Abstract
All solid malignancies share characteristic traits, including unlimited cellular proliferation, evasion of immune regulation, and the propensity to metastasize. The authors have previously described that a subnuclear structure, the perinucleolar compartment (PNC), is associated with the metastatic phenotype in solid tumor cancer cells. The percentage of cancer cells that contain PNCs (PNC prevalence) is indicative of the malignancy of a tumor both in vitro and in vivo, and thus PNC prevalence is a marker that reflects metastatic capability in a population of tumor cells. Although the function of the PNC remains to be determined, the PNC is highly enriched with small RNAs and RNA binding proteins. The initial chemical biology studies using a set of anticancer drugs that disassemble PNCs revealed a direct association of the structure with DNA. Therefore, PNC prevalence reduction as a phenotypic marker can be used to identify compounds that target cellular processes required for PNC maintenance and hence used to elucidate the nature of the PNC function. Here the authors report the development of an automated high-content screening assay that is capable of detecting PNC prevalence in prostate cancer cells (PC-3M) stably expressing a green fluorescent protein (GFP)–fusion protein that localizes to the PNC. The assay was optimized using known PNC-reducing drugs and non-PNC-reducing cytotoxic drugs. After optimization, the fidelity of the assay was probed with a collection of 8284 compounds and was shown to be robust and capable of detecting known and novel PNC-reducing compounds, making it the first reported high-content phenotypic screen for small changes in nuclear structure.
Keywords: high-content screening, nucleus, cancer, perinucleolar compartment, small molecules
INTRODUCTION
The perinucleolar compartment (PNC) is a nonmembrane-bound nuclear subdomain that is associated with but structurally distinct from the nucleolus. The PNC is a heritable trait, in which the number of PNCs per cell in daughter cells often mimics that of their mother cells. The PNC is heterogeneous in shape, is stable through interphase, disassembles during mitosis, and reassembles in early G1.1 The PNC is concentrated with newly synthesized RNA polymerase III (pol III) RNAs (MRP RNA, RNAse P H1 RNA, hY RNAs [hY1, 2, 5], Alu RNA, and SRP [7SL] RNA) and RNA binding proteins (nucleolin, PTB, CUG-BP, KSRP, Raver1, Raver2, and Rod12–9 and our unpublished data). Continuous transcription by pol III is necessary for the structural integrity of the PNC,6 and some protein components have been shown to bind RNAs within the PNC,10 implicating involvement of PNCs in pol III RNA metabolism. Chemical biology studies have demonstrated that PNC maintenance is dependent on the integrity of DNA, specifically DNA base pairing, and cell biology investigations demonstrated that the PNC is nucleated upon a DNA locus, indicating that the components of the PNC are most likely directly interacting with the DNA locus.11
Extensive in vitro studies have shown that the PNC is unique to tumor cells and preferentially forms in tumor cells derived from solid tissues.1,12 Examination of cancer cell lines from various origins and malignant capacities has shown that PNC prevalence correlates with the malignancy of tumors and has the potential to be developed as a pan-cancer prognostic marker.12 In vivo investigations using human breast tissue samples demonstrated that PNC prevalence was 0% in normal breast epithelium, increases in parallel with disease progression (as determined by staging), and reaches nearly 100% in distant metastases, demonstrating that PNC prevalence associates with the malignancy of breast cancer in vivo. Multivariate analysis further showed that a high PNC prevalence is associated with poor patient outcomes independent of current prognostic factors for stage I breast cancer patients.13 These studies demonstrate that the presence of the PNC reflects an advanced transformation state of cancer cells that are capable of metastasis.
Although the PNC is associated with a DNA locus and a handful of components have been identified, much remains to be investigated regarding the structure and function of the PNC and its mechanistic link to cancer. We have previously shown that PNC prevalence reduction is a valid strategy to identify compounds that can be used to study the structure and function of the PNC.12 It is necessary, however, to create a screening platform that is able to test a large number of compounds in an automated fashion. Here we report the development and robustness of an automated whole-cell phenotypic image-based screening assay that is capable of identifying such compounds. We have validated the assay by screening a collection of 8284 compounds. This is the first reported high-content screen that is able to detect small changes in nuclear structure, such as disassembly of the PNC, and thereby this strategy and technique are applicable to the discovery of compounds that can be used to study other nuclear substructures.
MATERIALS AND METHODS
Cell culture
PC-3M cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 1× penicillin/streptomycin, 10% fetal bovine serum, and 250 µg/mL Geneticin (all from Invitrogen, Carlsbad, CA). Cells were passaged when they reached 70% to 80% confluency, and all experiments were performed below passage number 25 (passage 1 started from the time the clonal population was first expanded to a 100-mm culture dish from a single cell).
Immunostaining
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min followed by 5 min permeabilization with 0.5% (vol/vol) Triton X-100 at room temperature. Primary antibodies (against PTB1 and CUG-BP8) were applied for 1 h at a dilution of 1:200. Cells were washed with PBS 3 times before incubations with secondary antibodies conjugated with FITC or Texas Red (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Signal was visualized using a Nikon Eclipse E800 microscope equipped with a SenSys cooled CCD camera (Photometrics, Tucson, AZ). Images were captured using Metamorph image acquisition software (Universal Imaging, Downingtown, PA).
