Abstract
Background/Aims
Wilson disease is characterized by hepatic copper overload and caused by mutations in the gene encoding the copper transporting P-type ATPase ATP7B. ATP7B interacts with COMMD1, a protein that is deleted in Bedlington terriers with hereditary copper toxicosis. Here we characterized the implications of the interaction between COMMD1 and ATP7B in relation to the pathogenesis of Wilson disease.
Methods
GST pull-down experiments, co-immunoprecipitations, immunofluoresensce microscopy, site-directed mutagenesis and biosynthetic labeling experiments were performed to characterize the interaction between COMMD1 and ATP7B and the effects of Wilson disease causing mutations.
Results
COMMD1 specifically interacted with the amino-terminal region of ATP7B. This interaction was independent of intracellular copper levels and of the expression of the copper chaperone ATOX1. Four Wilson disease patient derived mutations in this region of ATP7B significantly increased its binding to COMMD1. Two of these mutations also resulted in mislocalization and increased degradation rate of ATP7B. Although COMMD1 did not affect copper-induced trafficking of ATP7B, it markedly decreased the stability of newly synthesized ATP7B.
Conclusions
Our data implicate COMMD1 in the pathogenesis of Wilson disease and indicate that COMMD1 exerts its regulatory role in copper homeostasis through the regulation of ATP7B stability.
Introduction
The liver is essential for the maintenance of copper homeostasis as this organ plays a central role in the excretion of copper 1. Disorders associated with an overload of this essential yet toxic metal are therefore usually characterized by extensive liver damage, as can be observed in the autosomal recessive disorder Wilson disease (WD). The clinical presentation of WD is highly heterogeneous, and usually includes hepatic and/or neurological abnormalities due to toxic accumulation of copper in the liver and the brain 2. WD is caused by mutations in the ATP7B gene, which encodes a copper transporting P-type ATPase 3–6. ATP7B plays a key role in hepatic copper excretion, by virtue of its ability to transport copper across cellular membranes at the cost of ATP hydrolysis. Within its amino terminal region, ATP7B contains six copper binding sequences, through which it receives copper by a transient copper-dependent interaction with the copper chaperone ATOX17, 8. Under basal conditions, ATP7B resides in the trans Golgi network (TGN), where it facilitates the incorporation of copper in cuproenzymes such as ceruloplasmin. When the copper concentration in the cell exceeds a certain threshold, ATP7B translocates to a dispersed vesicular compartment in the periphery of the cell, an event that is generally believed to precede cellular excretion of copper 9.
Recently, COMMD1 was implicated as a novel regulator of hepatic copper excretion as a deficiency of this protein causes copper toxicosis in Bedlington terriers 10, 11, a canine disorder that shares many pathophysiological features with WD12. Subsequent identification of COMMD1 as an interacting partner of ATP7B further supports the role of COMMD1 in copper homeostasis 13, and suggests that these two proteins cooperate to facilitate excretion of copper from the hepatocyte into the bile canaliculus. Although COMMD1 has been postulated as a candidate gene for non-Wilsonian copper overload disorders, and as a modifier gene for the clinical presentation of WD, no disease associated mutations in COMMD1 have been detected in patients with these disorders, but a silent missense mutation in COMMD1 was possibly associated with an earlier onset of the disorder in patients with known ATP7B mutations 14–18. Recent studies have revealed that COMMD1 also functions in a variety of other cellular processes 19. Analysis of a COMMD1 knockout mouse implicated COMMD1 as a negative regulator of hypoxia-inducible factor 1 (HIF-1) 20. In addition, inhibitory functions for COMMD1 in sodium transport and NF-κB signaling have been identified 21–23. The role of COMMD1 in the HIF-1 and NF-κB pathways appear to be mediated through proteolytic regulation of key components of these pathways 20, 21, 24. Nevertheless, the function of COMMD1 within the regulation of copper homeostasis still remains elusive. To dissect the molecular function of COMMD1 in copper homeostasis, we here further characterized the interaction between COMMD1 and ATP7B and the role of this interaction in the pathogenesis of WD.
Materials and Methods
Constructs
COMMD1-GST, COMMD1-HA and IκBα-HA constructs were described previously 23, 25, 26. pEBB-ATP7B-Flag was generated by subcloning an ATP7B coding cDNA fragment from pEGFP-ATP7B (kindly provided by Dr. H. Roelofsen, University Medical Center Groningen, the Netherlands) into pEBB-Flag. cDNA sequence encoding the first 650 amino acids of ATP7B was amplified with PFU turbo (Stratagene, La Jolla, CA, USA) and subcloned in pEBB-Flag to obtain pEBB-ATP7B(1–650)-Flag. Mutations in pEBB-ATP7B(1–650)-Flag and in pEBB-ATP7B-Flag were introduced using the Quickchange site-directed mutagenesis method (Stratagene). Plasmids encoding short hairpin RNAs (shRNAs) were generated by cloning target sequences against COMMD1 (GTCTATTGCGTCTGCAGAC) or ATOX1 (GGTCTGCATTGAATCTGAC) in pRETRO-SUPER as previously described 20.
Cell culture and transfections
HEK293T and HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, L-glutamine, and penicillin/streptomycin. In some experiments, cells were incubated with media supplemented with 200 μM CuSO4, 50 μM bathocuproinedisulfonic acid (BCS) (Sigma), 15 μg/ml cycloheximide (Sigma), 3 μM MG132 (Calbiochem, Darmstadt, Germany), or 5 μM lactacystin (Sigma) for the indicated time periods. Standard calcium phosphate precipitation was used to transfect HEK293T cells. HepG2 cells were transfected using Fugene-6 (Roche, Basle, Switzerland). The generation of stable HEK 293T COMMD1 and ATOX1 knockdown cells has been described elsewhere 20.
Biosynthetic labeling of cell lines
After overnight transfection of subconfluent cultures, HEK293T cells were incubated in serum-free medium containing no methionine and cysteine for 30 min, and subsequently metabolically labeled with 50 μCi/ml redivue pro-mix L-[35S]-methionine and L-[35S]-cysteine (GE Healthcare, Diegem, Belgium) for 90 min at 37 °C. Following metabolic labeling, cells were chased with complete medium for different time periods as indicated in each experiment.
