Abstract
Variola virus (VARV) caused smallpox, one of the most devastating human diseases and the first to be eradicated, but its deliberate release represents a dangerous threat. Virulent orthopoxviruses infecting humans, such as monkeypox virus (MPXV), could fill the niche left by smallpox eradication and the cessation of vaccination. However, immunomodulatory activities and virulence determinants of VARV and MPXV remain largely unexplored. We report the molecular characterization of the VARV- and MPXV-secreted type I interferon-binding proteins, which interact with the cell surface after secretion and prevent type I interferon responses. The proteins expressed in the baculovirus system have been purified, and their interferon-binding properties characterized by surface plasmon resonance. The ability of these proteins to inhibit a broad range of interferons was investigated to identify potential adaptation to the human immune system. Furthermore, we demonstrate by Western blot and activity assays the expression of the type I interferon inhibitor during VARV and MPXV infections. These findings are relevant for the design of new vaccines and therapeutics to smallpox and emergent virulent orthopoxviruses because the type I interferon-binding protein is a major virulence factor in animal models, vaccination with this protein induces protective immunity, and its neutralization prevents disease progression.—Fernández de Marco, M. M., Alejo, A., Hudson, P., Damon, I. K., Alcami, A. The highly virulent variola and monkeypox viruses express secreted inhibitors of type I interferon.
Keywords: immune modulation, cytokine, infection, smallpox, vaccine
The Poxviridae are a family of large-dsDNA viruses that replicate in the cytoplasm of infected cells. Most members of the genera Orthopoxvirus (OPV), Parapoxvirus, Molluscipoxvirus, and Yatapoxvirus infect humans either exclusively, for example, variola virus (VARV) and molluscum contagiosum virus, or zoonotically, such as monkeypox virus (MPXV), vaccinia virus (VACV), or Yaba-like disease virus (YLDV). The consequences of these infections range from severe disease associated with high mortality to more benign localized infections such as seen with VACV infections of dairy cattle handlers in Brazil (1). VACV was the vaccine used to eradicate smallpox and is the prototypic member of the poxvirus family.
Two OPVs may cause severe disease in humans. VARV is the causative agent of smallpox, which was declared to be eradicated in 1980 as a result of the World Health Organization Smallpox Global Eradication Campaign, becoming the first and only viral disease eradicated by vaccination efforts (2). MPXV infects both humans and nonhuman primates, likely has a rodent reservoir, and is an emerging infectious disease, with cases observed in Africa and the United States (3). The deliberate release of VARV would have catastrophic consequences on global public health, considering that the majority of the human population has not been vaccinated or boosted in recent years, so there is a need to define the mechanisms of smallpox pathogenesis in order to develop intervention strategies (2). In addition, the reduced level of herd immunity against OPVs increases the possibility of infection with zoonotic OPVs, exemplified by VACV and cowpox virus infections in South America and Europe, respectively, and the more virulent MPXV, endemic in Central and West Africa, and the recent epidemic in the United States (3, 4).
Viral strategies to evade the immune response are likely pathogenesis determinants of smallpox and monkeypox (5, 6) and may also modulate an immunopathological reaction responsible for the toxemia reported in individuals suffering from severe smallpox and the adverse effects after smallpox vaccination (7).
The innate immune response is the first line of immune defense. One of its main effectors are interferons (IFNs), a family of multifunctional cytokines that are secreted from cells and inhibit virus replication via their direct antiviral and indirect immunoregulatory activities (8). Type I IFNs are induced by viral infection of almost any cell type and include various IFNα subtypes, IFNβ and IFNω among others. All type I IFNs bind to a common and widely expressed heterodimeric receptor and induce signaling through the Janus protein tyrosine-kinase and signal transducers and activators of transcription (STAT) pathway. Type I IFNs act by directly inducing an antiviral state in the cell (9) and have immunoregulatory activity (10). IFNγ, the only member of type II IFNs, is induced by antigen-stimulated lymphocytes and activates natural killer and cytotoxic T cells that destroy infected cells. Type III IFNs (IFNλ) are interleukin 10 (IL-10)-related cytokines with antiviral activity that are produced on cell infection by most cell types, including plasmacytoid dendritic cells (11). Although type III IFNs bind to a unique heterodimeric IFNλ receptor complex, they induce a type I IFN signaling pattern (12).
The central role of IFNs in antiviral defense is reinforced by the fact that most viruses interfere with IFN signaling pathways at different levels (8, 13). Poxviruses express intracellular proteins that target this pathway, such as the eIF-2 α homologue K3 (14) and the double-stranded RNA-binding protein E3 (15). An IFN evasion strategy particular to poxviruses is the expression of secreted IFN decoy receptors, including the IFNα/β-binding protein (IFNα/βBP) (16,17,18,19) and IFNγ receptor (20, 21), which bind IFNs with high affinity and prevent their interaction with cellular receptors. The IFNα/βBP, encoded by the VACV strain Western Reserve (WR) B18R gene, is an immunoglobulin (Ig) superfamily glycoprotein of sequence unrelated to type I IFN receptors (22, 23). It is secreted from infected cells and acts both in solution and when associated with the cell surface, preventing the establishment of an antiviral state in surrounding uninfected cells (17). Uniquely among IFN receptors, and indicative of their potential host range, the poxvirus-encoded IFN decoy receptors bind IFNs from a broad range of host species (19). Deletion of the IFNα/βBP gene from the VACV WR genome attenuates the virus in a mouse model of infection (19, 24), and it has been recently demonstrated that the IFNα/βBP is also essential for ectromelia virus (ECTV) virulence and that vaccination with the IFNα/βBP induces protective immunity (25).
