpentanoate (Map). The ninth and final moiety, deduced mainly from HMBC and comparisons with the literature, was constructed as 4-amino-2,2-dimethyl-3-oxo-pentanoate (Dmop).14
Table 1.
1H and 13C NMR Data for DMMC (1) Recorded in CDCl3 at 400 and 100 MHz, Respectively.
| Residue | Position | δC | δH, mult., J (Hz) | HMBCa |
|---|---|---|---|---|
| Map | 1 | 172.8 | ||
| 2 | 42.7 | 2.76, qd (7.0, 1.6) | 1, 3 | |
| 3 | 51.4 | 4.52, m | ||
| 4 (NH) | 7.07, d (10.1) | 3, 8 | ||
| 5 | 10.0 | 1.10, d (7.0) | 1, 2, 3 | |
| 6 | 26.3 | 1.56, m;1.48, m | 2, 3, 7 | |
| 7 | 11.2 | 0.93, t (7.4) | 3, 6 | |
| Ala | 8 | 173.0 | ||
| 9 | 48.5 | 4.45, m | 8, 11 | |
| 10 (NH) | 7.76, d (7.9) | 9, 11, 12 | ||
| 11 | 15.7 | 1.07, d (7.9) | 8, 9 | |
| Ibu | 12 | 172.3 | ||
| 13 | 55.1 | |||
| 14 | 210.3 | |||
| 15 | 52.0 | 4.92, m | 14, 19 | |
| 16 (NH) | 7.32, d (6.4) | 14, 15, 20 | ||
| 17 | 22.5 | 1.53, s | 12, 13, 14, 18 | |
| 18 | 21.7 | 1.48, s | 12, 13, 14, 17 | |
| 19 | 19.3 | 1.16, d (6.8) | 14, 15 | |
| N-Me Phe | 20 | 168.0 | ||
| 21 | 61.0 | 5.23, m | 20, 23, 24, 30 | |
| 22 (N) | ||||
| 23 | 35.7 | 3.31, dd (14.0, 6.3) | 20, 21, 24, 25/29 | |
| 2.89, dd (14.0, 8.2) | ||||
| 24 | 137.0 | |||
| 25/29 | 129.7 | 7.25, m | 23, 24, 26/28, 27 | |
| 26/28 | 129.0 | 7.28, m | 24, 25/29 | |
| 27 | 127.2 | 7.20, t (6.9) | 25/29 | |
| 30 | 29.5 | 2.99, s | 21, 31 | |
| N-MeVal | 31 | 170.4 | ||
| 32 | 58.3 | 4.79, d (10.6) | 31, 34, 35, 36, 37, 38 | |
| 33 (N) | ||||
| 34 | 27.3 | 2.22, m | 32, 35, 36 | |
| 35 | 18.7 | 0.33, d (6.4) | 32, 34, 36 | |
| 36 | 18.6 | 0.74, d (6.5) | 32, 34, 35 | |
| 37 | 29.5 | 2.97, s | 32, 38 | |
| Gly | 38 | 169.5 | ||
| 39 | 40.8 | 4.62, dd (17.6, 7.9) | 38, 41 | |
| 3.47, d (17.6) | ||||
| 40 (NH) | 7.58 d (7.9) | 41 | ||
| N-MeIle | 41 | 171.3 | ||
| 42 | 61.4 | 4.91, m | 41, 44, 45, 47, 48, 49 | |
| 43 (N) | ||||
| 44 | 33.0 | 2.11, m | ||
| 45 | 25.2 | 1.46, m;1.11, m | 44 | |
| 46 | 10.3 | 0.89, m | 44, 45 | |
| 47 | 15.8 | 1.03, d (7.3) | 42, 44, 45 | |
| 48 | 30.6 | 3.22, s | 42, 49 | |
| Gly | 49 | 170.2 | ||
| 50 | 41.0 | 4.44, m | 49, 52 | |
| 3.57, dd (15.9, 4.6) | ||||
| 51 (NH) | 7.36, t (5.5) | 49, 50 | ||
| Hmpa | 52 | 170.7 | ||
| 53 | 78.6 | 5.21, m | 1, 52, 54, 55, 57 | |
| 54 | 37.6 | 2.07, m | ||
| 55 | 24.0 | 1.47, m;1.24, m | 53, 54, 56 | |
| 56 | 11.9 | 0.90, m | 54, 55 | |
| 57 | 15.4 | 0.90, m | 53, 54 |
Proton showing correlation to indicated carbon resonance.
