Abstract
In Bacillus megaterium, the synthesis of vitamin B12 (cobalamin) and sirohaem diverges at sirohydrochlorin along the branched modified tetrapyrrole biosynthetic pathway. This key intermediate is made by the action of SirC, a precorrin-2 dehydrogenase that requires NAD+ as a cofactor. The structure of SirC has now been solved by X-ray crystallography to 2.8 Å (1 Å = 0.1 nm) resolution. The protein is shown to consist of three domains and has a similar topology to the multifunctional sirohaem synthases Met8p and the N-terminal region of CysG, both of which catalyse not only the dehydrogenation of precorrin-2 but also the ferrochelation of sirohydrochlorin to give sirohaem. Guided by the structure, in the present study a number of active-site residues within SirC were investigated by site-directed mutagenesis. No active-site general base was identified, although surprisingly some of the resulting protein variants were found to have significantly enhanced catalytic activity. Unexpectedly, SirC was found to bind metal ions such as cobalt and copper, and to bind them in an identical fashion with that observed in Met8p. It is suggested that SirC may have evolved from a Met8p-like protein by loss of its chelatase activity. It is proposed that the ability of SirC to act as a single monofunctional enzyme, in conjunction with an independent chelatase, may provide greater control over the intermediate at this branchpoint in the synthesis of sirohaem and cobalamin.
Keywords: chelatase, cobalamin (vitamin B12), dehydrogenase, precorrin-2, sirohaem, sirohydrochlorin
INTRODUCTION
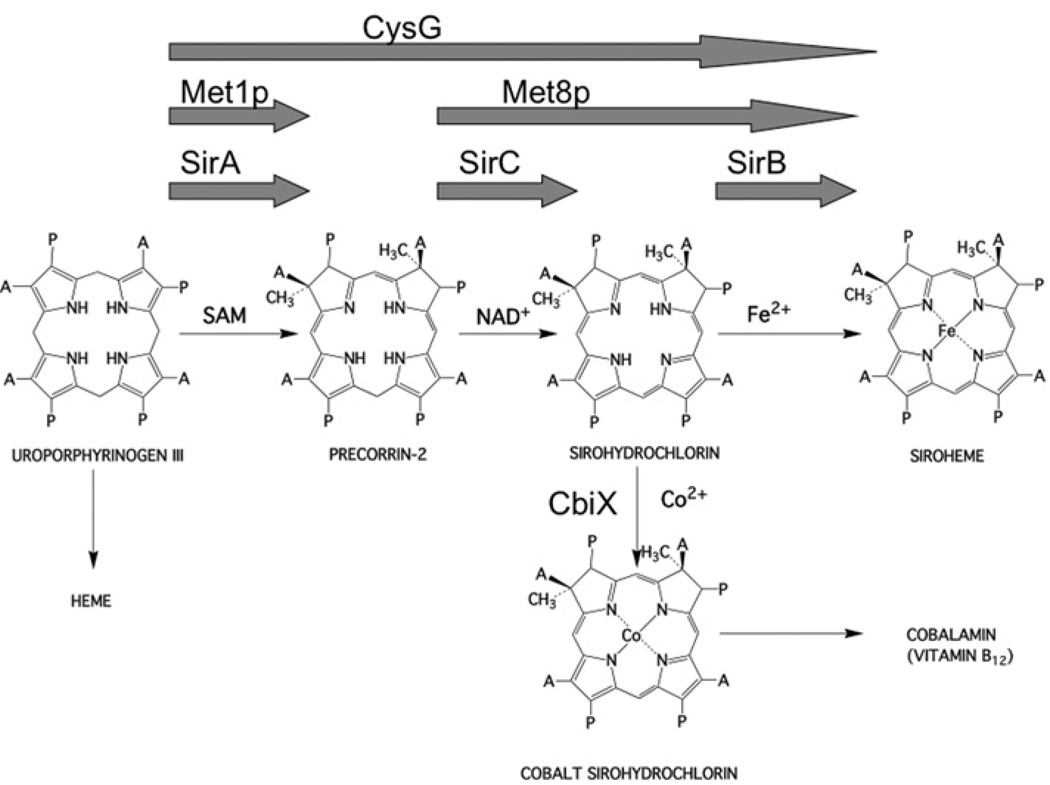
Sirohaem is a modified tetrapyrrole that is required as a prosthetic group in the six-electron reduction of both sulfite and nitrite [1–4]. As with all modified tetrapyrroles, sirohaem is synthesized via a branched biosynthetic pathway that also oversees the construction of molecules such as haem and cobalamin (vitamin B12) [5,6]. The first macrocyclic and branchpoint precursor in the pathway is uroporphyrinogen III. In the Bacilli, sirohaem is synthesized from uroporphyrinogen III in three steps (Figure 1) [7,8]. This transformation is initiated by the addition of two SAM (S-adenosyl-l-methionine)-derived methyl groups to positions 2 and 7 of the uroporphyrinogen III framework in a reaction catalysed by SirA (S-adenosyl-l-methionine uroporphyrinogen III methyltransferase; also known as SUMT, EC 2.1.1.107) to generate precorrin-2 [8]. Subsequently, this highly unstable dipyrrocorphin [9] is dehydrogenated by SirC (precorrin-2 dehydrogenase; EC 1.3.1.76) in an NAD+-dependent process to yield sirohydrochlorin [8]. Finally, ferrochelation by SirB (sirohydrochlorin ferrochelatase; EC 4.99.1.4) yields the end product sirohaem [8], which is duly incorporated into either the appropriate assimilatory sulfite or nitrite reductase.
Figure 1. Transformation of uroporphyrinogen III into sirohaem and cobalamin.
For sirohaem synthesis, three steps are involved. First, uroporphyrinogen III is methylated at positions 2 and 7 to yield precorrin-2, then this is oxidized to give sirohydrochlorin and finally iron insertion yields sirohaem. These steps are either catalysed by individual enzymes (e.g. SirA, SirB and SirC, as depicted by the three arrows), or by two enzymes (e.g. Met1p and Met8p, as depicted by the two arrows) or by a single multifunctional enzyme (e.g. CysG, the one large arrow). In the structures A=acetate side chain and P=propionate side chain.
