Abstract
Old and unneeded intracellular macromolecules are delivered through autophagy to lysosomes that degrade macromolecules into bioactive monomers such as amino acids. Autophagy is conserved in eukaryotes and is essential for the maintenance of cellular metabolism. Currently, more than 30 autophagy-related genes (Atgs) have been identified in yeast. Of these genes, the18 that are essential for autophagosome formation are also conserved in mammalian cells. Atg9 is the only transmembrane Atg protein required for autophagosome formation. Although the subcellular localization of the Atg9A protein (Atg9Ap) has been examined, little is known about its precise cell and tissue distribution. To determine this, we produced an antibody specific to mouse Atg9Ap. The antibody recognized both non-glycosylated and glycosylated Atg9Ap, which have molecular masses of ∼94 kDa and 105 kDa, respectively. Although Atg9Ap was ubiquitously detected, it was highly expressed in neurons of the central nervous system. In Purkinje cells, Atg9Ap immunoreactivity was localized in the endoplasmic reticulum (ER), trans-Golgi network (TGN), lysosomes/late endosomes, and in axon terminals. These results suggest that Atg9Ap may be involved in autophagosome formation in the ER and axon terminals of neurons, the TGN, and lysosomes/late endosomes. (J Histochem Cytochem 58:443–453, 2010)
Keywords: autophagy, mouse, Atg9A, neurons, Purkinje cells, axons
Autophagy is a eukaryotic self-degradation system that plays a pivotal role in the maintenance of cellular homeostasis (Mortimore and Poso 1987; Kuma et al. 2004; Meijer and Dubbelhuis 2004). Moreover, autophagy is induced by various stresses under various pathological conditions, such as starvation, inflammation, and ischemia. Once autophagy is induced in cells, old and unneeded substances, including organelles, together with part of the cytoplasm, are sequestered by double-membrane structures to form autophagosomes. These structures either receive lysosomal hydrolytic enzymes via transporting vesicles from the trans-Golgi network (TGN) or fuse with lysosomes to become autolysosomes (Klionsky and Emr 2000; Ohsumi 2001; Uchiyama et al. 2008). Many autophagy-related genes (Atgs) have been revealed in the yeast Saccharomyces cerevisiae, and mammalian homologs of these genes have also been identified (Klionsky et al. 2003; Mizushima 2007). Moreover, it has been shown that these Atg proteins are essential for autophagosome formation (Mizushima 2007; Uchiyama et al. 2008; Ravikumar et al. 2009).
The Atg9 protein (Atg9p), which is the only integral membrane Atg protein, is localized in the phagophore/pre-autophagosomal structure (PAS) (Suzuki et al. 2001; Yen and Klionsky 2007), although the structure in mammalian cells remains unknown. The PAS, in which several Atg proteins, including Atg9p, are localized (Krick et al. 2008; Sekito et al. 2009), might be the origin of the autophagosomal membranes, suggesting that Atg9p plays a crucial role in the formation of autophagosomes. It has been proposed that Atg9p is recruited to the PAS in an Atg11-dependent manner under nutrient-rich conditions, whereas during nutrient deprivation, recruitment of Atg9p depends on Atg17p (Sekito et al. 2009). Moreover, Atg9p cycles between the TGN and late endosomes in a ULK1-dependent manner (Young et al. 2006), whereas cycling between the PAS and peripheral regions of the cells occurs via several other mechanisms (Lang et al. 2000; Noda et al. 2000; Reggiori et al. 2005; Krick et al. 2008).
The role of Atg9A in the formation of autophagosomes remains to be identified, although subcellular localization of Atg9A protein (Atg9Ap) is clearly dependent on nutrient availability. Because autophagy is a highly conserved degradation system, it is expected that tissue distribution of Atg expression will be relatively uniform. However, the expression of human Atg9A mRNA is tissue dependent (Yamada et al. 2005). To examine in greater detail the precise cell and tissue distribution of Atg9Ap, we prepared an antibody specific to mouse Atg9Ap. Interestingly, Atg9Ap was predominantly expressed in neurons, including both axons and axon terminals.
Materials and Methods
Animals
The experiments described here were performed in compliance with the regulations of the Review Committee for Animal Experimentation of Juntendo University. C57BL/6J mice, 3 and 8 weeks old, were obtained from Charles River Laboratories of Japan (Yokohama, Japan), and were subsequently housed in specific pathogen-free conditions at Juntendo University.
Generation of an Antibody Against the Mouse Atg9Ap
The mouse Atg9A gene corresponding to an open reading frame was amplified by a PCR that used mouse brain cDNA as a template. The amplified Atg9A gene was subcloned into the EcoRI/HindIII site of the pGEM3Z vector (Promega; Madison, WI). The sequences of the sense and anti-sense primers are shown in Table 1. The DNA sequence that corresponds to the amino acid sequence of mAtg9A between aa 720 and aa 839 was generated by PCR (Table 1) that used mAtg9A/pGEM3Z as a template. The DNA was then subcloned into the BamHI/EcoRI site of the pGEX6P-1 vector (GE Healthcare UK, Ltd.; Little Chalfont, UK) to express the GST fusion protein. The GST–Atg9A fusion protein was purified using glutathione-Sepharose 4B and MacroPrep Ceramic Hydroxyapatite (Bio-Rad; Hercules, CA), and the GST was excised using PreScission proteinase (GE Healthcare UK). The antibody was prepared as previously described (Lu et al. 2005). Briefly, the purified Atg9Ap was emulsified with Titer Max Gold adjuvant (CytRx Corp.; Los Angeles, CA), and the emulsification was injected subcutaneously into female rabbits at intervals of 2–4 weeks. Polyclonal antibodies specific to Atg9Ap were purified from antisera, using an affinity column conjugated with the antigen.
