Abstract
Carnitine is a quaternary amine compound prevalent in animal tissues, and a potential carbon, nitrogen and energy source for pathogens during infection. Characterization of activities in Pseudomonas aeruginosa cell lysates has previously shown that carnitine is converted to 3-dehydrocarnitine (3-dhc) which is in turn metabolized to glycine betaine (GB), an intermediate metabolite in the catabolism of carnitine to glycine. However, the identities of the enzymes required for carnitine catabolism were not known. We used a genetic screen of the P. aeruginosa PA14 transposon mutant library to identify genes required for growth on carnitine. We identified two genomic regions and their adjacent transcriptional regulators that are required for carnitine catabolism. The PA5388–PA5384 region contains the predicted P. aeruginosa carnitine dehydrogenase homologue along with other genes required for growth on carnitine. The second region identified, PA1999–PA2000, encodes the α and β subunits of a predicted 3-ketoacid CoA-transferase, an enzymic activity hypothesized to be involved in the first step of deacetylation of 3-dhc. Furthermore, we confirmed that an intact GB catabolic pathway is required for growth on carnitine. The PA5389 and PA1998 transcription factors are required for growth on carnitine. PA5389 is required for induction of the PA5388–PA5384 transcripts in response to carnitine, and the PA1999–PA2000 transcripts are induced in a PA1998-dependent manner and induction appears to depend on a carnitine catabolite, possibly 3-dhc. These results provide important insight into elements required for carnitine catabolism in P. aeruginosa and probably in other bacteria.
INTRODUCTION
Carnitine is a quaternary amine compound required for transport of long-chain fatty acids into the mitochondria of animals (Bremer, 1983). This important physiological role results in a relatively large amount of carnitine present in fat-metabolizing tissues which could be utilized by micro-organisms during tissue necrosis or infection. Additionally, carnitine and acylcarnitine derivatives are found at high micromolar concentrations in circulating and extracellular fluids (Bremer, 1983). Most bacteria can use carnitine as an osmoprotectant, where it functions as a compatible solute (Peluso et al., 2000). Some bacteria, including Pseudomonas aeruginosa, can utilize carnitine as sole source of carbon and nitrogen (Aurich & Lorenz, 1959). The genes encoding the enzymes required for aerobic carnitine catabolism to glycine betaine (GB) have not been described for P. aeruginosa.
In Pseudomonas and Arthrobacter, carnitine has been shown to be converted to GB, which is then catabolized to glycine (Meskys et al., 2001; Strack et al., 1964). While a requirement for the Sinorhizobium dimethylglycine catabolic genes has been demonstrated (Burnet et al., 2000; Goldmann et al., 1991), it has not been experimentally verified with specific catabolic mutants in P. aeruginosa.
Published data provide insight into the catabolism of carnitine to GB (Lindstedt et al., 1967). The first step is the oxidation of the hydroxyl group on carbon 3, forming 3-dehydrocarnitine (3-dhc). The P. aeruginosa enzyme involved in the conversion of carnitine to 3-dhc, l-(−)-carnitine dehydrogenase (CDH) (EC 1 . 1 . 1 . 108), has been biochemically characterized (Aurich et al., 1968; Kleber & Aurich, 1967). The gene encoding a bacterial CDH was first cloned from Xanthomonas (Mori et al., 1988), and an available sequence from Alcaligenes (E05045) (Houriyou et al., 1991) allowed Uanschou and colleagues to identify a predicted conserved operon containing the CDH homologue in a variety of organisms known to utilize carnitine as sole carbon and nitrogen source (Uanschou et al., 2005). Based on this bioinformatics analysis, the putative P. aeruginosa CDH homologue is predicted to be encoded by PA5386, although the role of this gene has not been confirmed in any pseudomonad to date. The second predicted step in the catabolism of carnitine is the conversion of 3-dhc to GB. In P. aeruginosa cytosolic extracts, this reaction was shown to be CoA- and ATP-dependent, but the enzyme responsible was not characterized (Lindstedt et al., 1967), and no gene or protein associated with this activity has been described previously in any bacterium.
To identify the genes involved in carnitine catabolism, we have screened the PA14 non-redundant transposon mutant library (Liberati et al., 2006) for strains capable of growth on GB but incapable of growth on carnitine. From this screen, we identified genes in the predicted CDH operon and confirmed the role of these genes in P. aeruginosa carnitine catabolism. Our screen also enabled us to identify a two-gene operon encoding the two subunits of a predicted 3-ketoacid CoA-transferase, the enzymic activity of which is hypothesized to be required to for the conversion of 3-dhc to GB (Lindstedt et al., 1967). Based on our screen and the analysis of mutants defective in GB catabolism in P. aeruginosa (Wargo et al., 2008), we were able to demonstrate that, as predicted, mutant strains incapable of growth on GB were similarly unable to grow on carnitine. Finally, while conditions that induce P. aeruginosa CDH enzyme activity were previously determined, the molecular basis of this regulation was not known. Through these studies, we identified two transcriptional regulators that are essential for P. aeruginosa growth on carnitine due to regulation of the neighbouring catabolic genes, shedding light on the transcriptional control of the carnitine metabolism.
METHODS
Strains and growth conditions.
P. aeruginosa PA14 and PAO1 wild-type strains (Rahme et al., 1995; Stover et al., 2000), transposon mutants and deletion strains, as well as E. coli strains (Table 1) were maintained on LB medium. For experiments with single carbon sources, MOPS minimal medium (Neidhardt et al., 1974), with the addition of a specified carbon source at 20 mM, was used unless otherwise noted. When necessary, gentamicin was added to a final concentration of 10 μg ml−1 for E. coli, 25 μg ml−1 for P. aeruginosa in MOPS, and 50 μg ml−1 for P. aeruginosa in LB.
Table 1.