Sirohydrochlorin also acts as an intermediate in cobalamin biosynthesis, where cobalt insertion by enzymes such as CbiK [10,11] or CbiX [5,12] ensures that the intermediate is further manipulated along the anaerobic cobalamin biosynthesis branch of the pathway (Figure 1) [5]. In this respect, SirC plays an important role in both sirohaem and cobalamin biosynthesis [8].
In Saccharomyces cerevisiae, the transformation of uroporphyrinogen III into sirohaem requires just two enzymes, a methyltransferase (Metlp) for the synthesis of precorrin-2, and a bifunctional dehydrogenase/ferrochelatase (Met8p) (Figure 1) [13,14]. The sequence of the bifunctional Met8p displays similarity to the monofunctional SirC and thus Met8p would appear to be a dehydrogenase that has acquired chelatase activity [8]. Met8p has been crystallized and the structure of the enzyme solved to 2.2 Å (1 Å = 0.1 nm) resolution [15], revealing that the protein adopts a novel fold that bears no resemblance to the previously determined structures of cobalto- [10,16] or ferro-chelatases [17,18]. Analysis of mutant variants of Met8p suggests that the catalytic activities are accommodated within a single active site, and that Asp141 plays an essential role in both dehydrogenase and chelatase processes [15].
In some bacteria, the transformation of uroporphyrinogen III into sirohaem is mediated by a single multifunctional enzyme called CysG (Figure 1) [19,20]. In effect, CysG represents a fusion between a uroporphyrinogen III methyltransferase and a bifunctional dehydrogenase/ferrochelatase (such as Met8p). This interpretation has been confirmed by gene dissection and mutagenesis experiments that indicate that the N-terminal 200 amino acids of CysG house the dehydrogenase/ferrochelatase functionality, whereas the C-terminal 257 amino acids mediate the methyltransferase activity [21]. It has been assumed that the bifunctional catalytic activities found associated with Met8p and the N-terminal region of CysG have arisen by acquisition of metal binding and metal insertion activity into the preexisting framework of the dehydrogenase [8]. The previous structure elucidation of CysG also revealed that the enzyme is a phosphoprotein, where phosphorylation of Ser220 appears to control the activity of the enzyme [22]. A driving force for the appearance of the multifunctional enzymes could be to direct substrate channelling along a particular branch of the pathway. This would explain why Bacilli have separate enzymes for the sirohaem branch, since it allows sirohydrochlorin to be used not only for sirohaem synthesis but also for cobalamin production. In the present study we report on the structure and function of the Bacillus megaterium SirC, an NAD+-dependent precorrin-2 dehydrogenase, and compare it with the structure of Met8p, which is both a dehydrogenase and sirohydrochlorin ferrochelatase. In contrast with the previous idea that Met8p may have arisen from a precorrin-2 dehydrogenase by acquisition of chelatase activity, we suggest that SirC is more likely to have evolved from a protein such as Met8p by loss of its chelatase activity.
EXPERIMENTAL
Chemicals and reagents
Chemicals and reagents were purchased from Sigma–Aldrich unless otherwise stated. Bacterial strains were purchased from Novagen, Invitrogen or Promega.
Protein purification
SirC was produced recombinantly in Escherichia coli as described previously [8]. Briefly, sirC was cloned into pET14b and transformed into BL21(DE3)pLysS(codon+) cells. Upon induction with 0.4 mM IPTG (isopropyl β-d-thiogalactoside), SirC was produced with a N-terminal His6 (hexa-histidine) tag. The protein was purified using a Ni2+-chelating column followed by dialysis into 20 mM sodium citrate (pH 6.5), 100 mM NaCl and 1 mM DTT (dithiothreitol) and size-exclusion chromatography. Met8p was purified as described previously [15].
Mutagenesis
The SirC mutants N98A, S101A, S101D, S102A, D105A and S124A were generated using the Stratagene site-directed mutagenesis QuikChange® kit following the manufacturer’s protocol.
Precorrin-2 dehydrogenase assay
The precorrin-2 dehydrogenase assay was performed as described previously [15]. Briefly, activity was monitored by incubating 2.5 µM precorrin-2 with 10 µg of SirC in a reaction volume of 1 ml with 7.5 mM NAD+ in 0.05 M Tris buffer (pH 8.0) containing 0.1 M NaCl. Sirohydrochlorin was monitored by the appearance of an absorption peak at 376 nm. Initial rates were recorded on a Hewlett Packard 8452A photodiode array spectrophotometer and assays were performed in triplicate.
Crystallization
The protein was concentrated to 8 mg/ml for crystallization trials. Crystals of SirC were grown using the vapour-diffusion method using a hanging drop containing 2 µl of protein (8–10 mg/ml) and 2 µl of well solution comprising 2.3 M (NH3)4SO4, 0.1 M Mes and 0.5–5 mM LiCl (pH 6.0). Potassium tetrachloroplatinate(II) and thimerisol derivatives were generated by soaking crystals for 5–10 min with 1 mM concentrations of metal compound in the well solution. All crystals were cryoprotected by elevating the solution around the crystal to 4 M (NH3)4SO4 plus buffer and salt. All data were collected on a copper rotating anode X-ray source using CuKα radiation at 1.54 Å (4.5 kW, 100 mA), and processed with the HKL2000 suite [23].
Structure determination
The structure was determined to 2.8 Å by multiple isomorphous replacement with anomalous scattering using K2PtCl4 and thimerisol derivatives [24] (Table 1). Solvent flattening (solvent content of 50%) in RESOLVE [25] and subsequent automated model building generated a starting model. The model building was finished using the programs O [26], Coot [27] and CCP4 [28]. Refinement in REFMAC [29] using TLS [30] restraints generated a final model with an Rfactor = 19.5% and Rfree = 26.3%.
Table 1. Data collection and refinement statistics.