In this study we demonstrate that VARV and MPXV express a soluble IFNα/βBP, which binds to the cell surface and protects cells from the antiviral effects of IFN. A detailed characterization of the interaction of the VARV and MPXV IFNα/βBPs with a broad range of type I IFNs from different species and with type III IFNs was carried out to identify potential differences with VACV and adaptation to the human IFN system. The implications of these findings for the improvement of vaccines and therapeutics against smallpox and emergent virulent poxviruses is discussed.
MATERIALS AND METHODS
Cells and reagents
HeLa, A549, and BSC-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) with 5–10% fetal calf serum (FCS). Sf21 and High Five™ cells (Invitrogen, Carlsbad, CA, USA) were grown in TC-100 medium with 10% FCS. Vesicular stomatitis virus (VSV) strain Cocal was obtained from W. James (Oxford University, Oxford, UK). Recombinant hIFNα-G, hIFNα-J1, hIFNα-K, hIFNα-B2, hIFNα-F, hIFNω, mIFNα-A, mIFNβ, and Rhesus/cynomolgus IFNα-2 were from PBL InterferonSource (Piscataway, NJ, USA). hIFNα-A, hIL-10, mIL-10, and mIL28A were from PeproTech (Rocky Hill, NJ, USA). hIFNβ was from Calbiochem (San Diego, CA, USA) or PBL InterferonSource. hIFNα-2b (Intron A) was from Schering-Plough (Kenilworth, NJ, USA). Recombinant hIL-28A and hIL-29 were from R&D Systems (Minneapolis, MN, USA).
Cloning and expression of viral IFNα/βBP
The VACV strain WR B18R, MPXV strain MSF6 B16R, and VARV strain BSH1975 B17R genes were amplified by PCR. MPXV and VARV template DNAs were provided by H. Meyer (Bundeswehr Institute of Microbiology, Munich, Germany) and the U.S. Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA), respectively. The region amplified corresponded to the coding region without the predicted signal peptide. PCR products were cloned into pAL7, a modified pFastBac plasmid bearing the honeybee mellitin signal peptide at the 5′ region and a C-terminal V5-His tag. The plasmids generated were pMF1 (VARV B17), pMF2 (VACV B18), and pMF6 (MPXV B16). The sequence of the inserts was confirmed. The MPXV B16R gene from strain MSF6 was identical to the sequence published for MPXV strain Zaire 96 (26). Handling of VARV DNA was performed under World Health Organization (WHO) permission and in accordance with WHO guidelines. Recombinant baculoviruses were obtained using the Bac to Bac system (Invitrogen). Recombinant baculovirus expressing ECTV semaphorin homologue (SEMA) will be described elsewhere (unpublished results).
Generation of MPXV- and VARV-infected cell supernatants
BSC-40 cells were infected with MPXV West African (MPXV USA2003-WI-044) and Congo Basin (MPXV RCG2003-RCG-358) strains at 10 PFU/cell in Opti-MEM (Invitrogen), and media were harvested at 48 h postinfection. Media were centrifuged at 1000 g for 5 min and UV inactivated. VARV (Bangladesh 1975) infections were performed in a similar way under BSL4 laboratory conditions, and media were collected at 30 h postinfection. The supernatant was γ-irradiated (5×106 rad) on ice for 4 h and safety tested to demonstrate no viral growth in 2 sequential passages. VARV reagents were made in 2000, using a WHO-approved protocol. Supernatants were stored at −70°C. For all samples, 1 ml of supernatant corresponds to ∼1 × 106 cells.
Immunofluorescent cell staining
HeLa cells were incubated for 30 min at 37°C with supernatants from baculovirus-infected High5 cells. Unbound protein was removed with 5 washes in cold PBS, and cells were fixed with 4% paraformaldehyde for 20 min at room temperature. Immunofluorescent detection was performed without cell membrane permeabilization using a monoclonal anti-V5 antibody (Invitrogen), followed by Alexa Fluor® 488 donkey anti-mouse IgG (Invitrogen). Cell nuclei were stained with DAPI (Calbiochem).
Inhibition of type I and type III IFN-induced STAT-1 phosphorylation
HeLa (for IFN type I) or A549 (for IFN type III) cells were incubated with supernatants from baculovirus-infected cells and thoroughly washed to remove unbound material. Subsequently, cells were incubated with 1000 U of IFN for 15 or 30 min at 37°C. Cells were lysed in Laemmli buffer and analyzed by Western blot using the anti-phospho STAT1 (Tyr701) and anti-STAT1 antibodies from Cell Signaling and anti-V5 antibody.
Inhibition of type I IFN-induced transcription
HeLa cells grown in 96-well plates were cotransfected using FuGene HD (Roche) with 100 ng/well of pISRE-Luc, pCRE-Luc, or pTAL-Luc (Mercury Pathway Profiling System; Clontech, Palo Alto, CA, USA) and 5 ng/well of phRLnull (Promega, Madison, WI, USA) plasmid bearing the Renilla luciferase for in-well normalization of transfection efficiency. At 24 h post-transfection, 50 U of hIFNαA was added in the presence or absence of 20 ng of the indicated purified recombinant proteins, and cells were incubated for 16 h. Luciferase activity was detected using the Dual-Glo Luciferase assay system (Promega).
Neutralization of IFN antiviral activity
Recombinant IFNs were incubated with culture supernatants or purified proteins in 5% FCS DMEM for 30–90 min at 37°C and added to HeLa cells grown in 96-well plates for 16 h. Cells were then infected with 50 PFU/cell of VSV strain cocal for 48 h (16). Cell viability was determined using the Cell Titer 96 Aqueous One Solution cell proliferation assay (Promega).