Immunoprecipitations, GST pull-down assays, and immunoblotting
Cells were lysed in lysis buffer (1% Triton X-100, 25 mM HEPES; pH 7.9, 100 mM NaCl, 1 mM EDTA, 10% glycerol) supplemented with 1 mM Na3VO4, 1 mM PMSF, protease inhibitors (Roche) and 1 mM DTT. For copper or BCS treated cells, lysis buffer was also supplemented with 1mM CuCl2 and 5mM DTT instead of 1mM DTT, or with 1mM BCS, respectively, as previously described 7. Precipitations with GSH-sepharose, or immunoprecipitations with anti-HA (Sigma), were performed as previously described 25. Protein detection was performed by immunoblotting for Flag (Sigma), HA, GST (Santa Cruz, Santa Cruz, CA, USA), short-chain L-3-hydroxyacyl-CoA dehydrogenase (SCHAD) 11, COMMD1 11 or ATOX1 7 as described previously 11. To quantify immunoprecipitated proteins, relative densities of protein bands were measured with a Bio-Rad GS-700 imaging densitometer and analyzed with Quantity One software (Bio-Rad, Hercules, CA, USA). Biosynthetically labeled cells were lysed in Ripa lysis buffer (20 mM Tris/HCl, pH 7.4, 0.5% Triton X-100, 150 mM NaCl, 10 mM sodium EDTA, and 0.5% sodium deoxycholic acid), supplemented with 1 mM Na3VO4, 1 mM PMSF, and protease inhibitors. Cell lysates were subjected to immunoprecipitations using anti-Flag affinity gel (Sigma) after addition of 1.5 volumes buffer A (1% SDS, 1% Triton X-100, 0.5% sodium deoxycholic acid, 0.5% BSA and 1 mM PMSF, in PBS). Immunoprecipitates were washed with buffer A prior to SDS PAGE. Gels were soaked in Amplify (Amersham Pharmacia Biotech) prior to fluorographic exposure.
Confocal immunofluoresence microscopy
For fluorescence microscopy, cells were seeded and grown on cover slips (Marienfeld, Bad Mergentheim, Germany). Cells were fixed with 4% paraformaldehyde, and subsequently incubated with antibodies to ATP7B (kindly provided by Dr. S Lutsenko, Oregon Health & Science University, Portland, OR, USA), COMMD1 (Abnova, Taipei City, Taiwan), p230 (BD Biosciences, Alphen aan den Rijn, The Netherlands) or Calreticulin (Alexis Biochemicals, Lausen, Switzerland). Antibodies were visualized by incubation with Alexa Fluor 488- or Alexa Fluor 568-conjugated donkey anti-mouse, anti-rabbit, or anti-rat IgG (H+L) (Invitrogen, Breda, the Netherlands). Visualization of wild-type or mutant ATP7B-Flag in HepG2 cells was performed by incubation with FITC-conjugated anti-Flag antibodies (Sigma). The cover slips were mounted with FluorSave reagent (Calbiochem), and images were obtained utilizing a Zeiss Axiovert 100M confocal microscope equipped with a Zeiss LSM 510 Meta spectrometer.
Results
COMMD1 and ATP7B are localized in overlapping regions in HEK293T cells
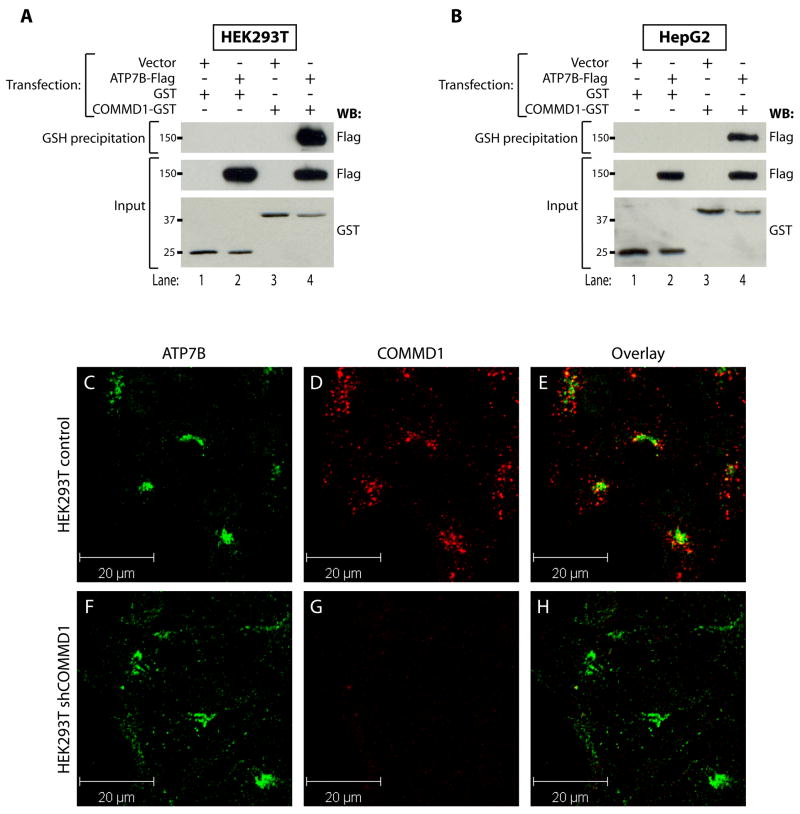
To study the interaction of COMMD1 with ATP7B, ATP7B-Flag was expressed in HEK293T or HepG2 cells together with either GST or a COMMD1-GST fusion protein. Analysis of glutathione sepharose precipitates from lysates of these cells confirmed a specific interaction of ATP7B-Flag with COMMD1-GST in both cell lines (Figures 1A–B). To determine if both proteins are localized in overlapping regions in the cell, double-label immunofluorescence microscopy experiments were performed in HEK293T cells. ATP7B displayed a dense perinuclear distribution, consistent with a localization in the TGN (Figure 1C and F). COMMD1 was localized in cytoplasmic vesicular compartments that were particularly profound in the perinuclear region (Figure 1D). The observed localization of both ATP7B and COMMD1 was consistent with previous reports 9, 11. Overlay of the images (Figure 1E) revealed that ATP7B and COMMD1 localization partially overlaps, supporting an endogenous interaction between these proteins. Immunoreactivity for COMMD1 was completely absent in HEK293T cells that were stably transduced with plasmids encoding short hairpin RNAs (shRNAs) directed against COMMD1 (Figure 1G), confirming the specificity of the antibody used to detect COMMD1.
Figure 1. COMMD1 and ATP7B interact and are localized in overlapping regions in the cells.
A+B. Glutathione-sepharose precipitation using cell lysates of HEK293T (A), or HepG2 (B) cells expressing ATP7B-Flag together with GST or COMMD1-GST. Precipitates were washed and separated by SDS-PAGE and immunoblotted as indicated. Input indicates direct analysis of cell lysates.
C-H. HEK293T cells stably transfected with an empty shRNA vector (control), or HEK293T cells with a stable knockdown of COMMD1 (shCOMMD1) were analyzed by double-label indirect confocal immunofluorescene using antibodies against ATP7B and COMMD1, visualized by Alexa488 conjugated donkey anti-rat and Alexa568 conjugated donkey anti-mouse IgG, respectively. ATP7B localization is presented in images C and F and COMMD1 localization is presented in images D and G. Overlap in staining is depicted in yellow in the overlay images (E and H).