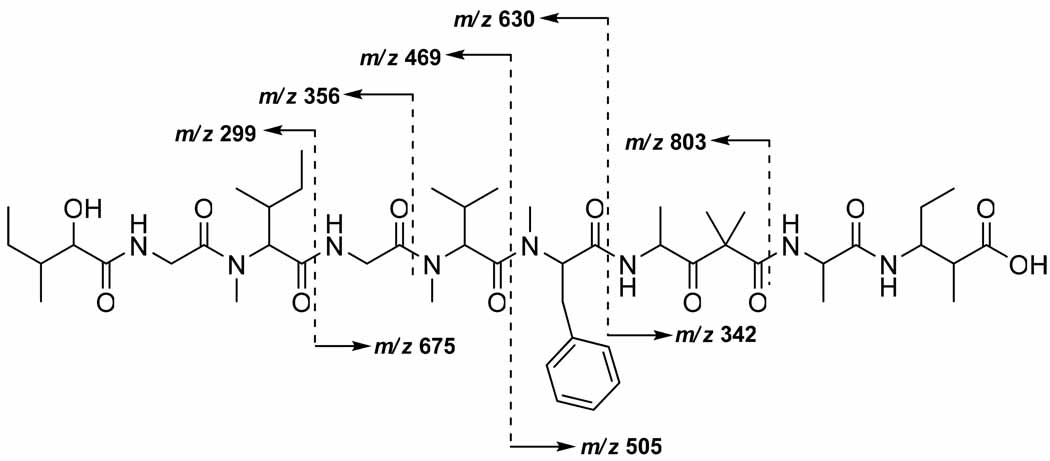
Sequencing of the amino and hydroxy acids in DMMC was accomplished by HMBC (Table 1), and supported by CID MS fragmentation of the base hydrolysis product of 1 (Figure 2). The configuration of the stereogenic centers found in 1 was determined by chiral HPLC and Marfey’s analysis. Fortunately, the doubling and broadening of several specific 1H- and 13C NMR signals, as observed for dolastatin 12, lyngbyastatin 1 and others, was not present in the spectra recorded for DMMC (1), thus suggesting the presence of a single C-15 epimer.12
Figure 2.
CID MS fragmentation pattern of derivative 2.
Cytotoxicity of cyclic (1) and linear DMMC (2)
The original extract (WHG-1338) was identified as solid tumor selective and very potent.17 The zone differential between the three human tumor cell lines HCT-116 human colon cancer, H125 human lung cancer, MCF-7 human breast cancer and the CCRF-CEM human leukemia was 500 zone units when 2.3 µg of extract was applied to the filter disk. As noted above, bioassay directed fractionation yielded DMMC (1) as the most active pure compound. The same zone value to the HCT-116 and H125 cell lines was achieved with 117 ng of 1, indicating an approximately 20-fold purification from crude extract to obtain DMMC. Interestingly, the linear form of DMMC (2) retained full potency in this assay and gave the same clearing zone at 35 ng/disk.
IC50 determinations were carried out for both 1 and 2 against 4 cells lines as shown in Table 2. HCT-116 was the most sensitive cell line with IC50 values of 20 nM and 16 nM, respectively. The H-460 cell line was about 5-fold less sensitive, the MDA-MB-435 cell line over 10-fold less sensitive, and Neuro-2A cells were resistant to both compounds.
Table 2.
Cytotoxicity of 1 and 2 Against a Panel of Cancer Cell Lines.
| IC50 (µM) | ||
|---|---|---|
| Cyclic DMMC (1) | Linear DMMC (2) | |
|
HCT-116 human colon carcinoma |
0.020 | 0.016 |
|
H-460 human large cell lung carcinoma |
0.063 | 0.094 |
|
MDA-MB-435 human carcinoma |
0.22 | 0.23 |
|
Neuro-2A murine neuroblastoma |
> 1.0 | > 1.0 |
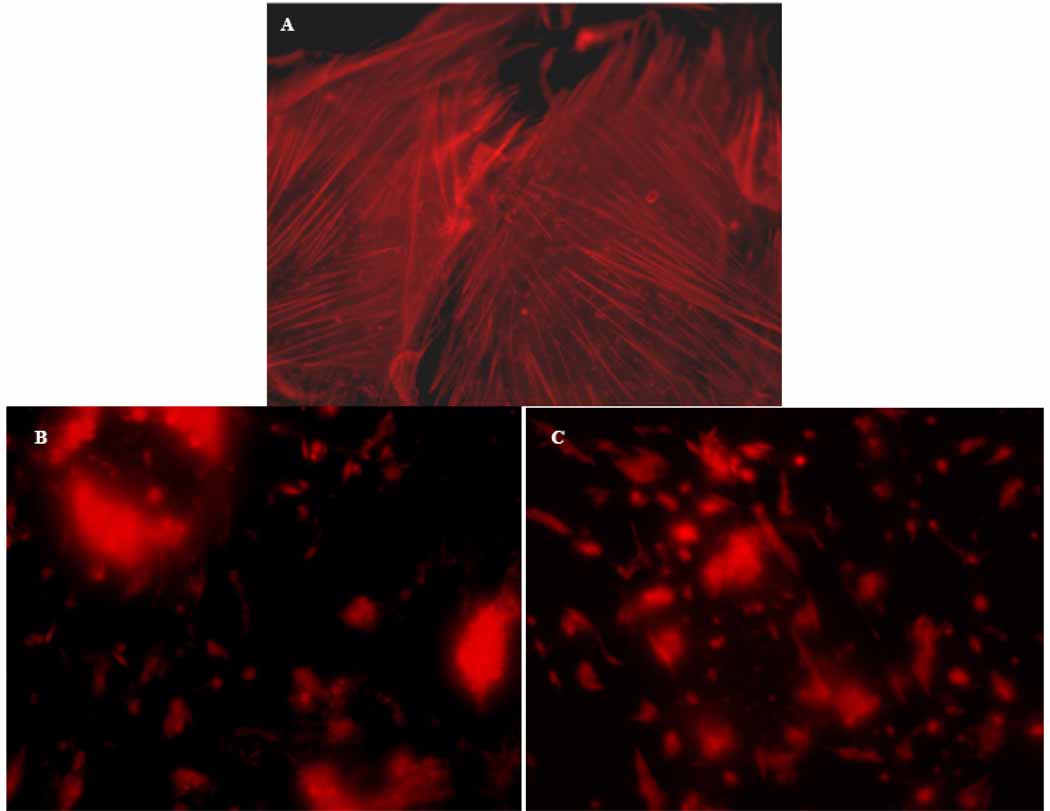
Actin microfilament disruption assays were conducted to further understand the mechanism of action for compounds 1 and 2. Cyclic (1) and linear (2) DMMC at 52 nM caused the complete loss of filamentous (F)-actin coincident with dramatic changes in cell morphology when tested against A-10 cells (Figure 4). The effects were specific for microfilaments as there was no evidence of microtubule loss at these drug concentrations. Binuclear cells were present, consistent with inhibition of the actin-dependent process of cytokinesis. Evidence of apoptosis and the breakdown of nuclei into apoptotic bodies were prevalent at 52 nM, and altered cellular morphology accompanied total disruption of the microfilament network at this concentration.