Strains and plasmids
| Strain (gene name*) | Strain† | Description |
|---|---|---|
| P. aeruginosa PAO1-derived strains | ||
| PAO1 | DH20 | P. aeruginosa WT |
| PA1997 : : Tn | PAO1 transposon mutant 478‡ | |
| PA1998 : : Tn (dhcR) | PAO1 transposon mutant 3 | |
| ΔPA1999–PA2000 (dhcAB) | MJ99 | Clean PA1999–PA2000 deletion |
| PA2001 : : Tn (atoB) | PAO1 transposon mutant 59 | |
| PA5384 : : Tn | PAO1 transposon mutant 47947 | |
| PA5385 : : Tn (cdhB) | PAO1 transposon mutant 10906 | |
| PA5386 : : Tn (cdhA) | PAO1 transposon mutant 52057 | |
| PA5387 : : Tn (cdhC) | PAO1 transposon mutant 14404 | |
| PA5388 : : Tn | PAO1 transposon mutant 37920 | |
| P. aeruginosa PA14-derived strains | ||
| PA14 | DH122 | P. aeruginosa WT |
| ΔgbcA-B | DH802 | PA5410–PA5411 deletion (Wargo et al., 2008) |
| ΔdgcA | DH848 | PA5398 deletion (Wargo et al., 2008) |
| PA1998 : : Tn (dhcR) | PA14 transposon mutant 26061§ | |
| PA1999 : : Tn (dhcA) | PA14 transposon mutant 30401 | |
| PA2000 : : Tn (dhcB) | PA14 transposon mutant 40012 | |
| ΔPA1999–PA2000 (ΔdhcAB) | MJ98 | Clean PA1999–PA2000 deletion |
| PA5385 : : Tn (cdhB) | PA14 transposon mutant 27032 | |
| ΔPA5386–PA5387 (ΔcdhCA) | MJ262 | In-frame PA5386–PA5387 deletion |
| ΔPA5388 | MJ192 | Clean PA5388 deletion |
| ΔPA5389 (cdhR) | MJ11 | Clean PA5389 deletion |
| Plasmids | ||
| pUCP22 | High-copy Pseudomonas stabilized vector; GmR (Schweizer, 1991) | |
| pEX18-Gm | Integrating vector in P. aeruginosa; GmR (Schweizer, 1993) | |
| pMQ30 | Suicide vector; GmR (Shanks et al., 2006) | |
| pMQ80 | High-copy Pseudomonas/yeast shuttle vector (Shanks et al., 2006) | |
| pMW5 | pUCP22 containing lacZYA from pRS415 (Wargo et al., 2009) | |
| pMW19 | 6×His-tagged PA5389 in pMQ80 | |
| pMW34 | PA1999 promoter fusion in pMW5 | |
| pMW37 | 6×His-tagged PA1998 in pMQ80 | |
| pMW40 | PA1998 deletion plasmid | |
| pMW41 | PA1999–PA2000 deletion plasmid | |
| pMW46 | PA5388 promoter fusion in pMW5 | |
| pMW50 | 6×His-tagged PA1999 and untagged PA2000 in pMQ80 | |
| pMW56 | PA5388 deletion plasmid | |
| pMW79 | PA14 genomic clone containing the PA5388–PA5384 region | |
| pMW85 | PA5389 deletion plasmid | |
| pMW89 | Subclone of pMW79 containing only the PA5388–PA5384 region | |
| pMW91 | PA5385 and partial PA5384 sequence in pUCP22 | |
| pMW95 | 6×His-tagged PA5386 and untagged PA5385 in pMQ80 | |
| pMW96 | PA5388 coding sequence in pMQ80 | |
| pMW99 | PA5386–PA5387 deletion plasmid | |
| pMW102 | PA5386 and PA5385 coding region subcloned from pMW79 |
*Gene names refer to the P. aeruginosa database nomenclature or are specified in this report.
†Strain numbers for non-transposon strains refers to the database designation used in our laboratory to facilitate strain requests.
‡PAO1 transposon mutants are numbered according to the two-allele library (Jacobs et al., 2003). All PAO1 transposon strains are tetracycline-resistant.
§PA14 transposon mutants are numbered according to the database identification numbers (Liberati et al., 2006). All PA14 strains are gentamicin-resistant.
Genetic screen.
The PA14 non-redundant transposon mutant library was replicated as described by Liberati et al. (2006). The library was replicated onto LB agar master plates overnight at 37 °C. Strains were subsequently replicated onto MOPS-carnitine plates and incubated at 37 °C for 24 h. Strains defective for growth on MOPS-carnitine were recovered from the LB master plates and a secondary screen was conducted for growth on MOPS agar plates with 20 mM carnitine, GB, glucose or pyruvate, and incubated at 37 °C for 24 h. Transposon insertion sites were confirmed using gene-specific primers and the PMFLGM.GB-4a primer (Liberati et al., 2006).
Growth assays.
Growth assays were conducted in 1.0 ml volumes in 24-well polystyrene plates inoculated using overnight cultures grown in MOPS medium containing 25 mM pyruvate and 5 mM glucose. These overnight cultures were then washed and resuspended in MOPS medium with the appropriate carbon source to an OD600 of 0.05 prior to inoculation of the plates. The plates were shaken at 37 °C, and growth was measured by OD600 every hour.
During the course of this study we noted significantly better growth of wild-type strains on MOPS-carnitine agar plates compared to MOPS-carnitine liquid medium. For the PA5386 rescue construct described below, cells showed no growth after 24 h in MOPS-carnitine liquid medium, but we did see robust growth of the same strain, but not the vector control, on MOPS-carnitine agar plates.
Construction of deletion strains and complementation constructs.
Gene numbers in this paper refer to the numbers from the PAO1 genome. There is at least 98 % identity at both the nucleotide and amino acid levels when PAO1 and PA14 sequences are compared for all the genes described in this report. Organization of both regions and the surrounding genomic context of the genes of interest were identical between the two strains.