Values in parentheses represent the highest-resolution shell. Rsym = 100 × Σ|I−|/Σ I, where is the average intensity from multiple observations of symmetry related reflections. Rcryst = 100 × Σ‖Fo|−|Fc‖/Σ|Fo| over 95% of the data. Rfree = Rcryst on 5% of the data not used in refinement.
| Space group H32 | Native | K2PtCl4 | Thimerisol |
|---|---|---|---|
| Unit cell (Å) | |||
| a=b= | 108.6 | 108.6 | 108.7 |
| c= | 201.03 | 200.7 | 199.9 |
| Number of observations | 192283 | 58434 | 175874 |
| Number of unique reflections | 20508 | 18841 | 30506 |
| Resolution range (Å) | 25–2.3 (2.38–2.3) | 20–2.9 (3.0–2.9) | 20–2.5 (2.59–2.5) |
| Completeness (%) | 99.4 (94.7) | 96.3 (99.2) | 99.7 (98.5) |
| I/σI | 35.6 (5.4) | 6.6 (1.2) | 23.3 (6.9) |
| Rsym | 6.2 (35.2) | 16.8 (42.7) | 7.2 (25.9) |
| Rcryst | 19.5 | ||
| Rfree | 26.3 | ||
| rmsd | |||
| Bonds (Å) | 0.013 | ||
| Angles (°) | 1.42 | ||
| Average B-factor (Å2) | |||
| Main chain | 43.9 | ||
| Side chain | 44.6 | ||
| Waters | 42.2 | ||
| Ligands:SO4 (7) GOL (1) | 57.9 |
EPR
To produce samples of SirC for EPR spectroscopy, the protein was purified by metal-chelate chromatography and cleaved by thrombin to remove the His6-tag. The protein was purified further by size-exclusion chromatography, as described above. The protein was taken into an anaerobic environment and exchanged into degassed buffer containing 50 mM Hepes (pH 8.0). The protein concentration was calculated as 47 µM on the basis of absorbance at 280 nm, from which EPR samples were generated containing 1:1 ratios of protein to Cu(II) or Co(II). The samples were sealed and frozen in liquid nitrogen. EPR spectra were obtained using a Bruker ELEXSYS E500/580 EPR spectrometer operating at X-band. Temperature control was effected using Oxford Instruments ESR900 and ESR935 cryostats interfaced with an ITC503 temperature controller. Experimental conditions were as given in the relevant Figure captions.
RESULTS AND DISCUSSION
Structure of SirC
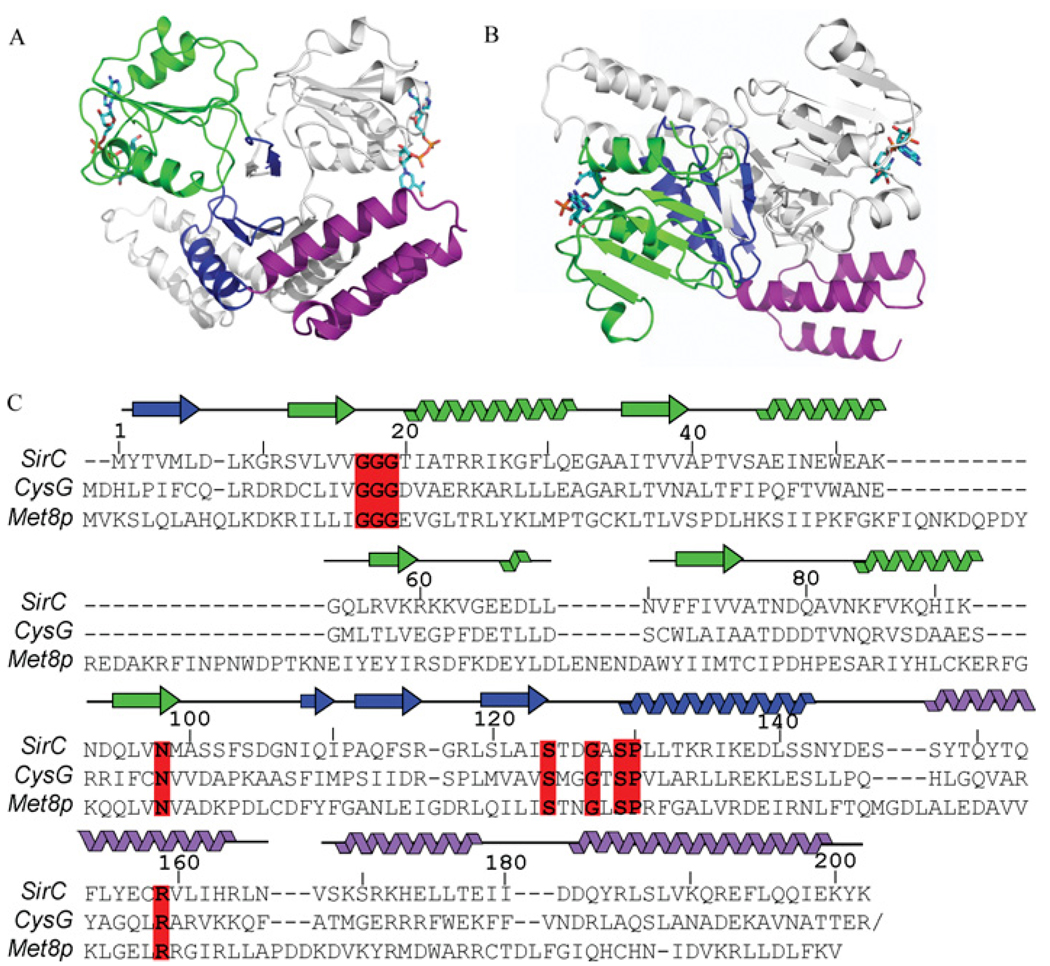
The crystal structure of His6-tagged Bacillus megaterium SirC was determined by isomorphous replacement with anomalous scattering, and refined to R/Rfree values of 19.5/26.3 with good geometry (Table 1). The SirC structure comprises three domains: an N-terminal NAD+-binding domain, a central dimerization domain and a C-terminal helical domain of unknown function (Figure 2). The structure resembles both the yeast sirohaem synthase, Met8p (PDB code: 1KYQ), with which it shares 11% identity, and the Salmonella enterica CysG (PDB code: 1PJS), with which it shares 25% sequence identity over the N-terminal region of the protein. SirC is the smallest of these three enzymes by virtue of its shorter loops and secondary structure elements. All three of the proteins are homodimers, in which the three domains cross each other in the form of an ‘X’ along a 2-fold axis of symmetry. The orientation of the paired NAD+-domains is almost perpendicular to that of the paired helical domains.
Figure 2. Structure of SirC.
