Abstract
Kaposi’s sarcoma (KS) and its causative agent, Kaposi’s sarcoma associated herpesvirus (KSHV/HHV-8), a gamma2 herpesvirus, have distinctive geographical distributions that are largely unexplained. We propose the “oncoweed” hypothesis to explain these differences, namely that environmental cofactors present in KS endemic regions cause frequent reactivation of KSHV in infected subjects, leading to increased viral shedding and transmission leading to increased prevalence of KSHV infection as well as high viral load levels and antibody titers. Reactivation also plays a role in the pathogenesis of KSHV-associated malignancies. To test this hypothesis, we employed an in vitro KSHV reactivation assay that measured increases in KSHV viral load in KSHV infected primary effusion lymphoma (PEL) cells and screened aqueous natural product extracts from KS endemic regions. Of 4,842 extracts from 38 countries, 184 (5%) caused KSHV reactivation. Extracts that caused reactivation came from a wide variety of plant families, and extracts from Africa, where KSHV is highly prevalent, caused the greatest level of reactivation. Time course experiments were performed using 28 extracts that caused the highest levels of reactivation. The specificity of the effects on viral replication was examined using transcriptional profiling of all viral mRNAs. The array data indicated that the natural extracts caused an ordered cascade of lytic replication similar to that seen after induction with synthetic activators. These in vitro data provide support for the “oncoweed” hypothesis by demonstrating basic biological plausibility.
Keywords: Kaposi’s sarcoma associated herpesvirus, natural products, reactivation
Much has been learned about the epidemiology and pathogenesis of Kaposi’s sarcoma (KS) since the discovery in 1994 of the causative agent, Kaposi’s sarcoma-associated Herpesvirus (KSHV), also known as Human Herpesvirus 8.1 The striking geographical differences in incidence of classic KS and the equally striking differences in incidence of AIDS KS, according to HIV risk group, can be partially explained by differences in the prevalence of KSHV. Prior to the AIDS epidemic, KS was most common in sub-Saharan Africa and Mediterranean countries but was rare in the US and northern Europe.2,3 Even now, in the US and Northern Europe, KS is rarely seen in patients who acquire HIV via blood products or heterosexual sex, but it is common in homosexual men.4
The prevalence of KSHV infection also shows considerable variation geographically and between HIV risk groups, largely mirroring the variations seen in the incidence of KS. Prevalence is high in sub-Saharan Africa and some Mediterranean countries and low in most northern European and Asian populations.5 In Italy, the incidence of KS is higher in the south than in the centre and north of the peninsula. The prevalence of KSHV accords with this geographical distribution.6 There are exceptions to this general rule, however. For instance, the high prevalence of KSHV in The Gambia where the incidence of KS is low is striking.7 The high prevalence of KSHV in South American Amerindians is also surprising. Although KS has not been reported in Amerindians, it is possible that this may be due to lack of ascertainment because of poor access to health care and the low life expectancy among these populations.8,9 In the US and Europe, KSHV is rare in the general population but relatively common in homosexual men, explaining the high risk of KS in HIV infected homosexual men.
Although the variation in the incidence of KS disease can thus be partially explained by differences in KSHV prevalence, no explanation exists for the variation in KSHV prevalence, since like other herpesviruses, KSHV is known to have coevolved with the human species.10 Variation in viral prevalence may be due to host genetic or environmental risk factors. Host genetic factors likely play an important part in the elevated risk for KS in elderly men of Jewish descent, since the prevalence of KSHV is not elevated in this population.11 Similarly, KS risk is 10-fold higher in transplant recipients from Saudi Arabia than from other areas, despite a modest KSHV prevalence in this population.12 In these instances, the high risk of KS is likely due to a genetically determined increased risk of disease amongst infected subjects.
We believe that environmental risk factors are more important than genetic predispositions in determining the variation in KSHV prevalence and KS risk in many areas and populations. Although KSHV prevalence is high in Africa, it is not elevated in subjects of African descent living in the Caribbean.13,14 In a study of STD clinic attendees in London, we found a high prevalence of KSHV in subjects born in Africa but not in those whose parents were born in Africa, suggesting that a single generation in a non-African environment is sufficient to reduce prevalence.14 Conversely, the prevalence of KSHV in South African blood donors of Northern European descent is 10% (Whitby and Sitas, unpublished data), compared with estimates of 1–2% in Northern European blood donors living in Northern Europe.
Unlike other viruses, such as Hepatitis B virus, the relationship between socioeconomic factors such as household crowding and KSHV prevalence is inconsistent. KSHV is common in sub-Saharan Africa but rare in Asian countries where similar, though less extreme, socioeconomic conditions exist. Moreover, it is also relatively common in affluent countries such as Italy. Therefore, the role of socioeconomic factors in determining KSHV transmission and prevalence is unclear.
To elucidate a role for environmental cofactors in KSHV infection and KS disease, we formulated the “oncoweed hypothesis”: Environmental cofactors present in KSHV endemic regions cause reactivation of KSHV in latently infected subjects. This leads to an increase in KSHV shedding and hence transmission and also results in elevated viral loads and antibody titres. Frequent reactivation and lytic replication not only increases the pool of infected individuals but lytic replication of KSHV may also be important in KS pathogenesis.15 High viral load and high antibody titres are known risk factors for KS.16,17 Furthermore, many of the KSHV genes shown to have transforming properties are expressed during viral reactivation and in primary KS lesions.18,19
To test the premise of this hypothesis, we developed an in vitro KSHV reactivation assay that measured increases in viral load in latently KSHV infected cells and used it to screen natural products for potential environmental cofactors of KSHV infection and disease. The Natural Products Branch of the National Cancer Institute maintains a repository of aqueous and organic extracts of land plants, marine invertebrates and fungi collected from numerous tropical and subtropical countries. These extracts are used to screen for antiviral and anticancer agents using a variety of techniques. In this study, we reversed the usual rationale and used the library to screen for agents that reactivate a virus that is associated with human cancer.
Methods
Selection of natural products extracts
Extracts for testing in the KSHV reactivation assay were selected from the NCI/DTP anticancer screening library of >2,200 unique microtiter plates, which include 130,000 aqueous and organic plant extracts. Plants and animals were collected from 1989 to the present under contract to the Developmental Therapeutics Program, National Cancer Institute, for anticancer screening. Organizations with expertise in the flora and fauna of specific regions were New York Botanical Garden for Central and South America, Missouri Botanical Garden for Africa and University of Illinois at Chicago Circle for southeast Asia. Collectors were responsible for taxonomic identification and preparation of a herbarium voucher. Detailed collection records and taxonomic information are permanently stored in an NCI database. Plants were air dried in the field and then transported to the NCI facility at Frederick, MD for storage (−20°C) until processed.
Dried plants were finely ground using a Holmes Brothers hammermill, extracted by percolation, first with dichloromethane/ methanol (1:1) and then with water. Organic solvent was removed by rotary evaporation. Aqueous extract was frozen and lyophilized. Additional detail of the extraction process can be found at http://npsg.ncifcrf.gov. For microtiter plate preparation, an aliquot of the lyophilized aqueous extract was solubilized in water and transferred into a single well of a 96-well plate by a robotic liquid handling system (TECAN Genesis 150, permanent tip), 88 different extracts per plate. The aqueous extracts in the plate were again vacuum dried using a Savant Speed-Vac 210 (Thermo), leaving a residue of 50 µg/well. All plants, extracts and microtiter plates were stored in −20°C freezers to prevent degradation.