Screening procedure
To maximize throughput and minimize reagent expenditure, we developed this assay in a 1536-well plate format. Cells were harvested from near-confluent T175 flasks using 2 mL of TrypLE and 5 mL of PBS (Invitrogen), pelleted, and resuspended in complete DMEM media (lacking Geneticin). The cells were seeded at a density of 1000 cells/well in 5 µL of media into black, micro-clear-bottom 1536-well low base plates (Aurora Discovery, San Diego, CA) using a Multidrop Combi 8 channel dispenser (Thermo Fisher Scientific, Inc., Waltham, MA). The plates were then incubated at 37 °C in 5% CO2 to allow attachment of cells to the plate. After the overnight incubation, a 1536-well pintool (Kalypsys, San Diego, CA) transferred 23 nL of compounds from library stock plates (all compounds solubilized in 100% DMSO) to the cell plates. The final volume of DMSO in the cell plates was 0.5%, which does not affect PNC prevalence.11 The cells were incubated with the compounds for 18 to 20 h11 and then fixed by the addition of 4 uL of 6% EM grade paraformaldehyde (Electron Microscopy Services, Hatfield, PA) containing 0.1% Triton X-100 (Sigma Aldrich, St. Louis, MO) directly to the media. The fix solution was removed using a Kalypsys aspirator station after a 20-min room temperature incubation. The fixed cells were washed twice with 5 µL Hank’s buffered salt solution (HBSS) per well with a final addition of HBSS containing 10 µg/mL Hoechst 33342 (Invitrogen). Plates were sealed with clear tape and stored in the dark at 4 °C until they were ready for imaging.
Automated microscopy and automated data analysis
The IN Cell Analyzer 1000 automated fluorescent imaging system (GE Healthcare, Piscataway, NJ) was used for automated image acquisition. Images were acquired with a 20× objective using a 475/20-nm excitation filter, a 535/50-nm HQ emission filter, a Q505LP dichroic filter, and an exposure time of 100 to 150 ms (adjusted to obtain a dynamic range of ~200 to 1750) with no camera binning. The instrument acquired images of each well in a 1536-well plate with a laser-based autofocus system. To score PNC prevalence in a high-content throughput, we used the Multi-Target Analysis (MTA) algorithm (GE Healthcare, Investigator v3.5) to identify individual cells and granules (PNCs) within these cells. The nucleus was segmented via a region growing method (50 µm2 minimum area) with light shading and noise removal to allow “touching” nuclei to be separated. Granules in the nucleus (PNCs) were segmented using a multiscale top hat method measuring granules of 1 to 2 µm in size and using a smart masking method to identify more precisely the boundaries of each segmented granule.
The algorithm was optimized and validated using positive (50 µM camptothecin) and negative controls (DMSO). In particular, the MTA algorithm allows for the identification of multiple subcellular compartments and organelles (or granules) within those compartments. In this instance, we took advantage of the algorithm’s capability to identify objects within the same color channel that only differ in size or fluorescent intensity. Also, the algorithm allows for building complex hierarchical classification systems, using output measures within the algorithm to filter and define subpopulations. For this particular assay, PNC-positive cells were scored when 1 to 3 PNC granules were detected per nucleus. Cells containing 0 granules were scored as PNC negative and/or cells with >3 granules were assumed to be false positives (very few cells have more than 3 PNCs in one focus plane) and also scored as PNC negative.
RESULTS
Creation of a stable cell line with a GFP marker of the PNC
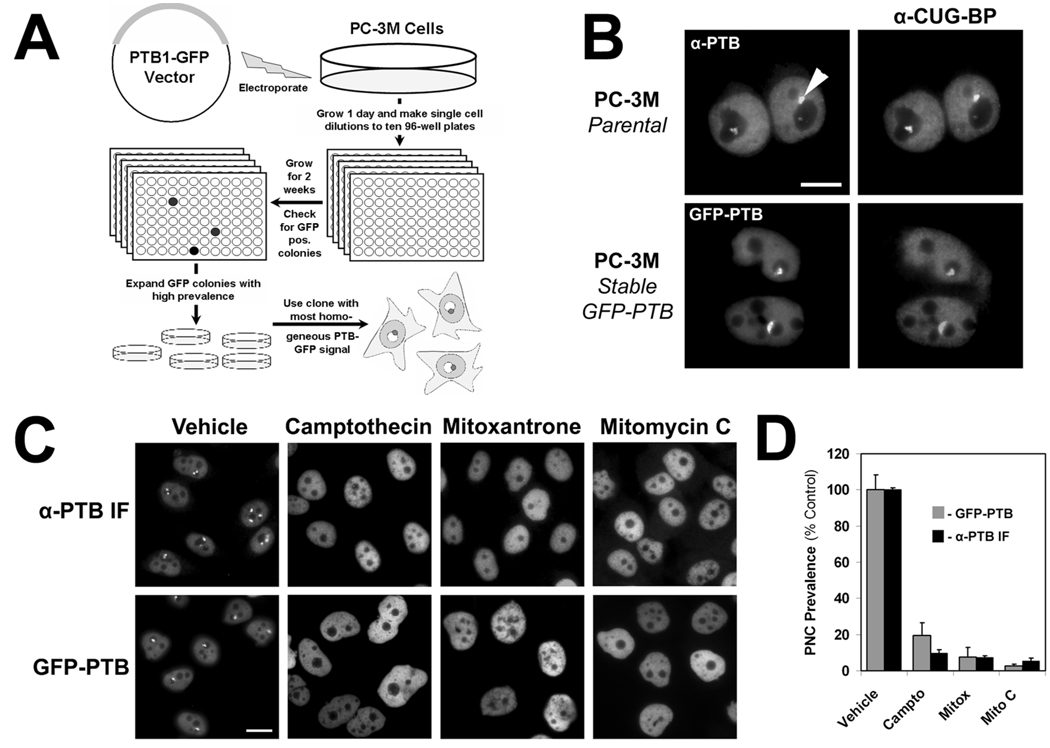
To detect the PNC without the need for immunofluorescent staining, we created a cell line stably expressing a fluorescent protein that localizes to the PNC. The polypyrimidine binding tract protein (PTB) localizes diffusely to the nucleoplasm but is highly enriched in the PNC if present,7 and its expression is essential for the maintenance of the PNC.9 PC-3M (metastatic prostate cancer cell line) cells, with a PNC prevalence of ~80%,12 were transfected with a vector expressing PTB isoform 1 with a C-terminus green fluorescent protein (GFP) tag (PTB-GFP). The PTB-GFP fusion protein has been previously shown to behave similar to endogenous PTB.9 PC-3M cells were chosen because the size of PNCs in PC-3M cells is among the largest we have observed (0.5 to 4 microns [data not shown] compared to only 0.25 to 1 micron in other cell lines).1 After transfection, the cells were allowed to grow for 1 day in a large culture dish, and then single cell dilutions were prepared and grown in ten 96-well plates for 2 weeks under G-418 selection. Wells were scanned with an inverted fluorescent microscope, and 5 wells were found that had stable PTB-GFP expression and high PNC prevalence. The PC-3M clone with the most homogeneous diffuse nuclear fluorescent pattern of PTB expression was chosen as the screening cell line and is referred to here on as the PTB-GFP cell line (Fig. 1A).