Table 1.
Primer sequences for cloning of DNA fragments of Atg9A (NM_001003917) and GAPDH (NM_008084)
| Gene | Use | Location (bp) | Primers | Sequences |
|---|---|---|---|---|
| Atg9A | Cloning, RT-PCR | 1–2,520 | Sense | CGGAATTCATGGCACAGTTTGACACTGA |
| Anti-sense | GGGAAGCTTCTATACCTTGTGCACTTG | |||
| GAPDH | Control, RT-PCR | 520–971 | Sense | ACCACAGTCCATGCCATCAC |
| Anti-sense | TCCACCACCCTGTTGCTGTA | |||
| Atg9A | Antigen | 2,158–2,520 | Sense | GCGGATCCCAGACCCAGGCTGAGCCCGA |
| Anti-sense | GGAATTCCTATACCTTGTGCACTTGAG | |||
| Atg9A | ECFP–Atg9A | 1–2,520 | Sense | ATCTCGAGCTATGGCACAGTTTGACACTGA |
| Anti-sense | TATCTAGACTATACCTTGTGCACTTGAG |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Antibodies
A rabbit polyclonal anti-Atg9Ap antibody was generated in this study. The commercially available antibodies used for immunohisto-/cytochemistry and Western blotting in the present study were as follows: a rabbit polyclonal antibody to green fluorescent protein (GFP) (MBL; Nagoya, Japan), a rat monoclonal antibody to GFP (Nacalai Tesuque; Kyoto, Japan), mouse monoclonal antibodies to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Ambion; Austin, TX), GM130 (a Golgi marker) (Becton, Dickinson and Company; Franklin Lakes, NJ), the glucose-regulated protein 78/BiP [an endoplasmic reticulum (ER) marker protein marker, BiP] (Becton, Dickinson and Company), early endosome marker (EEA1) (Becton, Dickinson and Company), γ-adaptin (a marker protein for the TGN) (Becton, Dickinson and Company), vesicle docking protein p115 (a marker protein for ER/cis-Golgi, p115) (Becton, Dickinson and Company), mouse glia fibrillary acidic protein (GFAP) (Sigma-Aldrich; St. Louis, MO), neurofilament-L (NF-L) (Novus Biologicals; Littleton, CO), synaptophysin (PROGEN Biotechnik; Heidelberg, Germany), a rat polyclonal anti-mouse lamp1 antibody (a lysosomal membrane protein marker) (the Developmental Studies Hybridoma Bank; Iowa city, IA), guinea pig polyclonal antibodies to mouse ubiquitous mitochondrial creatine kinase (CK-Mi) (Frontier Science; Ishikari, Japan), glutamate transporter (GLAST; Frontier Science), and a goat polyclonal antibody to mouse vesicular GABA transporter (VGAT; Frontier Science). Moreover, horseradish peroxidase-conjugated, species-specific secondary antibodies were used in Western blotting and immunostaining (Dako; Glostrup, Denmark). An Alexa Fluor 488–conjugated, species-specific secondary antibody and an Alexa Fluor 594–conjugated, species-specific secondary antibody were used in immunostaining (Invitrogen; Carlsbad, CA).
Cell Culture and Transfection
NIH3T3 cells were cultured in DMEM (Nacalai Tesque) supplemented with 100 U/ml penicillin, 100 μg/ml streptomycin (Nacalai Tesque), and 10% fetal bovine serum (10% FBS DMEM). Cells were transfected using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Plasmid Construction and Cell Transfection
An open reading frame of the mouse Atg9A gene was subcloned into the XhoI/XbaI site of the pECFP-c1 vector (Clontech; Mountain View, CA) (Table 1). The ECFP–Atg9A vector was transfected into NIH3T3 cells, and positive clones were selected by addition of G418 (Nacalai Tesuque) to the medium.
RNA Interference of Mouse Atg9A
RNA interference (RNAi) of Atg9A was performed as described previously (Yoshimura et al. 2006). Four oligonucleotides for mouse Atg9A#1 and Atg9A#2 (Table 2) were synthesized. Each oligonucleotide was annealed and subcloned into the pSUPER_neo vector (OligoEngine; Seattle, WA). RNAi vectors were transfected into NIH3T3 cells, and the transfected cells were cultured for 14 days, during which positive clones were selected by addition of G418 to the medium.
Table 2.
Oligonucleotide sequences for RNAi of Atg9A mRNA
| Use | Target sequences (bp) | Oligonucleotides | Sequences |
|---|---|---|---|
| Atg9A RNAi #1 | 3–23 | Sense | GATCCGGCACAGTTTGACACTGAATATTCAAGAGAT-ATTCAGTGTCAAACTGTGCCTTTTTTGGAAA |
| Anti-sense | AGCTTTTCCAAAAAAGGCACAGTTTGACACTGAATA-TCTCTTGAATATTCAGTGTCAAACTGTGCCG | ||
| Atg9A RNAi #2 | 428–448 | Sense | GATCCGGATCCACCGGCTTATCAAGTTTCAAGAGAA-CTTGATAAGCCGGTGGATCCTTTTTTGGAAA |
| Anti-sense | AGCTTTTCCAAAAAAGGATCCACCGGCTTATCAAGT-TCTCTTGAAACTTGATAAGCCGGTGGATCCG |
RNAi, RNAi interference.
RT-PCR of Atg9A and GAPDH
Total RNAs were extracted from wild-type and Atg9A knockdown NIH3T3 cells using ISOGEN (Nippon Gene; Tokyo, Japan). Total RNAs were then reverse transcribed using RevaTra Ace (Toyobo; Osaka, Japan). The primers used for GAPDH and Atg9A are shown in Table 1.