Figure 4.
Effects of cyclic (1) and linear (2) DMMC on the actin cytoskeleton of A-10 cells. After 24 h of drug exposure, the cells were fixed, permeabilized and exposed to the microfilament-staining reagent TRITC-phalloidin (visualized as red). (A) Control cells were treated with vehicle alone. (B) Cells treated with DMMC (1) at 52 nM caused complete loss of the cellular microfilament network and generated binucleated cells (not shown). At 10 nM DMMC, no microfilament loss was observed in A-10 cells. (C) Treatment of cells with linearized DMMC (2) at 52 nM also induced the complete disruption of cellular microfilament networks and generated binucleated cells (not shown).
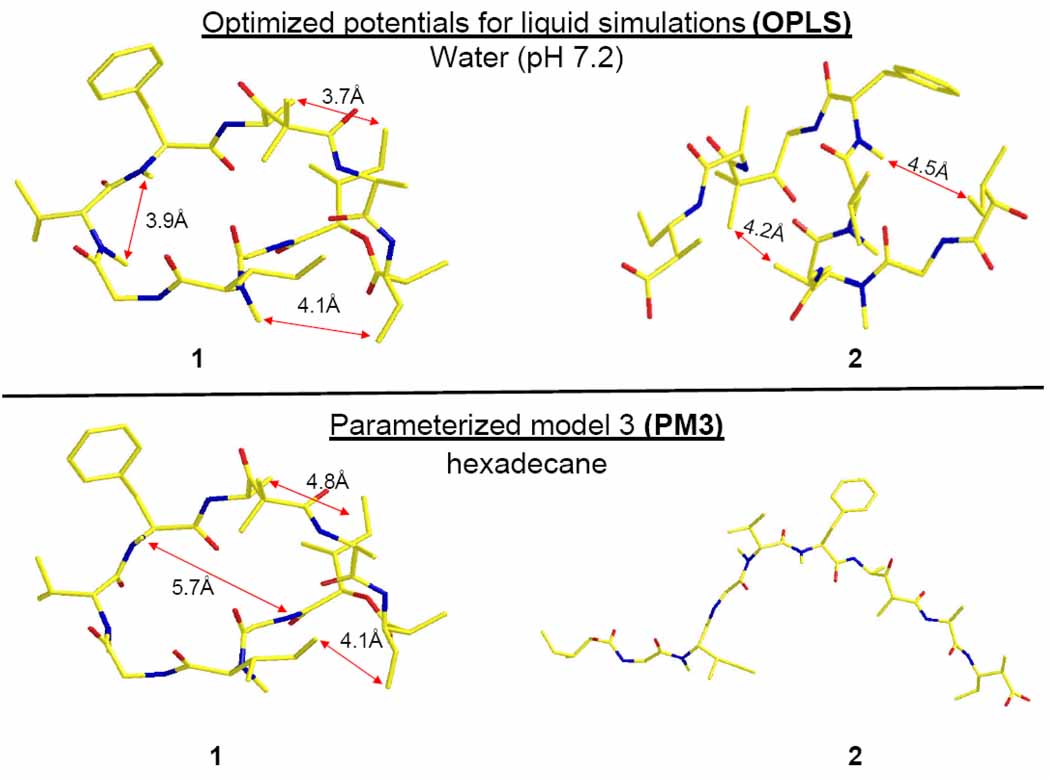
Energy minimized molecular modeling of both compounds 1 and 2 under aqueous conditions as well as under hydrophobic cell membrane conditions shows that the two molecules behave differently in these two environments. Using semi-empirical optimized potentials for liquid simulations (OPLS) modeled at pH 7.2, the linear molecule 2 takes on a ‘double hair-pin-like’ conformation while in the hexadecane parameterized model 3 (PM3) calculations, compound 2 ‘unwinds’ to a nearly linear conformation (Figure 5).18 By contrast, native DMMC (1) maintains a similar conformation under both modeling conditions. One possible interpretation of these data, based on the differences in the lowest energy conformations of 1 and 2 as well as their biological activity profiles, is that natural product 1 functions as a pro-drug which crosses the cell membrane and is then cleaved by cellular esterases found in the cytosol, to yield the highly bioactive linearized form of the molecule (2). Alternatively, it is possible that linear DMMC (2) is equally active to the cyclic form (1) because it adopts an active conformation similar to DMMC, but only on the surface of actin, its biomolecular target.