FIG. 1.
(A) Schematic for the creation of PC-3M cells stably expressing polypyrimidine binding tract protein–green fluorescent protein (PTB-GFP) to mark the perinucleolar compartment (PNC). (B) Immunofluorescent staining of PTB and CUG-BP in parental PC-3M cells to demonstrate colocalization within the PNC (arrowhead) and colocalization of PTB-GFP and CUG-BP to show that PTB-GFP marks the PNC. (C) Treatment of PTB-GFP cells and parental PC-3M cells (immunofluorescent staining to PTB) with 3 known PNC-disassembling chemotherapeutic drugs.11 (D) Quantification of experiments in section C (n = 3, error bars = SD).
The PNCs in the PTB-GFP cell line were compared to those in the parental PC-3M cell line. PNC number, size, and shape were similar in both cell lines, and PTB-GFP colocalized with CUG-BP, another protein component of the PNC (Fig. 1B). Our previous studies have shown that several anticancer drugs are able to disassemble the PNC in PC-3M cells and that their affect on the PNC is due to specific molecular actions, not as a result of cytotoxicity or growth inhibition.11 Therefore, the response of the PTB-GFP cell line to several of these drugs (camptothecin, mitoxantrone, and mitomycin C) was compared to the response of the parental cell line immunofluorescently stained with a PTB antibody. After a 20-h treatment, the PNC prevalence reduction by these drugs in the PTB-GFP cell line was similar to that of the parental cell line (Figs. 1C,D), validating the utility of the PTB-GFP cell line for the screening of PNC prevalence-reducing compounds.
Parameters for PNC detection and assay fidelity
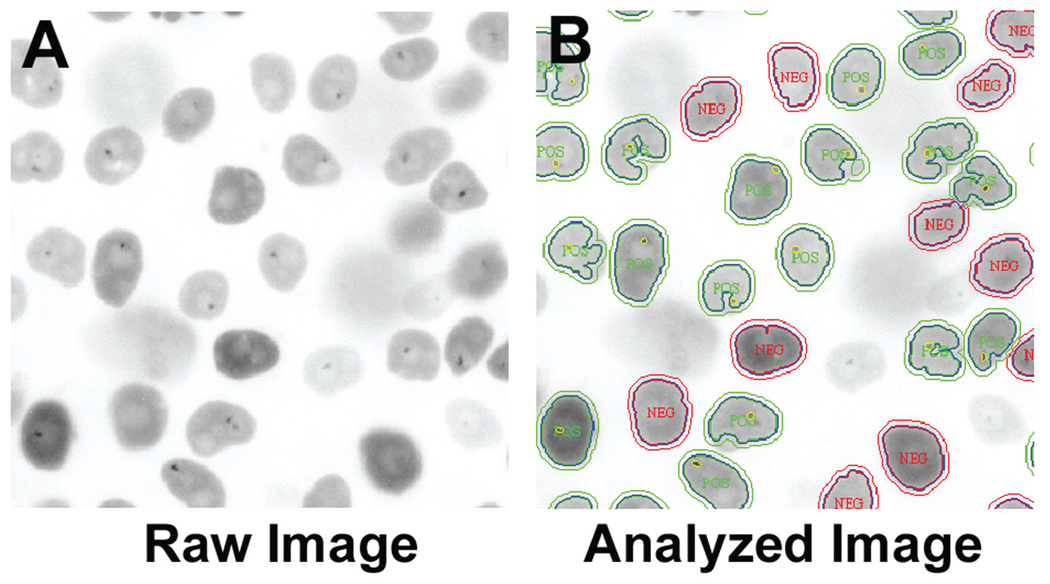
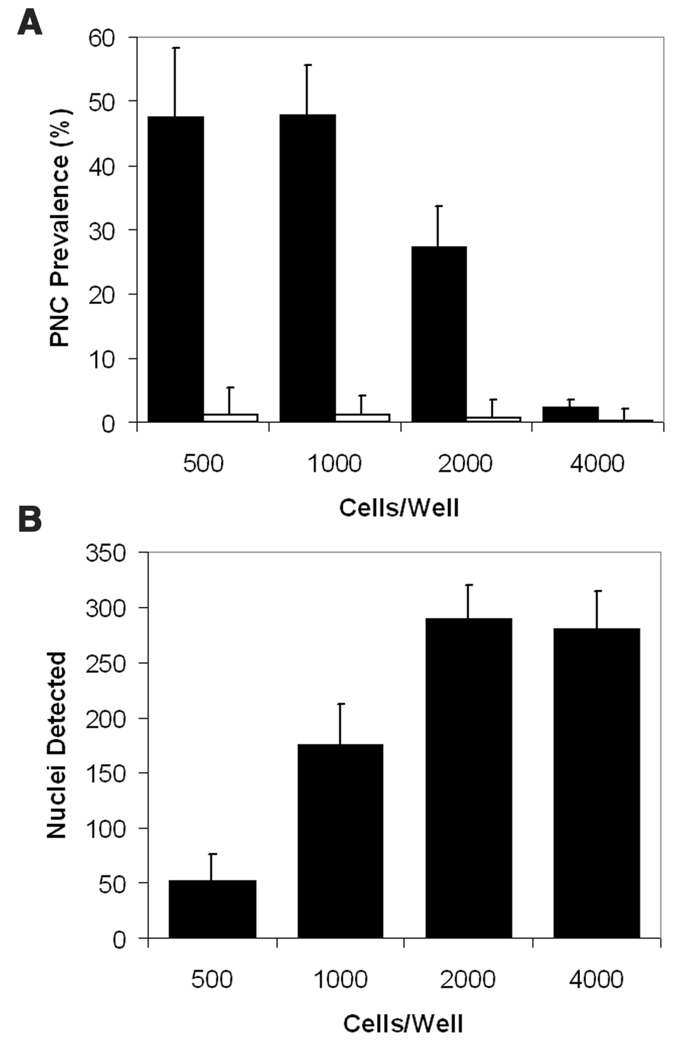
To score PNC prevalence in the high-content assay, we optimized an imaging analysis algorithm. The MTA is a preconfigured algorithm developed to identify subregions of interest within a larger segmented object and was chosen as the PNC detection algorithm because it allowed PNC segmentation and cell population filtering. PNC-positive cells were detected in the untreated controls as a small punctate perinucleolar GFP signal, which is easily visible amid a diffuse nuclear fluorescence (Fig. 2A). The resulting algorithm effectively detected PNCs in the PC-3M PTB-GFP cell line (Fig. 2B). After algorithm optimization, we determined the optimal number of cells per well to be detected by the software. Cells were plated at densities of 500 to 4000 cells/well in a 1536-well plate, allowed to adhere overnight, treated with either vehicle or 50 µM camptothecin