Protein expression and purification
Supernatants from baculovirus-infected High5 cells were concentrated on a stirred ultrafiltration cell (Amicon, Danvers, MA, USA) using YM-10 membranes (Millipore, Bedford, MA, USA), and buffer was exchanged into binding buffer (50 mM phosphate, 300 mM NaCl, and 10 mM imidazole, pH 7.4) on PD-10 desalting columns (GE Healthcare, Little Chalfont, UK). The His-tagged recombinant proteins were affinity-purified using Ni-NTA (Qiagen, Valencia, CA) columns, dialyzed into PBS, and concentrated using Vivaspin 500 columns (Sartorius Stedim Biotech, Aubagne, France). Protein purity was checked on Coomassie-blue-stained SDS-PAGE and quantitated by a BCA assay (Pierce Biotechnology, Rockford, IL, USA).
Biomolecular interaction analysis by SPR
Cytokine binding specificity and affinity constants were determined using a BIAcore X biosensor (BIAcore, Uppsala, Sweden). For ligand screening experiments, purified proteins were amine coupled to CM5 chips to ∼5000 RU (5000 pg/mm2). IFNs were injected at 10 or 100 nM in HBS-EP buffer [10 mM Hepes, 150 mM NaCl, 3 mM EDTA, and 0.005% (v/v) surfactant P20, pH 7.4] at a flow rate of 10 μl/min, and association and dissociation were monitored. The surface was regenerated using 10 mM glycine-HCl, pH 1.5. For kinetic analyses, proteins were immobilized at low densities (Rmax<200 RU). Different concentrations of the analyte were injected at a flow rate of 30 μl/min over 2 min and allowed to dissociate for 3–5 min. BIAcore sensorgrams were analyzed using BIAevaluation 3.2 software. Bulk refractive index changes were removed by subtracting the reference flow cell responses, and the average response of a blank injection was subtracted from all analyte sensorgrams to remove systematic artifacts. Kinetic data were globally fitted to a 1:1 Langmuir model.
RESULTS
VARV and MPXV encode secreted IFNα/βBPs that bind to the cell membrane
We expressed recombinant versions of VACV WR IFNα/βBP B18 and its orthologues VARV B17 and MPXV B16 proteins fused to V5 and 6xHis C-terminal tags (Supplemental Fig. 1). All 3 recombinant proteins were secreted from baculovirus-infected insect cells with molecular sizes of 52 kDa for VACV and MPXV proteins or 50 kDa for the VARV protein (Supplemental Fig. 2A and Fig. 2). Cell extracts obtained in the presence of tunicamycin, an inhibitor of N-glycosylation, showed a single band of 37 kDa in each case, corresponding to the predicted size of the unmodified proteins (Supplemental Fig. 2B).
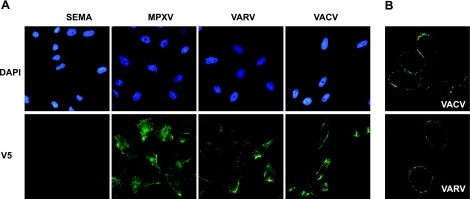
Figure 1.
Cell surface binding of VACV IFNα/βBP and its VARV and MPXV orthologues. A) Anti-V5 immunofluorescent staining of nonpermeabilized HeLa cells incubated with supernatants containing the indicated V5-tagged recombinant proteins. Cell nuclei were visualized using DAPI. B) Confocal microscopy of cells incubated as before, showing plasmatic membrane localization of the indicated proteins.
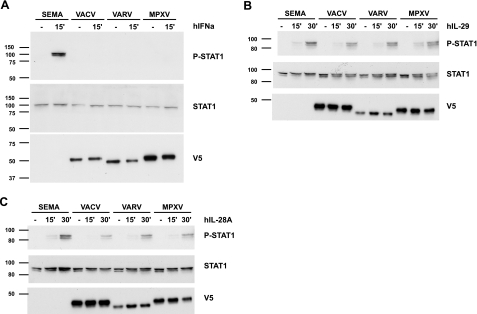
Figure 2.
Inhibition of type I but not type III IFN-induced signaling by the VACV B18 protein and its VARV and MPXV orthologues. HeLa or A549 cells were incubated with supernatants containing the indicated V5-tagged recombinant proteins and subsequently exposed to 1000 U of hIFNα (A), hIL-29 (B), or hIL-28A (C) for the indicated times. Samples were analyzed by Western blot using anti-phospho-STAT-1, anti-STAT-1, and anti-V5 antibodies as indicated. Molecular size markers (kDa) are at left.
VACV B18 binds to the plasma membrane of neighboring, uninfected cells after secretion from infected cells (17). Immunofluorescence of HeLa cell monolayers preincubated with supernatants from recombinant baculovirus-infected cells containing the mentioned proteins showed an intense staining indicative of protein binding to the cell surface for the 3 IFNα/βBPs but not with a control secreted protein, the SEMA encoded by ECTV (Fig. 1A). Confocal microscopy analysis showed a clear plasmatic membrane staining pattern for the VACV and VARV proteins (Fig. 1B) and the MPXV protein (not shown).
The VARV B17 and MPXV B16 proteins inhibit type I IFN-induced signaling
To examine whether cell surface-associated proteins prevented IFN-induced signaling, HeLa cells were incubated with recombinant proteins and washed to remove unbound protein. Addition of human IFNα (hIFNα) induced STAT-1 phosphorylation in HeLa cells preincubated with the control protein SEMA (Fig. 2A). However, cells preincubated with either VACV B18 protein or its VARV or MPXV orthologues did not show STAT-1 phosphorylation, indicating that the 3 IFNα/βBPs block hIFN-α-induced signaling. All 3 proteins, but not the control protein, were detected by Western blot analysis, confirming their binding to the plasma membrane. In a similar experiment, neither the VACV nor its VARV or MPXV orthologues blocked STAT-1 phosphorylation induced by type III IFNs (IL-29 and IL-28A), which was readily detected in the absence or the presence of the viral proteins (Fig. 2B, C).