Natural products were selected from all over the world but there was a definite bias towards geographical areas where KS is endemic. The vast majority of extracts (4,572) were from land plants, but extracts were included from 86 marine plants, 97 marine animals and 87 fungi. More than 50% of extracts selected were from African countries including Cameroon, Central Africa Republic, Gabon, Ghana, Madagascar and Tanzania. The remaining extracts were selected as being representative of the rest of the world, including regions where KS is rare. Plants were not selected by family or genus but rather represented a wide variety of groups. Extracts known to be highly toxic to cells in tissue culture (based upon previous anticancer testing) were excluded. Organic and aqueous extracts were available but screening was restricted to aqueous extracts, since aqueous components of plants were most likely to be present as environmental exposures. Aqueous extracts that caused greater than 25% growth inhibition in a panel of 60 transformed cell lines at a concentration of 100 µg/ml were also excluded.
KSHV quantitative PCR assay
We developed a KSHV quantitative real time PCR assay based on Applied Biosystems (ABI) technology. The KSHV genomic sequence used for assay design was GenBank record U75698 derived from the BC-1 cell line.20 Primers and probe were designed to target the KSHV K6 gene region, viral macrophage inflammatory protein α (v-mip-α), using Gene Runner software (Version 3.00, Hastings Software) according to ABI guidelines. The selected primers and probe sequence are as follows:
K6-10F: 5′-CGCCTAATAGCTGCTGCTACGG-3′ (nt 27330-27309)
K6-10R: 5′-TGCATCAGCTGCCTAACCCAG-3′ (nt 27159–27179)
p-K6-10: 5′-FAM-CACCCACCGCCCGTCCAAATTC-TAMRA-3′ (nt 27298-27277)
The primers and probe were synthesized and HPLC purified by Operon (Alameda, CA). The optimized KSHV K6 master mix consisted of 3 mM magnesium chloride, 0.05% EIA reagent grade gelatin (Sigma, St. Louis, MO), 0.01% Tween 20 (Sigma, St. Louis, MO), 200 µM each dATP, dCTP, dGTP, 400 µM dUTP, 1 µM forward and reverse primers, 200 nM probe, 0.1 U/µl AmpErase uracil-N-glycosylase and 1.25 U/µl AmpliTaq Gold polymerase in TaqMan buffer A (TaqMan PCR Core Reagents kit, Applied Biosystems, Foster City, CA). PCR reactions contained 40 µl of master mix and 10 µl of test material for a final volume of 50 µl. The cycling conditions were 2 min hold at 50°C, 95°C hold for 10 min and 45 cycles performed at 95°C for 15 sec, 55°C for 30 sec and an annealing step of 60°C for 1 min. All assays were performed using an ABI Prism 7700 automated detection system that captured and analyzed the data using the Sequence Detection System software version 1.6.
A 500-bp fragment of the KSHV K6 gene, which includes the 176-bp fragment amplified by the TaqMan primers, was cloned and used to prepare a plasmid based standard. The standard curve was prepared by making 10-fold serial dilutions ranging from 10 to 100 copies in TE buffer (nuclease-free 0.1 M Tris-EDTA buffer (Invitrogen, Carlsbad, CA)) containing 0.1 µg/ml of human placental DNA (Sigma, St. Louis, MO). The Sequence Detection software performs a linear regression and plots the standard curve. The KSHV K6 standard curve has a least-squares regression (R2) value of greater than 0.97, which reflects titration accuracy. The slope of the standard curve is indicative of 90–97% PCR efficiency and the Y intercept value is consistent with one copy template input. The estimated viral copy numbers for test samples are determined by interpolation of the sample Ct value onto the K6 standard curve. The lower limit of detection was 3 copies of KSHV genomic DNA per PCR. In practice, the KSHV K6 assay sensitivity is 1 copy of KSHV per 1 million cell equivalents in clinical samples.
KSHV reactivation screen
Lyophilized aqueous extracts were provided at a concentration of 50 µg per well in 96-well plates. BCP-1 cells21 latently infected with KSHV were added at a concentration of 2.5 × 105 cells per well and incubated for 3 days at 37°C, 5% CO2 in RPMI (Invitrogen) supplemented with 20% FBS (Hyclone, Logan, UT). A 96-well plate of uninduced BCP-1 cells served as the negative control and another plate of BCP-1 cells, induced with 0.3 mM sodium butyrate, was used as the positive control.
DNA was extracted using either the Qiagen QiAmp 96-well blood extraction kit, (Qiagen, Valencia CA) or Puregene 96-well kit (Gentra Systems, Minneapolis, MN). Real-time quantitative PCR assays were performed for KSHV K6 and the cell quantitation marker, ERV-3, as described above and previously.22,23 Viral load was calculated and expressed as the number of copies per 105 cells.
BCP-1 cells, like all PEL cell lines, undergo spontaneous reactivation and up to 5% of cells will be undergoing lytic infection at any time. Differences in KSHV viral load after 72 hr were used to classify the extracts as activators, inactive or suppressors. A reduction in viral load compared with the uninduced (negative control) cells therefore suggests that lytic replication is being inhibited, whereas increased viral load is indicative of reactivation. Extracts were considered activators if the log10 KSHV viral load was 2 standard deviations greater than the mean of the viral load in the sodium butyrate stimulated (positive control) cells (log10 KSHV copies = 7.77). The mean log10 viral load in the sodium butyrate induced cells was 7.05 (geometric mean = 1.12 × 107 KSHV copies/10 cells with a standard deviation of 0.36 log10 KSHV copies/ 105 compared with 7.03 (geometric mean = 1.07 × 107 KSHV copies/105 cells) with a standard deviation of 0.23 log10 KSHV copies/105 in the uninduced (negative control) BCP-1 cells. Extracts were considered inhibitory if the viral load was greater than 2 standard deviations less than the log10 mean of the uninduced BCP-1 cells (log10 KSHV copies = 6.57). All remaining extracts were classified as either inactive or indeterminate. Comparisons between African and non-African extracts were made using the Likelihood Ratio χ2 test.
Time course experiments
We selected the 28 extracts that showed the most potent activity for follow-up experiments. These extracts produced viral loads of >9 logs in the initial screen. Replicate natural product plates were created using the same concentration (50 µg/well) of extracts. BCP-1 cells were added to each well at a concentration of 2.5 × 105 cells in 200 µl of RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 20% FBS (Hyclone, Logan, UT). Plates were incubated at 37°C, 5% CO2 for 1 to 4 days. One plate was harvested each day for 4 consecutive days. From each well, 100 µl was added to 1 ml of Trizol (Invitrogen, Carlsbad, CA) and frozen at −80°C for later RNA extraction. DNA was extracted from the remaining 100 µl using the QiAmp 96-well blood extraction kit (Qiagen, Valencia, CA), according to the manufacturer’s instructions. Real-time quantitative PCR assays were performed for KSHV K6 as described earlier and the cell quantitation marker ERV-3 as previously described.22,23 Viral load was calculated and expressed as KSHV copies per 104 cells.
Negative controls consisted of unstimulated BCP-1 cells and positive controls were BCP-1 cells stimulated with a gradient of sodium butyrate concentrations ranging from 3 to 100 nM. The KSHV viral load estimates for each extract were normalized by subtracting the average of the negative control (unstimulated BCP-1) viral copy number and dividing by the corresponding 100 nM sodium butyrate positive control copy number for each day. The data measure the effect greater than that of butyrate and were thus expressed as standard sodium butyrate units.
Real-time PCR based KSHV viral gene expression array
RNA was isolated from 106 cells using RNAzol (Tel-Test, Friendswood, Texas) as previously described,19 and reverse-transcribed using Superscript-II reverse transcriptase (Invitrogen, Carlsbad, CA), according to the manufacturers’ recommendations. The cDNA was diluted to a final volume of 600 µl in distilled water and stored at −80°C The KSHV real-time QPCR array and analysis have been previously described.18 The final QPCR reaction contained 2.5 µl of primer mix (final concentration 166 nM), 7.5 µl of 2× SYBR PCR mix (Applied Biosystems, Foster City, CA) and 5 µl of cDNA.