Figure 5.
Lowest energy conformations of DMMC (1) and its base-opened seco-acid form (2): top, structures modeled for aqueous conditions at pH 7.2 (e.g. cytoplasm mimicking) using the optimized potentials for liquid simulations (OPLS) parameter set; bottom, structures modeled for hydrophobic conditions (e.g. cell membrane-mimicking) using the semi-empirical parameterized model 3 (PM3).18,19,20
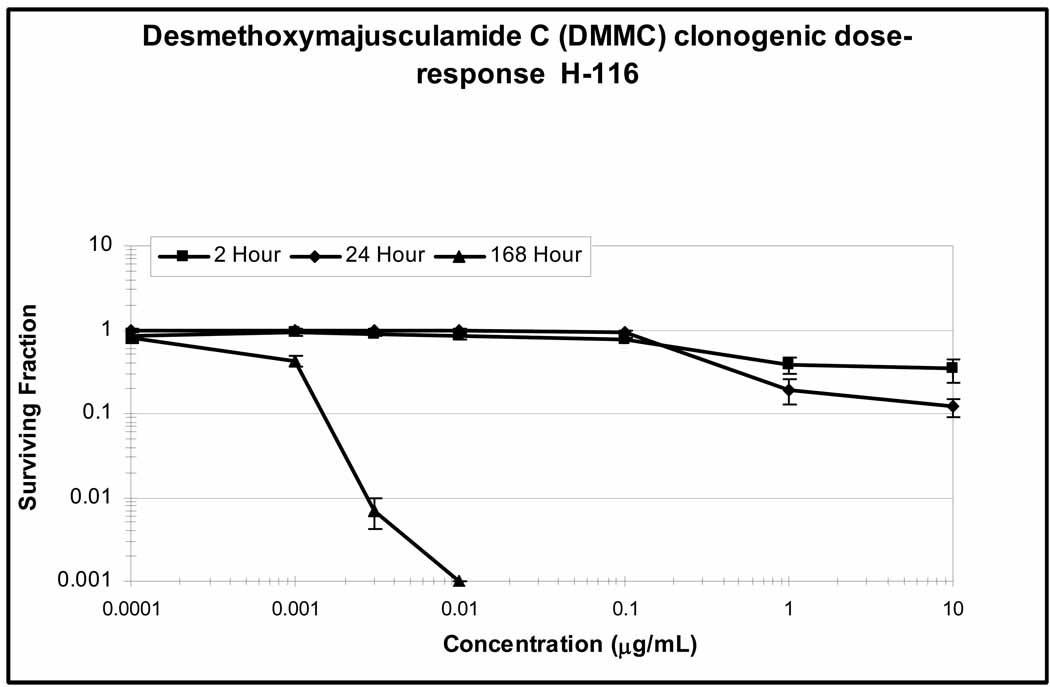
A clonogenic concentration-survival study was conducted for compound 1 to both define the effect of exposure duration on cytotoxicity and to provide a guide for determining the most effective dose schedule for the subsequent therapeutic assessment.17 As shown in Figure 6, clonogenic survival of HCT-116 cells was determined at three different exposure durations: 2 h, 24 h and continuous (168 h or 7 day), as a function of drug concentration. The concentration for an exposure duration that yields a surviving fraction of 10% (tS10) was determined: 2S10>10.5 µM; 24S10>10.5 µM; 168S10=2.1 nM. These results indicate that in order to observe a therapeutic effect, the concentration of DMMC with HCT-116 cells in vivo needed to be >10.5 µM at either the 2 h or 24 h time point if a single, bolus dose was administered, or continuously at >2.1 nM if a chronic (7 day) dosing was given.
Figure 6.
Clonogenic dose-response curve of HCT-116 human colon cancer cells exposed to DMMC (1) for 2 h (square), 24 h (diamond), and 168 h (triangle) in vitro.