Western Blot of Atg9Ap, GFP, and GAPDH
NIH3T3 cells that had been transfected with either the ECFP–Atg9A or Atg9A RNAi vector were harvested, lysed with lysis buffer [1% Triton X-100 and protease inhibitor cocktail (Nacalai Tesuque) in PBS], and incubated on ice for 30 min. The lysates were then centrifuged at 20,400 × g for 10 min, and the resultant supernatants were collected. Mouse tissues were pulverized in liquid nitrogen using a Cryo-Tec frozen tissue crusher (Microtec Co.; Funabashi, Japan). Pulverized tissues were homogenized with 5–10 vols of lysis buffer using a Polytron PT3100 (Kinematica; Littau, Switzerland), and the resultant homogenates were incubated on ice for 30 min. These lysates were then centrifuged at 20,400 × g for 10 min, and the resultant supernatants were collected. The lysates (20 μg) were subjected to SDS-PAGE, followed by Western blotting using antibodies to Atg9Ap (1:500), GFP (1:1000), or GAPDH (1:100,000). Quantification of protein bands was performed using an Image Gauge software program (Fuji Photo Film Co., Ltd.; Tokyo, Japan).
Immunohisto-/cytochemistry
Immunostaining was used to examine tissue and cellular distribution, as reported previously (Koike et al. 2000,2005; Yoshimura et al. 2006). Adult male and young C57BL/6J mice (8 and 3 weeks of age, respectively) were deeply anesthetized with pentobarbital (25 mg/kg intraperitoneal injection), and were fixed by cardiac perfusion with 4% paraformaldehyde buffered with 0.1 M phosphate buffer containing 4% sucrose. Because Atg9Ap was highly expressed in the central nervous system (CNS), the brains were excised and further fixed in the same fixative for 24 hr. Tissue samples for cryosections were cryoprotected by immersion in 15% and 30% sucrose solutions and then embedded in OCT compound (Miles; Elkhart, IN). The embedded tissues were cut into 10-μm sections with a cryostat (CM3050; Leica, Nussloch, Germany). The sections were placed on silane-coated glass slides and immunostained with the anti-Atg9A antibody (diluted 1:200) at 4C for 2 days, followed by further incubation with peroxidase-conjugated streptavidin (Vectastain ABC kit; Vector Laboratories, Burlingame, CA) for 1 hr at room temperature. After each step, the sections were rinsed thoroughly in 0.1 M phosphate-buffered 0.5 M saline (pH 7.2), containing 0.1% Tween 20 (TPBS). Staining for peroxidase was performed using 0.0125% 3,3-diaminobenzidine tetrahydrochloride (DAB) and 0.002% H2O2 in 0.05 M Tris-HCl buffer (pH 7.6) for 5 min (Vector Laboratories). For double-immunofluorescent staining, the cryosections were incubated with the anti-Atg9Ap antibody and an antibody to an intracellular marker such as BiP (1:50), p115 (1:100), GM130 (1:200), γ-adaptin (1:100), EEA1 (1:100), CK-Mi (1:200), and lamp1 (1:100), or GFAP (1:200), NF-L (1:50), or synaptophysin (1:100), VGAT (1:500), and GLAST (1:300) at 4C for 2 days. The cryosections were then incubated with a mixture of an Alexa Fluor 488–coupled goat or donkey anti-rabbit antibody and either an anti-mouse, anti-rat, anti-donkey or anti-guinea pig IgG coupled with Alexa Fluor 594 (Invitrogen) for 1 hr at room temperature. The sections were then coverslipped using Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and viewed under a confocal laser scanning microscope (LSM) (FV1000; Olympus, Tokyo, Japan).
Wild-type (mock-transfected), ECFP–Atg9A-transfected and Atg9A mRNA knockdown NIH3T3 cells were fixed with 4% paraformaldehyde buffered with 0.1 M phosphate buffer containing 4% sucrose. The cells were then subjected to either single immunostaining with the anti-Atg9A antibody (1:200) or double immunostaining with the anti-GFP (1:200) and anti-Atg9A antibodies at 4C for 2 days. The cells were then incubated with either Alexa Fluor 488–coupled, goat anti-rabbit IgG or a mixture of Alexa Fluor 488–coupled, goat anti-rabbit and Alexa Fluor 594–coupled, anti-mouse IgG (Invitrogen), respectively, for 1 hr at room temperature. The sections were then coverslipped using Vectashield mounting medium with DAPI (Vector Laboratories) and viewed under an LSM.
Results
Generation of the Specific Antibody Against Mouse Atg9Ap
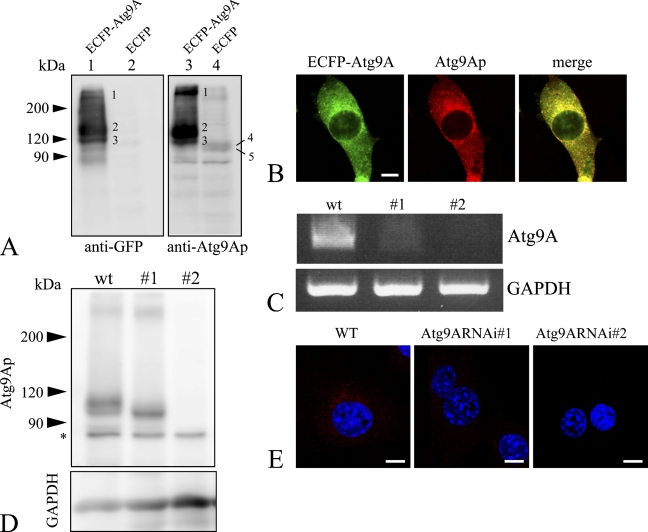
To examine both the tissue distribution and subcellular localization of endogenous Atg9Ap, a specific antibody against the carboxyl terminal region of the mouse Atg9Ap was prepared. Specificity of the prepared antibody was confirmed using NIH3T3 cells expressing the Atg9Ap fused with the carboxyl terminal end of ECFP (ECFP–Atg9). The cell lysates were subjected to Western blot analysis for the Atg9A fusion protein using antibodies to GFP and Atg9Ap, and its positive bands were found at molecular masses of 94, 105, 123, 134, and >200 kDa (Figure 1A). Based on its amino acid sequence, the molecular mass of Atg9Ap is deduced to be 94 kDa. Thus, it is reasonable to assume that the 94-kDa protein band (Figure 1A; the band marked number 5 in Lanes 3 and 4) was Atg9Ap that was synthesized in the rough ER. Glycosylation of human Atg9Ap at Asn99 results in the molecular mass of 105 kDa (Young et al. 2006). Therefore, the 94-kDa and 105-kDa bands correspond to endogenous, non-glycosylated and glycosylated, Atg9Ap (Figure 1A, marked numbers 5 and 4 in Lanes 3 and 4, respectively). This result is consistent with the 123-kDa and 134-kDa bands (marked numbers 3 and 2, respectively), which were detected by the anti-GFP antibody (Figure 1A, Lane 1). Because the molecular mass of ECFP is 29 kDa, the 123-kDa and 134-kDa protein bands were non-glycosylated and glycosylated ECFP–Atg9A, respectively.