Some internal segments of the PA5388–PA5384 region proved difficult to amplify from genomic DNA using various primer sets with a large range of melting temperatures. To bypass this issue we constructed a library containing partial Sau3AI digest fragments of P. aeruginosa PA14 genomic DNA, size-selected for 4–15 kb, cloned into BamHI-digested pUCP22. The library contained approximately 11 000 clones. We pooled these clones and isolated the plasmid DNA. This pooled plasmid library was used to transform the PA5386 : : Tn strain and we selected for growth on carnitine as sole carbon source. After secondary verification, this selection yielded a single clone containing the genomic region corresponding to the PAO1 genome base pairs 6 057 482–6 067 212, as verified by sequencing the ends of the clone. This clone is referred to as pMW79 and served as a cloning template for this genomic region. This clone also rescues the growth defects of all deletions and transposons in the PA5388–PA5385 region (data not shown).
Deletion constructs for PA1998, PA1999–PA2000, PA5386–PA5387 and PA5388 were made using plasmid pMQ30 (Shanks et al., 2006), and the PA5389 deletion construct was made using plasmid pEX18Gm. Deletions in P. aeruginosa were made by recombination as described previously (Schweizer, 1993; Wargo et al., 2008). With the exception of the PA5386–PA5387 deletion construct, upstream and downstream regions of the genes of interest were amplified by PCR from genomic DNA or from the pMW79 construct. The splice overlap extension PCR products were cut and ligated into a suicide vector. After transformation into E. coli DH5α and verification of the correct sequence, constructs in pMQ30 were transformed into E. coli S17/λpir. This E. coli strain was then mated with the recipient P. aeruginosa strain and single crossover mutants were selected for growth on gentamicin. Recombinants were screened by PCR after selecting for double crossover events by growth on 5 % sucrose LB plates with no NaCl. Primers for the deletion constructs are listed in Table S1 (available with the online version of this paper). Construction of the PA5386–PA5387 deletion construct was done by cloning the appropriate SacI–EcoRI fragment from pMW79 into pUCP22, generating pMW91 (Fig. 1a), followed by cloning of the appropriate HindIII–BamHI fragment into pMW91. The HindIII–EcoRI fragment from the resulting construct was ligated into a similarly cut pMQ30 to generate pMW99.
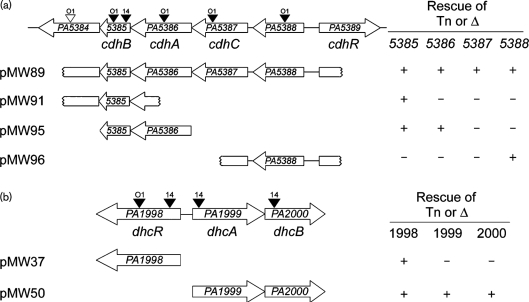
Fig. 1.
Two regions of the P. aeruginosa genome contain genes required for carnitine catabolism to GB. (a) The PA5384–PA5389 region contains genes involved in the first step of carnitine catabolism, conversion to 3-dhc. (b) The PA1998–PA2000 region contains genes predicted to be involved in conversion of 3-dhc to GB. Transposon insertions are marked with triangles above the corresponding gene with a label to specify the strain carrying the transposon; O1=PAO1, 14=PA14. Black triangles represent transposon insertions that could not grow on carnitine and white triangles represent insertions that did not alter growth on carnitine. Complementation constructs are shown below each genomic region with notation at the left denoting capability (+) or failure (−) to rescue a deletion or transposon insertion into the gene at the top of the column. Growth rates for strains summarized here can be seen in Table S2. The gene names below each genomic region are based on the naming scheme proposed in this report.
Complementation plasmids for PA1998, PA1999–PA2000, PA5386–PA5385 and PA5389 were constructed in the pMQ80 vector using the primers listed in Table S1. Each of these constructs incorporates an N-terminal 6×His tag on the first gene in the construct. The PA5385 : : Tn insertion was rescued by pMW91, which contains the entire PA5385 sequence and part of the PA5384 sequence (Fig. 1a). The PA5388 rescue construct, pMW96, was generated by cloning the appropriate KpnI–BamHI fragment from pMW79 into pUCP22 (Fig. 1a). In addition to these individual gene rescue constructs, we subcloned pMW79 to generate a plasmid containing only the PA5388–PA5385 region intact, named pMW89 (Fig. 1a).
Cytoplasmic extracts and enzyme assays.
Strains PA14, ΔPA1999–PA2000 and ΔPA5387–PA5386 were grown overnight in 3 ml LB broth and transferred to 50 ml MOPS medium with 20 mM pyruvate, 5 mM carnitine and 0.1 mg tryptone ml−1 in 250 ml flasks. These cultures were grown for ∼8 h until late-exponential phase at 37 °C, shaking at 220 r.p.m. The centrifuged cell pellet was washed with 3 pellet volumes of 10 mM Tris/HCl, pH 8.0, 1 mM EDTA (TE) and resuspended in 600 μl TE. Cells were lysed by addition of lysozyme to 3 mg ml−1. After the lysis reaction, NaCl and MgCl2 were added to a final concentration of 50 and 4 mM, respectively, followed by DNA digestion with 100 units DNase I (Invitrogen) for 30 min. The extract was clarified by centrifugation at maximum speed in a microcentrifuge for 10 min at 4 °C. The clarified extract was dialysed in a 10 000 molecular weight cut-off Slide-A-Lyzer (Pierce Biotechnology) cassette against reaction buffer (RB; 100 mM HEPES, pH 7.3, 50 mM NaCl, 4 mM MgCl2) for 16 h at 4 °C. The dialysed extract was centrifuged for 10 min at 4 °C and the protein concentration of this supernatant was measured by the Bradford assay (Bio-Rad).
We measured carnitine-dependent NADH formation in cytoplasmic extracts by monitoring A340 (Aurich et al., 1967). Reaction conditions were: RB plus 1.25 mM NAD+, 25 μg cytoplasmic extract ml−1, and either 50 mM l-carnitine (stock solution of 0.5 M in 0.5 M HEPES, pH 7.3) or an equivalent volume of 0.5 M HEPES, pH 7.3. The addition of carnitine or the HEPES blank was defined as the initiation of the reaction and A340 was measured every 30 s for 10 min using an extinction coefficient of 6220 M−1 cm−1 to calculate specific activity. As described by Aurich et al. (1967), we also noted a linear reaction for only 4 min.