(A) Front view of the SirC homodimer, with the three structural domains highlighted; NAD+-binding domain (green/light green), central dimerization domain (blue/light blue) and C-terminal helical domain (purple/light purple). (B) SirC homodimer viewed from the top looking down on the NAD+-binding domain with the C-terminal helical domains below. (C) Structure- and sequence-based alignment of SirC, Met8p and the N-terminal part of CysG. The SirC secondary structure is mapped to the sequence. Invariant residues, between these three sequences and a larger assortment of SirC precorrin-2 dehydrogenases, including Staphylococcus aureus (YP_042039.1), Geobacillus kaustophilus (YP_146257.1), Bacillus cereus (NP_977872.1), Bacillus anthracis (NP_843903.1), Staphylococcus epidermidis (NP_765732.1), Oceanobacillus iheyensis (NP_692578.1), Pelotomaculum thermopropionicum (GAA01341.1), Heliobacillus mobilis (ABH04863.1), Desulfotomaculum reducens (ZP_01148319.1), Bacillus halodurans (NP_242363.1), Bacillus licheniformis (YP_078957.1), delta proteobacterium MLMS-1 (ZP_01287446.1), Moorella thermoacetica (YP_430108.1) and Herpetosiphon aurantiacus (ZP_01424816.1) are boxed in red.
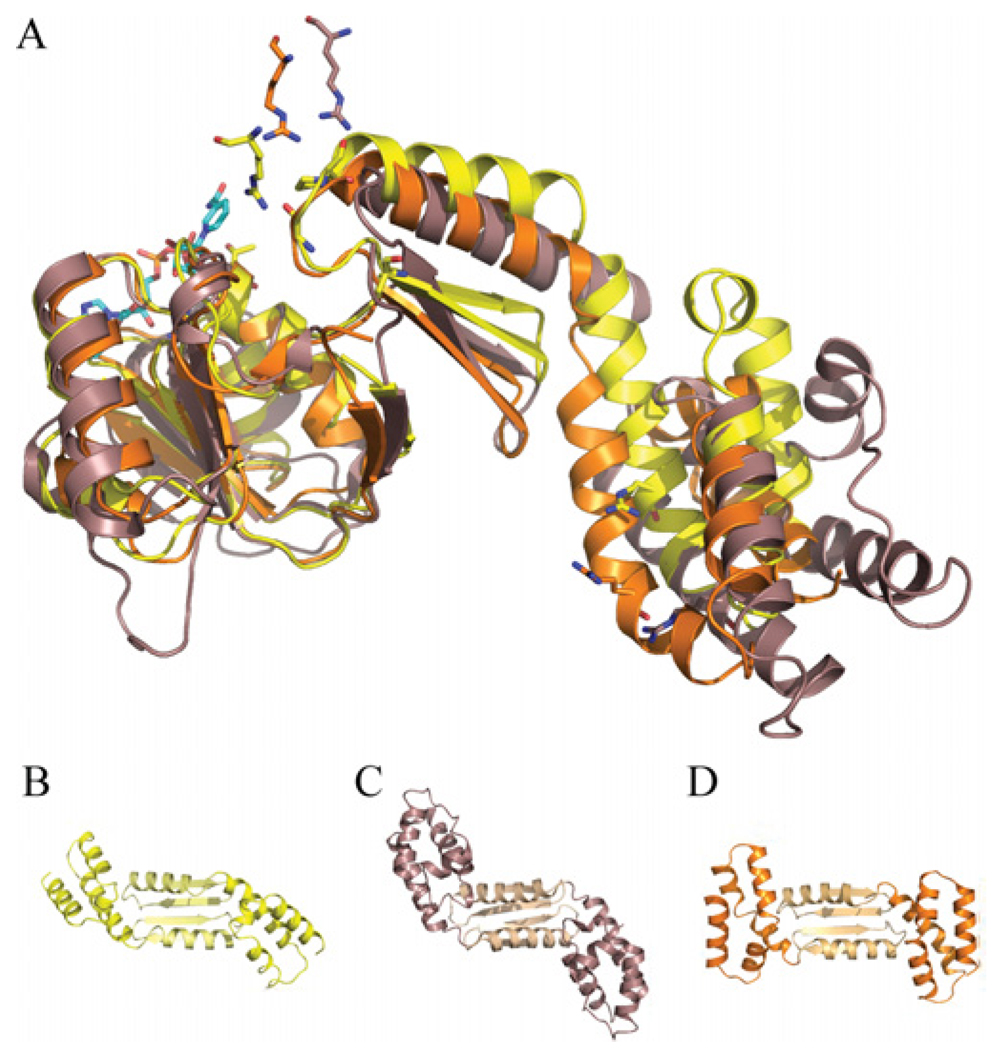
Optimized alignments between dimeric SirC, CysG and Met8p over their first ~190 residues gives rmsds (root mean square deviations) of 3.0–4.8 Å, with the two bacterial proteins aligning more closely with each other than with the yeast Met8p structure. If NAD+-binding domains alone are aligned, the rmsd between CysG and SirC is 1.1 Å over 100 Cα’s. When monomers from each structure are aligned on their NAD+-binding domains, a significant relative twist in the orientation of the C-terminal helical domains is revealed (Figure 3). The structural deviation begins immediately after the NAD+-binding domain where the β-strands in the dimerization domain twist with respect to the 2-fold axis of the homodimer but also a tilt away from the central core of the dimer.
Figure 3. Conformational variation in SirC homologues.
(A) Alignment of three structures (monomers) based on their NAD+-binding domains, SirC (yellow), Met8p (puce; purplish/brown), and CysG (orange). SirC active-site residues are shown in stick representation. The residues equivalent to SirC Arg159 appear above the active site because they are presented by the other molecule of the homodimer (not shown). Using this alignment, the C-terminal helical domains (dark colour) and central dimerization domains (light colour) are as viewed from the bottom of the structure. (B) SirC. (C) CysG. (D) Met8p.