COMMD1 interacts with the copper-binding amino terminal region of ATP7B independent of ATOX1 and copper concentration
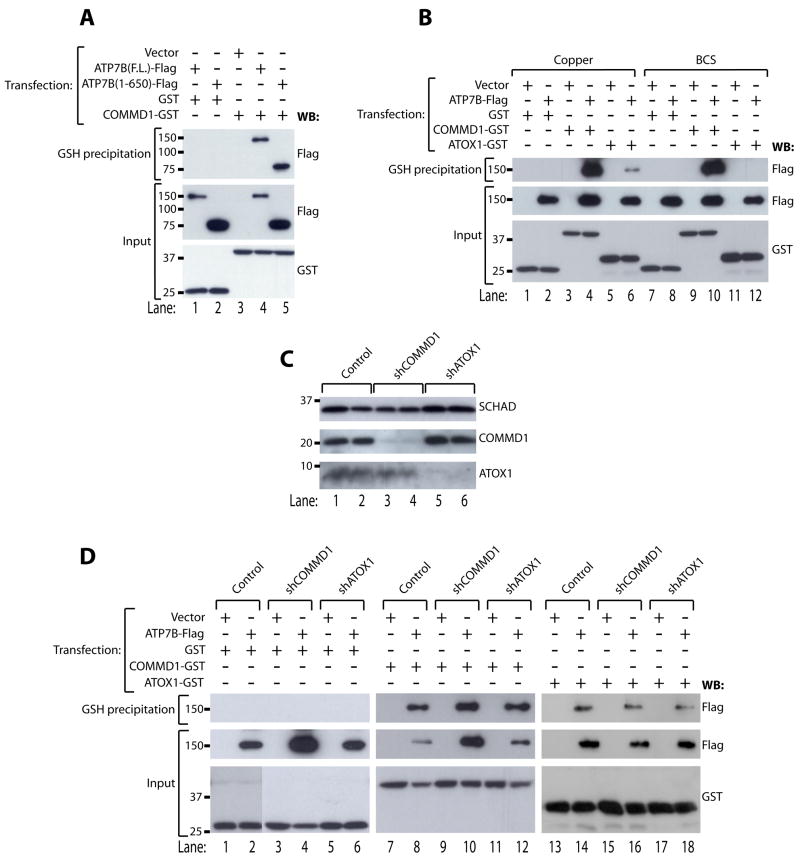
Next, full length ATP7B-Flag or a truncated variant of ATP7B consisting of the amino-terminal 650 amino acids of the full length protein (ATP7B(1–650)-Flag), were expressed in HEK293T cells together with GST or COMMD1-GST. Analysis of glutathione-sepharose precipitates from lysates of these cells (Figure 2A) indicated that the amino terminal region of ATP7B is sufficient for interaction with COMMD1, as shown previously 13. Within its amino terminal region, ATP7B contains six copper binding sequences, through which it receives copper by a transient copper-dependent interaction with the copper chaperone ATOX17, 8. Whereas incubation with the copper chelator BCS efficiently prevented interaction of ATP7B-Flag and ATOX1-GST (Figure 2B, compare lanes 6 and 12), no reproducible effects of CuSO4 or BCS incubation on the interaction of COMMD1-GST with ATP7B-Flag were observed (Figure 2B, compare lanes 4 and 10). These data suggest that COMMD1 binds to the amino terminus of ATP7B, independently of cellular copper levels. To determine if ATOX1 is essential for the interaction of COMMD1 with ATP7B, HEK293T cells were stably transfected with plasmids encoding short hairpin RNAs (shRNAs) directed against ATOX1, resulting in an efficient knockdown of ATOX1 (4% residual expression compared to control cells, figure 2C). This knockdown of ATOX1 did not affect the interaction between COMMD1-GST and ATP7B-Flag (Figure 2D). In the reciprocal experiment, efficient knockdown of endogenous COMMD1 (12% residual expression compared to control cells, figure 2C) did not result in impairment of the interaction between ATOX1-GST and ATP7B-Flag (Figure 2D). These data indicate that COMMD1 and ATOX1 bind to the same region of ATP7B, but they do so independent of each other.
Figure 2. COMMD1 interacts with the amino terminus of ATP7B in a copper- and ATOX1-independent manner.
A. Glutathione-sepharose precipitation was performed as described under figure 1A using cell lysates of HEK293T cells expressing ATP7B(F.L.) or ATP7B(1–650) coupled to Flag epitopes together with GST or COMMD1-GST.
B. HEK293T cells were transfected with cDNA constructs encoding full length ATP7B-Flag, GST, COMMD1-GST or ATOX1-GST as indicated. Cells were treated overnight with 200 μM CuSO4 or BCS (bathocuproinedisulfonic acid), prior to glutathione-sepharose precipitation as described under figure 1A
. C. Western blot analysis of COMMD1 and ATOX1 expression in cell lysates of HEK293T cells after a stable knockdown of COMMD1 (shCOMMD1) or ATOX1 (shATOX1). HEK293T cells stably transfected with an empty shRNA vector were used as a negative control (Control). SCHAD was probed as loading control.
D. HEK293T control cells, or HEK293T cells stably expressing shRNAs targeting COMMD1 (shCOMMD1) or ATOX1 (shATOX1) were transfected with cDNA constructs encoding ATP7B-Flag together with GST (D), COMMD1-GST (E), or ATOX1-GST (F). Lysates were used for glutathione-sepharose precipitation, as described under figure 1A.
COMMD1 does not regulate copper-induced trafficking of ATP7B
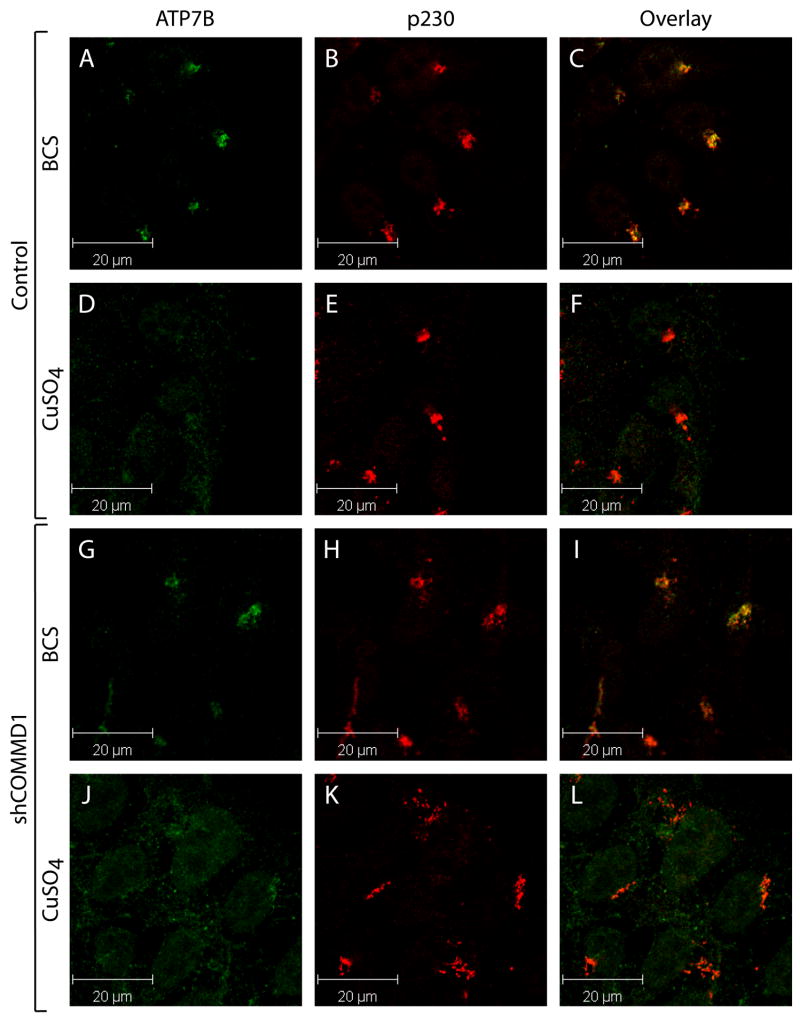
To assess if COMMD1 has a regulatory role in copper-dependent trafficking of ATP7B, the localization of ATP7B in response to copper was determined in COMMD1-deficient and control HEK293T cells. Incubation with BCS resulted in localization of ATP7B in dense perinuclear regions overlapping with the TGN marker p230 in both cell lines (Figure 3A–C and G–I). Treatment with 200 μM CuSO4 for 1 hour resulted in relocalization of ATP7B towards the cell periphery, as appreciated by loss of overlap between ATP7B and p230 staining (Figure 3 D–F). This relocalization was not impaired in COMMD1 deficient cells, also not when cells were treated with 200 μM CuSO4 for different time intervals, or with different concentrations of CuSO4 (Figure 3J–L and data not shown). Similarly, overexpression of COMMD1 did not interfere with the copper-dependent relocalization of ATP7B (data not shown). Taken together, these data suggest that COMMD1 does not have an essential regulatory role in the copper-induced relocalization of ATP7B.