for 18 h, and were then fixed and analyzed. When cell density was at 500 and 1000 cells/well (Fig. 3A), the maximum PNC prevalence of untreated cells determined by the algorithm was about 50%. Cells plated at 2000 were nearly confluent, and 4000 were overly confluent, which prevented proper PNC detection by the software. The number of nuclei per 20× field as detected by the software increased as cell densities increased but peaked at 2000 cells/well due to inappropriate software segmentation of nuclei in the wells with 4000 cells (Fig. 3B). To balance PNC detection with maximal nuclear counts, we selected 1000 cells/well as the optimal plating density, at which the PNC prevalence was about 50%. Table 1 depicts the final plate preparation and screening protocol.
FIG. 2.
(A) Inverted gray-scale image of polypyrimidine binding tract protein–green fluorescent protein (PTB-GFP) PC-3M cells, treated with 0.5% DMSO and imaged with a 20× objective, GFP filter, and 100 ms exposure. Dark dots in the nucleus are perinucleolar compartments (PNCs). (B) Raw image with the superimposed postprocessing masks created by the IN Cell Analyzer 1000 Workstation MTA algorithm. POS, cells with PNCs; NEG, cells without PNCs as determined by the algorithm. Only a portion of the entire field of view captured is displayed.
FIG. 3.
The effect of (A) cell density on perinucleolar compartment (PNC) prevalence detection in 0.5% DMSO-treated cells (black bars) and in 50 µ camptothecin-treated cells (white bars) by the IN Cell Analyzer 1000 Workstation MTA algorithm and of (B) cell density on the number of nuclei detected per field (n = 3 and error bars = SD).
Table 1.
Protocol Outline for Plate Preparation with Automation
| Sequence | Parameter | Value | Description |
|---|---|---|---|
| 1 | Reagent | 5 µL | 1000 cells/well in 1536 TC-treated plate |
| 2 | Time | 18–20 h | 37 °C 5% CO2 |
| 3 | Reagent | 23 nL | Compounds |
| 4 | Time | 18 h | 37 °C 5% CO2 |
| 5 | Reagent | 4 µL | 6% PFA with 0.1% Triton X-100 |
| 6 | Time | 20 min | Room temperature—fume hood |
| 7 | Decant | 8 µL | Kalypsys pipettor removes media + PFA |
| 8 | Reagent | 5 µL | HBSS buffer |
| 9 | Decant | 4 µL | Kalypsys pipettor with HBSS |
| 10 | Reagent | 5 µL | HBSS buffer |
| 11 | Decant | 4 µL | Kalypsys pipettor removes HBSS |
| 12 | Reagent | 5 µL | HBSS buffer with 10 µg/mL Hoechst 33342 |
| 13 | Detector | GFP filter set | IN Cell Analyzer 1000 measurement |
GFP, green fluorescent protein; HBSS, Hank’s buffered salt solution; PFA, paraformaldehyde; TC, tissue culture.