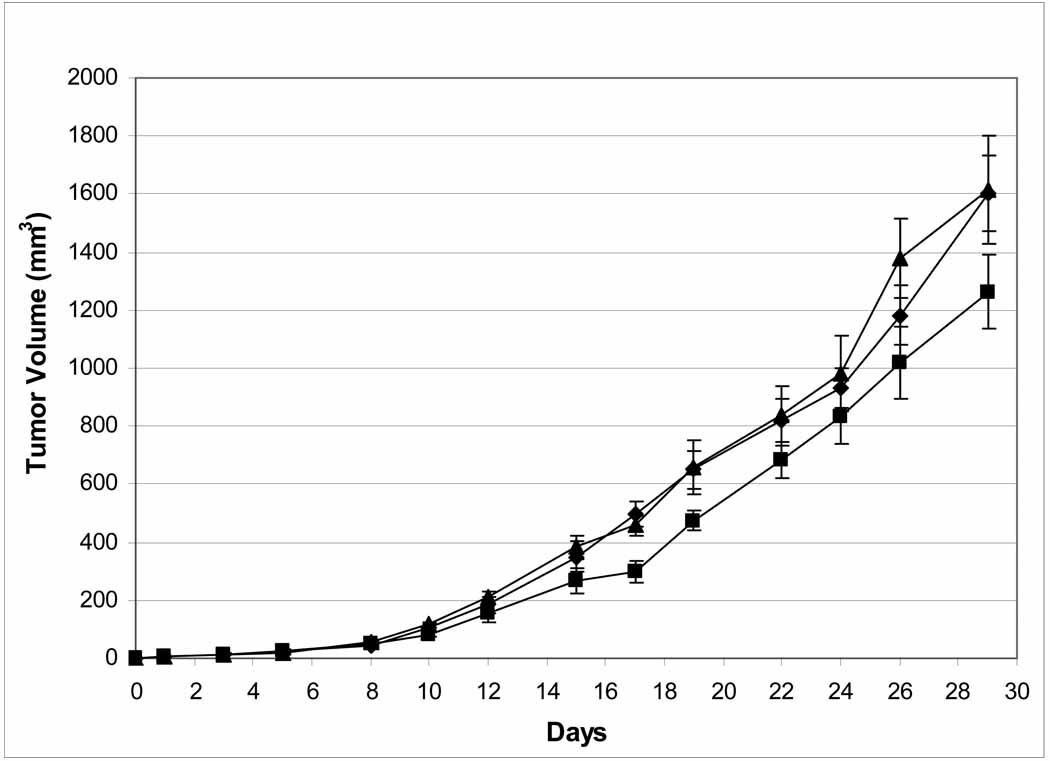
The clonogenic results were sufficiently encouraging to design a therapeutic efficacy trial of DMMC (1) using HCT-116 tumor cells in SCID mice. We first determined the maximum tolerated dose (MTD) to be approximately 2.5 mg/kg for SCID mice. Unfortunately, we did not have sufficient drug to carry out a 5-day therapeutic trial at 2.5 mg/kg/day. Because we needed two doses of drug to carry out the trial, and given the limitation of DMMC availability, we chose doses of 0.62 and 0.31 mg/kg/day. The results are presented in Figure 7 and show that the tumor growth rate for the lower dose schedule was identical to the untreated control, while the higher dose yielded a %T/C value of 60% at 17 days, and indicates that efficacy was found at about one-fourth of the maximum tolerated dose. A further 30 mg of DMMC (1) is needed for an adequate trial against HCT-116 at doses closer to the MTD; if found effective, evaluations should be repeated in both a multiple treatment schedule and against other tumor types.
Figure 7.
Therapeutic assessment of DMMC (1) against HCT-116 human colon cancer cells growing subcutaneously in SCID mice. Drug was administered daily for 5 days intravenously [untreated control (diamonds), 0.31 mg/kg/day (triangles), 0.62 mg/kg/day (squares)].
Conclusions
DMMC (1), a new cyclic depsipeptide from the marine cyanobacterium Lyngbya majuscula, was isolated through a cancer cell cytotoxicity assay directed process. As such, it represents the newest member of the majusculamide/lyngbyastatin natural product group, and extends our knowledge of the range of methylation patterns possible within this skeletal class. DMMC displays both a mechanism and potency of biological activity that is consistent with the most active members in this structural class. Remarkably, we observed a similar high level of cytotoxic activity in the ring-opened, linear form of this compound with IC50 values equivalent to those of the parent structure. Thus, in vitro cellular and limited in vivo therapeutic studies indicate the potential for DMMC, and possibly its linearized form, in cancer treatment.
The majusculamide/lyngbyastatin structure class displays interesting features from a biosynthetic perspective. The first and most obvious is the variable degree of methylation on the mixed PKS/NRPS backbone. As illustrated in Figure 3, there are five sites of variable C-, N- and O-methylation which give rise to the metabolite diversity found in this structural class. While some of the variability can be attributed to promiscuity of amino acid incorporation during NRPS chain extension (e.g. Tyr/Phe, Ile/Leu, Map/Ampa), additional variability derives from variable methylation of the tyrosine oxygen, as well as the amide nitrogen of several amino acid residues. As such, a growing appreciation of structure-activity relationships in this metabolite class is developing from these naturally-occurring analogs, and combined with the antitumor efficacy studies performed herein, should stimulate additional synthetic efforts to more completely evaluate this group of natural product for useful properties.
Methods
General Experimental
Optical rotation was measured on a Perkin-Elmer 243 polarimeter, UV recorded on a Waters Millipore Lambda-Max model 480 LC spectrophotometer and IR recorded on a Nicolet 510 Fourier transform IR spectrophotometer. All NMR data were recorded on Bruker AM400 (Table 1) and DRX300 MHz (Supporting Information) spectrometers, with the solvent CDCl3 used as an internal standard (δC 77.0, δH 7.26). Chemical shifts are reported in ppm and coupling constants (J) are reported in Hz. The FAB mass spectrum and CID mass spectrum were recorded on Kratos MS50TC and Perkin Elmer Sciex API3 mass spectrometers, respectively. HPLC isolation of 1 was performed using Waters 515 HPLC pumps, and all solvents were either freshly distilled or purchased as HPLC grade.