The VARV B17 and MPXV B16 proteins block the antiviral activity of several human type I IFNs
We next determined the ability of the IFNα/βBPs from VARV and MPXV to inhibit the antiviral activity of different human type I IFNs: hIFNα-A, hIFNα-2b, hIFNα-K, hIFNα-G, hIFNα-J1, hIFNα-F, hIFNα-B2, hIFNβ, and hIFNω (Fig. 3). IFN treatment completely abrogated VSV-induced cell killing in the presence of the control protein SEMA. By contrast, preincubation of IFNs with supernatants containing the IFNα/βBP from VACV, VARV, or MPXV resulted in complete cell death, demonstrating that all 3 proteins inhibit the antiviral effect of all the human IFNs tested. This effect was dose dependent for all the IFNs tested (not shown).
Figure 3.
Inhibition of type I IFN antiviral activity by VACV IFNα/βBP and its VARV and MPXV orthologues. Supernatants containing the indicated recombinant proteins were preincubated with 50 U of each type I IFN and subsequently added to HeLa cell monolayers. After a 16 h incubation, cells were infected with VSV, and cell viability was determined after 48 h. Controls of nontreated, uninfected (−VSV), and infected (+VSV) cells are at left, separated by a dashed line. Results are means ± sd of triplicate samples from one representative experiment.
Differential binding properties of the VARV B17 protein as compared to the VACV B18 protein
To further characterize the properties of VARV B17 in vitro in comparison to VACV B18, we purified both by affinity chromatography (Supplemental Fig. 2A). Both proteins bound similarly to all the human type I IFNs tested (hIFNα-A, hIFNα-2b, hIFNα-K, hIFNα-G, hIFNα-J1, hIFNα-F, hIFNα-B2, hIFNβ, and hIFNω) as well as to Rhesus/cynomolgus IFNα (not shown), as determined by surface plasmon resonance (SPR). Although both proteins bound the murine IFNα (mIFNα) equally well, the VARV B17 protein showed a much weaker binding to mIFNβ, with a very marked difference in the dissociation time (Fig. 4, top panels). In addition, we tested human and murine type III IFNs (hIL-28A, mIL-28A, and hIL-29) as well as the more distantly related human and murine IL-10 for binding to both proteins. Although VARV B17 was not able to bind strongly to any of these molecules, VACV B18 bound to hIL-28A, mIL-28A, hIL-29, and mIL-10 (Fig. 4, bottom panels). To further evaluate these differences, the binding affinites for several of these ligands were calculated by SPR (Table 1 and Supplemental Fig. 3). The affinites of VARV B17 and VACV B18 for hIFNα-Α, hIFNβ, and mIFNα were found to be high (ranking from 0.1 to 5 nM), and the VARV protein bound the human ligands with ∼5-fold higher affinity than the VACV orthologue. However, although VACV B18 showed comparatively weaker affinites (ranking from 3 to 15 nM) for mIFNβ and the type III IFNs, the VARV B17 protein showed no detectable binding to any of these ligands under the conditions used.
Figure 4.
Differential binding of VACV B18 and VARV B17 to human and murine IFNs. Purified recombinant VACV B18 or VARV B17 were coupled to a sensor chip and screened for interactions with the indicated molecules by SPR. Top panels: binding curves of each protein to indicated human and murine type I IFNs. Bottom panels: binding curves of each protein to human and murine type III IFNs and the related human and murine IL-10. Curves correspond to injections of 10 nM hIFNα-A, 10 nM hIFNβ, 10 nM mIFNα, and 100 nM of all other analytes. All curves are normalized to an arbitrary value of 100 RU for comparative purposes; hIFNα-A curve is shown as a reference. Data correspond to one representative experiment.
TABLE 1.
Comparison of kinetic parameters and derived affinity constants for the binding of VACV IFN type I binding protein and its VARV orthologue to IFNs
| IFN | VACV B18
|
VARV B17
|
||||
|---|---|---|---|---|---|---|
| ka (M−1s−1) | kd (s−1) | Kd (M) | ka (M−1s−1) | kd (s−1) | Kd (M) | |
| hIFNα | 3.89 × 105 | 2.25 × 10−4 | 5.8 × 10−10 | 1.22 × 106 | 1.32 × 10−4 | 1.08 × 10−10 |
| hIFNβ | 5.98 × 105 | 3.16 × 10−4 | 5.28 × 10−9 | 2.52 × 105 | 3.13 × 10−4 | 1.24 × 10−9 |
| mIFNα | 1.72 × 106 | 7.84 × 10−4 | 4.55 × 10−10 | 1.34 × 106 | 3.84 × 10−4 | 2.6 × 10−10 |
| mIFNβ | 1.04 × 105 | 3.33 × 10−4 | 3.2 × 10−9 | – | – | – |
| hIL28A | 2.45 × 105 | 3.64 × 10−3 | 1.49 × 10−8 | – | – | – |
| hIL29 | 1.93 × 105 | 2.6 × 10−3 | 1.35 × 10−8 | – | – | – |
| mIL28A | 8.32 × 105 | 1.12 × 10−2 | 1.34 × 10−8 | – | – | – |
In a luciferase reporter assay, preincubation of hIFNα-A with either VACV B18 or VARV B17 completely abrogated the IFN-induced activation of transcription from an IFN-stimulated response enhancer element (Supplemental Fig. 4A). In addition, both purified proteins blocked the antiviral activity of all the type I hIFNs tested (hIFNα-A, hIFNα-2b, hIFNα-K, hIFNα-G, hIFNα-J1, hIFNα-F, hIFNα-B2, hIFNβ, and hIFNω) in a dose-dependent manner (Supplemental Fig. 4B), with the concentration of protein needed to inhibit the IFN activity by 50% estimated at 10–40 ng/ml for all cases (Supplemental Fig. 4C).