Dehydrogenase active site
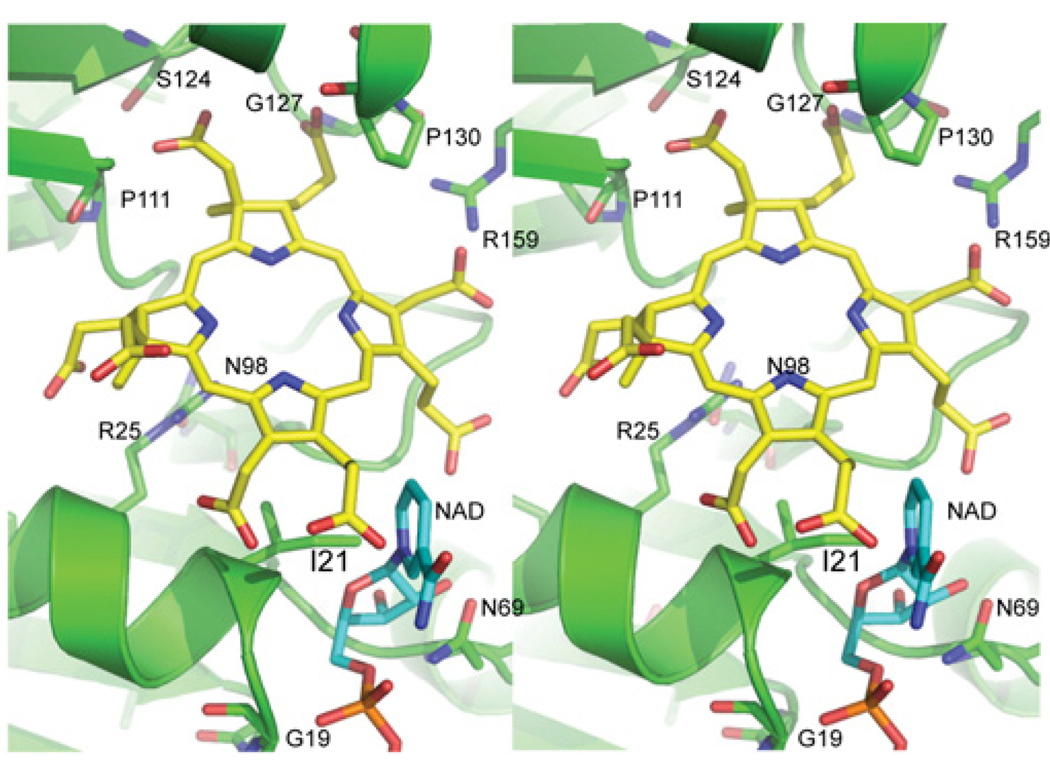
The NAD+-dependent dehydrogenase active site lies between the NAD+ domain and the dimerization domain. The NAD+-binding site is empty in the SirC structure, although the high level of similarity with other NAD+-binding domains [31] and with the NAD+-bound structures of Met8p and CysG facilitates the modelling of an NAD+ molecule to SirC (Figure 4). Canonical NAD+-binding domains contain two Rossmann folds where the adenosine portion of the NAD+ binds above the first β-strand, the phosphates sit on top of an invariant Gly–Gly–Gly sequence, and the nicotinamide portion of the coenzyme points toward the active-site cleft. The NAD+-bound structures of Met8p and CysG have poorly defined electron density for the nicotinamide portion of the ligand, which is seen to adopt a variety of conformations in other related structures. We are therefore reasonably confident about the model presented here for the SirC-bound NAD+ adenine and phosphates, but more tentative in our suggestion that the nicotinamide resembles that of the Met8p-bound NAD+ and forms an edge of the putative active-site cleft, rather than that of the CysG-NAD+ where the β-nicotinamide is located further away from the cleft.
Figure 4. Stereodiagram of the SirC active site and putative substrate binding location of precorrin-2 and β-nicotinamide-binding sites.
Conserved and invariant residues are highlighted. Arg159 is presented from the alternate monomer of the SirC homodimer. The location of the bound SO4 2− group in the SirC structure, and the phosphorylated threonine residue in the CysG structure is just above the C-ring near Gly127 and Ser124. β-Nicotinamide is shown in cyan and SirC is shown in green.
A comparison of precorrin-2 dehydrogenase primary sequences from 18 species, all with greater than 55% identity with B. megaterium SirC, provides a list of nine invariant residues (results not shown). Owing to the low overall sequence similarity, a combination of structure-based and amino acid sequence-based alignments is necessary to obtain an accurate alignment of SirC, Met8p and CysG (Figure 2C).
The active-site cleft contains all of the invariant residues. Gly17, Gly18 and Gly19 lie within the NAD+-binding pocket, whereas the other seven invariant residues are predicted to interact with the precorrin-2 substrate. Asn98 lies at the base of the binding pocket pointing up toward the open cleft (Figure 4). Ser124, Gly127, Ser129 and Pro130 (SxxGxSP) form a cluster of invariant residues on the opposite side of the active-site cleft from the NAD+, within the central dimerization domain. Finally, Arg159 resides on the C-terminal helical domain and contributes to the active site from the alternate molecule of the homodimer. Differences in the relative positions of the domains found in SirC, Met8p and the N-terminal region of CysG are clear (Figure 4), but the significance of these differences in terms of catalytic mechanism is unclear. Curiously, the location of the invariant Arg159 with respect to the active sites differs dramatically; it is clearly positioned within the active-site cleft of SirC (Figure 4), whereas in the Met8p and CysG structures it is found 8–11 Å away from the active site. Sequence similarity is quite low in the C-terminal helical domain and Arg159 is only identified as an invariant residue after using secondary structural alignments of the three proteins (Figure 2C).
Modelling of a precorrin-2 substrate and site-directed mutagenesis
We have built a plausible model of precorrin-2 bound to the active-site cleft between the β-nicotinamide moiety and the cluster of invariant residues (Figure 4). The bond between C-14 and C-15 is oxidized during the dehydrogenase reaction with a concomitant loss of the proton from the C-ring pyrrole N-3. The modelled precorrin-2 substrate places the C-15 atom nearest to the β-nicotinamide, while pointing the A- and B-rings toward the SxxGxSP residue cluster (Figure 4). In this position, the methylated A- and B-rings pack against conserved residues in the dimerization domain, and their acetate and propionate side chains extend further into the cleft, where they could form hydrogen bonds with the main-chain carbonyl oxygens and amide nitrogens of the invariant residues and the side chain of Ser124. Ser124 is located on the back of the active site among a number of other conserved residues, opposite to the NAD+-binding pocket. The equivalent residues in Met8p and CysG are largely glycine residues and proline residues, and so it had originally been proposed that these residues would be involved in domain motions or substrate binding [15]. An S124A variant was found to have 33% of the wild-type activity, supporting a role for Ser124 in substrate binding and maximizing enzyme activity (Table 2). In our apoenzyme crystal structure, Ser124 points away from the active site, although a minor conformational change upon substrate binding might induce it to face the active site.