Cyanobacterial Collection and Identification
The marine cyanobacterium Lyngbya majuscula was collected from the Kauviti Reef of Yanuca Island, Fiji on February 16, 2000. The specimen was identified morphologically by WHG (voucher specimen available as collection number VKR-16/Feb/00-05). The material was stored in 2-propanol at reduced temperature until extraction.
Isolation of DMMC (1)
Approximately 300 g dry weight of the cyanobacterium was repetitively extracted with 2:1 CH2Cl2/CH3OH to yield 5.5 g of crude extract. A portion of the extract (5.3 g) was fractionated by vacuum liquid chromatography (VLC) over silica gel. The fraction eluting with 100% EtOAc was further chromatographed by C18 SPE (7:3 CH3OH/H2O) followed by RP HPLC (Phenomenex Sphereclone 5 µm ODS column, 17:3 CH3OH/H2O) to yield 27.5 mg of pure 1.
Desmethoxymajusculamide C (1)
Colorless glassy oil; [α]22D -104 (c 1.86, CH2Cl2); IR (neat) 3315, 2966, 2934, 2877, 1739, 1641, 1519, 1460, 1410, 1286 cm−1; 1H and 13C NMR data, see Table 1 and Supporting Information; HR FAB MS m/z [M + H]+ 955.5867 (calculated for C49H79N8O11, 955.5868).
Base Hydrolysis of 1
Approximately 6 mg of 1 was suspended in 2 mL of a 1:1 CH3OH/0.5 M NaOH solution and allowed to stand overnight at room temperature. The CH3OH was removed by evaporating under N2 and the mixture neutralized with HCl and then extracted with EtOAc. The organic layer was dried under N2 and purified using HPLC (17:3 CH3OH/H2O) to give the base hydrolysis product 2 at > 98% yield and gave the following FAB MS (3-nitrobenzylalcohol) m/z 973 (35), 803 (25), 675 (41), 630 (52), 505 (30), 469 (100), 356 (66), 342 (54), 299 (82), 244 (31).
Absolute Configuration of the Standard Amino Acid Residues in 1
Approximately 1.0 mg of 1 was hydrolyzed with 6N HCl (Ace high pressure tube, microwave, 1.0 min), and the hydrolysate was evaporated to dryness and resuspended in H2O (50 µL). A 0.1% 1-fluoro-2,4-dinitrophenyl-5-l-alaninamide (l-Marfey's reagent) solution in acetone (50 µL) and 20 µL of 1N NaHCO3 were added, and the mixture was heated at 40°C for 1 h. The solution was cooled to room temperature, neutralized with 10 µL of 2N HCl and evaporated to dryness. The residue was resuspended in H2O (50 µL) and analyzed by reversed-phase HPLC (LiChrospher 100 C18, 5 µm, UV detection at 340 nm) using a linear gradient of 9:1 50 mM triethylammonium phosphate (TEAP) buffer (pH 3.1)/CH3CN to 1:1 TEAP/CH3CN over 60 min. The retention times (tR, min) of the derivatized residues in the hydrolysate of 1 matched l-Ala (22.1; d-Ala, 26.9), N-Me-l-Val (36.5; N-Me-d-Val, 39.7), and N-Me-l-Phe (38.5; N-Me-d-Phe, 39.2). Given that only N-Me-l-Ile and N-Me-l-allo-Ile were commercially available, the d-Marfey's reagent was used to make N-Me-d-Ile and N-Me-d-allo-Ile chromatographic equivalents. The retention time of the Ile derivative from the hydrolysate matched that of N-Me-l-Ile (40.7 min; N-Me-l-allo-Ile, 41.2 min; N-Me-d-Ile, 44.2 min; N-Me-d-allo-Ile, 44.7 min).
Absolute configuration of the non-standard amino- and hydroxy-acid residues in 1
Determination of the absolute configuration for the 3-amino-2-methylpentanoic (Map), 2-hydroxy-3-methylpentanoic (HMPA), and 4-amino-2,2-dimethyl-3-oxopentanoic acid (Ibu) residues was accomplished using a combination of Marfey’s method and chiral HPLC. Authentic chromatographic standards for the Map and Ibu residues were obtained as gifts from the laboratory of R.E. Moore, Department of Chemistry, University of Hawaii. In both cases, approximately 1.0 mg of 1 was hydrolyzed with 6N HCl (Ace high pressure tube, microwave, 1.0 min), and the hydrolysate was evaporated to dryness and resuspended in H2O (50 µL). A 0.1% 1-fluoro-2,4-dinitrophenyl-5-L-alaninamide (Marfey's reagent) solution in acetone (50 µL) and 20 µL of 1N NaHCO3 were added, and the mixture was heated at 40°C for 1 h. The solution was cooled to room temperature, neutralized with 10 µL of 2N HCl and evaporated to dryness. The residue was resuspended in H2O (50 µL) and analyzed by reversed-phase HPLC (LiChrospher 100 C18, 5µm, UV detection at 340 nm) using a linear gradient of 9:1 50 mM triethylammonium phosphate (TEAP) buffer (pH 3.1)/CH3CN to 4:6 TEAP/CH3CN over 60 min. Retention times, under these conditions, for the hydrolysate of 1 indicated 2S,3R-Map (47.0 min; 2R,3S-Map, 36.5 min; 2S,3S-Map, 37.0 min; 2R,3R-Map, 28.9 min); 4S-Ibu (34.2 min; 4R-Ibu, 32.8 min, respectively).