Figure 1.
Characterization of the anti–Atg9A protein (anti-Atg9Ap) antibody. (A) Western blot analysis. Lysates from NIH3T3 cells that express ECFP–Atg9A (Lanes 1 and 3) or ECFP (Lanes 2 and 4) were stained with anti-green fluorescent protein (anti-GFP) (Lanes 1 and 2) or anti-Atg9Ap antibodies (Lanes 3 and 4). Respective numbers on bands indicate the following molecular masses: 1: ∼200 kDa; 2: 134 kDa; 3: 123 kDa; 4: 105 kDa; and 5: 94 kDa. The 105-kDa and 94-kDa bands correspond to endogenous Atg9Aps that are weakly, but distinctly, detected in Lanes 3 and 4, but not in Lanes 1 and 2. (B) Immunostaining using the anti-Atg9Ap antibody. NIH3T3 cells in which ECFP–Atg9A was expressed were immunostained with anti-GFP (green) and anti-Atg9Ap antibodies (red). Positive signals for the two proteins show significant overlap (yellow). (C) Suppression of Atg9A expression by RNA interference. mRNAs obtained from wild-type (wt) and Atg9A knockdown (#1 and #2) NIH3T3 cells were reverse transcribed, and the Atg9A gene was amplified by PCR. Upper panel shows Atg9Ap, and lower panel shows glyceraldehyde-3-phosphate dehydrogenase (GAPDH). (D) Western blot analysis of endogenous Atg9Ap in wild-type and Atg9A mRNA knockdown (#1 and #2) NIH3T3 cells. Upper panel shows protein bands immunopositive for Atg9Ap, and lower panel shows a protein band immunopositive for GAPDH. Expression of the Atg9Ap is significantly suppressed in the knockdown cells. Asterisk indicates a nonspecific band. (E) Immunofluorescent staining for the Atg9Ap in wild-type (WT) and Atg9A mRNA knockdown (Atg9ARNAi#1 and Atg9ARNAi#2) NIH3T3 cells using the anti-Atg9Ap antibody (red). Nuclei are counterstained with DAPI. Bar = 10 μm.
To further examine the antibody specificity, ECFP–Atg9A-expressing cells were stained with the Atg9Ap antibody and observed by confocal microscopy. Intense immunosignals for Atg9Ap were detected in the perinuclear regions and peripheral dotted structures. GFP-positive signals indicative of ECFP–Atg9A were largely coincident with Atg9Ap-positive signals (Figure 1B). To verify the antibody specificity by a genetic method, we established cells in which Atg9Ap expression was suppressed by RNAi. RNAi vectors were introduced into NIH3T3 cells, some of which showed suppressed Atg9A expression. Decreased levels of Atg9A mRNA and protein expression were confirmed in these cells by RT-PCR and Western blotting, respectively (Figures 1C and 1D). As shown in Figure 1E, immunoreactivity for Atg9Ap was detected in the perinuclear region of the control cells, but was significantly reduced in or had largely disappeared from the cytoplasm of the Atg9Ap knockdown cells. Moreover, the Atg9A-positive bands with the molecular mass of >200 kDa were not evident in the Atg9A mRNA knockdown cells. This result indicates that these large molecules may correspond to dimerized forms of Atg9p. Therefore, the results of these experiments confirmed that the antibody was specific for Atg9Ap and could be used in subsequent experiments to study the tissue and cellular distribution of Atg9Ap. Furthermore, we also confirmed that the antibody generated could recognize human Atg9Ap and could be used for both Western blotting and immunohistochemistry (data not shown).