Table 2. Activities of SirC and protein variants.
The values for specific dehydrogenase activity are means ± S.D.
| Enzyme form | Specific dehydrogenase activity (nmol/min per mg) | Percentage of wild-type activity |
|---|---|---|
| Wild-type | 20.98 ( ± 2.10) | 100 |
| N98A | 4.98 ( ± 1.39) | 23.7 |
| S101A | 30.10 ( ± 1.87) | 143.5 |
| S101D | 58.55 ( ± 7.42) | 279.1 |
| S102A | 48.77 ( ± 3.48) | 232.5 |
| D105A | 66.84 ( ± 7.43) | 318.6 |
| S124A | 6.92 ( ± 1.40) | 33.0 |
A side chain extending from position 127 would protrude into the cleft, suggesting that invariant Gly127 may have been selected not only to facilitate a turn in secondary structure, but also to permit tight interactions with the substrate. The hydroxy side chain of Ser129 points away from the active site and forms a hydrogen bond with the amide nitrogen of Ile132. This linkage may support the tight turn between strand-8 and helix-5. The adjacent Pro130 sits slightly above the B-ring of the tetrapyrrole-derived substrate and may contribute both a structural role in the initiation of helix-5 and hydrophobic interactions with the B-ring. Arg159 enters the active site near Pro130 and potentially contributes both hydrogen bonds and a compensatory positive charge to the precorrin-2 carboxylates, most likely to the C-ring acetate.
A phosphorylated serine residue was identified in the structure of CysG at position Ser128 [22]. This residue is equivalent to Thr125 in SirC and, intriguingly, SirC electron density that we interpret as a free sulfate group is adjacent to Thr125 at the same position as the phospho-group in CysG. A regulatory role for CysG phosphorylation was proposed based on the observation that mutation of Ser128 to alanine resulted in a variant that was fully active whereas mutation to aspartate was inhibitory [22]. The sulfate moiety found in the SirC structure is one of many sulfates modelled in the structure due to the high concentration of ammonium sulfate in the crystallization solution. There is no evidence for a covalent interaction as would be expected if the threonine residue was actually phosphorylated in our structure, and the hydrogen bond distance between a sulfate oxygen and the Thr125 hydroxy group is 2.6 Å. The presence of the ion in both structures suggests a readiness of this pocket on the surface of the protein to accept negatively charged ions such as the precorrin-2 carboxylates, and further substantiates a role for this pocket in binding the substrate.
Characterization of other active-site residues
The Met8p active site includes Asp141, which has been proposed to function as a general base for catalysis [15]. Interestingly, SirC displays a serine residue (Ser101) at the equivalent position, although there is a nearby aspartate residue (Asp105) displaced by ~6 Å from the corresponding Met8p site. These residues, and the intervening Ser102 residue, whose density is poorly defined, were investigated by mutagenesis. Surprisingly, mutation of either serine to alanine resulted in a 1.4- (S101A) and 2.3- (S102A) fold increase in activity (Table 2).
Guided by the Met8p structure and the importance of Met8p Asp141 for metal chelation [15], SirC Ser101 was changed to aspartate to see whether this generated a protein that had chelatase activity. Although this variant did not display any metal-inserting activity, the SirC S101D protein did show enhanced dehydrogenanse activity (Table 2). Interestingly, mutation of Asp105 to alanine also resulted in an increase in activity of SirC (Table 2), demonstrating that this residue does not have an essential role in catalysis and that SirC Asp105 is not equivalent to Met8p Asp141.
These studies indicate that the SirC active site does not house a conserved nucleophile that is used to help promote the dehydrogenation, and we favour a model in which catalysis results from the proximity of the tetrapyrrole-derived substrate with bound NAD+. Surprisingly, many of the mutant variants had increased catalytic activity in comparison with wild-type SirC. In part, this may be due to the instability of the wild-type enzyme, which has a tendency to aggregate over time. It may be that the various mutations enhanced the stability of the enzyme, permitting a higher apparent activity. Without an accurate active-site titrant it is not possible to make further comment. It would have been useful to have characterized many of the mutants made in this study further, especially with respect to Km and kcat values. However, the difficulty of making the precorrin-2 substrate and performing the assays under strict anaerobic conditions precluded the generation of such data.
Metal-binding studies of Met8p and SirC from B. megaterium
It is a logical assumption to make that the chelatase activity of Met8p is likely to have evolved by acquisition of metal-binding ability within a protein such as SirC. With this in mind we thought that it may be possible to identify how the metal ion that is required for ferrochelation activity is bound in Met8p by comparing it with SirC. However, neither sequence alignments nor crystal soaks indicated a conserved metal-binding site in Met8p. We thus thought that it may be possible to identify a metal-binding site in Met8p by EPR and distinguish some of the ligands involved.
Met8p was subject to EPR spectroscopy in the presence of either Co(II) or Cu(II) because it is known to possess cobaltochelatase activity during anaerobic cobalamin biosynthesis [14] and because Cu(II) can often substitute for Co(II) and has a more readily interpretable EPR spectrum. Co(II) and Cu(II) EPR spectra are sensitive reporters of the ion environment because the spin and orbital energies of the ion’s unpaired electrons are affected by the arrangement and nature of the atoms ligating the ion. SirC from B. megaterium was originally deployed as a negative control in these studies, but surprisingly it became apparent that SirC was also able to bind both Co(II) and Cu(II) in the same configuration as seen for Met8p.