Viral CT values obtained by real-time QPCR were normalized to gapdh according to dCT = CT (viral mRNA) – CT (gapdh). The dCT values were imported into ArrayMiner v5.2 (Optimal Design, Brussels, Belgium) and clustered based on standard correlation metric. Fold induction was calculated by ddCT method (fold = 1.7 exp(dCTmock-Maximum (dCTinduced)).
Western blotting for K8.1 A/B viral glycoproteins
Supernatants from untreated BCP-1 cells and cells stimulated for 96 hr with extracts from C. dependens, E. coccinea, C. polystachyus and D.fragrans (top activators identified in screen) were filtered and then concentrated through a 20% sucrose gradient at 17,000 rpm, 4°C for 2 hr. Virus was resuspended in 300 µl (concentrated 100× original volume). Isolated KSHV (1.5 µl) from each condition was subjected to electrophoresis in a 7% Tris-Acetate polyacrylamide gel (Invitrogen, Carlsbad, CA) under reducing conditions. After transfer to Immobilon-P PVDF membrane (Millipore, Billerica, MA), the blot was probed with mouse anti-K8.1A/B (Advanced Biotechnologies, Columbia, MD), washed and probed with goat anti-mouse secondary antibody (Invitrogen, Carlsbad, CA). Chemiluminescence using Immunstar HRP (Biorad, Hercules, CA) was detected on Lumi-Film (Roche, Indianapolis, IN).
Electron microscopy
Preparation of cultured cells for EM ultrastructural analysis was described previously.24 Briefly, BCP-1 cells stimulated for 96 hr with extracts from C. dependens, E. coccinea, C. polystachyus and D. fragrans as well as 0.3 mM sodium butyrate (about 1 × 106/ ml) were centrifuged at 100g, supernatant aspirated and gently overlaid with 2% glutaraldehyde in cacodylate buffer (0.1 M, pH. 7.4) followed by post fixation in 1% osmium in same buffer. The cell pellet was stained en bloc in 0.5% uranyl acetate in acetate buffer (0.1 M, pH 4.0), dehydrated in graded ethanol and propylene oxide and infiltrated in an equal volume of epoxy resin and propylene oxide overnight. The sample was embedded in epoxy resin and cured for 48 hr in 60°C The cured block was thin-sectioned, stained in uranyl acetate and lead citrate and examined with an EM equipped with a digital camera and operated at 75 kV.
Results
A total of 4,842 extracts from 38 countries were tested for their ability to induce KSHV reactivation in BCP-1 cells. Most were from land plants but extracts from 86 marine plants, 97 marine animals and 87 species of fungi were included. Usable results were obtained from 3,703 extracts. A total of 2,958 extracts were inactive, 561 reduced levels of spontaneous KSHV replication and 184 (5.0%) induced viral reactivation above the level of sodium butyrate, which is the most potent chemical inducer of KSHV reactivation. The degree of viral reactivation by natural products was significantly higher than that obtained by host biological response modifiers such as interferon-gamma.25,26
More than half (2,477) of the extracts with usable results were from African land plants and 126 (5.1%) of these were activators, compared with 58 (4.7%) from other or unknown geographical regions (Table I). Activators included 3 marine plants, 1 marine animal and 1 fungal extract.
TABLE I.
Natural Products Causing KSHV Reactivation by Geographical Region
| Region | Total products tested |
Inactive (%) | Activators (%) | Inhibitors (%) |
|---|---|---|---|---|
| Africa | 2,477 | 2,010 (81.1) | 126 (5.1) | 341 (13.8) |
| Non-Africa and unknown: | 1226 | 948 (77.3) | 58 (4.7) | 220 (17.9) |
| Asia and Europe | 40 | 33 (82.5) | 0 | 7 (17.5) |
| Central America and Caribbean | 245 | 193 (78.7) | 14 (5.6) | 38 (15.5) |
| South America | 284 | 210 (73.9) | 9 (3.2) | 65 (22.9) |
| Philippines and Indonesia | 415 | 311 (74.9) | 30 (7.2) | 74 (17.8) |
| Unknown | 242 | 201 (83.4) | 5 (2.1) | 36 (14.9) |
Using likelihood ratio χ2 tests, African plants were more likely to cause reactivation than non-African plants (p = 0.024: likelihood ratio χ2 = 7.442, 2 df). Of the activating extracts, the magnitude of activation was greater for those from Africa (log10 mean = 8.41) compared with those from elsewhere (log10 mean = 8.21, p = 0.044, t-test). These associations were only weakly statistically significant. There were no activators from Europe or Asia, although only a small number of extracts from these regions were tested. Somewhat surprisingly, the proportion of extracts from Central America and the Caribbean and from The Philippines and Indonesia causing reactivation was higher than for African plants (5.6% and 7.2%, respectively vs. 5.1%), although the numbers of extracts were relatively small.
Viral activators belonged to a wide variety of plant families (Table II), although consistent with sampling based only on geography; for many families only a few extracts were tested. For example, 2 out of 3 samples from the Hypericaceae and 3 out of 7 samples from the Pittosporaceae were classified as activators. Between 7 and 15% of the extracts from the Acanthaceae, Boraginaceae, Celastraceae, Leguminosae, Rutaceae and Solanaceae were classified as activators compared with 5% or less for other families. The 28 natural products that had the highest reactivation levels in the screening experiment are listed in Table III, along with the family to which they belong, country of origin and the induced KSHV level expressed as log10 KSHV copies per 105 cells.
TABLE II.
Plant Families from which 5% or More Extracts Produced KSHV Reactivation
| Plant family | Number of extracts screened |
Activators (%) |
|---|---|---|
| Acanthaceae | 27 | 4 (14.8) |
| Apocynaceae | 121 | 6 (5.0) |
| Asteraceae | 79 | 4 (5.0) |
| Boraginaceae | 24 | 2 (8.3) |
| Caesalpiniaceae | 163 | 8 (4.9) |
| Celastraceae | 57 | 4 (7.0) |
| Hypericaceae | 3 | 2 (67.0.) |
| Leguminosae | 31 | 3 (9.7) |
| Mimosaceae | 66 | 3 (5.0) |
| Moraceae | 109 | 6 (5.5) |
| Pittosporaceae | 7 | 3 (43.0) |
| Rutaceae | 42 | 4 (9.5) |
| Solanaceae | 32 | 3 (9.4) |
TABLE III.