We measured carnitine-dependent CoA utilization in cytoplasmic extracts using the 5,5-dithiobis(2-nitrobenzoic acid) (DTNB) method (Engeser et al., 1979). Reaction conditions were: RB plus 4 mM NAD+, 4 mM ATP, 1 mM CoA, 25 μg cytoplasmic extract ml−1, and with 50 mM l-carnitine or an equivalent volume of 0.5 M HEPES, pH 7.3. Aliquots were taken every 2 min and treated with DTNB at a final concentration of 75 μM. The thiophenalate-CoA complex was measured by A412 and an extinction coefficient of 13 600 M−1 cm−1 to calculate specific activity.
RNA isolation and quantitative real-time RT-PCR (qRT-PCR).
For RNA samples, cells were grown overnight in MOPS with 25 mM pyruvate and 5 mM glucose as described for the growth assays, harvested by centrifugation, resuspended in MOPS with 25 mM pyruvate and 2 mM of the listed carbon source, and grown for 2 h at 37 °C. RNA was isolated and cDNA created as described previously (Wargo et al., 2008). Primers are listed in Table S1. qRT-PCR was conducted with SYBR Green and AmpliTaq Gold DNA polymerase, according to the manufacturer's instructions (Applied Biosystems). The amplification conditions were: 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, 56 °C for 30 s, and 72 °C for 30 s using an Applied Biosystems 7500 instrument. Only one PCR product was obtained for all primers and all samples based on melting curve analysis. Transcript levels were normalized to the rplU transcript (Kuchma et al., 2005).
Reporter constructs.
We generated promoter fusions to lacZYA in pMW5 (Wargo et al., 2009) by amplification with promoter-specific primers listed in Table S1. Numbering from the translational start, the PA5388 promoter fusion contained the −422 to −21 segment, and the PA1999 promoter fusion contained the −258 to −11 segment. Briefly, each PCR product was gel-purified, digested with HindIII and KpnI, and ligated into similarly digested pMW5. Ligation mixtures were transformed into E. coli DH5α cells (Invitrogen) by chemical transformation. Plasmid preparations from E. coli clones were transformed into P. aeruginosa by electroporation (Choi et al., 2006) and selected for growth on gentamicin. After overnight growth in MOPS-pyruvate-glucose medium, as described for the growth assays, cells were pelleted and resuspended in MOPS with 25 mM pyruvate and 2 mM of the inducing compound, unless otherwise specified. Cells were grown under inducing conditions for 3 h at 37 °C. β-Galactosidase assays were conducted according to Miller (1972).
RESULTS
Identification of mutants defective for growth on carnitine
The P. aeruginosa PA14 non-redundant library was screened to find mutants unable to grow in MOPS minimal medium with carnitine as sole carbon source. To sort our mutants into functional categories and eliminate auxotrophs, we performed secondary screens for growth on glucose, pyruvate and GB as sole carbon sources. Mutants that failed to grow on either pyruvate or glucose were not studied further. The 14 mutants unable to grow on carnitine could be separated into two groups, consisting of strains that grew on neither carnitine nor GB and those that were only defective in growth on carnitine. The strains defective for growth on both carnitine and GB were found to have transposon insertions in genes required for GB catabolism in P. aeruginosa, including gbcA, gbcB, dgcAB, soxBDAG and gbdR (Wargo et al., 2008), indicating that GB is an essential intermediate in the catabolism of carnitine, consistent with data from previous studies of carnitine catabolism (Lindstedt et al., 1967).
Mutants defective for growth on carnitine, but not GB, included mutants with transposon insertions in a predicted thioesterase (PA5385), a predicted periplasmic component of an ABC transporter (PA5388), two genes of a predicted operon (PA1999–PA2000) and a putative transcriptional regulatory protein (PA1998). The two regions identified as being involved in carnitine catabolism are shown in Fig. 1(a, b).
The roles of the PA5388–PA5384 genes in carnitine catabolism
P. aeruginosa strain PA14 mutants with transposon insertions in PA5385 and PA5388 resulted in the inability of P. aeruginosa to grow on carnitine, while retaining the ability to grow on pyruvate (Fig. 1a and Table S2, available with the online version of this paper). Because strains PA14 and PAO1 have high homology within this region and both strains grow well on carnitine, we also used mutants from the PAO1 transposon collection in our analyses. The P. aeruginosa strain PAO1 transposon library contained strains with transposon insertions in each ORF of this region (Jacobs et al., 2003). PAO1 mutants with transposon insertions in each of the PA5388, PA5387, PA5386 and PA5385 genes were unable to grow on carnitine (Fig. 1a and Table S2). The measured growth rate for one representative transposon mutant strain from each gene is shown in Table S2. PA14 and PAO1 strains with transposon insertions in PA5384 were capable of normal growth on carnitine, suggesting that this gene is not necessary for growth on free carnitine.
Deletion of PA5388 was sufficient to abrogate growth on carnitine, supporting a role for this ProX homologue in carnitine uptake. This deletion could be rescued by the pMW96 construct (Fig. 1a and Table S2). Studies to further characterize the PA5388 protein and its role in transport are on-going (G. Beattie, personal communication).