Tissue Distribution of Atg9Ap
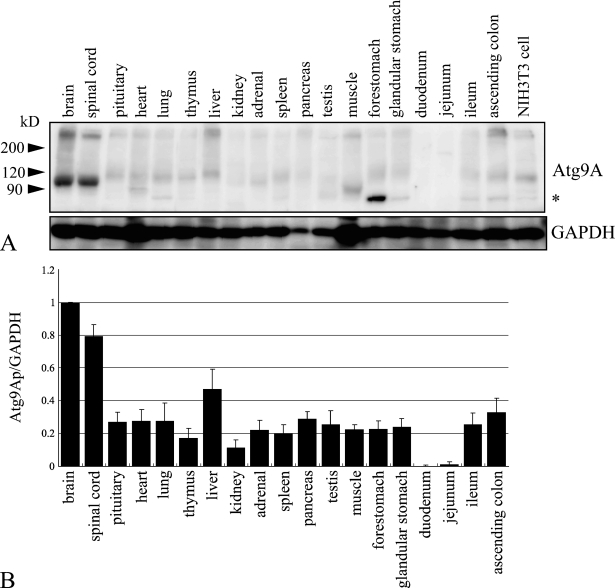
As stated above, the antibody recognized endogenous Atg9Ap in NIH3T3 cells. To examine the levels of Atg9Ap expression in various mouse tissues, extracts from each tissue were subjected to Western blotting using the antibody. As shown in Figure 2A, Atg9Ap was found to be ubiquitous in mouse tissues, although the level of Atg9Ap expression was tissue dependent. The main bands observed in each tissue had molecular mass that ranged from ∼95 kDa to 120 kDa. Only Asn99 of Atg9Ap is known to be glycosylated in HeLa cells, but the molecule has four possible N-glycosylation sites (Young et al. 2006). Thus, it is possible that the glycosylation of mouse Atg9Ap varies with cell type (Figure 2A). The level of Atg9Ap expression was compared among tissues using the intensity of the major bands positive for Atg9Ap in the Western blots (Figure 2B). The highest expression level was detected in the brain and spinal cord, and the expression was lowest in the duodenum and jejunum. These results indicate that expression of Atg9A is tissue dependent.
Figure 2.
Distribution of Atg9Ap in various mouse tissues. (A) Western blot analysis of various mouse tissue lysates using anti-Atg9Ap (upper panel) or anti-GAPDH (lower panel) antibodies. Asterisk indicates a nonspecific band. (B) Protein levels of the Atg9A relative to those of GAPDH in various mouse tissues. The intensity of each band in A was measured, and the ratio of the amount of the Atg9Ap to GAPDH protein was calculated, using a brain ratio equal to 1. The mean values ± standard errors were calculated from the results of three independent experiments.
Localization of Atg9Ap in CNS Tissues
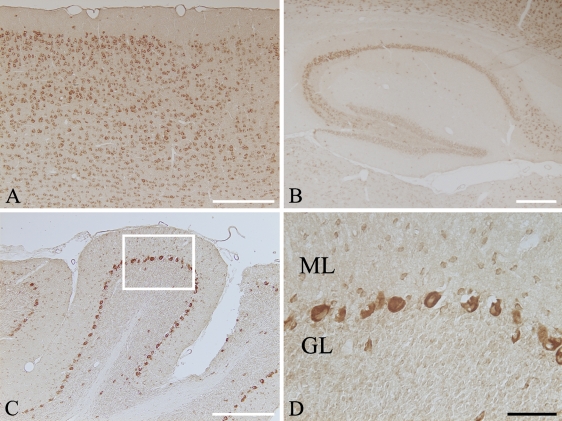
Because the expression of Atg9Ap was high in CNS tissues, we used immunohistochemistry to further examine Atg9Ap localization in mouse brain. As shown in Figure 3, immunoreactivity for Atg9Ap was mostly localized in CNS neurons, but the signal intensity varied with the brain region; the immunoreactivity was highly detected in neurons of the cerebral cortex from the external granule cell layer (Layer II) to the multiform layer (Layer VI) (Figure 3A), pyramidal neurons in the hippocampus (Figure 3B), and Purkinje cells in the cerebellum (Figures 3C and 3D). In addition to Purkinje cells in the cerebellum, the immunoreactivity was also found in other neuronal cells that were adjacent to Purkinje cells or in the molecular layer of the cerebellum (Figure 3D). Based on the distribution pattern of the cells, these immunopositive cells might be basket cells. The immunosignal in these neurons was weaker than that observed in Purkinje cells, but was much more intense than the weak signal detected in granule cells (Figure 3D).
Figure 3.
Immunohistochemical staining for the Atg9Ap in mouse brain. Cryosections from the cerebral cortex (A), hippocampus (B), and cerebellum (C) were immunostained with the anti-Atg9A antibody and visualized with DAB. Enlargement of boxed area in C is shown in D. Immunoreactivity for Atg9Ap is localized in various types of neurons. In cerebellum, the immunosignal is intensely detected in the perikaryal region of Purkinje cells, and it is also seen in the small neurons that are dispersed in the molecular layer. ML, molecular layer; GL, granular cell layer. Bars: A–C = 100 μm; D = 20 μm.
Localization of Atg9Ap in Purkinje Cells
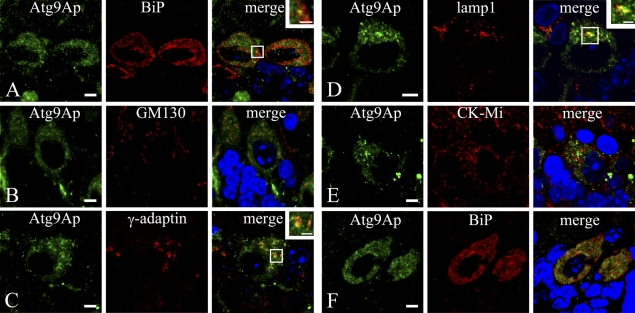
Although Atg9Ap was intensely localized in Purkinje cells, its precise role in neurons remains to be determined. To uncover the significance of Atg9Ap in neuronal cells, its colocalization with various organell markers was examined using immunocytochemistry as follows: BiP for an ER marker; p115 for an ER/cis-Golgi marker; GM130 for a Golgi marker; γ-adaptin mainly for a TGN marker; EEA1 for an early endosome marker; lamp1 for a lysosomal/late endosomal marker; and ubiquitous CK-Mi for a mitochondrial marker (Figure 4). The immunosignal for Atg9Ap was colocalized largely with γ-adaptin and lamp1, which were used as markers of the TGN and lysosomes/late endosomes, respectively (Figures 4C and 4D). The Atg9A immunosignal was colocalized in part with BiP in the ER (Figure 4A). In particular, costaining of Atg9Ap and BiP was more pronounced in cerebellar Purkinje neurons of 3-week-old mice (Figure 4F). Other markers, including p115, GM130 (Figure 4B), and EEA1, were stained well in Purkinje cells, but they were not colocalized with Atg9Ap (data for p115 and EEA1 are not shown). In addition, the CK-Mi mitochondrial marker was not colocalized with Atg9Ap (Figure 4E).