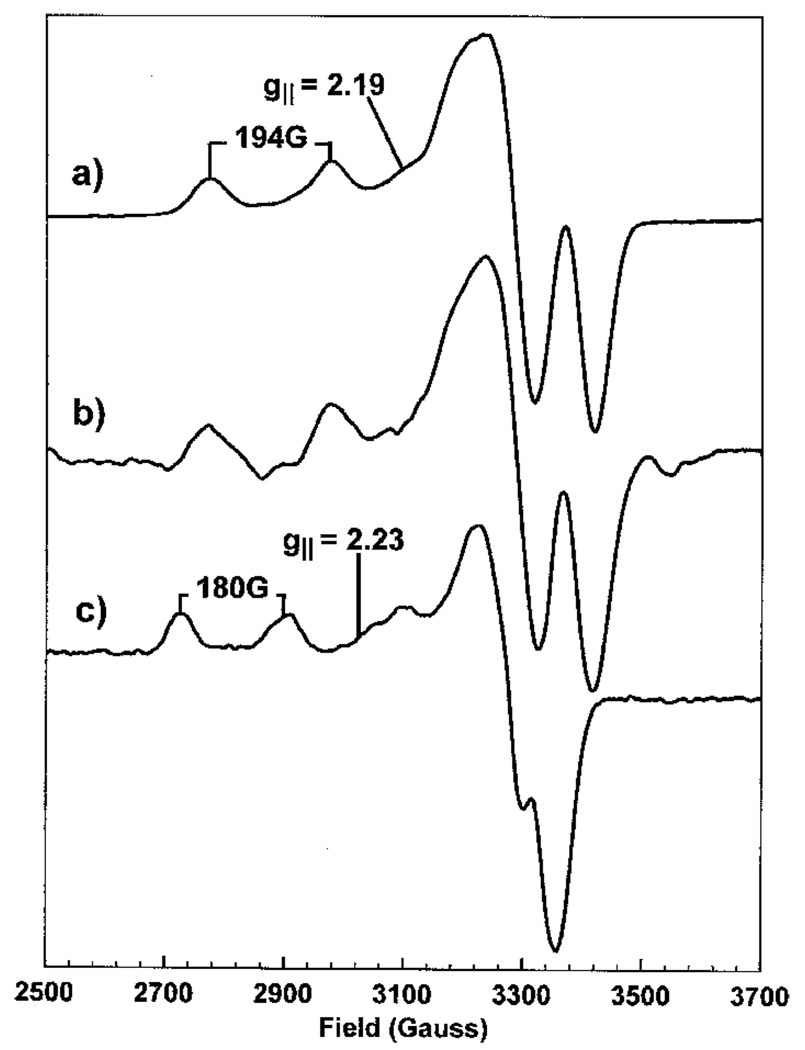
The Cu(II) EPR spectra (Figure 5) are clearly different in the presence (Figure 5a) and the absence (Figure 5c) of stoichiometric SirC. Cu(II) EPR spectra are axial in symmetry, having a large first-derivative g‖ feature to high field often overlapping four smaller g‖ features having absorption line shapes which arise from the hyperfine splitting (A‖) of the spectrum by the I = 3/2 Cu nuclear spin [32]. The single unpaired electron of the Cu(II) ion is located in the dz2 orbital. Therefore the aforementioned hyperfine splitting and the g‖ value are indicative of the ligand arrangement around the Cu(II) ion. For Cu(II) bound to SirC these values are g‖ = 2.19 and A‖ = 194 G. However, the Cu(II) ions in the experiment with buffer alone, g‖ = 2.23 and A‖ = 180 G, are clearly distinguishable from the SirC-bound form. In the presence of Met8p, again at a 1:1 protein to metal ion stoichiometry, the Cu(II) spectrum (Figure 5b) displays identical g‖ and A‖ values with those observed in the presence of SirC. These parameters can be interpreted based on the knowledge of the EPR parameters of Cu(II) complexes of known structure [33]. They suggest that the Cu(II)-binding site on SirC, and Met8p, consists of at least two nitrogen ligands, together with two oxygen ligands. The geometries of these ligating groups are identical in both SirC and Met8p.
Figure 5. X-band EPR spectra of Cu(II).
Cu(II) in a 1:1 stoichiometry with SirC (a), in a 1:1 stoichiometry with Met8p (b) and in buffer alone (c). Microwave power 0.5 mW, modulation amplitude 5 G for (a) and (c) and 8 G for (b), temperature 20 K.
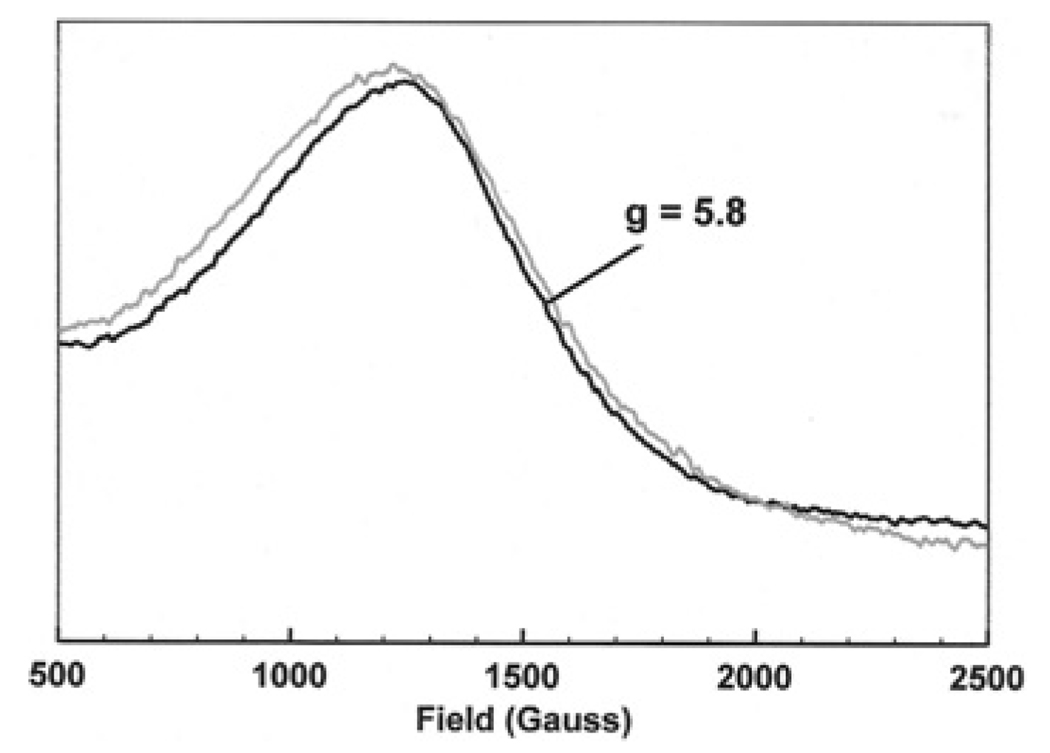
Co(II) ions in the high-spin state have three unpaired electrons (S = 3/2) and consequently give rise to a broad signal at a high g value (g = 5.8) with unresolved hyperfine splitting [32]. This makes such spectra difficult to analyse, but small qualitative differences are evident in linewidth and g value on binding Co(II) to SirC (Figure 6). This suggests that SirC is also able to bind to a metal ion that is a known substrate for Met8p, i.e. Co(II). An octahedral environment containing several strong ligands such as nitrogens would be expected to effect a spin-state change in the Co(II) ion, leading to pairing of some electrons and a large change in the spectrum (including a change in g value to below g = 3) [32]. Since Figure 6 shows no evidence of such a change, the CoII/CuII-binding site on SirC is probably tetrahedral or tetragonal and includes no more than two nitrogens.