An IFN inhibitory activity is secreted by MPXV- and VARV-infected cells
To address whether an active IFNα/βBP is expressed and secreted during MPXV or VARV infection, BSC-1 cells were infected with strains USA or RCG of MPXV or strain BSH1975 of VARV, and supernatants were collected at 48 or 30 h postinfection. Virus present in the supernatants was inactivated to allow safe handling of the samples. Western blot analyses using a polyclonal anti-VACV B18 protein antibody showed a band of ∼50 kDa in supernatants from cells infected with either RCG or USA MPXV strains as well as VARV, but not in mock-infected cells, and likely corresponds to the MPXV B16 protein and VARV B17 protein, respectively (Fig. 5A). Supernatants from cells infected with each MPXV strain, but not from mock-infected cells, efficiently inhibited hIFNα-2b, hIFNα-A, or hIFNβ-induced antiviral activity in a dose-dependent manner (Fig. 5B). Similarly, supernatants from VARV-infected cells contained a specific type I IFN inhibitory activity (Fig. 5C).
Figure 5.
Expression and activity of the IFNα/βBP in supernatants from MPXV- or VARV-infected cells. A) Supernatants from VACV-, MPXV-, or VARV-infected cells were analyzed by Western blot using an anti-B18 protein antibody along with negative (media) and purified recombinant B18 protein (VACV B18) as indicated. B, C) Increasing amounts of supernatants from MPXV strain USA- or RCG-infected cells (B) or VARV strain BSH75 X or mock-infected cells (media) (C) were preincubated with either 5 or 50 U of type I hIFNs as indicated and added to HeLa cell monolayers for 16 h. As a control, 10 ng of purified VACV B18 protein was preincubated with IFN and added to HeLa cell monolayers as before. Cells were infected with VSV, and cell viability was determined after 48 h. Controls of nontreated, uninfected (−VSV) and infected (+VSV) cells were included in each case. Results are means ± sd of triplicate samples from one representative experiment.
DISCUSSION
Viral immunomodulatory mechanisms are key determinants of pathogenesis. Currently the mechanisms of immune evasion in VARV and MPXV are largely unknown and mostly inferred from data from other poxviral species. Given the relevance of both viruses as human pathogens, it is important to study the properties of the VARV and MPXV proteins with immunomodulatory potential. In this report we have characterized the secreted type I IFN inhibitors from both viral species, comparing their activity to their VACV orthologue. Moreover, we have shown the expression of both viral proteins during infection with VARV and MPXV in cell culture.
The genomes of several VARV and MPXV strains have been sequenced and proteins with immunomodulatory potential predicted. However, very few such proteins have been characterized so far (reviewed in refs. 27, 28). The IFNα/βBP has been shown to contribute to poxvirus pathogenesis in VACV infections (18, 19), and more recently this has been confirmed using the mousepox model, considered the best surrogate mouse model for human smallpox. In the absence of the ECTV IFNα/βBP, the LD50 in susceptible mice was raised ≥107-fold, and this attenuation was accompanied by an increased innate and adaptive immune response (25). Therefore, it is reasonable to speculate that the VARV and MPXV IFNα/βBPs may play a role in pathogenesis. A rapid activation of IFN-associated genes was observed in peripheral blood mononuclear cells from VARV-infected cynomolgus macaques (29). In addition, the inability to induce a strong IFN response appeared to correlate with more severe disease. This is consistent with the hypothesis that VARV regulation of IFN responses is a key determinant of smallpox pathogenesis. Here we demonstrate that VARV B17 is expressed and active as a type I IFN inhibitor during infection. To our knowledge, this is the first demonstration of expression and activity of a native-secreted immunomodulatory VARV protein. Similarly, we have shown expression of the active protein in both RCG and USA strains of MPXV during infection in cell culture. However, we cannot formally rule out the possibility that VARV or MPXV may encode additional secreted antitype I IFN activity distinct from the IFNα/βBP characterized in this report. Previously, transcription of the B16R gene from MPXV strain Zaire96 was described to occur early during tissue culture infection, but protein expression was not reported (30). Note that orthologues of both intracellular poxviral IFN-resistance proteins E3 and K3 are present in VARV but truncated or absent in MPXV (28). This may make MPXV far more reliant for the control of the host’s immune response on the secreted type I IFN inhibitor than other poxviruses.
VACV B18 is a decoy-type I IFN receptor with broad species specificity and blocks the antiviral activity of human IFNα1, IFNα2, IFNα7, IFNα8, IFNβ, IFNω, IFNκ, and IFNε (18, 19, 31). Our results show that the VARV and MPXV orthologues block the antiviral activity of 8 different human type I IFNs, with an inhibitory profile similar to that of the VACV protein. In addition, both the VARV and MPXV proteins block monkey IFN activity, indicating that its activity should be evident in the macaque model of infection.
A unique property of VACV B18 is its ability to bind to the cell surface and prevent IFN signaling at this location (17, 18). It was suggested that secretion is the best way to transport this protein to uninfected cells to coat a surrounding uninfected tissue, which would be its most relevant site of action physiologically. We show that this property is shared by the VARV and MPXV IFNα/βBPs. Interestingly, the more distantly related secreted IFN inhibitor protein from the yatapoxvirus YLDV also binds to the cell surface (31).
The antiviral activity of type III IFN is restricted to viruses that enter through epithelia, such as those lining the lung (32). Type III IFN impairs VACV replication in vivo (33) and, more importantly, the YLDV orthologue of VACV B18 inhibits human type III IFN activity in cell culture (31). Because VARV and MPXV enter their host mainly through the respiratory system, it was important to determine whether the VARV or MPXV IFNα/βBP may also block type III IFN signaling. As shown before (31), we found that the VACV protein does not block type III IFN-induced signaling. Similarly, neither its MPXV nor its VARV orthologues blocked type III IFN activity. The importance of type III IFN in response against human poxviral infections is not known. YLDV is a primate virus that can be transmitted to humans causing a mild illness clearly distinct from monkeypox and smallpox, and perhaps type III IFN is more important in the local control of yatapoxvirus infections. Alternatively, other OPVs may encode different secreted immunomodulators targeting type III IFNs. It is also conceivable that the intracellular blockade of type I IFN-induced responses mediated by E3L, K3L, and the VH1 phosphatase (34) is also efficient against type III IFN signaling and sufficient to prevent its antiviral activity in vivo. Surprisingly, we found that the VACV, but not the VARV IFNα/βBP, did bind human and murine type III IFNs. The inability of this protein to block their activity may be due to its relatively low binding affinity. This lower affinity or the nature of the recombinant proteins used may be the reason why Huang et al. (31) did not detect this interaction. However, this finding has important consequences for structure-function studies, especially for the determination of the motifs involved in type I vs. type III IFN binding in poxviral orthologues of VACV B18.