Preparation and chiral analysis of HMPA
l-Ile (100 mg, 0.75 mmol) was dissolved in 50 mL 0.2 N perchloric acid (0°C). To this was added a cold (0°C) solution of NaSO3 (1.4 g, 20 mmol) in 20 mL H2O with rapid stirring. With continued stirring the reaction mixture was allowed to reach room temperature until evolution of N2 subsided (~ 30 min). The solution was then brought to boil for 3 min, cooled to room temperature, and saturated with NaCl. The mixture was then extracted with Et2O and dried under vacuum. The three other stereoisomers 2R,3R-HMPA, 2R,3S-HMPA, and 2S,3R-HMPA were synthesized in a similar manner from d-Ile, d-allo-Ile, and l-allo-Ile, respectively.21 A portion of the resultant oil was dissolved in aq 2 mM CuSO4 buffer for HPLC. The retention time [Chirex-D, isocratic system (85:15) 2 mM CuSO4: MeCN] of the natural product hydrolysate matched that for 2S,3S-HMPA (17.1 min; 2R,3S-HMPA, 11.8 min; 2S,3R-HMPA, 9.2 min and 2R,3R-HMPA, 21.2 min, respectively).
Determination of IC50 values for 1 and 2 against HCT-116 cells
Concentration-cell number studies (IC50 assay) were carried out against HCT-116 cells. These cells were grown in 5 mL culture medium (RPMI-1640 + 15% FBS containing 1% penicillin-streptomycin, and 1% Glutamine) at 37 °C and 5% CO2 at a starting concentration of 5×104 cells/T25 flask. On day 3, cells were exposed to different concentrations of the drug. Flasks were incubated for 120 h (5 d) in a 5% CO2 incubator at 37 °C and the cells harvested with trypsin, washed once with HBSS and then resuspended in HBSS and counted using a hemocytometer. The results were normalized to an untreated control and IC50 values determined using Prism 4.0.
DMMC (1) Clonogenic Dose-Response Analysis against HCT-116 Cells
Concentration- and time-survival studies were carried out with HCT-116 cells seeded at 200 or 20,000 cells in 60 mm dishes. DMMC (1) was added to the medium (RPMI + 10% FBS) to a final concentration of 10 µg/mL (= 10.5 µM) and 10-fold dilutions thereof. At either 2 or 24 h, the drug containing medium was removed and fresh medium without drug was added. For continuous exposure to drug, the medium containing the compounds remained in contact with the cells for the entire incubation period (168 h). The dishes were incubated for 7 days, the medium removed and the colonies stained with methylene blue. Colonies containing 50 cells or more were counted. The results were normalized to an untreated control. Plating efficiency for the untreated cells was about 90%.
Therapeutic Assessment of 1 and 2
The in vivo therapeutic assessment trial was carried out using the HCT-116 human colon tumor model as previously described.22 Individual mouse body weights for each experiment were within 5 g, and all mice were over 17 g at the start of therapy. The mice were supplied food and water ad libidum. SCID mice were pooled, implanted subcutaneously with 106 tissue-derived tumor cells, and pooled again before distribution to treatment and control groups (5 mice per group). Treatment with 1 started 1 day after tumor inoculation. Mice were sacrificed after 30 days had elapsed from tumor inoculation. Tumor weights were estimated using two-dimensional caliper measurements done three times per week using the following formula: tumor weight (mg) (a × b2)/2, where a and b are the tumor length and width, respectively, in mm. The median tumor weight was calculated as an indication of antitumor effectiveness. The parameter, % T/C, was determined after each measurement and the minimum value reported as therapeutic efficacy. Compound 1 was prepared as a stock solution in DMSO, diluted 1:1 (v/v) with Cremophor-propylene glycol (40:60 v/v) and further diluted at least 10-fold with saline before injection. Drug was prepared at 0.05 and 0.25 mg/mL for intravenous administration as a bolus injection, daily for 5 days, in 0.25 mL volumes via the tail vein, corresponding to 0.62 and 0.31 mg/kg/day, respectively.
Microfilament Disrupting Activity of Compounds 1 and 2
Cyclic (1) and linear (2) DMMC were tested for microfilament-disrupting activity using rhodamine-phalloidin dye visualization. A-10 cells were grown on glass cover slips in Basal Medium Eagle (BME) containing 10% fetal calf serum. The cells were incubated with the test compounds for 24 h and then fixed with 3% paraformaldehyde for 20 min, permeabilized with 0.2% Triton X-100 for 2 min, and chemically reduced with NaBH4 (1 mg/mL in PBS) three times for 5 min each. Following a 45 min incubation with 100 nM TRITC-phalloidin in phosphate buffered saline, the cover slips were washed, stained with 4,6-diamidino-2-phenylindole (DAPI), mounted on microscope slides, and examined using a Nikon Eclipse ES800 fluorescence microscope with a digital camera.