Name, Family and Country of Origin of Natural Products from which Aqueous Extracts Produced the Highest Levels of KSHV Reactivation in Initial Screen
| Name | Family | Country of origin | Log copies per 105 cells |
|---|---|---|---|
| Chasmanthera dependens | Menispermaceae | Tanzania | 10.36 |
| Lepistemon owariensis | Convolvulaceae | Tanzania | 10.32 |
| Chenopodium fasciculosum | Chenopodiaceae | Tanzania | 10.08 |
| Acacia amythethophylla | Mimosaceae | Tanzania | 10.00 |
| Vepris simplicifolia | Rutaceae | Tanzania | 9.96 |
| Dalbergia ecastaphyllum | Fabaceae | Ghana | 9.94 |
| Not identified1 | Leguminoseae | Papua New Guinea | 9.88 |
| Pittosporum verticillatum | Pittosporaceae | Madagascar | 9.83 |
| Dracaena fragrans | Agavaceae | Tanzania | 9.74 |
| Cleistanthus polystachyus | Euphorbiaceae | Cameroon | 9.71 |
| Astripomoea grantii | Convolvulaceae | Tanzania | 9.70 |
| Not identified2 | Not Identified | Philippine Islands | 9.57 |
| Eugenia soyauxii | Myrtaceae | Gabon | 9.51 |
| Psorospermum febrifugum | Hyperiaceae | Tanzania | 9.51 |
| Maesa lanceolata | Myrsinaceae | Tanzania | 9.45 |
| Emilia coccinea | Asteraceae | Tanzania | 9.30 |
| Erythrina dominguezii | Leguminosae | Bolivia | 9.30 |
| Salacia whytei | Celastraceae | Gabon | 9.28 |
| Salvadora persica | Salvadoraceae | Tanzania | 9.28 |
| Brucea antidysenterica | Simaroubaceae | Tanzania | 9.26 |
| Chlorophytum blepharophyllum | Liliaceae | Tanzania | 9.26 |
| Ekebergia benguelensis | Meliaceae | Tanzania | 9.23 |
| Cnestis ferruginea | Connaraceae | Cameroon | 9.20 |
| Stericulia tragacantha | Sterculiaceae | Gabon | 9.18 |
| Testulea gabonensis | Ochnaceae | Gabon | 9.15 |
| Datura stramonium | Solanaceae | Tanzania | 9.15 |
| Canavalia rosea | Fabaceae | Ghana | 9.04 |
| Aspilia bussei | Asteraceae | Ghana | 9.04 |
Unidentified legume.
Unidentified marine animal.
Apparent inhibition of KSHV replication was observed with 555 extracts (15%).These extracts included 525 land plants, 11 marine plants, 9 marine animals and 10 fungi. We considered it unlikely that these extracts were specifically inhibiting KSHV replication. More likely, the reduction in KSHV load that was observed with these extracts was probably caused by toxic effects of the extracts specific to BCP-1 cells, even though we had excluded extracts known to cause cytotoxicity in other cell lines. Some of these extracts may cause specific inhibition of KSHV replication but, since this was not the focus of this study, we did not attempt to characterize these further, with the exception of 3 extracts selected for the viral transcription array experiment.
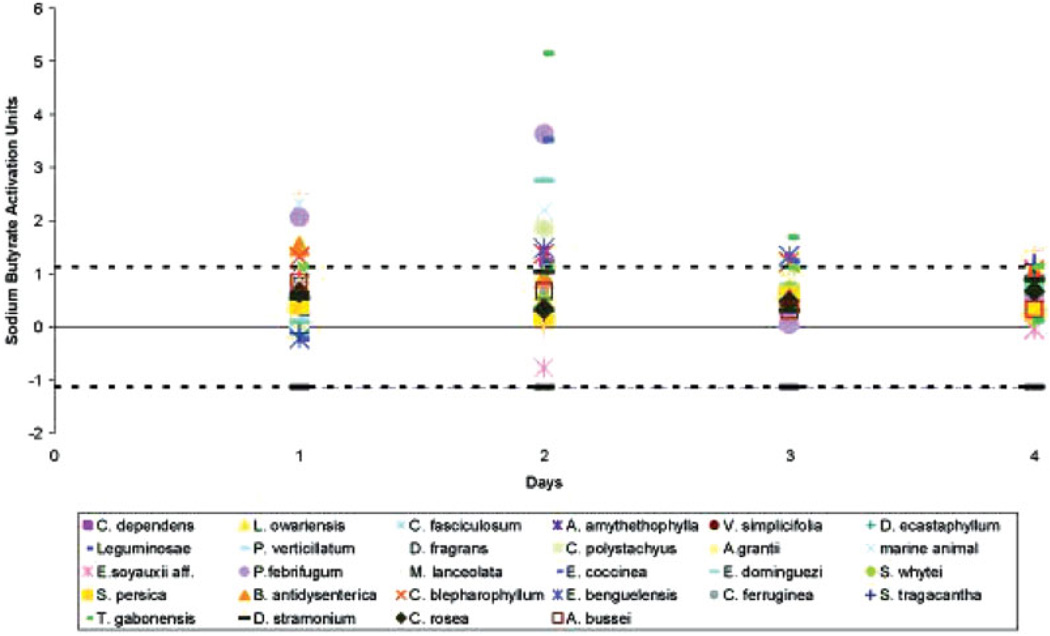
We further characterized the dynamics of reactivation by retesting the 28 most potent extracts from the initial screen in a 4-day time course experiment over 4 days. Using the K6 real-time quantitative PCR assay, we confirmed the initial screening results for 22 of the extracts by showing KSHV viral load increases over time (Fig. 1). To further elucidate the mechanism of reactivation, RNA was extracted from duplicate cell pellets for viral gene expression analysis using a previously reported viral array18 that queries the entire KSHV transcriptome by real time quantitative PCR. Four extracts causing high levels of reactivation, Dracaena fragrans, Chasmanthera dependens, Cleistanthus polystachyus and Emilia coccinea, were chosen for RNA analysis as well as 3 causing apparent inhibition: Uvaria angalensia, Keetia carmichailli and Garcinia species.
FIGURE 1.
Time course experiment with 28 extracts that caused the highest increases in KSHV viral load in the initial screen. The names and country of origin of the extracts are listed in Table III. The results are shown relative to the viral load observed in sodium butyrate induced cells. The standard deviation was calculated from the results of day 1 for all samples. Black dashed lines indicate one and a half standard deviations above and below the normalized average unstimulated control values for each day.
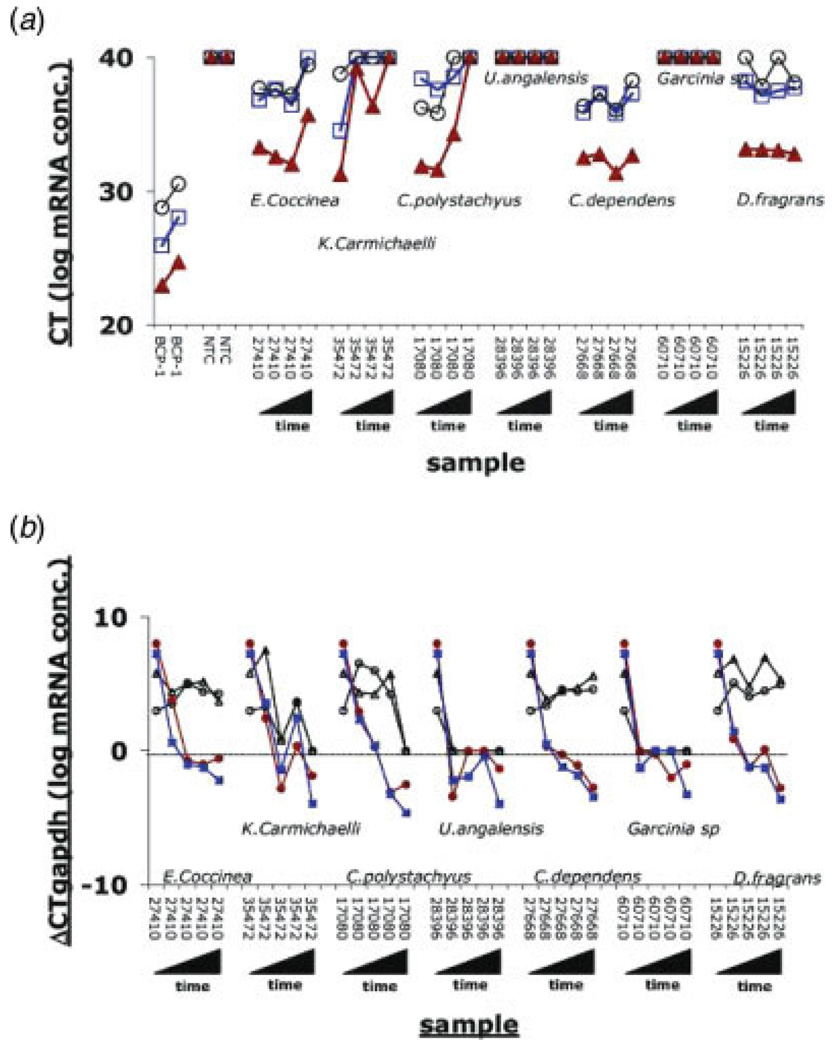
No mRNAs could be isolated from pellets in experiments using extracts, U. angolensia and Garcinia species, which caused apparent inhibition, consistent with our assumption that the natural product extract killed the cells early on. The other results fell into 2 categories; E. coccinea, C. dependens and D. fragrans caused only minimal changes in levels of 3 different cellular mRNAs over time, suggesting that the extract did not affect cell viability or basic cellular transcription (Fig. 2a). In contrast, extracts from K. carmichailli and C. polystachyus caused a rapid decrease in cellular mRNA levels between 24 and 48 hr post exposure, suggesting a deleterious effect on viability or down regulation of basic cellular transcription. On the basis if this result, we normalized all data to the level of gapdh for each sample at each time point.