Figure 4.
Localization of Atg9Ap in Purkinje cells of the cerebellum in mice at 8 (A–E) and 3 (F) weeks of age. (A–F) Mouse Purkinje cells were coimmunostained with the anti-Atg9A antibody (green) and with an antibody for various organelle markers (red): BiP for endoplasmic reticulum (A,F); GM130 for Golgi apparatus (B); γ-adaptin for TGN (C); lamp1 for lysosome/late endosome (D); and mitochondrial creatine kinase (CK-Mi) for mitochondria (E). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Bars: A–F = 5 μm; insets = 2 μm.
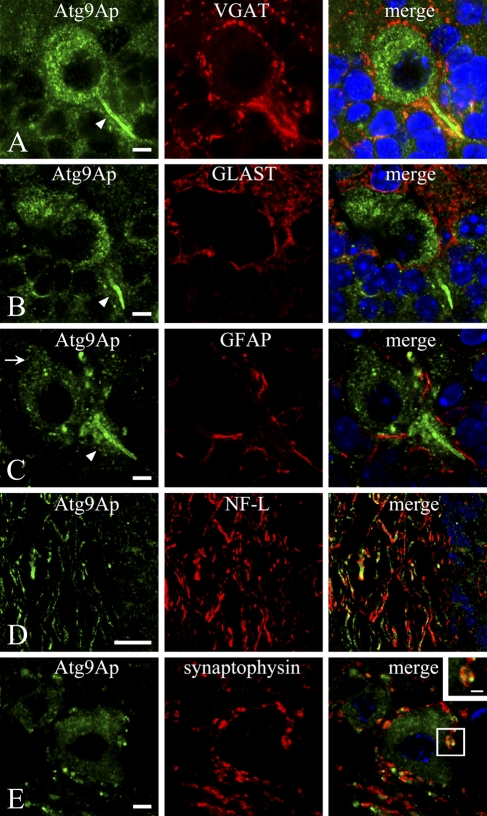
In addition to perinuclear localization of Atg9Ap in Purkinje cells, intense staining was observed in the initial portion of the axon (Figures 5A–5C), and weak but distinct staining was detected in dendrites (Figure 5C). Because it has been shown that around the initial axon segments of Purkinje cells the axons and terminals of basket cells form a dense plexus called a pinceau that is positive for VGAT (Chaudhry et al. 1998; Miura et al. 2006), we also performed double staining of Atg9Ap with VGAT (Figure 5A). Moreover, Atg9Ap was costained for GLAST or GFAP for astroglial cells (Figures 5B and 5C), NF-L for axons (Figure 5D), and synaptophysin for axon terminals (Figure 5E). As stated above, VGAT immunoreactivity was densely present in axons and terminals around the Atg9Ap-positive initial segment of Purkinje cells and colocalized with the immunosignal for Atg9Ap in them (Figure 5A). However, VGAT-positive axons and terminals were largely negative for Atg9Ap immunoreactivity when they were present around Atg9Ap-positive Purkinje cell bodies (Figure 5A). GLAST- and GFAP-positive astroglial cells were observed around Purkinje cells (Figures 5B and 5C). However, these GLAST- and GFAP-positive cells were largely negative for the immunosignal for Atg9Ap in the Purkinje cell layer, although punctate staining for Atg9Ap was in some cases discerned in the GFAP- or GLAST-positive astroglial cells (data not shown). Moreover, the immunosignal for Atg9Ap coexisted with that for NF-L in axons that ran through the white matter in the cerebellum, although it was relatively weak and discontinuous in these axons (Figure 5D). Furthermore, the immunoreactivity for synaptophysin was localized in axon terminals around neurons of the deep cerebellar nuclei, and the Atg9Ap-immunopositive signal was also detected in such synaptophysin-positive axon terminals (Figure 5E). In comparison to the dense distribution of VGAT-positive axon terminals of basket cells around Atg9Ap-positive initial axon terminals of Purkinje cells (Figure 5A), the synaptophysin-positive axon terminals were loosely distributed around neurons of deep cerebellar nuclei (Figure 5E). Because Atg9Ap-positive dots were small in number in the synaptophysin-positive axon terminal regions, there were some synaptophysin-positive terminals that lacked Atg9Ap-positive dots (Figure 5E). These axon terminals were often confirmed by confocal laser scanning microscopy to have small Atg9Ap-positive dots in other sectional planes of the doubly stained cerebellar sections (data not shown). These results suggest that Atg9Ap might be involved in autophagosome formation in axon terminals.
Figure 5.
Double immunostaining for Atg9Ap and vesicular GABA transporter (VGAT) in the Purkinje cell layer (A), GLAST (B), and glia fibrillary acidic protein (GFAP) (C) in the Purkinje cell layer, neurofilament-L (NF-L) in the white matter (D), and synaptophysin in the dentate nucleus (E) of the cerebellum. Immunostaining for Atg9Ap (green) is intensely detected in the initial segment of the Purkinje cell (arrowheads) (A–C), whereas it is weak but distinct in a dendrite (arrow) (C). Immunoreactivity for VGAT (red) is densely detected in axons and axon terminals of basket cells around Atg9Ap-positive initial axon terminals of Purkinje cells to form a pinceau, in which the immunosignal for Atg9Ap is colocalized (A). Moreover, the immunoreactivity for Atg9Ap is not colocalized in GLAST- or GFAP-positive astroglial cells (red) (B,C). In the white matter, Atg9Ap-immunopositive staining is longitudinally running in axons that are immunopositive for NF-L (red) (D). In the dentate nucleus, Atg9Ap-immunopositive large neuronal cell bodies are surrounded by synaptophysin-positive axon terminals (red), in which punctate signals for Atg9Ap overlap or are closely associated with synaptophysin (red) (D). Bars: A–C,E = 5 μm; D = 20 μm; inset in E = 2 μm.