Complementation analysis of transposon and deletion mutants provided insight into the genes necessary for growth on carnitine. The growth defect of PA5385 : : Tn strains was rescued by complementation with PA5385 on a plasmid (pMW91) (Fig. 1a and Table S2), indicating the importance of this gene for carnitine catabolism. However, this gene is not sufficient as none of the other transposon insertions in PA5386, PA5387 or PA5388 were rescued by this plasmid. The rescue of growth of the PA5386 : : Tn strain using pMW95, which contains 6×His-tagged PA5386 and untagged PA5385 under control of an arabinose-inducible promoter was observed on solid medium with carnitine as sole source of carbon (see Methods). This plasmid, pMW95, was not able to rescue the growth phenotype resulting from transposon insertion in the PA5387 gene, suggesting that PA5387 is also required for growth on carnitine (Fig. 1a). While we were unable to construct deletion strains of either PA5386 or PA5387 via multiple independent constructs for reasons that remain unclear, we were able to generate a strain with a deletion eliminating the majority of the PA5386–PA5387 coding sequences, which resulted in loss of growth on carnitine (Table S2). Plasmid pMW95, which contains PA5385 and PA5386, was not able to rescue this growth phenotype.
The requirement for rescue of the PA5386 : : Tn strains using MOPS-carnitine plates was also noted for a non-His-tagged construct driven by an arabinose-inducible promoter, and a construct containing a portion of the PA5387 gene in addition to PA5386–PA5385 and driven by the lac promoter (pMW102, data not shown). The pMW89 construct, however, rescued all of the PA5386 and PA5387 mutant strains to near wild-type growth levels in both liquid and plate-based assays, suggesting the possibility that spatial co-transcription or co-translation of this region is important for the wild-type level of enzymic activity.
We next tested the requirement for the adjacent, divergently transcribed, AraC-family transcription factor gene, PA5389. Deletion of PA5389 prevents growth on carnitine (Fig. 1a and Table S2). The carnitine growth defect of the ΔPA5389 mutant was rescued by complementation with a 6×His-tagged PA5389 construct (pMW19) (Table S2).
Based on the reported induction conditions for CDH enzymic activity (Aurich et al., 1967), we predicted that the PA5388–PA5384 transcripts would be induced upon growth in carnitine, but not pyruvate or GB. The transcripts for these genes were induced in carnitine only, with levels of induction measured by quantitative qRT-PCR, as reported in Table 2.
Table 2.
qRT-PCR quantification of transcript induction by carnitine
| Transcript | Fold induction* (sd) |
|---|---|
| PA1998 | 1.1 (0.57) |
| PA1999 | 178.8 (7.75) |
| PA2000 | 128.5 (24.8) |
| PA2001 | 2.2 (0.37) |
| PA5384 | 158.1 (24.5) |
| PA5385 | 266.1 (14.9) |
| PA5386 | 399.5 (71.9) |
| PA5387 | 286.9 (30.6) |
| PA5388 | 412.4 (12.6) |
| PA5389 | 2.2 (0.16) |
*Fold induction was calculated by dividing carnitine-induced transcript levels by levels of transcript in pyruvate-grown cells after normalization to the rplU transcript.
To validate the function of the PA5387–PA5386 genes, we measured carnitine-dependent NADH formation in cytoplasmic extracts of PA14 wild-type and ΔPA5387–PA5386, by monitoring A340 as a measure of CDH activity (Aurich et al., 1967). PA14 wild-type extracts had a measured CDH activity of 1.89±0.27 μmol NADH min−1 mg−1, while extracts from the ΔPA5387–PA5386 strain had a CDH activity of only 0.21±0.02 μmol NADH min−1 mg−1. Background NADH formation in the absence of carnitine was similar in both strains.
The genes of the PA5384–PA5388 region are highly homologous to the CDH-containing region of other Gram-negative bacteria (Uanschou et al., 2005). In addition, the N-terminal sequence of a purified P. putida CDH (Goulas, 1988) aligns with the predicted N terminus of PA5386, matching 12 of 18 residues identically. Our screen, subsequent growth analyses and CDH enzymic activity assay confirms the predictions that this is the cdh region in P. aeruginosa, and that PA5386 is the cdh homologue in P. aeruginosa that is required for the first step in carnitine degradation (Fig. 2). We therefore designate PA5386 as cdhA. In some bacterial species, the proteins encoded by PA5386 and PA5385 are fused and encoded by one gene responsible for CDH activity (Uanschou et al., 2005). Transposon insertion into the PA5385 gene, downstream of the canonical cdh homologue, also results in loss of growth on carnitine and this mutant is rescued by the PA5385 coding sequence (pMW91) provided in trans (Fig. 1a and Table S2). These data support a role for PA5385 in carnitine catabolism, and therefore PA5385 is here named cdhB. The PA5387 gene, based on the growth analyses presented here, is required for growth on carnitine but has no homology to proteins with known function. Based on the genetic data presented here, PA5387 is here named cdhC. Deletion of the PA5388 gene via an in-frame non-polar excision results in lack of growth on carnitine. The protein encoded by PA5388 shows high similarity to ProX homologues and we predict it may function as part of a carnitine ABC-family transport system; however, we will refrain from naming this gene until the range of PA5388 ligands are identified. The PA5389 gene, required for induction of the PA5388–PA5384 genes, is renamed cdhR based on its regulatory role described here.
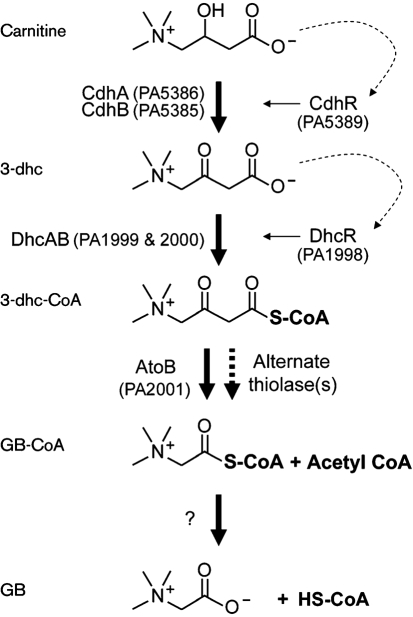
Fig. 2.