FIGURE 2.
(a) Raw CT values for three cellular mRNAs, namely actin (blue squares), gapdh (red triangles) and c-myc (black circles). CT values are depicted on the vertical axis and correspond to the level of mRNA on a logarithmic scale. Higher CT values indicate lower mRNA levels. Hence, a CT = 40, for the negative nontemplate control (NTC) lanes indicated no amplification. We used BCP-1 cells as a positive control. As expected, gapdh mRNA is more abundant than actin, which in turn is more abundant than c-myc mRNA, as indicated by their relative CT values. Moreover, a 1:2 dilution of the input cDNA pool resulted in the same, reproducible and significant increase in CT for each mRNA, affirming the quantitative nature of our analysis. (b) dCT values for 2 KSHV mRNAs, K14 (red circles) and ORF57 (blue squares) and 2 cellular mRNAs, actin (black open circles) and c-myc (black triangles) at days 0, 1, 2, 3 and 4 after exposure to natural product extracts. Lower CT values correspond to higher mRNA levels on a log 2 scale, and 0 indicates mRNA levels equivalent to gapdh mRNA.
To determine whether the extracts were specifically causing KSHV reactivation, we first quantified the levels of 2 early viral mRNAs: K14 and ORF57 (Fig. 2b). All 3 of the extracts causing apparent decrease of KSHV replication exhibited significant signs of cellular toxicity. We were therefore unable to demonstrate any specific effects on KSHV replication by these extracts. In contrast, for extracts causing activation, E. coccinea, C. polystachyus, C. dependens and D. fragrans, the 2 viral mRNAs showed a different pattern than the cellular mRNAs. Levels for K14 and ORF57 increased over time up to and beyond the level of gapdh, which is indicative of KSHV lytic reactivation. By comparison, the 2 cellular mRNAs, c-myc and actin showed little or no change, indicating that the effect was specific for viral transcripts. Table IV shows fold induction relative to mock treated cells for selected mRNAs, representing the different kinetic classes of genes. All classes were induced relative to mock treatment.
TABLE IV.
Maximum Fold Induction by Natural Product Extracts for Selected mRNAS
| Gene | E. coccinea | C. polystachyus | C. dependens | D. fragrans |
|---|---|---|---|---|
| ORF 26 | 214 | 614 | 586 | 658 |
| ORF 29 | 740 | 2602 | 1231 | 1155 |
| ORF 50 | 19 | 127 | 25 | 36 |
| ORF 57 | 103 | 294 | 332 | 363 |
| K8.1 | 61 | 63 | 103 | 93 |
| c-myc | 2 | 2 | 2 | 1.8 |
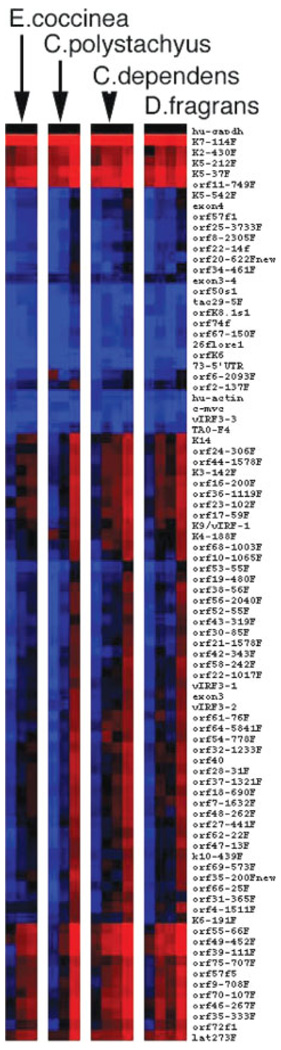
To evaluate the effects of E. coccinea, C. polystachyus, C. dependens and D. fragrans extracts on all KSHV mRNAs, we performed cluster analysis of the viral array data as shown in Figure 3. Most KSHV mRNAs were induced upon exposure to the natural products and with similar kinetics, as shown by the gradual shift from blue (low levels) to black (median level) to red (high levels) over time. This suggested that the transcriptional changes were the result of an ordered cascade of lytic reactivation rather than activation of random viral promoters by the extract. The RNA transcription data indicate that the increases in viral load observed with our real time PCR assay are a consequence of specific viral reactivation as a result of exposure to the natural product extracts.
FIGURE 3.
Heatmap representation of hierarchical clustering of dCT values for 73 KSHV mRNAs. Gapdh mRNA is present at an intermediate level, indicated by the black color. High abundance mRNAs are in red. The topmost group of viral mRNAs was present at high levels (indicated by red color in each sample) and changed very rapidly and significantly. A second group changed from low abundance (blue) to medium abundance (black). The third group of KSHV mRNAs increased slowly over time and only at the last time point exhibited levels above the median (red). In this depiction, black indicates equivalency to the level of gapdh, because individual genes are not normalized, which preserves information on the relative levels of each mRNA to one another. Primer pairs that amplified splice site specific mRNAs were excluded because the relative PCR efficiency was not comparable.