Figure 3. COMMD1 does not regulate copper-induced trafficking of ATP7B.
HEK293T cells stably transfected with an empty shRNA vector (Control, A–F), or with shCOMMD1 (G–L) were incubated with 50 μM BCS or 200 μM CuSO4. Cells were subsequently analyzed using antibodies against ATP7B and p230, visualized by Alexa488 conjugated donkey anti-rat and Alexa568 conjugated donkey anti-mouse IgG respectively. ATP7B localization is presented in images A, D, G, and H, and p230 localization is presented in images B, E, H, and K. Overlap in staining is depicted in yellow in the overlay images (C, F, I and K).
Several mutations in ATP7B associated with WD result in increased binding of ATP7B to COMMD1-GST
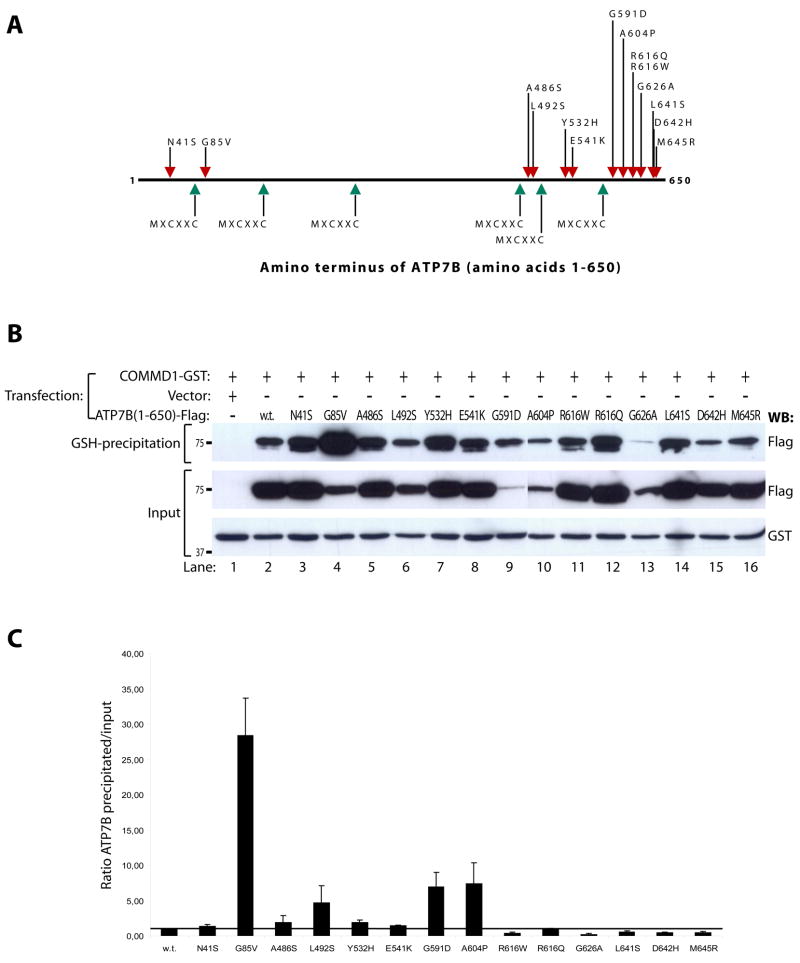
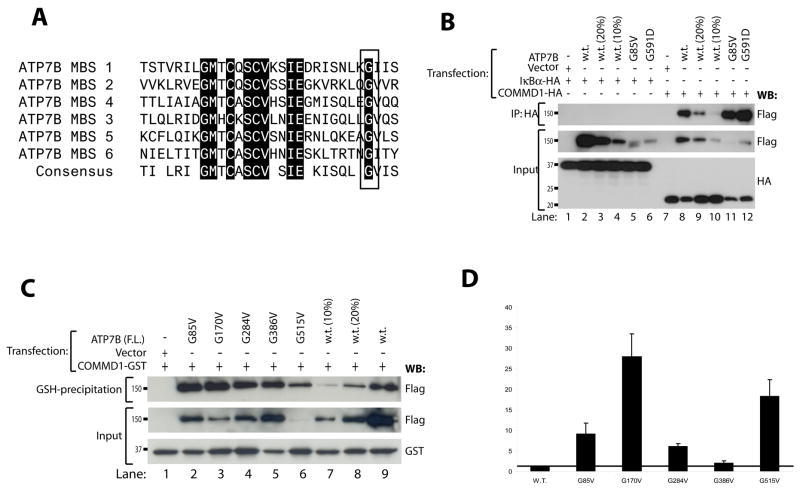
The interaction between COMMD1 and ATP7B suggests that these two proteins cooperate to maintain copper homeostasis. We therefore hypothesized that alteration of this interaction underlies the development of copper overload in WD. As COMMD1 directly binds to the amino terminus of ATP7B, this hypothesis predicts that WD-causing mutations in this region could interfere with the interaction of ATP7B with COMMD1. To assess this possibility, all the missense mutations causing WD in this region and deposited in the WD mutation database (as of February 2005) 6 were introduced in ATP7B(1–650)-Flag, yielding a total of 14 mutant cDNAs (Figure 4A). Glutathione precipitation using lysates of HEK293T cells transfected with these mutant cDNAs revealed that all mutants retained their ability to interact with COMMD1-GST (Figure 4B). Quantitation of precipitated ATP7B(1–650)-Flag relative to the amount present in the lysate revealed that several mutations; G85V, L492S, G591D, and A604P resulted in a marked increase in the efficiency of ATP7B(1–650)-Flag precipitation by COMMD1-GST (Figure 4C). A slight, but not reproducible, increase of this interaction was also observed for the A486S and Y532H mutations.
Figure 4. Several WD-causing mutations are associated with increased binding to COMMD1.