The proposed pathway for carnitine catabolism to GB in P. aeruginosa. Dashed arrows represent ligand activation of the respective transcription factor, with the solid arrows leading from the transcription factors representing transcriptional induction. Step 1: using NAD+, CDH catalyses the oxidation at carbon 3. Step 2: the predicted 3-ketoacid CoA-transferase, encoded by dhcAB, along with a source of activated CoA, catalyses the sulfur attack of carbon 1. Step 3: a thiolase, possibly encoded by atoB, catalyses the deacetylation reaction. Step 4: an unknown enzyme catalyses the removal of CoA, leaving GB.
The role of the PA1999–PA2000 operon in carnitine catabolism
Transposon insertion into PA1999 and PA2000 resulted in the inability of P. aeruginosa to grow on carnitine, while retaining the ability to grow on pyruvate (Fig. 1b and Table S2). We constructed a deletion of PA1999 and PA2000 (ΔPA1999–PA2000) in both PA14 and PAO1. Deletion of this predicted operon resulted in failure to grow on carnitine (Fig. 1b and Table S2). This deletion could be rescued by pMW50, encoding a 6×His-tagged PA1999 and untagged PA2000. Transposon insertion into, or deletion of, the adjacent, divergently transcribed LysR-family transcription factor, PA1998, also resulted in carnitine growth deficiency (Fig. 1b and Table S2). The deletion of PA1998 could be rescued by providing 6×His-tagged PA1998 on a plasmid (pMW37) as well as by arabinose-induced overexpression of PA1999–PA2000 (pMW50) (Fig. 1b).
We verified the operonic nature of PA1999–PA2000 by RT-PCR from carnitine-induced RNA using primers spanning these two reading frames. Based on this analysis (data not shown), we will now refer to the region as the PA1999–PA2000 operon.
Like the PA5388–PA5384 genes, the transcripts for PA1999 and PA2000 were induced in carnitine only, with levels of induction measured by qRT-PCR, as reported in Table 2.
The PA1999 and PA2000 genes encode the α and β subunits of a predicted 3-ketoacid CoA-transferase, which is the enzymic function predicted to be required as the first step in deacetylation of 3-dhc (a 3-ketoacid) to GB and acetyl-CoA (Fig. 2). To test the function of PA1999–PA2000, we measured carnitine-dependent utilization of CoA by measuring the loss of free CoA over time using the DTNB assay. PA14 wild-type cytoplasmic extracts had a carnitine-dependent CoA utilization rate of 1.88±0.42 μmol NADH min−1 mg−1, while extracts from ΔPA1999–PA2000 cells showed barely detectable carnitine-dependent utilization of CoA (0.05±0.03 μmol NADH min−1 mg−1). The background CoA utilization rate in the absence of carnitine was comparable in both strains. Combining the growth phenotype and induction data of this operon, together with the lack of carnitine-dependent utilization of CoA, there is strong support for the role of PA1999 and PA2000 in conversion of 3-dhc to GB. Therefore, we designate PA1999 and PA2000 as dhcA and dhcB, respectively. PA1998, required for induction of the dhcAB operon, is renamed dhcR based on its regulatory role described here.
Transcription factors required for growth on carnitine
In our screen and subsequent analyses we identified two transcription factors required for P. aeruginosa growth on carnitine; CdhR, encoded by PA5389, and DhcR, encoded by PA1998. We predicted that CdhR would be required for induction of the PA5388 promoter and DhcR would be required for induction of the dhcAB promoter. To investigate these hypotheses, we used reporter gene constructs of the upstream regions from PA5388 and dhcA fused to lacZYA (Fig. 3).
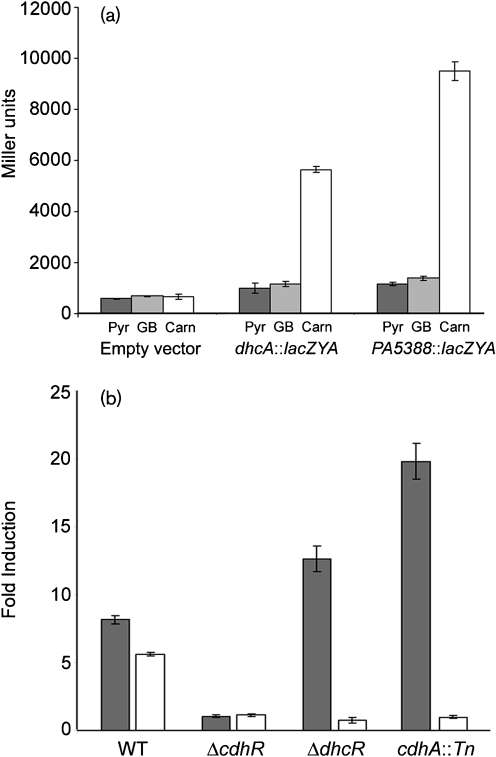
Fig. 3.
CdhR (PA5389) and DhcR (PA1998) are transcriptional regulators of carnitine catabolic genes. (a) lacZYA reporter fusions to the PA5388 (pMW46) and dhcA (pMW34) promoters are both induced in response to carnitine (Carn) and not in the presence of pyruvate (Pyr) or GB. (b) chdR is required for the induction of both the PA5388 : : lacZYA (grey bars) and dhcA : : lacZYA fusions (white bars). dhcR is only required for induction of the dhcA : : lacZYA fusion. When P. aeruginosa is unable to convert carnitine to 3-dhc (cdhA : : Tn), carnitine does not lead to induction of the dhcA : : lacZYA fusion. Error bars show sd.
Both the PA5388 : : lacZYA (pMW46) and dhcA : : lacZYA (pMW34) promoter fusions were induced strongly in response to carnitine in wild-type cells, whereas pyruvate and GB did not result in induction (Fig. 3a). In the ΔcdhR strain, neither the PA5388 : : lacZYA nor the dhcA : : lacZYA constructs were induced in response to carnitine (Fig. 3b). We hypothesize that regulatory control of carnitine catabolism is a two-step process, where carnitine induces transcription from the PA5388 promoter via CdhR, and 3-dhc induces transcription from the PA1999 promoter via DhcR. This model of carnitine catabolic regulation is shown in Fig. 2. Based on this model, the inability to catabolize carnitine to 3-dhc would prevent induction of the dhcA : : lacZYA construct in the presence of carnitine. To test this we transformed the cdhA : : Tn strain of PAO1 with the PA5388 : : lacZYA and dhcA : : lacZYA reporter fusions. As predicted, without the CDH activity to convert carnitine to 3-dhc, encoded by cdhA, the dhcA reporter was not induced in response to carnitine, while this mutation did not alter transcription from the PA5388 : : lacZYA fusion (Fig. 3).