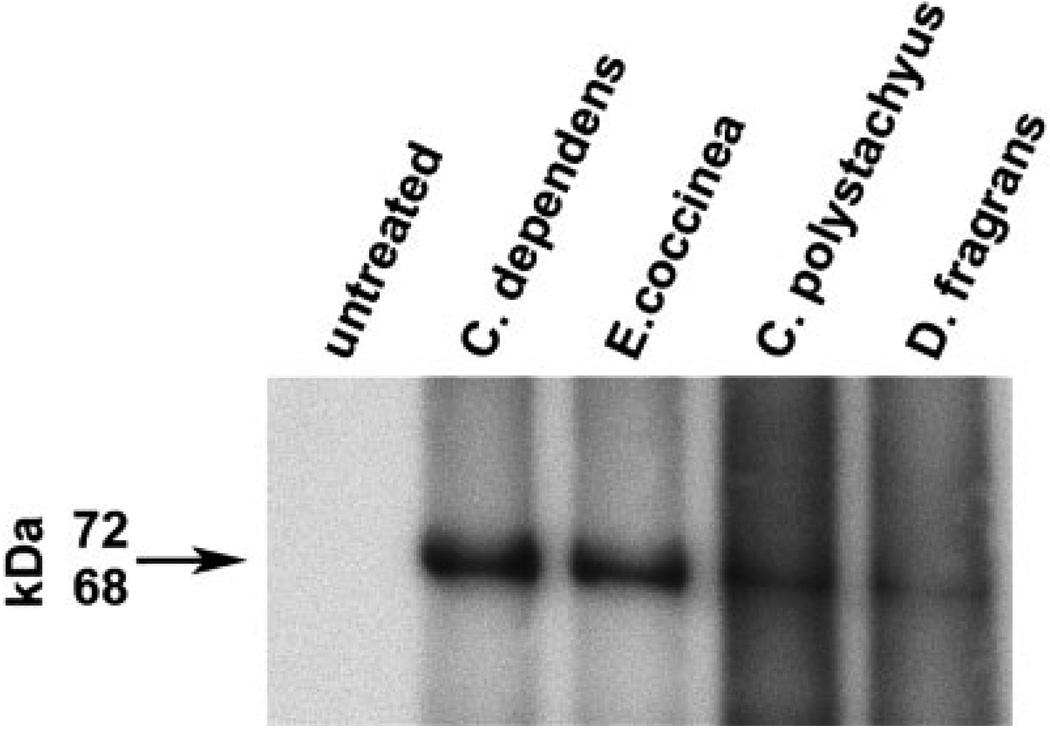
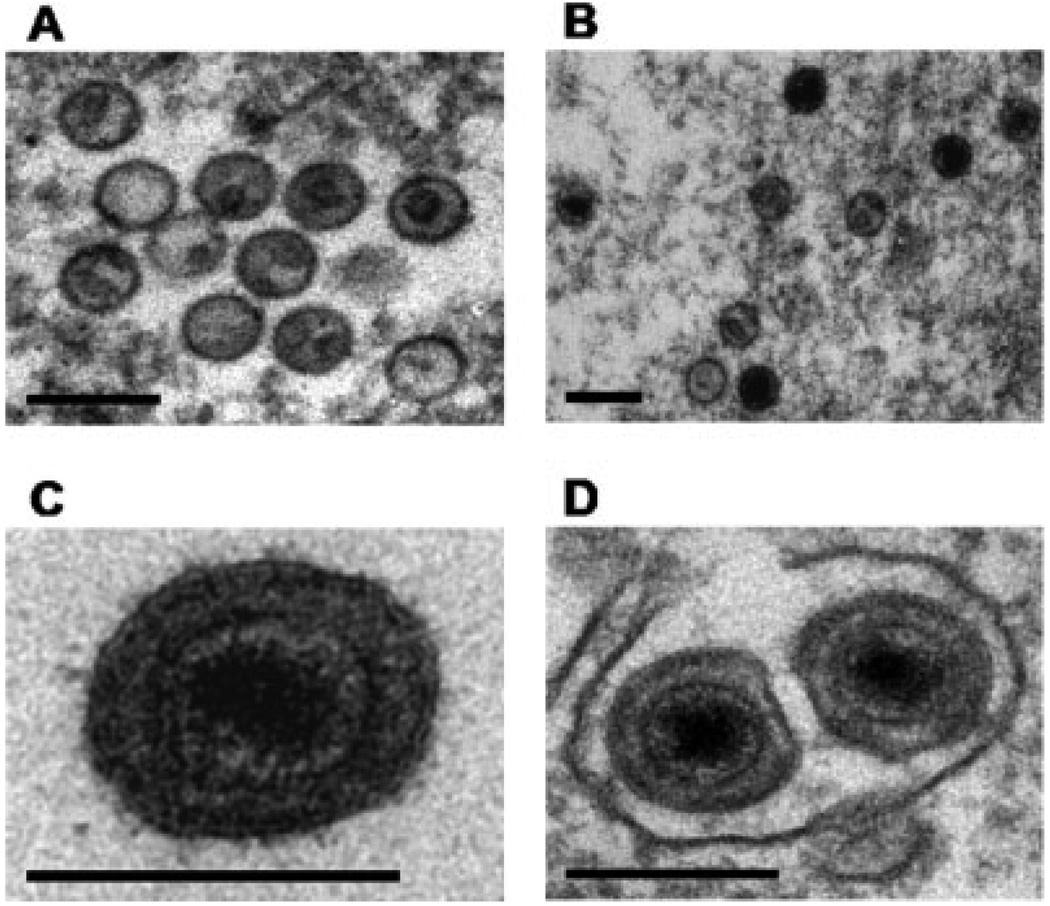
To determine whether viral reactivation by natural products resulted in virion production, we filtered and concentrated supernatants from untreated BCP-1 cells and cells treated with C. dependens, E. coccinea, C. polystachyus and D. fragrans for 96 hr. Immunoblotting of the KSHV late-lytic viral protein K8.1 A/B detected the expected 68–72 kDa broad band of virion-associated glycoprotein27 in samples from cells exposed to the 4 extracts, suggesting natural product reactivation induced a full lytic cycle of the virus (Fig. 4).K8.1 was not detected in the supernatant from the untreated BCP-1 cells. Cells from this experiment were processed for electron microscopy. Immature and mature virions were present in cells treated with all 4 extracts as well as the positive control. Representative virions are shown in Figure 5.
FIGURE 4.
KSHV virion production in supernatant from BCP-1 cells stimulated with extracts from C. dependens, E. coccinea, C. polystachyus and D. fragrans. Concentrated cell supernatants from untreated BCP-1 cells and extract exposed cells were run on a denaturing polyacrylamide gel and the proteins were transferred to a PVDF membrane. Immunoblotting for reactivity of the late lytic glycoprotein K8.1A/B demonstrated KSHV virion production in the cells treated with all 4 extracts but not in the untreated BCP-1 cells.
FIGURE 5.
Electron microscopy of BCP-1 cells stimulated with extracts from C. dependens, E. coccinea, C. polystachyus and D. fragrans demonstrated the production of typical herpesvirus virions. Immature and mature particles were observed in cells treated with all 4 extracts as well as the positive control (sodium butyrate). Panels A and B show immature particles assembled in the nucleus. Panel C shows mature particles in a cytoplasmic vacuole. Panel D shows an extracellular mature particle. The scale bar represents 200 nm.
Discussion
We have demonstrated that aqueous extracts of natural products collected from African countries known to have a high prevalence of KSHV infection and endemic KS can cause reactivation of KSHV in latently infected PEL cells. This supports the “oncoweed” hypothesis by providing a biological mechanism that may help to explain geographic variations in KSHV reactivation rates. Higher reactivation rates may explain higher prevalence, viral loads and transmission frequency in KS endemic regions.
Some caution must be applied to the interpretation of these data. The plants in the NCI repository were not collected to be representative of the flora of specific geographical areas but were selected for possibly having therapeutic potential against common cancers. Few extracts were available from regions with very low KSHV prevalence. The percentage of extracts inducing activation was only slightly higher if they were collected from African versus non African countries. In addition, a surprisingly high proportion of extracts from Central American and Caribbean plants, as well as plant extracts from The Philippines and Indonesia caused KSHV reactivation. These results warrant further study. However, among the activating extracts, the level of viral reactivation by African extracts was significantly higher than that from non-African extracts. This is evident from Table III which shows that 25 of the 28 most potent extracts were from Africa. The viral loads after reactivation with natural products exceeded those obtained after reactivation with known chemical inducers and host response modifiers. This supports the “oncoweed” hypothesis, since highly active extracts are more likely to have a biological effect on exposed subjects.