Discussion
The main findings of the present study can be summarized as follows: (1) Western blotting and immunohisto-/cytochemistry confirmed that the anti-Atg9Ap antibody recognized both non-glycosylated and glycosylated forms of Atg9Ap; (2) although the level of Atg9Ap expression in mice was tissue dependent, Atg9Ap expression was ubiquitous and was high in CNS tissue, particularly in neurons; and (3) in addition to TGN-lysosomal cycling, Atg9Ap was detected in part in the ER of Purkinje cell bodies and distinctly in axon terminals of the cells.
Although the PAS, in which Atg proteins are localized, has been shown in yeast, its existence in mammalian cells has not been confirmed. Nevertheless, like the PAS membrane in yeast, identification of the membrane origin of autophagosome formation in mammalian cells is very important. Because Atg9Ap is the only Atg membrane protein, it has been suggested that Atg9Ap transports membranes during autophagosome formation; the protein is localized to TGN and late endosomes, and it dynamically cycles between these organelles under starvation conditions (Young et al. 2006; Webber et al. 2007). In fact, generation of Atg9A-deficient mice has shown that the protein is essential for survival during neonatal starvation (Saitoh et al. 2009).
However, there are no studies on cell and tissue distribution in vivo, except for studies that have shown ubiquitous expression of human Atg9A mRNA by Northern blot analysis (Yamada et al. 2005; Young et al. 2006). To examine the distribution of Atg9Ap in mouse tissues, we generated an anti-Atg9Ap antibody that was confirmed to specifically recognize several forms of endogenous Atg9Ap by Western blotting and immunohisto-/cytochemistry of mouse tissue samples and ECFP–Atg9A-transfected and Atg9A mRNA knockdown NIH3T3 cells. Expression of human Atg9A mRNA is ubiquitous, but it is highest in skeletal muscle (Yamada et al. 2005; Young et al. 2006). Measurement of Atg9A mRNA using a real-time PCR method confirmed this result (data not shown). However, the expression of mouse Atg9Ap was tissue dependent and highest in the brain. These results indicate that the protein expression of Atg9A in mouse tissues may be regulated in a posttranslational manner.
To our knowledge, this is the first reported evidence that Atg9Ap is widely distributed in the neurons of the brain. In particular, immunocytochemistry showed that Atg9Ap is colocalized in γ-adaptin–positive TGN and lamp1-positive lysosomes/late endosomes in Purkinje neurons. These results are consistent with those of Young et al. (2006), who reported that the Atg9Ap dynamically cycles between the TGN and late endosomes in cultured cells. Although it has been suggested that the Atg9p is localized in part to mitochondria in yeast (Reggiori et al. 2005), endogenous and overexpressed mouse and human Atg9p were not present in the mitochondria of cultured cells (Yamada et al. 2005; Young et al. 2006; Webber et al. 2007). In the present study, Atg9Ap was not localized in CK-Mi–positive mitochondria in CNS neurons. In addition to TGN/lysosomal localization, the protein was also localized in the BiP-positive ER, particularly in the brains of young mice. Interestingly, recent studies have shown that the ER provides important components for autophagosome formation in relation to double FYVE domain protein 1 (Axe et al. 2008). In addition, electron microscopic tomography showed that the ER membrane has direct continuity with the autophagosomal membrane (Hayashi-Nishino et al. 2009; Ylä-Anttila et al. 2009). Because Atg9Ap is the only Atg membrane protein, its localization in the ER strongly suggests that the ER membrane is involved in the autophagosome formation in CNS neurons.
Although the immunosignal for Atg9Ap was predominantly detected in neuronal bodies, axons, and axon terminals, some neurons, such as granule cells in the cerebellum, were faint or negative for the Atg9Ap signal. Our present data concerning VGAT-positive axon terminals that formed a dense network around the initial axon terminal of Purkinje cells were consistent with results reported previously (Chaudhry et al. 1998; Miura et al. 2006). Atg9Ap-positive dots were found in VGAT-positive axon terminals of basket cells. Because astroglial cells are present in the Purkinje cell layer, we also examined double immunostaining of Atg9Ap and GFAP or GLAST for astroglial cell markers (Shibata et al. 1997), and found that the Atg9Ap immunosignal was largely negative in GFAP- or GLAST-positive astroglial cells in the Purkinje cell layer. Because autophagy plays a pivotal role in the maintenance of cellular homeostasis (Mortimore and Poso 1987; Kuma et al. 2004; Meijer and Dubbelhuis 2004), we could not exclude the possibility that Atg9Ap-positive dots are present in astroglial cells.
We have previously shown that autophagosomes with double membranes that are positive for LC3, an autophagosome marker, accumulate in axons in the corpus callosum of cathepsin D–deficient or cathepsins B– and L–double-deficient mouse brains (Koike et al. 2005), indicating that autophagosome formation occurs within axons. Moreover, it has been shown that Atg5/Atg7 are required for the maintenance of metabolism in axon terminals, and loss of either gene results in axonopathy that leads to neuronal degeneration (Komatsu et al. 2007; Nishiyama et al. 2007). In addition to these data, our present results showing the presence of Atg9Ap in axons and axon terminals of Purkinje cells are a valid argument for the involvement of Atg9Ap in autophagosome formation in these regions. Furthermore, it is very interesting that the immunosignal for Atg9Ap was intensely present in both VGAT-positive axon terminals of basket cells and the initial axon segment of Purkinje cells (Figure 5A). The dense localization of Atg9Ap in these pre- and postsynaptic regions, where metabolic turnover is thought to be active, may further support our hypothesis that Atg9Ap is involved in autophagosome formation.