DISCUSSION
Despite significant work on the enzymology of carnitine catabolism, the aerobic carnitine catabolic pathway to GB in Gram-negative bacteria has yet to be fully described. We have used a genetic screen to identify both the CDH homologue and the genes predicted to be involved in conversion of 3-dhc to GB. This screen also led to the discovery of the transcriptional regulators of both sets of catalytic genes, shedding light on the regulation of carnitine catabolism in P. aeruginosa.
While the genes encoding CDH in P. aeruginosa have been predicted as such (Uanschou et al., 2005), the genes encoding the enzymes for conversion of 3-dhc to GB have not been described in any bacterium. Here we identified the dhcAB operon as required for carnitine catabolism and propose that these genes encode subunits of the enzyme required to convert 3-dhc to GB. The dhcAB genes encode a predicted 3-ketoacid CoA-transferase, which gives us insight into the possible mechanism for deacetylation of 3-dhc.
Based on the enzymic activities and general reaction mechanisms, the enzymic steps between 3-dhc and GB are similar to the catabolism of β-hydroxybutyrate and acetoacetate in ketone bodies. Based on that analogy, the catabolism of carnitine to GB would follow the pathway shown in Fig. 2. According to this scheme, we have identified the genes encoding enzymes required for the first and second steps. The thiolase predicted to be required for the third step may be encoded by atoB (PA2001) which is predicted to encode the P. aeruginosa acetoacyl-CoA-thiolase involved in phospholipid metabolism. The PA2001 transcript is slightly induced in response to carnitine, and transposon insertion into atoB results in slow growth on carnitine (data not shown). This slow growth, versus lack of growth, may be explained by the presence of at least two alternate acetyl-CoA thiolases in P. aeruginosa encoded by PA3589 and PA3925. If AtoB functions as the primary thiolase in this reaction scheme, the remaining unidentified enzyme is that required for step 4; conversion of GB-CoA to GB and CoA. Similar to the predicted situation with AtoB, there may be multiple thiolases that could function to cleave CoA from GB. This may explain our inability to identify this gene in our genetic screen.
The CDH-encoding genomic region, as described by Uanschou et al. (2005), is remarkably conserved in a variety of bacteria, and contains three core components: the CDH-encoding enzyme (cdhA), the predicted thioesterase (cdhB), and a protein of unknown function, DUF849 (cdhC). We have determined the necessity of these three genes in carnitine degradation and for measured CDH activity – experimental verification of the hypothesis that all three of these coding regions are required for carnitine catabolism (Uanschou et al., 2005). In P. aeruginosa, as described in Methods, the cdhC region is particularly problematic to amplify by PCR; however, it is possible that the cdhC homologue in other organisms will be more amenable to molecular techniques.
The cdhR homologue is located adjacent to the CDH region of nearly all Gram-negative bacteria that catabolize carnitine (Uanschou et al., 2005). CdhR, an AraC-family transcription factor, has high similarity with the ArgR and GbdR transcription factors of P. aeruginosa, which induce transcription of target genes in response to the charged amine-containing compounds arginine and GB, respectively (Park et al., 1997; Wargo et al., 2008). The data presented here show that CdhR induces transcription in response to carnitine, a specificity that is likely to be conserved in the CdhR homologues of other bacteria.
CDH activity is required for carnitine-dependent induction of the dhcAB operon, as a transposon insertion into cdhA (PA5386) abrogates this induction. We hypothesize that DhcR binds 3-dhc or a derivative, which causes induction of the dhcAB operon. Additionally, mutations in either cdhA or dhcR result in hyperinduction of the PA5388 : : lacZYA reporter fusion (Fig. 3), as one would predict due to backup of the catabolic pathway leading to higher concentrations of free carnitine in the cytosol.
The dhcAB genes, encoding a predicted carnitine CoA-transferase, show similarity to a number of different 3-ketoacid CoA-transferases, suggesting that either the similarity is due to conserved structure unrelated to the binding site residues or that these enzymes are somewhat promiscuous. These two possibilities are not mutually exclusive but would have impacts on the study of dhcAB transcriptional regulation. If DhcAB and their homologues are capable of acting on a variety of 3-ketoacids, we would predict that the DhcR homologue has a similar range of inducing ligands. Understanding the regulation of this catabolic step could shed light on control of flux through this step in the presence of multiple 3-ketoacids.
This study provides a significant step forward in understanding carnitine catabolism in P. aeruginosa. The identification of DhcAB (PA1999 and PA2000) as the predicted 3-ketoacid CoA-transferase involved in 3-dhc catabolism gives support to the hypothesis that 3-dhc deacetylation proceeds in a manner similar to ketone body metabolism. Future studies will focus on biochemical examination of the reaction steps as well as exploration of the transcriptional regulation of the cdh and dhcAB genes.
Acknowledgments
This work was supported by the National Institutes of Health grant P20-RR018787 from the IDeA Program of the National Center for Research Resources (to D. A. H.), by the Ruth Kirchstein NRSA institutional fellowship awarded to the Department of Microbiology and Immunology, Dartmouth Medical School (grant T32 AI07519 supporting M. J. W.), and by the Cystic Fibrosis Foundation Research Development Program (grant STANTO07R0) (D. A. H. and M. J. W.).
Abbreviations
3-dhc, 3-dehydrocarnitine
CDH, l-(−)-carnitine dehydrogenase
GB, glycine betaine
Footnotes
Supplementary tables of primers used in this study, and doubling times of P. aeruginosa strains on pyruvate and carnitine are available with the online version of this paper.