To study the potential adaptation of the VARV IFNα/βBP to its human host, we compared the binding and specific activity of the purified protein with those of VACV B18. The binding affinites of VARV B17 for hIFNα-A and hIFNβ were slightly better (5-fold higher) than those of VACV B18. Although the specific activity of both proteins in IFNα and IFNβ inhibition assays in vitro did not show major differences, a potential advantage in vivo in the human host cannot be excluded. Interestingly, while VARV B17 binds mIFNα with an affinity comparable to that of VACV B18, it has lost the capacity to bind murine IFNβ as well as type III IFNs and IL-10. It should be noted that binding of VACV B18 to these cytokines does not block their activity (refs. 16, 31 and the present study) and is therefore probably not relevant in vivo. It is surprising that the OPV IFNα/βBPs have lost the ability to block mIFNβ activity while retaining hIFNβ-inhibitory activity since IFNβ-deficient mice are more susceptible to VACV (35).
Apparent adaptation of poxvirus immunomodulatory proteins to their specific hosts has been shown for the smallpox inhibitor of complement enzyme (SPICE), a VARV orthologue of the VACV complement control protein, which contributes to pathogenesis in vivo. SPICE is a highly specific and potent inhibitor of human complement as compared to the VACV protein (36). This enhanced function against human complement has been ascribed to 4 amino acid changes (37), illustrating that adaptation to the human host may not require many mutations. However, the VARV immunomodulators studied to date do not necessarily have higher affinities for their human ligands. Thus, the VARV IL-18BP showed similar affinities for its human and murine ligand (38), while VARV CrmB is a better inhibitor of murine than human tumor necrosis factor (39). This suggests the relative importance of the binding affinities of various virally encoded immune response modifiers for various human immune effectors in the maintenance of VARV in a human host system and perhaps indicates that VARV has only recently (in evolutionary time) adapted to a human host. Yatapoxviruses, on the contrary, show a marked species specificity in their secreted receptors for TNF and IFN (31, 40).
Because of the risk of emerging OPV infections and weaponization, and because the live VACV-based vaccine is contraindicated for portions of the global population, there is a considerable interest in developing novel anti-OPV vaccines (41, 42). It was previously shown that monoclonal antibodies against the secreted VACV/VARV growth factor could enhance protective immunity in a VACV intranasal mouse model of infection (43), which led the authors to propose that blockade of secreted viral immunomodulators represents an alternative to the traditional approach that prevents infection by virus neutralization. The IFNα/βBP encoded by ECTV, with properties similar to those of VARV and MPXV (16), has been shown to be a major contributor to virus virulence in mousepox (25), considered one of the best mouse models of human smallpox. The unprecedented attenuation of an ECTV mutant in the IFNα/βBP gene, with an increase of more than 107-fold in the LD50, clearly demonstrated that virulent OPVs require this protein to establish a smallpox-like disease and strongly suggested that neutralization of this viral IFN inhibitor is a therapeutic strategy to control disease severity. Consistent with this, vaccination with recombinant ECTV IFNα/βBP could effectively prevent fatal mousepox (25).
Our demonstration that the IFNα/βBP is actively expressed by cells infected with VARV and MPXV identifies this viral immunomodulatory protein as both a component of a protective vaccine and a target for therapeutics against virulent human poxviruses. Subunit vaccines against poxviruses are composed of structural proteins present in the virus particle and neutralize the infectivity of the virions, and the inclusion of viral-secreted virulence factors may serve as a complementary approach (25, 43). It should be noted that the smallpox vaccines Dryvax and modified virus Ankara encode truncated versions of the IFNα/βBP, which in the case of vaccinia virus Dryvax has been shown to be expressed and to bind IFN with 70-fold lower affinity than the full-length protein (17, 44). The inclusion of full-length IFNα/βBP, or, even better, a mutagenized version of the protein lacking IFN-binding activity, in the current smallpox vaccines may induce better anti-IFNα/βBP response and enhance their protective effect. In addition, monoclonal antibodies against the MPXV or VARV proteins could be helpful as a phamacological compound in exposed individuals to prevent disease in human monkeypox and smallpox cases. This would represent an alternative to the large proportion of individuals at high risk of serious adverse effects when receiving smallpox vaccination, due to immune deficiency, atopic dermatitis, or at the extremes of age (45).
In summary, we report the expression of an active IFNα/βBP by VARV and MPXV, and the characterization of their binding properties against a broad range of IFNs. This is the first demonstration of the expression of a soluble cytokine binding protein by VARV. The critical role of the IFNα/βBP in the pathogenesis of virulent poxviruses (25) emphasizes the relevance of these findings to understand the mechanisms of pathogenesis of smallpox and monkeypox. Furthermore, our findings identify the VARV and MPXV IFNα/βBPs as a target for vaccination and therapeutic treatment of infected individuals.
Supplementary Material
Acknowledgments
The authors thank R. Martín for excellent technical assistance and Sylvia Gutiérrez Erlandsson for help with microscopy. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. This work was funded by the Wellcome Trust and the European Union. A. Alejo was supported by a Ramon y Cajal contract (Spanish Ministry of Science and Innovation). This article is dedicated to the memory of Riccardo Wittek.