A. Schematic representation of the amino terminal 650 amino acids of ATP7B. Locations of metal-binding sequences are indicated by green arrows. Red arrows indicate locations of WD-causing mutations introduced in ATP7B(1–650)-Flag and ATP7B-Flag for experiments in Figures 4B, 4C and 5B.
B. Glutathione-sepharose precipitation using cell lysates of HEK293T cells expressing wild-type or mutant ATP7B(1–650)-Flag and COMMD1-GST, as described under figure 1A.
C. Quantification of the ratio of precipitated wild-type and mutant ATP7B(1–650)-Flag relative to input levels. Wild-type ratio was set to 1, and indicated by the black horizontal line. Shown is the average of at least three independent experiments. Error bars indicate standard errors of the mean.
G85 and G591 residues are conserved residues present in all six metal binding regions of ATP7B
Further inspection of the four residues affecting interaction with COMMD1 revealed that the G85 and G591 residues are located in the same conserved position in metal binding regions one and six of ATP7B, respectively (Figure 5A). Interestingly, the G85V mutation most dramatically affected the interaction of ATP7B-Flag with COMMD1-GST, resulting in a relative increase of ATP7B-Flag interacting with COMMD1-GST by factor 28.3. Co-immunoprecipitation of full length ATP7B-Flag harboring either G85V or G591D mutations with COMMD1-HA confirmed the effects of these mutations (Figure 5B). To assess if the other conserved glycine residues in metal binding regions 2-5 of ATP7B equally affect interaction of ATP7B with COMMD1, each of these residues were individually mutated to valines. Glutathione precipitation using lysates of HEK293T cells transfected with these mutant cDNAs revealed that mutation of the glycine residues in metal binding regions two, three and five also resulted in a dramatic increase of ATP7B-Flag interaction with COMMD1-GST (Figure 5C–D). A slight, but not reproducible, increase for this interaction was also observed for mutation of the glycine in metal binding region four. Similar results were obtained when these glycine residues were mutated to aspartic acid (data not shown).
Figure 5. G85 and G591 are conserved residues in all metal binding regions of ATP7B and are important for COMMD1-ATP7B interaction.
A. Sequence alignment of all six metal binding regions of ATP7B. The box indicates position of G85 and G591 and conserved glycine residues at the same position in other metal binding regions.
B. Full length ATP7B-Flag, ATP7B-G85V-Flag and ATP7B-G591D-Flag were expressed in HEK293T cells together with COMMD1-HA or IκBα-HA. To equalize wild-type ATP7B-Flag expression to mutant ATP7B-Flag expression, HEK293T cells were also transfected with less cDNA (20% and 10%) encoding wild-type ATP7B-Flag. Cells lysates were used for immunoprecipitation with anti-HA. Precipitates were immunoblotted as indicated. Input indicates direct analysis of cell lysates.
C. Wild type ATP7B-Flag and glycine to valine mutants of different metal binding regions in ATP7B-Flag were expressed in HEK293T cells together with GST or COMMD1-GST. To equalize wild-type ATP7B-Flag expression to mutant ATP7B-Flag expression, HEK293T cells were also transfected with less cDNA (20% and 10%) encoding wild-type ATP7B-Flag. Cell lysates were used for glutathione-sepharose precipitation as described under figure 1A
. D. Quantification of the ratio of precipitated wild-type and mutant ATP7B(1–650)-Flag relative to input levels presented in C. Wild-type ratio was set to 1, and indicated by the black line. Shown is the average of three independent experiments except for the G170V (n=2). Error bars indicate standard errors of the mean.
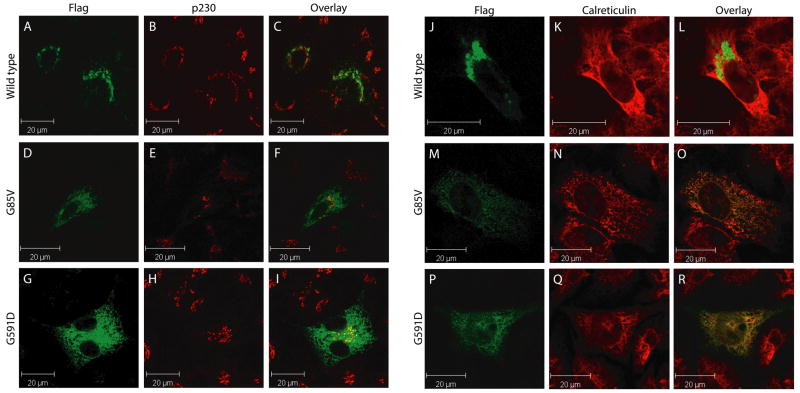
The G85V and G591D mutations lead to defective localization of ATP7B to the endoplasmic reticulum
Next, we set out to determine the effects of the G85V and G591D mutations on ATP7B expression and localization. HepG2 cells were transfected with wild-type or mutant ATP7B-Flag. Staining of these cells with FITC-conjugated anti-Flag antibodies revealed an overlapping localization of wild type ATP7B-Flag with the TGN marker p230 (Figure 6A–C). In contrast, both the G85V and G591D mutant ATP7B-Flag lost co-localization with p230 (Figure 6D–I), and displayed a more dispersed reticular localization pattern overlapping with the ER marker calreticulin (Figure 6M–R). These experiments suggest that the G85V and G591D mutations lead to mislocalization of ATP7B to the ER. Mislocalization of the G85V and G591D mutants could not be rescued by growing the cells at 30 °C, or by treatment with CuSO4. While COMMD1 staining partially overlapped with ATP7B G85V and G591D, expression of these mutants did not result in detectable alteration of overall COMMD1 localization (data not shown).
Figure 6. The G85V and G591D mutations lead to defective localization of ATP7B to the endoplasmic reticulum.
HEK293T cells were transfected with ATP7B-Flag (A–C and J–L), ATP7B-G85V-Flag (D–F and M–O) or ATP7B-G591D-Flag (G–I and P–R) and analyzed using antibodies against ATP7B (A, D, G, J, M, and P; green), p230 (B, E, and H; red) or Calreticulin (K, N, and Q; red), visualized by Alexa488 conjugated donkey anti-rat, Alexa568 conjugated donkey anti-mouse, Alexa568 conjugated donkey anti-rabbit IgG respectively. Overlap in staining is depicted in yellow in the overlay images (C, F, I, L, O, and R).