The plants causing activation included many that are used in traditional medicines, for basket making or as food. For example, C. dependens, P. febrifugum, M. lanceolota, E. coccinacea and B. antidysenterica are all used in traditional medicines. S. persica twigs are used for teeth cleaning and V. simplicifolia is used to make wooden bowls and spoons. Consequently, exposure of KSHV infected individuals to the active ingredients contained in these plants is plausible. Wojcicki recently described traditional practices associated with saliva exchange in sub-Saharan Africa. Many of these involved chewing of herbs by traditional healers and by mothers before a mixture of herbs and saliva were used for treatment.28 If the herbs used caused KSHV reactivation, this might be a very efficient route of KSHV transmission, since we know that viral shedding occurs in saliva.29–31
A role for environmental cofactors has been proposed in the etiology of endemic Burkitt’s lymphoma (BL), a monoclonal B cell malignancy of children that is caused by Epstein Barr virus and has a distinct geographical distribution in sub-Saharan Africa. One well defined cofactor is holoendemic malaria but it has also been proposed that exposure to sap from the plant Euphorbia tirucalli, a member of the Euphorbiaceae family may be an additional environmental cofactor.32–35 Laboratory studies have shown that E. tirucalli extracts or latex enhance EBV induced transformation,36 reduce Epstein-Barr virus (EBV) specific cellular immunity37 and cause reactivation of EBV lytic replication.35 These studies demonstrate the biological plausibility of a role for E. tirucalli as a cofactor in endemic Burkitt’s lymphoma, although definitive epidemiological studies have yet to be reported. There are additional reports of similar activities in related Euphorbiaceae extracts36,38 and in extracts from the unrelated Aleurites fordii,39 indicating that additional environmental factors may be important in EBV pathology. E. tirucalli, related Euphorbiaceae and Aleurites fordii are known to contain high levels of phorbol esters which are known inducers of EBV lytic reactivation.
There is considerable overlap between the regions of sub-Saharan Africa known to be endemic for KS and those where endemic BL occurs, although there are also important differences in the local distribution of the 2 diseases.2 We did not investigate reactivation of EBV in this study but it is plausible that some natural products may exert effects on EBV as well as KSHV. Of interest, C. polystachyus, an extract that caused high levels of KSHV reactivation in this study, is a member of the Euphorbiaceae. This family includes E. tirucalli, the plant studied as a potential cofactor for endemic BL. It is unlikely, however, that the biological activity of the extracts identified in this study was related to the presence of phorbol esters, since we used only aqueous extracts and phorbol esters would be found in organic extracts. Chemical fractionation of the most potent extracts to determine the active components is planned.
We conclude that environmental factors may play an important role in the epidemiology of KSHV. Such factors may be involved in enhancing transmission of KSHV in KS endemic areas and in accelerating progression to KS disease in KSHV latently infected individuals by increasing the viral reactivation frequency. Our extensive screening data provide support for this “oncoweed” hypothesis by demonstrating for the first time biological plausibility. Further epidemiological and laboratory studies are needed to fully understand the role of environmental cofactors in KSHV infection and disease.
Acknowledgements
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. This study was funded in part with federal funds from the National Institutes of Health to D.P.D. We thank Mr. Robert Shoemaker for helpful discussions, Mr. John Britt for help with natural products, Mr. Jason de la Cruz for help with electron microscopy, Mr. Doug Powell for help with statistics and Mr. Thomas Parks for help with figures.
Research funded by: National Cancer Institute, NIH; Contract number: NO1-CO-12400; Grant sponsor: National Institutes of Health; Grant number: CA109232; Grant sponsor: Intramural Research Program, NIH.
References
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Cook-Mozaffari P, Newton R, Beral V, Burkitt DP. The geographical distribution of Kaposi’s sarcoma and of lymphomas in Africa before the AIDS epidemic. Br J Cancer. 1998;78:1521–1528. doi: 10.1038/bjc.1998.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franceschi S, Geddes M. Epidemiology of classic Kaposi’s sarcoma, with special reference to mediterranean population. Tumori. 1995;81:308–314. doi: 10.1177/030089169508100502. [DOI] [PubMed] [Google Scholar]
- 4.Beral V, Peterman TA, Berkelman RL, Jaffe HW. Kaposi’s sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–128. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 5.Whitby D, Boshoff C. Kaposi’s sarcoma herpesvirus as a new paradigm for virus-induced oncogenesis. Curr Opin Oncol. 1998;10:405–412. doi: 10.1097/00001622-199809000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Whitby D, Luppi M, Barozzi P, Boshoff C, Weiss RA, Torelli G. Human herpesvirus 8 seroprevalence in blood donors and lymphoma patients from different regions of Italy. J Natl Cancer Inst. 1998;90:395–397. doi: 10.1093/jnci/90.5.395. [DOI] [PubMed] [Google Scholar]
- 7.Ariyoshi K, Schim van der Loeff M, Cook P, Whitby D, Corrah T, Jaffar S, Cham F, Sabally S, O’Donovan D, Weiss RA, Schulz TF, Whittle H. Kaposi’s sarcoma in the Gambia, West Africa is less frequent in human immunodeficiency virus type 2 than in human immunodeficiency virus type 1 infection despite a high prevalence of human herpesvirus 8. J Hum Virol. 1998;1:193–199. [PubMed] [Google Scholar]
- 8.Biggar RJ, Whitby D, Marshall V, Linhares AC, Black F. Human herpesvirus 8 in Brazilian Amerindians: a hyperendemic population with a new subtype. J Infect Dis. 2000;181:1562–1568. doi: 10.1086/315456. [DOI] [PubMed] [Google Scholar]
- 9.Whitby D, Marshall VA, Bagni RK, Wang CD, Gamache CJ, Guzman JR, Kron M, Ebbesen P, Biggar RJ. Genotypic characterization of Kaposi’s sarcoma-associated herpesvirus in asymptomatic infected subjects from isolated populations. J Gen Virol. 2004;85((Part 1)):155–163. doi: 10.1099/vir.0.19465-0. [DOI] [PubMed] [Google Scholar]
- 10.McGeoch DJ, Davison AJ. The descent of human herpesvirus 8. Semin Cancer Biol. 1999;9:201–209. doi: 10.1006/scbi.1999.0093. [DOI] [PubMed] [Google Scholar]
- 11.Engels EA, Clark E, Aledort LM, Goedert JJ, Whitby D. Kaposi’s sarcoma-associated herpesvirus infection in elderly Jews and non-Jews from New York City. Int J Epidemiol. 2002;31:946–950. doi: 10.1093/ije/31.5.946. [DOI] [PubMed] [Google Scholar]
- 12.Almuneef M, Nimjee S, Khoshnood K, Miller G, Rigsby MO. Prevalence of antibodies to human herpesvirus 8 (HHV-8) in Saudi Arabian patients with and without renal failure. Transplantation. 2001;71:1120–1124. doi: 10.1097/00007890-200104270-00019. [DOI] [PubMed] [Google Scholar]