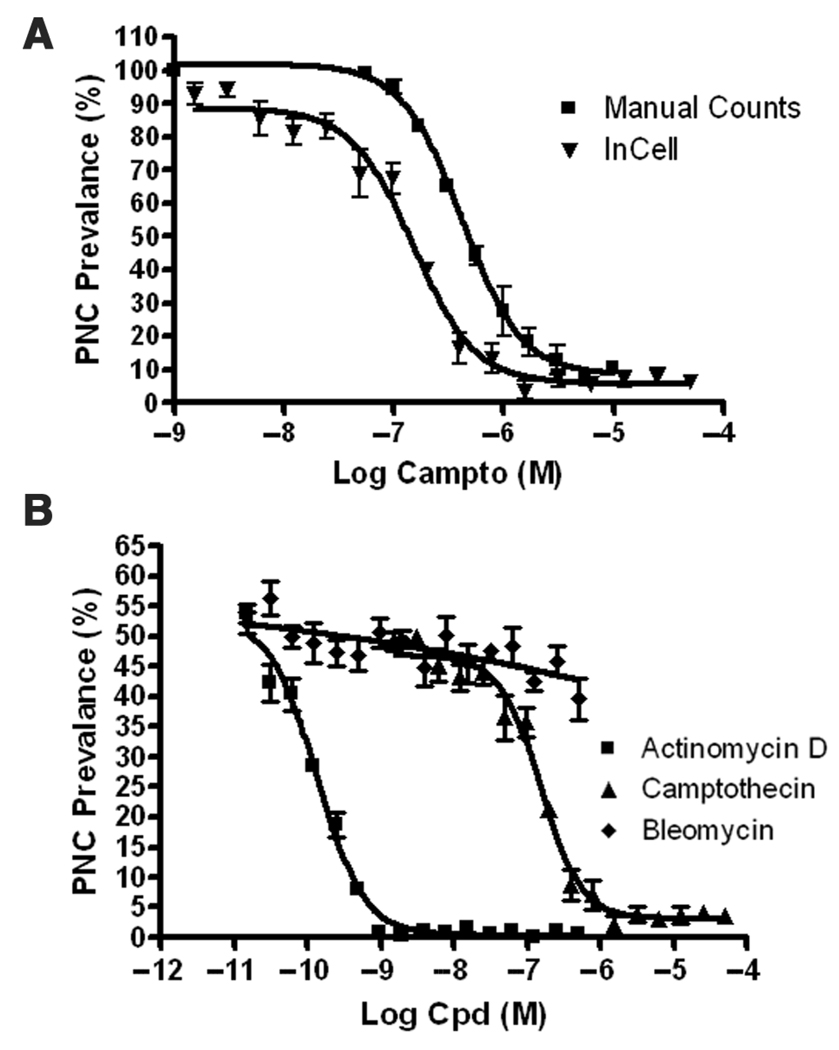
To validate the assay parameters, we manually determined a concentration response curve for camptothecin in the PTB-GFP (cells scored by eye with a microscope). The PNC prevalence in the PTB-GFP PC-3M determined manually using a 60× objective was 85% ± 6.4%, whereas it was 60.5% ± 7.0% as determined by the automated image analysis. This discrepancy can be attributed to the lower power magnification and the fact that not all PNCs are in the single focus plane when viewed by automated imaging; however, when both sets of data are normalized to the vehicle control, there is a correlation of the concentration response curves (Fig. 4A), validating the assay parameters for automated image acquisition and analysis. Although the automated assay is slightly less sensitive than the manual detection for PNC-positive cells, it is sufficient for accurate hit determination. PNC prevalence reduction to 25% of the control (minimum PNC prevalence to be considered a hit for the system reported here) as determined by the automated system corresponds to an approximate 45% reduction in PNC prevalence by the manual method (Fig. 4A), which is considered a substantial reduction.11 In addition to camptothecin, a concentration curve was created for actinomycin D (Fig. 4B), which was able to potently disassemble the PNC as shown in previous results.11 To ensure that cell death induced by cytotoxic compounds would not be falsely counted as hits by the automated system, we conducted a concentration-dependent study with a non-PNC-reducing and highly cytotoxic molecule, bleomycin. Even at the most cytotoxic concentrations tested, bleomycin did not cause a false decrease in PNC prevalence in the automated assay, in accordance with previous results using manual scoring.11
FIG. 4.
(A) Dependence of perinucleolar compartment (PNC) prevalence on camptothecin concentration for manual scoring with a 60× objective compared to that determined by the IN Cell Analyzer 1000 MTA algorithm (normalized to vehicle-treated cells). The calculated EC50% values for camptothecin were 419 nM and 152 nM for manual counts and automated counts, respectively. (B) Dependence of PNC prevalence on actinomycin D concentration compared to camptothecin by the IN Cell Analyzer 1000 MTA algorithm. PNC prevalence determination by the algorithm does not show dependence on bleomycin concentration (a highly cytotoxic non-PNC-reducing compound). The EC50% value for actinomycin D was 133 pM (n=4 and error bars = SD).