References
- De Souza Trindade G, Drumond B P, Guedes M I, Leite J A, Mota B E, Campos M A, da Fonseca F G, Nogueira M L, Lobato Z I, Bonjardim C A, Ferreira P C, Kroon E G. Zoonotic vaccinia virus infection in Brazil: clinical description and implications for health professionals. J Clin Microbiol. 2007;45:1370–1372. doi: 10.1128/JCM.00920-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G L, McFadden G. Smallpox: anything to declare? Nat Rev. 2002;2:521–527. doi: 10.1038/nri845. [DOI] [PubMed] [Google Scholar]
- Di Giulio D B, Eckburg P B. Human monkeypox: an emerging zoonosis. Lancet. 2004;4:15–25. doi: 10.1016/S1473-3099(03)00856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalca A, Rimoin A W, Bavari S, Whitehouse C A. Reemergence of monkeypox: prevalence, diagnostics, and countermeasures. Clin Infect Dis. 2005;41:1765–1771. doi: 10.1086/498155. [DOI] [PubMed] [Google Scholar]
- Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- Seet B T, Johnston J B, Brunetti C R, Barrett J W, Everett H, Cameron C, Sypula J, Nazarian S H, Lucas A, McFadden G. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- Stanford M M, McFadden G, Karupiah G, Chaudhri G. Immunopathogenesis of poxvirus infections: forecasting the impending storm. Immunol Cell Biol. 2007;85:93–102. doi: 10.1038/sj.icb.7100033. [DOI] [PubMed] [Google Scholar]
- Randall R E, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- Samuel C E. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- Biron C A. Role of early cytokines, including alpha and beta interferons (IFN-alpha/beta), in innate and adaptive immune responses to viral infections. Semin Immunol. 1998;10:383–390. doi: 10.1006/smim.1998.0138. [DOI] [PubMed] [Google Scholar]
- Ank N, Iversen M B, Bartholdy C, Staeheli P, Hartmann R, Jensen U B, Dagnaes-Hansen F, Thomsen A R, Chen Z, Haugen H, Klucher K, Paludan S R. An important role for type III interferon (IFN-lambda/IL-28) in TLR-induced antiviral activity. J Immunol. 2008;180:2474–2485. doi: 10.4049/jimmunol.180.4.2474. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hamming O J, Ank N, Paludan S R, Nielsen A L, Hartmann R. Type III interferon (IFN) induces a type I IFN-like response in a restricted subset of cells through signaling pathways involving both the Jak-STAT pathway and the mitogen-activated protein kinases. J Virol. 2007;81:7749–7758. doi: 10.1128/JVI.02438-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Yanai H. Interferon signalling network in innate defence. Cell Microbiol. 2006;8:907–922. doi: 10.1111/j.1462-5822.2006.00716.x. [DOI] [PubMed] [Google Scholar]
- Davies M V, Elroy-Stein O, Jagus R, Moss B, Kaufman R J. The vaccinia virus K3L gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor 2. J Virol. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H W, Watson J C, Jacobs B L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V P, Alcami A. Inhibition of interferons by ectromelia virus. J Virol. 2002;76:1124–1134. doi: 10.1128/JVI.76.3.1124-1134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcami A, Symons J A, Smith G L. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol. 2000;74:11230–11239. doi: 10.1128/jvi.74.23.11230-11239.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colamonici O R, Domanski P, Sweitzer S M, Larner A, Buller R M. Vaccinia virus B18R gene encodes a type I interferon-binding protein that blocks interferon alpha transmembrane signaling. J Biol Chem. 1995;270:15974–15978. doi: 10.1074/jbc.270.27.15974. [DOI] [PubMed] [Google Scholar]
- Symons J A, Alcami A, Smith G L. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Alcami A, Smith G L. Soluble interferon-gamma receptors encoded by poxviruses. Comp Immunol Microbiol Infect Dis. 1996;19:305–317. doi: 10.1016/0147-9571(96)00013-6. [DOI] [PubMed] [Google Scholar]
- Upton C, Mossman K, McFadden G. Encoding of a homolog of the IFN-gamma receptor by myxoma virus. Science. 1992;258:1369–1372. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- Smith G L, Chan Y S. Two vaccinia virus proteins structurally related to the interleukin-1 receptor and the immunoglobulin superfamily. J Gen Virol. 1991;72:511–518. doi: 10.1099/0022-1317-72-3-511. [DOI] [PubMed] [Google Scholar]
- Alcami A, Smith G L. A soluble receptor for interleukin-1 beta encoded by vaccinia virus: a novel mechanism of virus modulation of the host response to infection. Cell. 1992;71:153–167. doi: 10.1016/0092-8674(92)90274-g. [DOI] [PubMed] [Google Scholar]
- Verardi P H, Jones L A, Aziz F H, Ahmad S, Yilma T D. Vaccinia virus vectors with an inactivated gamma interferon receptor homolog gene (B8R) are attenuated in vivo without a concomitant reduction in immunogenicity. J Virol. 2001;75:11–18. doi: 10.1128/JVI.75.1.11-18.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R H, Cohen M, Tang Y, Lazear E, Whitbeck J C, Eisenberg R J, Cohen G H, Sigal L J. The orthopoxvirus type I IFN binding protein is essential for virulence and an effective target for vaccination. J Exp Med. 2008;205:981–992. doi: 10.1084/jem.20071854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchelkunov S N, Totmenin A V, Babkin I V, Safronov P F, Ryazankina O I, Petrov N A, Gutorov V V, Uvarova E A, Mikheev M V, Sisler J R, Esposito J J, Jahrling P B, Moss B, Sandakhchiev L S. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett. 2001;509:66–70. doi: 10.1016/S0014-5793(01)03144-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop L R, Oehlberg K A, Reid J J, Avci D, Rosengard A M. Variola virus immune evasion proteins. Microbes Infect. 2003;5:1049–1056. doi: 10.1016/s1286-4579(03)00194-1. [DOI] [PubMed] [Google Scholar]