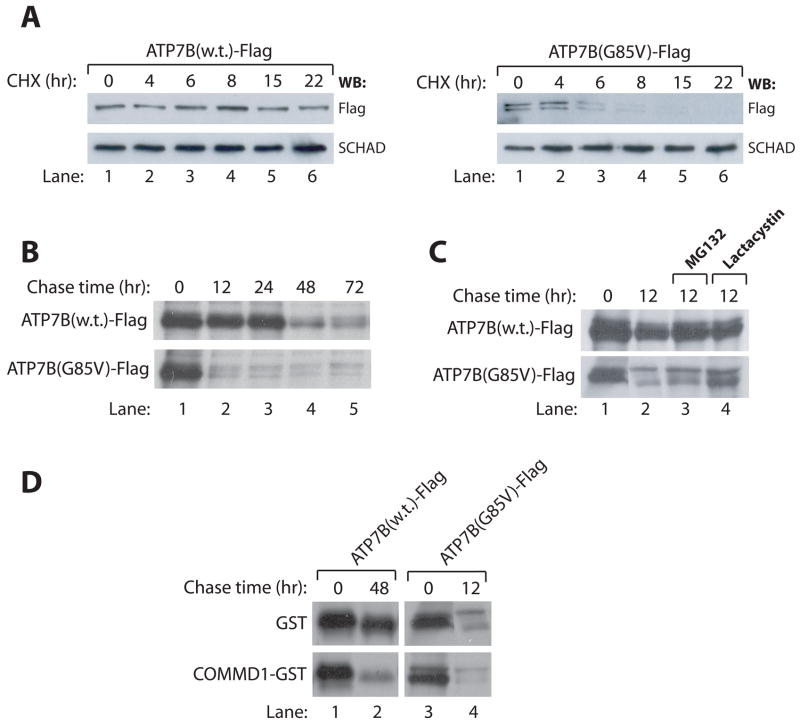
The G85V mutation leads to increased degradation of ATP7B
ER mislocalization of proteins is often associated with increased proteolysis, as has been demonstrated before for the common WD causing mutation H1069Q in ATP7B 27. Therefore we sought to determine if the G85V and G591D mutations result in an increase in proteolysis associated with the mislocalization to the ER.The low steady state expression level of the G591D mutant (figure 4B) prohibited reliable analysis of its stability. HEK293T cells were transfected with cDNA constructs encoding wild-type or G85V mutant ATP7B-Flag, and subsequently incubated with protein synthesis inhibitor cycloheximide for different time intervals. Western blot analysis demonstrates that wild-type ATP7B-Flag is relatively stable over a period of 22 hours, but the G85V mutant ATP7B-Flag is clearly degraded within 8 hours after addition of cycloheximide (Figure 7A). As a complementary approach, metabolic pulse-chase labeling was undertaken. This experiment revealed that, although the biosynthesis rates of wild-type and G85V mutant ATP7B-Flag were similar, the G85V mutant is subject to a dramatically increased rate of proteolysis (Figure 7C). Incubation of cells with the proteasomal inhibitors MG132 or lactacystin partially inhibited the degradation of G85V mutant ATP7B-Flag, suggesting that the increased proteolysis was at least partially mediated by the proteasome (Figure 7C).
Figure 7. ATP7B-G85V is subject to increased degradation, partially facilitated by COMMD1.
A: HEK293T cells were transfected with cDNA constructs encoding for ATP7B-Flag or ATP7B-G85V-Flag and subsequently incubated with cycloheximide for the indicated time intervals. Cell lysates were generated and analyzed for expression of ATP7B-Flag and ATP7B-G85V-Flag. Equal loading was confirmed by immunoblotting for SCHAD.
B: HEK293T cells were transfected with cDNA constructs encoding for ATP7B-Flag or ATP7B-G85V-Flag and biosynthetically labeled with 35S-labeled methionine and cysteine for 90 minutes. After labeling, cells were chased with cold medium for the indicated time intervals. Cell lysates were subjected to immunoprecipitation using anti-Flag affinity gel before analysis by SDS-PAGE. Proteins were visualized with fluorography.
C. HEK293T cells expressing ATP7B-Flag or ATP7B-G85V-Flag were subjected to biosynthetic labeling and immunoprecipitation as described in B. During 12 hr chase with cold media, cells were incubated with the proteasome inhibitors MG132 or Lactacystin as indicated.
D. HEK293T cells expressing ATP7B-Flag or ATP7B-G85V-Flag together with GST or COMMD1-GST were subjected to biosynthetic labeling and immunoprecipitation as described in B.
Previous studies revealed that COMMD1 associates with several proteins to regulate the stability of such interacting proteins 20, 21, 24. Therefore, we hypothesized that COMMD1 could also regulate the stability of ATP7B. To address this hypothesis, HEK293T cells were transfected with cDNA constructs encoding wild-type or G85V mutant ATP7B-Flag together with GST or COMMD1-GST. Pulse-chase analysis of ATP7B-Flag stability on these cells revealed that overexpression of COMMD1 results in increased proteolysis of wild-type ATP7B-Flag and, to a lesser extent also of G85V mutant ATP7B-Flag (Figure 7D).
Discussion
The hepatic copper overload in Bedlington terriers with a homozygous deletion of the second exon of COMMD1 implies a critical role of COMMD1 in the regulation of copper excretion 10. In addition, knockdown of COMMD1 by RNA interference in several cell lines results in increased cellular copper levels 25, 28. COMMD1 interacts with the copper transport protein ATP7B, which is mutated in WD, suggesting that these two proteins co-operate in the excretion of copper 13. Consequentially, these observations suggest that COMMD1 could play a role in the development of WD. Consistent with this notion, heterozygosity for a silent missense mutation in COMMD1 was possibly associated with an earlier onset of the disorder in patients with known ATP7B mutations 15. Because COMMD1 appears to depend on protein-protein interactions for its functions (reviewed in 19), we have characterized the effects of mutations in ATP7B associated with WD on its interaction with COMMD1. Several WD causing mutations markedly increased the amount of ATP7B that interacted with COMMD1, suggesting that deregulation of this interaction is associated with the development of WD. In addition to previously reported effects of WD causing mutations on ATP7B-mediated copper transport, cuproenzyme biosynthesis, ATP-binding and –hydrolysis, localization and copper-induced trafficking, post-translational modifications, and protein-protein interactions, this observation provides a novel molecular mechanism in the pathogenesis of WD.
Further characterization of the mutations that de-regulate the interaction of ATP7B with COMMD1 has led to valuable insights in the functionality of this interaction. The G85 and G591 residues in ATP7B are highly conserved at a position were a glycine is present in all six amino-terminal metal binding regions, indicating that these residues are of structural importance, which is underscored by our observation that mutation of other glycine residues also leads to increased binding to COMMD1. In addition, previous studies indicated that the G85V and G591D mutations markedly impair the interaction of ATP7B with ATOX1 7. Analysis of the localization of the G85V and G591D mutations demonstrated that both mutations result in defective localization of ATP7B to the ER in the cell. A similar defect has been demonstrated for several other WD causing mutations (e.g. see refs 27, 29). ER mislocalization of proteins is often due to misfolding and associated with proteasomal degradation 30. A well known example of this process in human disease development is the ER associated degradation of ΔF508 CFTR mutant in cystic fibrosis 31. In addition, it has previously been shown that the common WD causing mutation H1069Q results in increased proteolysis, associated with ER mislocalization of ATP7B 27. Two complementary approaches indicated that the G85V mutant ATP7B protein has a dramatically shortened half-life. Incubation with the proteasome inhibitor lactacystin partially inhibited the degradation of the G85V mutant, indicating that this degradation is at least in part mediated by the proteasome.