References
- Aurich, H. & Lorenz, I. (1959). On the catabolism of carnitine by Pseudomonas pyocyanea. Acta Biol Med Ger 3, 272–275. [PubMed] [Google Scholar]
- Aurich, H., Kleber, H. P. & Schopp, W. D. (1967). An inducible carnitine dehydrogenase from Pseudomonas aeruginosa. Biochim Biophys Acta 139, 505–507. [DOI] [PubMed] [Google Scholar]
- Aurich, H., Kleber, H. P., Sorger, H. & Tauchert, H. (1968). Purification and properties of carnitine dehydrogenase from Pseudomonas aeruginosa. Eur J Biochem 6, 196–201. [DOI] [PubMed] [Google Scholar]
- Bremer, J. (1983). Carnitine – metabolism and functions. Physiol Rev 63, 1420–1480. [DOI] [PubMed] [Google Scholar]
- Burnet, M. W., Goldmann, A., Message, B., Drong, R., El Amrani, A., Loreau, O., Slightom, J. & Tepfer, D. (2000). The stachydrine catabolism region in Sinorhizobium meliloti encodes a multi-enzyme complex similar to the xenobiotic degrading systems in other bacteria. Gene 244, 151–161. [DOI] [PubMed] [Google Scholar]
- Choi, K.-H., Kumar, A. & Schweizer, H. P. (2006). A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J Microbiol Methods 64, 391–397. [DOI] [PubMed] [Google Scholar]
- Engeser, H., Hubner, K., Straub, J. & Lynen, F. (1979). Identity of malonyl and palmitoyl transferase of fatty acid synthetase from yeast. Functional interrelationships between the acyl transferases. Eur J Biochem 101, 407–412. [DOI] [PubMed] [Google Scholar]
- Goldmann, A., Boivin, C., Fleury, V., Message, B., Lecoeur, L., Maille, M. & Tepfer, D. (1991). Betaine use by rhizosphere bacteria: genes essential for trigonelline, stachydrine, and carnitine catabolism in Rhizobium meliloti are located on pSym in the symbiotic region. Mol Plant Microbe Interact 4, 571–578. [DOI] [PubMed] [Google Scholar]
- Goulas, P. (1988). Purification and properties of carnitine dehydrogenase from Pseudomonas putida. Biochim Biophys Acta 957, 335–339. [DOI] [PubMed] [Google Scholar]
- Houriyou, K., Takahashi, M., Mizoguchi, J. & Imamura, S. (1991). Essentially pure microorganism capable of producing carnitine dehydrogenase. Patent no. 1993161492-A/1 (Japan).
- Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C. & other authors (2003). Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 100, 14339–14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleber, H. P. & Aurich, H. (1967). Evidence for an inducible active transport of carnitine in Pseudomonas aeruginosa. Biochem Biophys Res Commun 26, 255–260. [DOI] [PubMed] [Google Scholar]
- Kuchma, S. L., Connolly, J. P. & O'Toole, G. A. (2005). A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol 187, 1441–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati, N. T., Urbach, J. M., Miyata, S., Lee, D. G., Drenkard, E., Wu, G., Villanueva, J., Wei, T. & Ausubel, F. M. (2006). An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103, 2833–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt, G., Lindstedt, S., Midtvedt, T. & Tofft, M. (1967). The formation and degradation of carnitine in Pseudomonas. Biochemistry 6, 1262–1270. [Google Scholar]
- Meskys, R., Harris, R. J., Casaite, V., Basran, J. & Scrutton, N. S. (2001). Organization of the genes involved in dimethylglycine and sarcosine degradation in Arthrobacter spp.: implications for glycine betaine catabolism. Eur J Biochem 268, 3390–3398. [DOI] [PubMed] [Google Scholar]
- Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory.
- Mori, N., Shirota, K., Kitamoto, Y. & Ichikawa, Y. (1988). Cloning and expression in Escherichia coli of the carnitine dehydrogenase gene from Xanthomonas translucens. Agric Biol Chem 52, 851–852. [Google Scholar]
- Neidhardt, F. C., Bloch, P. L. & Smith, D. F. (1974). Culture medium for enterobacteria. J Bacteriol 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S. M., Lu, C. D. & Abdelal, A. T. (1997). Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol 179, 5300–5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso, G., Barbarisi, A., Savica, V., Reda, E., Nicolai, R., Benatti, P. & Calvani, M. (2000). Carnitine: an osmolyte that plays a metabolic role. J Cell Biochem 80, 1–10. [DOI] [PubMed] [Google Scholar]
- Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G. & Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. P. (1991). Escherichia–Pseudomonas shuttle vectors derived from pUC18/19. Gene 97, 109–112. [DOI] [PubMed] [Google Scholar]
- Schweizer, H. D. (1993). Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15, 831–834. [PubMed] [Google Scholar]
- Shanks, R. M., Caiazza, N. C., Hinsa, S. M., Toutain, C. M. & O'Toole, G. A. (2006). Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J. & other authors (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964. [DOI] [PubMed] [Google Scholar]
- Strack, E., Aurich, H. & Gruener, E. (1964). On the degradation capacity of certain Pseudomonas species for (−)-carnitine. Z Allg Mikrobiol 61, 154–160. [DOI] [PubMed] [Google Scholar]
- Uanschou, C., Frieht, R. & Pittner, F. (2005). What to learn from a comparative genomic analysis of l-carnitine dehydrogenase. Monatsh Chem 136, 1365–1381. [Google Scholar]
- Wargo, M. J., Szwergold, B. S. & Hogan, D. A. (2008). Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190, 2690–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo, M. J., Ho, T. C., Gross, M. J., Whittaker, L. A. & Hogan, D. A. (2009). GbdR regulates Pseudomonas aeruginosa plcH and pchP transcription in response to choline catabolites. Infect Immun 77, 1103–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]