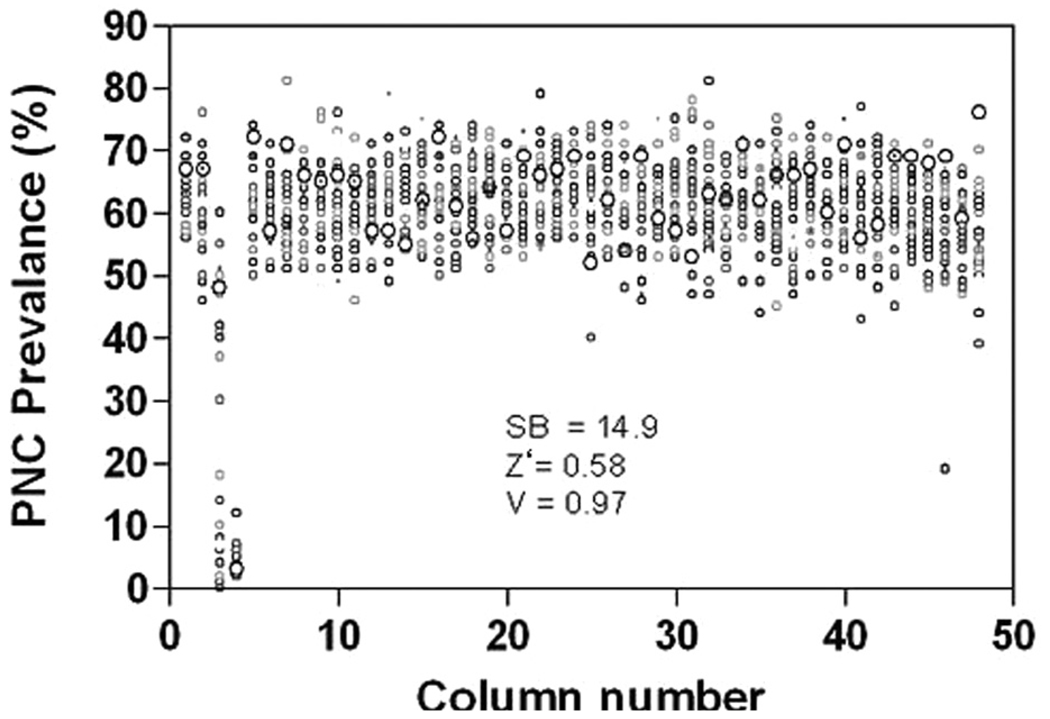
To statistically validate the functional assay and image analysis algorithm, we analyzed the data from camptothecin and vehicle-treated cells in 2 ways. First, the Z′ score, a general measure of robustness for high-throughput and high-content assays that considers fidelity based on only complete or no responses (in our assay: vehicle-treated and 50-µM camptothecin-treated cells), was 0.58 out of 1.00 (Fig. 5). This Z′ score is considered acceptable for cell-based assays, further validating the functional assay and the image analysis algorithm.14 The V score, unlike the Z′ score, takes into account data points that are not complete positives or negatives (i.e., concentration-dependent response).15 Using the camptothecin concentration response curves generated with this automated assay, we obtained a V score of 0.97 out of 1.00 (Fig. 5), which again indicates the robustness of the assay parameters. The higher V score can be explained by several factors. First, the variation in PNC prevalence is fairly consistent among the concentrations of camptothecin (Fig. 4). In addition, the Z′ score is more sensitive to variations in the standard deviation of positive and negative controls, which are introduced during imaging acquisition and analysis, whereas the V score is less susceptible to this variation because multiple points across the concentration response curve are included in its calculation. A signal-to-background ratio of 14.9 was observed in 50 µM camptothecin treatment compared to vehicle-treated cells. These results demonstrate that the automated system we have designed and optimized is sensitive and specific enough for utilization in large screens.
FIG. 5.
DMSO plate values from a 1536-well plate. Columns 1, 2, and 5 to 48 received DMSO (0.5% final concentration). Column 3 received a titration of camptothecin (50 µM down by 1:2 dilutions), and column 4 received 50 µM camptothecin. The cells were plated at 1000 per well and were treated for 18 h. The signal-to-background ratio (SB, n = 3, with 32 wells for 50 µM camptothecin and n = 1408 wells for DMSO, per trial), Z′ score (Z′, n = 3 with 32 wells for positive and 1408 wells for negative), and V score (V, n = 3 per 12 concentration dilution series) are listed in the figure.
To probe the robustness of this assay, we prepared and analyzed plates consisting of 1388 vehicle-treated wells and 10 random wells treated with 50 µM camptothecin and 10 wells treated with 10 µM actinomycin D (both these treatments disassemble most PNCs) using this system. The experiments were run in duplicate, and the results show that all 20 positive control wells were identified as reducing PNC prevalence to below 5% of the control, giving a false-negative rate of 0%. In the first run, there were 6 vehicle-treated wells that came up as false positives, and there were 18 wells in the second run; however, only 1 well came up as a false positive in both runs, giving an initial false-positive rate of 0.08%. After reviewing the images of wells that were false positives, it was determined that they were usually caused by a missed autofocus of the instrument, which resulted in blurred images, leading to falsely deflated PNC prevalences. Therefore, all experiments for this assay were run in duplicate to eliminate technical error/variability, and only compounds that were positive in both trials were considered hits. Approximately 0.5% to 1% of the wells missed autofocus (data not shown), so the chance of a false positive in duplicate plates from a missed autofocus was about 0.025% to 0.01%.
Screening libraries of known drugs and biologically active compounds
The robustness and utility of the optimized assay were first tested by screening the Library of Pharmacologically Active Compounds (LOPAC) from Sigma. For this screen, a hit compound had to reduce PNC prevalence to below 25% of the control value at 25 µM in both of 2 independent trials. A less stringent hit selection was used in this trial to determine if hits that were slightly less effective, which may belong to novel mechanistic classes, were able to be discovered and later be used to probe the function of the PNC. In addition, this less stringent hit rate would allow us to test fidelity of the assay by retesting the hits manually and determining if there any were false positives. With the hit cutoff of 25% PNC prevalence compared to control in 2 independent trials, 11 hits were discovered from the 1280 compounds in this library, giving a hit rate of ~0.8%. Five previously known PNC prevalence-reducing compounds—camptothecin, ellipticine, mitoxantrone, etoposide,11 and genistein (our unpublished data)—were identified as hits, further validating this assay. The other 6 compounds were retested manually and scored by eye to confirm their activity, and it was found that 3 were able to reduce PNC prevalence significantly compared to vehicle-treated cells (Table 2) and 3 could not (data not shown), giving a true hit rate of 0.6%. Images of these hits were then examined, and it was found that the 3 false-positive hits did not appear to reduce the PNC but drastically altered nuclear morphology (i.e., severe nuclear fragmentation or mitotic arrest), which likely caused the algorithm to report a falsely deflated PNC prevalence. This observation was confirmed by manual counting of PNC prevalence in nonapoptotic and nonmitotic cells in follow-up assays (data not shown). Therefore, all hits should be verified by manual or semiautomated inspection of the images obtained by the instrument prior to follow-up assays. The mechanisms of action for the LOPAC hits prove that this assay is capable of detecting previously known compounds and discovering new classes of compounds that disassemble the PNC.