- 13.Ablashi D, Chatlynne L, Cooper H, Thomas D, Yadav M, Norhanom AW, Chandana AK, Churdboonchart V, Kulpradist SA, Patnaik M, Liegmann K, Masood R, et al. Seroprevalence of human herpesvirus-8 (HHV-8) in countries of Southeast Asia compared to the USA, the Caribbean and Africa. Br J Cancer. 1999;81:893–897. doi: 10.1038/sj.bjc.6690782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith NA, Sabin CA, Gopal R, Bourboulia D, Labbet W, Boshoff C, Barlow D, Band B, Peters BS, de Ruiter A, Brown DW, Weiss RA, et al. Serologic evidence of human herpesvirus 8 transmission by homosexual but not heterosexual sex. J Infect Dis. 1999;180:600–606. doi: 10.1086/314926. [DOI] [PubMed] [Google Scholar]
- 15.Grundhoff A, Ganem D. Inefficient establishment of KSHV latency suggests an additional role for continued lytic replication in Kaposi sarcoma pathogenesis. J Clin Invest. 2004;113:124–136. doi: 10.1172/JCI200417803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engels EA, Biggar RJ, Marshall VA, Walters MA, Gamache CJ, Whitby D, Goedert JJ. Detection and quantification of Kaposi’s sarcoma-associated herpesvirus to predict AIDS-associated Kaposi’s sarcoma. AIDS. 2003;17:1847–1851. doi: 10.1097/00002030-200308150-00015. [DOI] [PubMed] [Google Scholar]
- 17.Sitas F, Carrara H, Beral V, Newton R, Reeves G, Bull D, Jentsch U, Pacella-Norman R, Bourboulia D, Whitby D, Boshoff C, Weiss R. Antibodies against human herpesvirus 8 in black South African patients with cancer. N Engl J Med. 1999;340:1863–1871. doi: 10.1056/NEJM199906173402403. [DOI] [PubMed] [Google Scholar]
- 18.Fakhari FD, Dittmer DP. Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J Virol. 2002;76:6213–6223. doi: 10.1128/JVI.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmer DP. Transcription profile of Kaposi’s sarcoma-associated herpesvirus in primary Kaposi’s sarcoma lesions as determined by realtime PCR arrays. Cancer Res. 2003;63:2010–2015. [PubMed] [Google Scholar]
- 20.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boshoff C, Gao SJ, Healy LE, Matthews S, Thomas AJ, Coignet L, Warnke RA, Strauchen JA, Matutes E, Kamel OW, Moore PS, Weiss RA, et al. Establishing a KSHV+ cell line (BCP-1) from peripheral blood and characterizing its growth in Nod/SCID mice. Blood. 1998;91:1671–1679. [PubMed] [Google Scholar]
- 22.de Sanjose S, Marshall V, Sola J, Palacio V, Almirall R, Goedert JJ, Bosch FX, Whitby D. Prevalence of Kaposi’s sarcoma-associated herpesvirus infection in sex workers and women from the general population in Spain. Int J Cancer. 2002;98:155–158. doi: 10.1002/ijc.10190. [DOI] [PubMed] [Google Scholar]
- 23.Yuan CC, Miley W, Waters D. A quantification of human cells using an ERV-3 real time PCR assay. J Virol Methods. 2001;91:109–117. doi: 10.1016/s0166-0934(00)00244-5. [DOI] [PubMed] [Google Scholar]
- 24.Gonda MA, Aaronson SA, Ellmore N, Zeve VH, Nagashima K. Ultrastructural studies of surface features of human normal and tumor cells in tissue culture by scanning and transmission electron microscopy. J Natl Cancer Inst. 1976;56:245–263. doi: 10.1093/jnci/56.2.245. [DOI] [PubMed] [Google Scholar]
- 25.Chang J, Renne R, Dittmer D, Ganem D. Inflammatory cytokines and the reactivation of Kaposi’s sarcoma- associated herpesvirus lytic replication. Virology. 2000;266:17–25. doi: 10.1006/viro.1999.0077. [DOI] [PubMed] [Google Scholar]
- 26.Zoeteweij JP, Eyes ST, Orenstein JM, Kawamura T, Wu L, Chandran B, Forghani B, Blauvelt A. Identification and rapid quantification of early-and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J Virol. 1999;73:5894–5902. doi: 10.1128/jvi.73.7.5894-5902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu L, Puri V, Chandran B. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology. 1999;262:237–249. doi: 10.1006/viro.1999.9900. [DOI] [PubMed] [Google Scholar]
- 28.Wojcicki JM. Traditional behavioural practices, the exchange of saliva and HHV-8 transmission in sub-Saharan African populations. Br J Cancer. 2003;89:2016–2017. doi: 10.1038/sj.bjc.6601390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mbulaiteye SM, Pfeiffer RM, Engels EA, Marshall V, Bakaki PM, Owor AM, Ndugwa CM, Katongole-Mbidde E, Goedert JJ, Biggar RJ, Whitby D. Detection of kaposi sarcoma-associated herpesvirus DNA in saliva and buffy-coat samples from children with sickle cell disease in Uganda. J Infect Dis. 2004;190:1382–1386. doi: 10.1086/424489. [DOI] [PubMed] [Google Scholar]
- 30.Vieira J, Huang ML, Koelle DM, Corey L. Transmissible Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi’s sarcoma. J Virol. 1997;71:7083–7087. doi: 10.1128/jvi.71.9.7083-7087.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster-Cyriaque J. Development of Kaposi’s sarcoma in a surgical wound. N Engl J Med. 2002;346:1207–1210. doi: 10.1056/NEJMoa012125. [DOI] [PubMed] [Google Scholar]
- 32.Mizuno F, Osato T, Imai S, Koizumi S, Aya T, Kinoshita T, Hirai N, Hirota M, Ohigashi H, Koshimizu K, et al. Epstein-Barr virus-enhancing plant promoters in east Africa. AIDS Res. 1986;2 Suppl. 1:S151–S155. [PubMed] [Google Scholar]
- 33.Osato T, Mizuno F, Imai S, Aya T, Koizumi S, Kinoshita T, Tokuda H, Ito Y, Hirai N, Hirota M, et al. African Burkitt’s lymphoma and an Epstein-Barr virus-enhancing plant Euphorbia tirucalli. Lancet. 1987;1:1257–1258. doi: 10.1016/s0140-6736(87)92700-0. [DOI] [PubMed] [Google Scholar]
- 34.van den Bosch C, Griffin BE, Kazembe P, Dziweni C, Kadzamira L. Are plant factors a missing link in the evolution of endemic Burkitt’s lymphoma? Br J Cancer. 1993;68:1232–1235. doi: 10.1038/bjc.1993.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacNeil A, Sumba OP, Lutzke ML, Moormann A, Rochford R. Activation of the Epstein-Barr virus lytic cycle by the latex of the plant Euphorbia tirucalli. Br J Cancer. 2003;88:1566–1569. doi: 10.1038/sj.bjc.6600929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mizuno F, Koizumi S, Osato T, Kokwaro JO, Ito Y. Chinese and African Euphorbiaceae plant extracts: markedly enhancing effect on Epstein-Barr virus-induced transformation. Cancer Lett. 1983;19:199–205. doi: 10.1016/0304-3835(83)90155-6. [DOI] [PubMed] [Google Scholar]
- 37.Imai S, Sugiura M, Mizuno F, Ohigashi H, Koshimizu K, Chiba S, Osato T. African Burkitt’s lymphoma: a plant, Euphorbia tirucalli, reduces Epstein-Barr virus-specific cellular immunity. Anticancer Res. 1994;14:933–936. [PubMed] [Google Scholar]
- 38.Ohigashi H, Koshimizu K, Tokuda H, Hiramatsu S, Jato J, Ito Y. Epstein-Barr virus-inducing activity of Euphorbiaceae plants commonly grown in Cameroon. Cancer Lett. 1985;28:135–141. doi: 10.1016/0304-3835(85)90068-0. [DOI] [PubMed] [Google Scholar]
- 39.Ito Y, Yanase S, Tokuda H, Kishishita M, Ohigashi H, Hirota M, Koshimizu K. Epstein-Barr virus activation by tung oil, extracts of Aleurites fordii and its diterpene ester 12-O-hexadecanoyl-16-hydroxy-phorbol-13-acetate. Cancer Lett. 1983;18:87–95. doi: 10.1016/0304-3835(83)90121-0. [DOI] [PubMed] [Google Scholar]