Supplementary Material
Figure 1.
Molecular structures of cyclic DMMC (1) and its linear hydrolysis product (2).
Acknowledgments
We thank B. Marquez and T. Okino for collection of the source cyanobacterium and the country of Fiji for permission to make these collections. We thank the Environmental Health Sciences Center at OSU for mass spectrometry and the Chemistry Department, OSU for NMR spectroscopy. This work was supported by NIH grant CA100851.
Footnotes
Supporting Information Available: FT-IR, MALDI-TOF-MS, 1H NMR, 13C NMR, 1H-1H COSY, 1H-13C HMBC NMR, and 1H-13C meHSQC NMR of DMMC (1) in CDCl3 at either 300 MHz (1H) or 75 MHz (13C). This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.Papakonstani EA, Stournaras C. Methods Enzymol. 2007;428:227–240. doi: 10.1016/S0076-6879(07)28012-7. [DOI] [PubMed] [Google Scholar]
- 2.Janmey PA, Chaponnier C. Curr. Opin. Cell Biol. 1995;7:111–117. doi: 10.1016/0955-0674(95)80052-2. [DOI] [PubMed] [Google Scholar]
- 3.Jordan MA, Wilson L. Curr. Opin. Cell. Biol. 1998;10:123–130. doi: 10.1016/s0955-0674(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 4.Sahai E. Nat. Rev. Cancer. 2007;7:737–749. doi: 10.1038/nrc2229. [DOI] [PubMed] [Google Scholar]
- 5.Simmons TL, Andrianasolo E, McPhail KL, Flatt P, Gerwick WH. Mol. Cancer Ther. 2005;4:333–342. [PubMed] [Google Scholar]
- 6.Simmons TL, Gerwick WH. Anticancer Drugs of Marine Origin. In: Walsh PJ, Smith S, Fleming L, Solo-Gabriele H, Gerwick WH, editors. Oceans and Human Health: Risks and Remedies from the Seas. UK: Elsevier Science and Technology; 2008. p. 431. [Google Scholar]
- 7.Bubb MR, Senderowicz AMJ, Sausville EA, Duncan KLK, Korn ED. J. Biol. Chem. 1994;269:14869–14871. [PubMed] [Google Scholar]
- 8.Marquez BL, Watts KS, Yokochi A, Roberts MA, Verdier-Pinard P, Jimenez JI, Hamel E, Scheuer PJ, Gerwick WH. J. Nat. Prod. 2002;65:866–871. doi: 10.1021/np0106283. [DOI] [PubMed] [Google Scholar]
- 9.Bai R, Covell DG, Liu C, Ghosh AK, Hamel E. J. Chem. Biol. 2002;277:32165–32171. doi: 10.1074/jbc.M205076200. [DOI] [PubMed] [Google Scholar]
- 10.Morton WM, Ayscough KR, McLaughlin PJ. Nat. Cell Biol. 2000;2:376–378. doi: 10.1038/35014075. [DOI] [PubMed] [Google Scholar]
- 11.Klehchin VA, Allingham JS, King R, Tanaka J, Marriott G, Rayment I. Nat. Struct. Biol. 2003;10:1058–1063. doi: 10.1038/nsb1006. [DOI] [PubMed] [Google Scholar]
- 12.Harrigan GG, Yoshida WY, Moore RE, Nagle DG, Park PU, Biggs J, Paul VJ, Mooberry SL, Corbett TH, Valeriote FA. J. Nat. Prod. 1998;61:1221–1225. doi: 10.1021/np9801211. [DOI] [PubMed] [Google Scholar]
- 13.Bia R, Verdier-Panard P, Gangwar S, Stessman CC, McClure KJ, Sausville EA, Pettit GR, Bates RB, Hamel E. Mol. Pharmacol. 2001;59:462–469. doi: 10.1124/mol.59.3.462. [DOI] [PubMed] [Google Scholar]
- 14.Carter DC, Moore RE, Mynderse JS, Niemczura WP, Todd JS. J. Org. Chem. 1984;49:236–241. [Google Scholar]
- 15.Pettit GR, Kamano Y, Kizu H, et al. Heterocycles. 1989;28:553–558. [Google Scholar]
- 16.Mynderse JS, Hunt AH, Moore RE. J. Nat. Prod. 1988;51:1299–1301. [Google Scholar]
- 17.Subramanian BN, Tenney K, Crews P, Gunatilaka L, Valeriote FA. J. Exp. Ther. Oncol. 2006;5:195–204. [PMC free article] [PubMed] [Google Scholar]
- 18.Spartan ’02 (Linux) Irvine, CA: Wavefunction Inc; 92612. [Google Scholar]
- 19.Jorgenson WL, Tirado-Rives J. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 20.Stewart JJP. J. Comput. Chem. 1989;10:221–264. [Google Scholar]
- 21.Mamer OA. Meth. Enzymol. 2000;324:3–10. doi: 10.1016/s0076-6879(00)24213-4. [DOI] [PubMed] [Google Scholar]
- 22.Corbett T, Valeriote F, LoRosso P, et al. J. Pharmacog. 1995;S33:102–122. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.