Table 2.
List of Compounds Discovered from the Screening of 5 Libraries (8284 compounds) and Whether the Compound or Its Mechanistic Class Was Previously Known to Disassemble the PNC
| Library | Compound | Previously Discovered | Mechanistic Class | Mechanistic Class Known to Reduce PNC |
|---|---|---|---|---|
| LOPAC | (S)-(+)-camptothecin | [10] | Topo I inhibitor | Yes |
| 7-Chloro-4-hydroxy-2-phenyl-1,8-naphthyridine | No | Adenosine receptor antagonist | No | |
| Diphenyleneiodonium chloride | No | Nitric oxide synthase inhibitor | No | |
| Ellipticine | [10] | Intercalator | Yes | |
| Etoposide | [10] | Topo II inhibitor | Yes | |
| Genistein | Yes (our unpublished data) | Topo II inhibitor at high doses | Yes | |
| Mitoxantrone | [10] | Intercalator | Yes | |
| Methotrexate | [10] | Inhibits RNA synthesis | Yes | |
| Quinacrine dihydrochloride | No | Intercalator | Yes | |
| NCGC in house | Acriflavinium | Yes (our unpublished data) | Intercalator | Yes |
| 9-Aminoacridine | Yes (our unpublished data) | Intercalator | Yes | |
| Daunorubicin | [10] | DNA intercalator and Topo II inhibitor | Yes | |
| 3,6-Diaminoacridine | Yes (our unpublished data) | Intercalator | Yes | |
| Doxorubicin | [10] | DNA intercalator and Topo II inhibitor | Yes | |
| Mofetil | No | Nucleoside analog | Yes | |
| Topotecan | [10] | Topo I inhibitor | Yes | |
| Zidovudine | No | Nucleoside analog | Yes | |
| Tocris/Timtec | Actinomycin D | [10] | DNA intercalator and Topo II inhibitor | Yes |
| Camptothecin | [10] | Topo I inhibitor | Yes | |
| Chelidonine | No | Tubilin inhibitor | No | |
| Prestwick | Camptothecin | [10] | Topo I inhibitor | Yes |
| Daunorubicin | [10] | DNA intercalator and Topo II inhibitor | Yes | |
| Doxorubicin | [10] | DNA intercalator and Topo II inhibitor | Yes | |
| Biomol | Actinomycin D | [10] | DNA intercalator and Topo II inhibitor | Yes |
| (S)-(+)-Camptothecin | [10] | Topo I inhibitor | Yes | |
| Diphenyleneiodonium chloride | No | Nitric oxide synthase inhibitor | No | |
| Hydroxy-camptothecin | No | Topo I inhibitor | Yes |
LOPAC, Library of Pharmacologically Active Compounds; NCGC, National Institutes of Health Chemical Genomics Center; PNC, perinucleolar compartment.
To identify potent PNC prevalence-reducing compounds, we used a stringent hit criteria based on p-value scoring (Bonferroni correction was used to account for multiple hypothesis testing effects). The p-values were calculated using a t-test for the differences in PNC prevalence of compound-treated and DMSO-treated cells, and p-values were considered significant if they were less than 0.05/1536 (the multiple hypothesis correction value used). This stringent hit selection criterion was used to screen an in-house National Institutes of Health Chemical Genomics Center (NCGC) library of 2816 known bioactive drugs and diagnostic compounds. The screen was performed in duplicate at a final concentration of 50 and 25 µM (4 plates total). Eight hits were produced from this screen, 6 of which reduced PNC prevalence to below 6% of the control in all 4 trials and 2 of which reduced PNC prevalence to below 6% of the control in 3 of the trials (Table 2). Topotecan, doxorubicin, and daunorubicin have been identified in a previous chemical biology study in which PNC prevalence was determined through manual scoring,11 validating the ability of this assay to detect PNC prevalence-reducing compounds. Acriflavinium, 9-aminoacridine, and 3,6-diaminoacridine are all acridines that are potent DNA and RNA intercalators, and previous studies have shown that DNA intercalators drastically reduce PNC prevalence.11 Mofetil and zidovudine (positive in 3 of 4 trials) are both nucleoside analogs, a class of molecule that can modestly reduce prevalence PNC via pol III inhibition.11 We also screened (in duplicate at 25 µM with a hit cutoff of 6% PNC prevalence of the control) Timtec’s Natural Product Library (480 compounds), Tocris’ Tocriscreen library of biologically active molecules (1180 compounds), Prestwick’s library of marketed drugs and biologically active molecules (1120 compounds), and 1 plate of molecules sampled from all of Biomol’s compound libraries (1408 compounds). The hits from these libraries overlapped with hits found in the NCGC and LOPAC libraries and provided a hit, chelidonine, whose mechanism is not known to affect the PNC (Table 2). Therefore, the screening of 5 different collections of biologically active compounds validated our assay because previously known PNC-reducing compounds were detected, several hits were found in more than 1 library, and we were able to discover compounds whose mechanism is not known to affect the PNC.
DISCUSSION
The PNC has been shown to be specifically associated with metastatic potential of cancer cells.12,13 Identification of the mechanism of PNC disassembly by small molecules could help elucidate the structure and function of the PNC and its possible role in metastatic cells. Initial identification of a subset of anticancer compounds and analyses of their targets have led to the finding that the PNC is associated with a DNA locus,11 demonstrating the effectiveness of chemical biology approaches to dissect the molecular function of the PNC. Because PNCs form in only highly metastatic cells, understanding the function of PNCs can provide insight into the biology of metastatic behavior. Therefore, we have developed and reported here an automated high-content screening assay for the discovery of PNC-reducing compounds, which is the first such screen for changes in subnuclear structures. Using the PTB-GFP cell line and positive and negative control treatments and a rage of camptothecin concentrations, we found this assay to be robust with a Z′ score of 0.58 and a V score of 0.97. Screening of 5 libraries consisting of 8284 compounds produced several hits, which confirmed the robustness of this assay as the hits consisted of novel compounds and previously known compounds, which often overlapped among the libraries.