- Weaver J R, Isaacs S N. Monkeypox virus and insights into its immunomodulatory proteins. Immunol Rev. 2008;225:96–113. doi: 10.1111/j.1600-065X.2008.00691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubins K H, Hensley L E, Jahrling P B, Whitney A R, Geisbert T W, Huggins J W, Owen A, Leduc J W, Brown P O, Relman D A. The host response to smallpox: analysis of the gene expression program in peripheral blood cells in a nonhuman primate model. Proc Natl Acad Sci U S A. 2004;101:15190–15195. doi: 10.1073/pnas.0405759101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubins K H, Hensley L E, Bell G W, Wang C, Lefkowitz E J, Brown P O, Relman D A. Comparative analysis of viral gene expression programs during poxvirus infection: a transcriptional map of the vaccinia and monkeypox genomes. PLoS ONE. 2008;3:e2628. doi: 10.1371/journal.pone.0002628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Smirnov S V, Lewis-Antes A, Balan M, Li W, Tang S, Silke G V, Putz M M, Smith G L, Kotenko S V. Inhibition of type I and type III interferons by a secreted glycoprotein from Yaba-like disease virus. Proc Natl Acad Sci U S A. 2007;104:9822–9827. doi: 10.1073/pnas.0610352104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mordstein M, Kochs G, Dumoutier L, Renauld J C, Paludan S R, Klucher K, Staeheli P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008;4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett N W, Buttigieg K, Kotenko S V, Smith G L. Murine interferon lambdas (type III interferons) exhibit potent antiviral activity in vivo in a poxvirus infection model. J Gen Virol. 2005;86:1589–1596. doi: 10.1099/vir.0.80904-0. [DOI] [PubMed] [Google Scholar]
- Mann B A, Huang J H, Li P, Chang H C, Slee R B, O'Sullivan A, Anita M, Yeh N, Klemsz M J, Brutkiewicz R R, Blum J S, Kaplan M H. Vaccinia virus blocks Stat1-dependent and Stat1-independent gene expression induced by type I and type II interferons. J Interferon Cytokine Res. 2008;28:367–380. doi: 10.1089/jir.2007.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deonarain R, Alcami A, Alexiou M, Dallman M J, Gewert D R, Porter A C. Impaired antiviral response and alpha/beta interferon induction in mice lacking beta interferon. J Virol. 2000;74:3404–3409. doi: 10.1128/jvi.74.7.3404-3409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengard A M, Liu Y, Nie Z, Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc Natl Acad Sci U S A. 2002;99:8808–8813. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav V N, Pyaram K, Mullick J, Sahu A. Identification of hot spots in the variola virus complement inhibitor (SPICE) for human complement regulation. J Virol. 2008;82:3283–3294. doi: 10.1128/JVI.01935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteban D J, Nuara A A, Buller R M. Interleukin-18 and glycosaminoglycan binding by a protein encoded by variola virus. J Virol. 2004;85:1291–1299. doi: 10.1099/vir.0.79902-0. [DOI] [PubMed] [Google Scholar]
- Alejo A, Ruiz-Arguello M B, Ho Y, Smith V P, Saraiva M, Alcami A. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc Natl Acad Sci U S A. 2006;103:5995–6000. doi: 10.1073/pnas.0510462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti C R, Paulose-Murphy M, Singh R, Qin J, Barrett J W, Tardivel A, Schneider P, Essani K, McFadden G. A secreted high-affinity inhibitor of human TNF from Tanapox virus. Proc Natl Acad Sci U S A. 2003;100:4831–4836. doi: 10.1073/pnas.0737244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P L, Americo J L, Wyatt L S, Eller L A, Whitbeck J C, Cohen G H, Eisenberg R J, Hartmann C J, Jackson D L, Kulesh D A, Martinez M J, Miller D M, Mucker E M, Shamblin J D, Zwiers S H, Huggins J W, Jahrling P B, Moss B. Immunogenicity of a highly attenuated MVA smallpox vaccine and protection against monkeypox. Nature. 2004;428:182–185. doi: 10.1038/nature02331. [DOI] [PubMed] [Google Scholar]
- Earl P L, Americo J L, Wyatt L S, Espenshade O, Bassler J, Gong K, Lin S, Peters E, Rhodes L, Jr, Spano Y E, Silvera P M, Moss B. Rapid protection in a monkeypox model by a single injection of a replication-deficient vaccinia virus. Proc Natl Acad Sci U S A. 2008;105:10889–10894. doi: 10.1073/pnas.0804985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Yang H, Kim S K, Reche P A, Tirabassi R S, Hussey R E, Chishti Y, Rheinwald J G, Morehead T J, Zech T, Damon I K, Welsh R M, Reinherz E L. Biochemical and functional analysis of smallpox growth factor (SPGF) and anti-SPGF monoclonal antibodies. J Biol Chem. 2004;279:25838–25848. doi: 10.1074/jbc.M400343200. [DOI] [PubMed] [Google Scholar]
- Waibler Z, Anzaghe M, Frenz T, Schwantes A, Pohlmann C, Ludwig H, Palomo-Otero M, Alcami A, Sutter G, Kalinke U. Vaccinia virus-mediated inhibition of type I interferon responses is a multifactorial process involving the soluble type I interferon receptor B18 and intracellular components. J Virol. 2009;83:1563–1571. doi: 10.1128/JVI.01617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs B L, Langland J O, Kibler K V, Denzler K L, White S D, Holechek S A, Wong S, Huynh T, Baskin C R. Vaccinia virus vaccines: past, present and future. Antiviral Res. 2009;84:1–13. doi: 10.1016/j.antiviral.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.