The increased interaction of this mutant with COMMD1 led us to hypothesize that COMMD1 facilitates the degradation of ATP7B. This hypothesis is supported by several recent studies that have implicated COMMD1 as a negative regulator of the stability of a number of proteins in the HIF-1 and NF-κB pathways 20, 21, 24. Within the NF-κB pathway, COMMD1 specifically promotes the ubiquitination and subsequent proteasomal degradation of RelA, whereas it protects IκBα from the same fait 21, 24. COMMD1 has also been shown to inhibit the activity of the epithelial sodium channel (ENAC), through association with the αENaC and δENaC subunits 22. The mechanism of this inhibition possibly involves ubiquitination and degradation, since it is well known that the ubiquitin-proteasome pathway plays an important role in the regulation of ENaC 32. Furthermore, COMMD1 interacts with a number of subunits of E3 ubiquitin-ligase complexes, including Cullin1, Cullin2, Cullin3, Cullin5, and the X-linked inhibitor of apoptosis (XIAP), suggesting a direct role for COMMD1 in ubiquitin modifications 20, 21, 24, 25. Taken together, these studies strongly implicate COMMD1 as a general negative regulator of protein stability in a number of cellular processes. Consistent with this proposed function, we have observed that overexpression of COMMD1 resulted in an increase in proteolytic turnover of newly synthesized ATP7B. The increased interaction with ATP7B as a result of several WD causing mutations suggests that COMMD1 is involved in the quality control of ATP7B. The importance of quality control mechanisms is underscored by estimates that, in general, 30% of all newly synthesized proteins are targeted for degradation as a result of misfolding 33. In this context, failure to maintain proper quality control of ATP7B in Bedlington terriers as a result of COMMD1 deficiency might be the mechanism that underlies the development of copper toxicosis. High copper intake through modern commercial dog food over the years has resulted in hepatic copper concentrations in healthy dogs that are over 10-fold higher than in humans 34. This might predispose dogs to develop copper toxicosis, as evidenced by the occurrence of copper associated liver disease in a variety of dog breeds other then Bedlington terriers 12, 35. We hypothesize that absence of COMMD1 in affected Bedlington terriers disrupts ATP7B quality control, resulting in a gradual build-up of suboptimally functioning ATP7B, which perturbs the ability of the hepatocyte to cope with such high copper concentrations. This hypothesis is supported by the observation that, although serum ceruloplasmin levels appear normal, the incorporation of copper in ceruloplasmin is greatly reduced in a manner that inversely correlates with hepatic copper levels consistent with a gradual impairment of the function of ATP7B 36. In conclusion, we have presented biochemical evidence that COMMD1, a protein identified through genetic studies in dogs with copper toxicosis, shows a markedly increased interaction with ATP7B affected by distinct mutations. These data suggest that this increased interaction contributes to the molecular basis of WD in patients harboring such mutations. This observation might partially explain the clinical heterogeneity observed in WD. In addition, this study indicates that COMMD1 regulates ATP7B stability, rather then copper-induced trafficking or interactions with ATOX1. Recently, nine human homologues of COMMD1 have been identified that share a common functional domain known as the COMM domain. These proteins have a similar inhibitory function in the NF-κB pathway as COMMD1 23, 37. These structural and functional similarities among COMMD proteins suggest that they might have a complementary function in copper homeostasis. This is supported by our observation that three COMMD proteins, other then COMMD1, also interact with ATP7B (P. de Bie et al., unpublished data). Further studies on the molecular functions of COMMD1 and its homologues will provide valuable and novel insights into the regulation of ATP7B, and consequentially, copper homeostasis.
Acknowledgments
This work has been funded by the Dutch Digestive Diseases Foundation grant WS 02–34. We thank Olivier van Beekum and Eric Kalkhoven for assistance in generating the COMMD1 and ATOX1 knockdown cell lines, Eric Kalkhoven and Jackie Senior for critically reading the manuscript, and Thomas Müller and members of the Klomp and Wijmenga laboratories for helpful discussions.
Abbreviations
- ER
endoplasmic reticulum
- GST
glutathione-S-transferase
- HEK293T
human embryonic kidney 293T cells
- HIF1
hypoxia inducible factor 1
- NF-κB
nuclear factor κB
- NF-κB
nuclear factor κB
- shRNA
short hairpin RNA
- TGN
trans Golgi network
- WD
Wilson disease
Footnotes
Author conflict of interest statement: The Authors declare that they have no conflicts of interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wijmenga C, Klomp LW. Molecular regulation of copper excretion in the liver. Proc Nutr Soc. 2004;63:31–39. doi: 10.1079/pns2003316. [DOI] [PubMed] [Google Scholar]
- 2.Gitlin JD. Wilson disease. Gastroenterology. 2003;125:1868–77. doi: 10.1053/j.gastro.2003.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Bull PC, Thomas GR, Rommens JM, Forbes JR, Cox DW. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet. 1993;5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 4.Tanzi RE, Petrukhin K, Chernov I, Pellequer JL, Wasco W, Ross B, Romano DM, Parano E, Pavone L, Brzustowicz LM. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet. 1993;5:344–350. doi: 10.1038/ng1293-344. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi Y, Heiny ME, Gitlin JD. Isolation and characterization of a human liver cDNA as a candidate gene for Wilson disease. Biochem Biophys Res Commun. 1993;197:271–277. doi: 10.1006/bbrc.1993.2471. [DOI] [PubMed] [Google Scholar]
- 6.Kenny S, Cox DW. Wilson Disease Mutation Database. http://www.medicalgenetics.med.ualberta.ca/wilson/index.php.