The automated PNC reduction assay is capable of screening a large number compounds in a short time. It is possible to fix 10 plates (15,360 wells) in a 60-min window due to the PTB-GFP cell line and our 1-step fixation method. This process would take an entire day to complete 5 plates if the cells had to be fixed and immunofluorescently stained and the variability arising from several wash steps required for antibody usage could easily make the assay impossible to conduct in a 1536-well format. The throughput of this assay on the IN Cell Analyzer 1000 is approximately 14 plates (21,504 wells) per an 8-h day, assuming a 35-min plate read time and accounting for instrument setup.
From the diffuse nuclear distribution of the PTB-GFP, the image analysis software was able to detect nuclear boundaries comparable to nuclei stained with a DNA dye, Hoechst 33342 (data not shown). The advantages of using the GFP channel for nuclear detection are reduced read time (single-channel GFP reading takes ~30 min/1536-well plate vs. 45 min for a 2-color acquisition at 100-ms exposure in both channels), and the data file is 4.2 gigabytes as opposed to 8.4 GB per plate. The main disadvantage of using GFP for detection is that mitotic or apoptotic cells cannot always be excluded from analysis as with nuclear stains and thereby occasionally causes a falsely deflated PNC prevalence. Because Hoechst is included in the final HBSS solution, manual or automated reimaging of wells of interest is an easy option for hit validation; however, after extensive analysis, we have found that the most efficient method for secondary validation of hit criteria is manual examination of the image obtained by the IN Cell Analyzer 1000 because the low hit rate observed in this preliminary screening is expected to translate to future screens. Greater than 100 hits can be confirmed or excluded in less than 1 h by comparing the image of the hits to an image of vehicle-treated control cells to ensure there is an obvious decrease in PNC prevalence. In addition, false positives due to a missed autofocus, high toxicity (low cell number), mitotic block, fluorescent compounds, or nuclear fragmentation are excluded from the hits during this manual inspection process. The false positives can automatically be eliminated in future screens by creation of filters in the software for the Hoechst 33342 channel to identify the aforementioned false-positive phenotypes (data not shown); however, because the hit rate is low and manual inspection of the images is rapid and decreases the chance of exclusion of true positives by the software filters, we believe that manual inspection provides satisfactory efficiency for hit confirmations prior to follow-up assays.
In summary, we have reported the development and validation of a robust high-content screening assay for PNC prevalence reduction. This assay will be used to screen large libraries of compounds, with the goal of discovering novel hits that can be used to elucidate the function of the PNC and discover novel biology driving metastasis and also potentially be developed as anticancer drugs. This assay can be adapted to screen for siRNAs that affect the PNC and can also serve as the basis for screens that cause subtle changes in nuclear structure other than PNC disassembly with minimal modifications.
ACKNOWLEDGMENTS
We thank Robert Graves of GE Healthcare for technical assistance and discussion and Noel Southall, Ajit Jadhav, and Trung Nguyen of the NIH Chemical Genomics Center for guidance.
REFERENCES
- 1.Huang S, Deerinck TJ, Ellisman MH, Spector DL. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall MP, Huang S, Black DL. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol Biol Cell. 2004;15:774–786. doi: 10.1091/mbc.E03-09-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang S, Deerinck TJ, Ellisman MH, Spector DL. The perinucleolar compartment and transcription. J Cell Biol. 1998;143:35–47. doi: 10.1083/jcb.143.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huttelmaier S, Illenberger S, Grosheva I, Rudiger M, Singer RH, Jockusch BM. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J Cell Biol. 2001;155:775–786. doi: 10.1083/jcb.200105044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kleinhenz B, Fabienke M, Swiniarski S, Wittenmayer N, Kirsch J, Jockusch BM, et al. Raver2, a new member of the hnRNP family. FEBS Lett. 2005;579:4254–4258. doi: 10.1016/j.febslet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Kopp K, Huang S. Perinucleolar compartment and transformation. J Cell Biochem. 2005;95:217–225. doi: 10.1002/jcb.20403. [DOI] [PubMed] [Google Scholar]
- 7.Matera AG, Frey MR, Margelot K, Wolin SL. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, et al. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Politz JC, Pederson T, Huang S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell. 2003;14:2425–2435. doi: 10.1091/mbc.E02-12-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lorenz M. Visualizing protein-RNA interactions inside cells by fluorescence resonance energy transfer. RNA. 2009;15:97–103. doi: 10.1261/rna.1307809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norton JT, Wang C, Gjidoda A, Henry RW, Huang S. The perinucleolar compartment is directly associated with DNA. J Biol Chem. 2009;284:4090–4101. doi: 10.1074/jbc.M807255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Norton JT, Pollock CB, Wang C, Schink JC, Kim JJ, Huang S. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer. 2008;113:861–869. doi: 10.1002/cncr.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamath RV, Thor AD, Wang C, Edgerton SM, Slusarczyk A, Leary DJ, et al. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005;65:246–253. [PubMed] [Google Scholar]
- 14.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 15.Ravkin I, Temov V, Nelson A, Zarowitz MA, Hoopes M, Verhovsky Y, et al. Multiplexed high-throughput image cytometry using encoded carriers. Proc SPIE. 2004;5322:52–63. [Google Scholar]