Collectively, the results of the present study, which were obtained using a specific antibody to Atg9Ap, revealed that the protein was localized in the ER, TGN, and lysosomes/late endosomes in Parkinje cells of the mouse cerebellum. Moreover, localization of Atg9Ap both in the axon terminals of Purkinje and basket cells and in the ER of Purkinje cells suggests that Atg9Ap, which is the only membrane Atg protein, might play an essential role in the initiation of autophagosome formation.
Acknowledgments
This work was supported by a grant-in-aid for Creative Scientific Research from the Japan Society for the Promotion of Science.
This article is distributed under the terms of a License to Publish Agreement (http://www.jhc.org/misc/ltopub.shtml). JHC deposits all of its published articles into the U.S. National Institutes of Health (http://www.nih.gov/) and PubMed Central (http://www.pubmedcentral.nih.gov/) repositories for public release twelve months after publication.
References
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, et al. (2008) Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182:685–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry FA, Reimer RJ, Bellocchio EE, Danbolt NC, Osen KK, Edwards RH, Storm-Mathisen J (1998) The vesicular GABA transporter, VGAT, localizes to synaptic vesicles in sets of glycinergic as well as GABAergic neurons. J Neurosci 18:9733–9750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A (2009) A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11:1433–1437 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Cregg JM, Dunn WA Jr, Emr SD, Sakai Y, Sandoval IV, Sibirny A, et al. (2003) A unified nomenclature for yeast autophagy-related genes. Dev Cell 5:539–545 [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD (2000) Autophagy as a regulated pathway of cellular degradation. Science 290:1717–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, Schulz-Schaeffer W, et al. (2000) Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci 20:6898–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E, Gotow T, et al. (2005) Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid-lipofuscinoses (Batten disease). Am J Pathol 167:1713–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr, Iwata J, Kominami E, Chait BT, et al. (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA 104:14489–14494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R, Muehe Y, Prick T, Bremer S, Schlotterhose P, Eskelinen EL, Millen J, et al. (2008) Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol Biol Cell 19:4492–4505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, et al. (2004) The role of autophagy during the early neonatal starvation period. Nature 432:1032–1036 [DOI] [PubMed] [Google Scholar]
- Lang T, Reiche S, Straub M, Bredschneider M, Thumm M (2000) Autophagy and the cvt pathway both depend on AUT9. J Bacteriol 182:2125–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Dono K, Gotoh K, Shibata M, Koike M, Marubashi S, Miyamoto A, et al. (2005) Participation of autophagy in the degeneration process of rat hepatocytes after transplantation following prolonged cold preservation. Arch Histol Cytol 68:71–80 [DOI] [PubMed] [Google Scholar]
- Meijer AJ, Dubbelhuis PF (2004) Amino acid signalling and the integration of metabolism. Biochem Biophys Res Commun 313:397–403 [DOI] [PubMed] [Google Scholar]
- Miura E, Fukaya M, Sato T, Sugihara K, Asano M, Yoshioka K, Watanabe M (2006) Expression and distribution of JNK/SAPK-associated scaffold protein JSAP1 in developing and adult mouse brain. J Neurochem 97:1431–1446 [DOI] [PubMed] [Google Scholar]
- Mizushima N (2007) Autophagy: process and function. Genes Dev 21:2861–2873 [DOI] [PubMed] [Google Scholar]
- Mortimore GE, Poso AR (1987) Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu Rev Nutr 7:539–564 [DOI] [PubMed] [Google Scholar]
- Nishiyama J, Miura E, Mizushima N, Watanabe M, Yuzaki M (2007) Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy 3:591–596 [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 148:465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y (2001) Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2:211–216 [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Futter M, Jahreiss L, Korolchuk VI, Lichtenberg M, Luo S, Massey DC, et al. (2009) Mammalian macroautophagy at a glance. J Cell Sci 122:1707–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Shintani T, Nair U, Klionsky DJ (2005) Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 1:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, et al. (2009) Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA 106:20842–20846 [DOI] [PMC free article] [PubMed]
- Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y (2009) Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells 14:525–538 [DOI] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, Ikenaka K, Wada K, Tanaka K, Inoue Y (1997) Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci 17:9212–9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 20:5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, Shibata M, Koike M, Yoshimura K, Sasaki M (2008) Autophagy-physiology and pathophysiology. Histochem Cell Biol 129:407–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Young AR, Tooze SA (2007) Atg9 trafficking in mammalian cells. Autophagy 3:54–56 [DOI] [PubMed] [Google Scholar]
- Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW (2005) Endothelial nitric-oxide synthase anti-sense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 280:18283–18290 [DOI] [PubMed] [Google Scholar]
- Yen WL, Klionsky DJ (2007) Atg27 is a second transmembrane cycling protein. Autophagy 3:254–256 [DOI] [PubMed] [Google Scholar]
- Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL (2009) 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5:1180–1185 [DOI] [PubMed] [Google Scholar]
- Yoshimura K, Shibata M, Koike M, Gotoh K, Fukaya M, Watanabe M, Uchiyama Y (2006) Effects of RNA interference of Atg4B on the limited proteolysis of LC3 in PC12 cells and expression of Atg4B in various rat tissues. Autophagy 2:200–208 [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, et al. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119:3888–3900 [DOI] [PubMed] [Google Scholar]