- 7.Hamza I, Schaefer M, Klomp LW, Gitlin JD. Interaction of the copper chaperone HAH1 with the Wilson disease protein is essential for copper homeostasis. Proc Natl Acad Sci U S A. 1999;96:13363–8. doi: 10.1073/pnas.96.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker JM, Tsivkovskii R, Lutsenko S. Metallochaperone Atox1 transfers copper to the NH2-terminal domain of the Wilson's disease protein and regulates its catalytic activity. J Biol Chem. 2002;277:27953–9. doi: 10.1074/jbc.M203845200. [DOI] [PubMed] [Google Scholar]
- 9.Hung IH, Suzuki M, Yamaguchi Y, Yuan DS, Klausner RD, Gitlin JD. Biochemical characterization of the Wilson disease protein and functional expression in the yeast Saccharomyces cerevisiae. J Biol Chem. 1997;272:21461–6. doi: 10.1074/jbc.272.34.21461. [DOI] [PubMed] [Google Scholar]
- 10.Van de Sluis B, Rothuizen J, Pearson PL, van Oost BA, Wijmenga C. Identification of a new copper metabolism gene by positional cloning in a purebred dog population. Hum Mol Genet. 2002;11:165–173. doi: 10.1093/hmg/11.2.165. [DOI] [PubMed] [Google Scholar]
- 11.Klomp AE, van de Sluis B, Klomp LW, Wijmenga C. The ubiquitously expressed MURR1 protein is absent in canine copper toxicosis. J Hepatol. 2003;39:703–709. doi: 10.1016/s0168-8278(03)00380-5. [DOI] [PubMed] [Google Scholar]
- 12.Fuentealba IC, Aburto EM. Animal models of copper-associated liver disease. Comp Hepatol. 2003;2:5. doi: 10.1186/1476-5926-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tao TY, Liu F, Klomp L, Wijmenga C, Gitlin JD. The copper toxicosis gene product Murr1 directly interacts with the Wilson disease protein. J Biol Chem. 2003;278:41593–41596. doi: 10.1074/jbc.C300391200. [DOI] [PubMed] [Google Scholar]
- 14.Muller T, van de Sluis B, Zhernakova A, van Binsbergen E, Janecke AR, Bavdekar A, Pandit A, Weirich-Schwaiger H, Witt H, Ellemunter H, Deutsch J, Denk H, Muller W, Sternlieb I, Tanner MS, Wijmenga C. The canine copper toxicosis gene MURR1 does not cause non-Wilsonian hepatic copper toxicosis. J Hepatol. 2003;38:164–168. doi: 10.1016/s0168-8278(02)00356-2. [DOI] [PubMed] [Google Scholar]
- 15.Stuehler B, Reichert J, Stremmel W, Schaefer M. Analysis of the human homologue of the canine copper toxicosis gene MURR1 in Wilson disease patients. J Mol Med. 2004;82:629–634. doi: 10.1007/s00109-004-0557-9. [DOI] [PubMed] [Google Scholar]
- 16.Wu ZY, Zhao GX, Chen WJ, Wang N, Wan B, Lin MT, Murong SX, Yu L. Mutation analysis of 218 Chinese patients with Wilson disease revealed no correlation between the canine copper toxicosis gene MURR1 and Wilson disease. J Mol Med. 2006:1–5. doi: 10.1007/s00109-005-0036-y. [DOI] [PubMed] [Google Scholar]
- 17.Coronado VA, Bonneville JA, Nazer H, Roberts EA, Cox DW. COMMD1 (MURR1) as a candidate in patients with copper storage disease of undefined etiology. Clin Genet. 2005;68:548–51. doi: 10.1111/j.1399-0004.2005.00524.x. [DOI] [PubMed] [Google Scholar]
- 18.Weiss KH, Merle U, Schaefer M, Ferenci P, Fullekrug J, Stremmel W. Copper toxicosis gene MURR1 is not changed in Wilson disease patients with normal blood ceruloplasmin levels. World J Gastroenterol. 2006;12:2239–42. doi: 10.3748/wjg.v12.i14.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Bie P, van de Sluis B, Klomp L, Wijmenga C. The Many Faces of the Copper Metabolism Protein MURR1/COMMD1. J Hered. 2005;96:803–811. doi: 10.1093/jhered/esi110. [DOI] [PubMed] [Google Scholar]
- 20.van de Sluis B, Muller P, Duran K, Chen A, Groot AJ, Klomp LW, Liu PP, Wijmenga C. Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice. Mol Cell Biol. 2007 doi: 10.1128/MCB.01932-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ganesh L, Burstein E, Guha-Niyogi A, Louder MK, Mascola JR, Klomp LW, Wijmenga C, Duckett CS, Nabel GJ. The gene product Murr1 restricts HIV-1 replication in resting CD4+ lymphocytes. Nature. 2003;426:853–857. doi: 10.1038/nature02171. [DOI] [PubMed] [Google Scholar]
- 22.Biasio W, Chang T, McIntosh CJ, McDonald FJ. Identification of Murr1 as a regulator of the human delta epithelial sodium channel. J Biol Chem. 2003;279:5429–5434. doi: 10.1074/jbc.M311155200. [DOI] [PubMed] [Google Scholar]
- 23.Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW, Duckett CS. COMMD Proteins, a Novel Family of Structural and Functional Homologs of MURR1. J Biol Chem. 2005;280:22222–32. doi: 10.1074/jbc.M501928200. [DOI] [PubMed] [Google Scholar]
- 24.Maine GN, Mao X, Komarck CM, Burstein E. COMMD1 promotes the ubiquitination of NF-kappaB subunits through a cullin-containing ubiquitin ligase. Embo J. 2007;26:436–47. doi: 10.1038/sj.emboj.7601489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burstein E, Ganesh L, Dick RD, Van De Sluis B, Wilkinson JC, Klomp LW, Wijmenga C, Brewer GJ, Nabel GJ, Duckett CS. A novel role for XIAP in copper homeostasis through regulation of MURR1. Embo J. 2004;23:244–254. doi: 10.1038/sj.emboj.7600031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duckett CS, Perkins ND, Kowalik TF, Schmid RM, Huang ES, Baldwin AS, Jr, Nabel GJ. Dimerization of NF-KB2 with RelA(p65) regulates DNA binding, transcriptional activation, and inhibition by an I kappa B-alpha (MAD-3) Mol Cell Biol. 1993;13:1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payne AS, Kelly EJ, Gitlin JD. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper-dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci U S A. 1998;95:10854–9. doi: 10.1073/pnas.95.18.10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spee B, Arends B, van Wees AM, Bode P, Penning LC, Rothuizen J. Functional consequences of RNA interference targeting COMMD1 in a canine hepatic cell line in relation to copper toxicosis. Anim Genet. 2007;38:168–70. doi: 10.1111/j.1365-2052.2007.01580.x. [DOI] [PubMed] [Google Scholar]
- 29.Harada M, Sakisaka S, Terada K, Kimura R, Kawaguchi T, Koga H, Kim M, Taniguchi E, Hanada S, Suganuma T, Furuta K, Sugiyama T, Sata M. A mutation of the Wilson disease protein, ATP7B, is degraded in the proteasomes and forms protein aggregates. Gastroenterology. 2001;120:967–74. doi: 10.1053/gast.2001.22543. [DOI] [PubMed] [Google Scholar]
- 30.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 31.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–34. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 32.Malik B, Price SR, Mitch WE, Yue Q, Eaton DC. Regulation of epithelial sodium channels by the ubiquitin-proteasome proteolytic pathway. Am J Physiol Renal Physiol. 2006;290:F1285–94. doi: 10.1152/ajprenal.00432.2005. [DOI] [PubMed] [Google Scholar]
- 33.Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–4. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 34.Su LC, Owen CA, Jr, Zollman PE, Hardy RM. A defect of biliary excretion of copper in copper-laden Bedlington terriers. Am J Physiol. 1982;243:G231–6. doi: 10.1152/ajpgi.1982.243.3.G226. [DOI] [PubMed] [Google Scholar]
- 35.Thornburg LP. A perspective on copper and liver disease in the dog. J Vet Diagn Invest. 2000;12:101–10. doi: 10.1177/104063870001200201. [DOI] [PubMed] [Google Scholar]
- 36.Su LC, Ravanshad S, Owen CA, Jr, McCall JT, Zollman PE, Hardy RM. A comparison of copper-loading disease in Bedlington terriers and Wilson's disease in humans. Am J Physiol. 1982;243:G226–30. doi: 10.1152/ajpgi.1982.243.3.G226. [DOI] [PubMed] [Google Scholar]
- 37.De Bie P, van de Sluis B, Burstein E, Duran KJ, Berger R, Duckett CS, Wijmenga C, Klomp LW. Characterization of COMMD protein-protein interactions in NF-kappaB signalling. Biochem J. 2006;398:63–71. doi: 10.1042/BJ20051664. [DOI] [PMC free article] [PubMed] [Google Scholar]