Figure 6. X-band EPR spectra of Co(II) bound to SirC (black line) and free in buffer solution (grey line).
Microwave power 0.5 mW, modulation amplitude 5 G, temperature 20 K.
Both spectra illustrate that SirC binds metal ions with the same geometry and type of ligands as Met8p. It is not possible to establish which residues are responsible for metal binding using sequence and structural comparisons because there is insufficient sequence conservation and structural similarities. Thus, rather than Met8p being an enzyme that has acquired metal binding and chelation activity, it seems more likely that SirC is an enzyme that has lost its chelation activity.
In summary, we have solved the crystal structure of the B. megaterium precorrin-2 dehydrogenase (SirC) and shown that it has a very similar structure to that of Met8p and the N-terminal region of CysG. Mutagenesis studies have revealed that the enzyme does not appear to have a residue within the active site that acts as a general base during the catalytic cycle of the enzyme. Indeed, individually changing a number of residues generated enzymes that have significantly more activity than the wild-type enzyme. For biosynthetic enzymes involved in the synthesis of small quantities of natural products such as sirohaem and vitamin B12, there is no pressing reason for them to be highly active. Unexpectedly, SirC was found to bind metal ions such as Co(II) and Cu(II) in the same manner as Met8p, which is required to bind metal for its chelatase activity. This may reflect the evolutionary history of SirC as a bifunctional enzyme that has lost an earlier chelatase activity. The proposed loss of ferrochelation activity in SirC results in production of the sirohydrochlorin product, which can be acted upon by specific ferro- and cobalto-chelatases for transformation into either sirohaem or cobalamin.
Acknowledgments
The research was supported by a grant from the BBSRC (Biotechnology and Biological Sciences Research Council) and from NIH (National Institutes of Health) RO1 GM56775 and P30 DK072437.
Abbreviations used
- rmsd
root mean square deviation
- SAM
S-adenosyl-l-methionine
- SirA
S-adenosyl-l-methionine uroporphyrinogen III methyltransferase
- SirB
sirohydrochlorin ferrochelatase
- SirC
precorrin-2 dehydrogenase.
Footnotes
Protein co-ordinates and structure factor amplitudes have been submitted to the Protein Databank with the PDB code 3DFZ.
REFERENCES
- 1.Murphy MJ, Siegel LM. Siroheme and sirohydrochlorin. The basis for a new type of porphyrin-related prosthetic group common to both assimilatory and dissimilatory sulfite reductases. J. Biol. Chem. 1973;248:6911–6919. [PubMed] [Google Scholar]
- 2.Murphy MJ, Siegel LM, Tove SR, Kamin H. Siroheme: a new prosthetic group participating in six-electron reduction reactions catalyzed by both sulfite and nitrite reductases. Proc. Natl. Acad. Sci. U.S.A. 1974;71:612–616. doi: 10.1073/pnas.71.3.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vega JM, Garrett RH. Siroheme: a prosthetic group of the Neurospora crassa assimilatory nitrite reductase. J. Biol. Chem. 1975;250:7980–7989. [PubMed] [Google Scholar]
- 4.Crane BR, Getzoff ED. The relationship between structure and function for the sulfite reductases. Curr. Opin. Struct. Biol. 1996;6:744–756. doi: 10.1016/s0959-440x(96)80003-0. [DOI] [PubMed] [Google Scholar]
- 5.Leech HK, Raux-Deery E, Heathcote P, Warren MJ. Production of cobalamin and sirohaem in Bacillus megaterium: an investigation into the role of the branchpoint chelatases sirohydrochlorin ferrochelatase (SirB) and sirohydrochlorin cobalt chelatase (CbiX) Biochem. Soc. Trans. 2002;30:610–613. doi: 10.1042/bst0300610. [DOI] [PubMed] [Google Scholar]
- 6.Schubert HL, Raux E, Warren MJ, Wilson KS. Optimization of Met8p crystals through protein-storage buffer manipulation. Acta Crystallogr. D Biol. Crystallogr. 2001;57:867–869. doi: 10.1107/s0907444901004619. [DOI] [PubMed] [Google Scholar]
- 7.Johansson P, Hederstedt L. Organization of genes for tetrapyrrole biosynthesis in gram-positive bacteria. Microbiology. 1999;145:529–538. doi: 10.1099/13500872-145-3-529. [DOI] [PubMed] [Google Scholar]
- 8.Raux E, Leech HK, Beck R, Schubert HL, Santander PJ, Roessner CA, Scott AI, Martens JH, Jahn D, Thermes C, et al. Identification and functional analysis of enzymes required for precorrin-2 dehydrogenation and metal ion insertion in the biosynthesis of sirohaem and cobalamin in Bacillus megaterium. Biochem. J. 2003;370:505–516. doi: 10.1042/BJ20021443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren MJ, Stolowich NJ, Santander PJ, Roessner CA, Sowa BA, Scott AI. Enzymatic synthesis of dihydrosirohydrochlorin (precorrin-2) and of a novel pyrrocorphin by uroporphyrinogen III methylase. FEBS Lett. 1990;261:76–80. doi: 10.1016/0014-5793(90)80640-5. [DOI] [PubMed] [Google Scholar]
- 10.Schubert HL, Raux E, Wilson KS, Warren MJ. Common chelatase design in the branched tetrapyrrole pathways of heme and anaerobic cobalamin synthesis. Biochemistry. 1999;38:10660–10669. doi: 10.1021/bi9906773. [DOI] [PubMed] [Google Scholar]
- 11.Raux E, Thermes C, Heathcote P, Rambach A, Warren MJ. A role for Salmonella typhimurium cbiK in cobalamin (vitamin B12) and siroheme biosynthesis. J. Bacteriol. 1997;179:3202–3212. doi: 10.1128/jb.179.10.3202-3212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brindley AA, Raux E, Leech HK, Schubert HL, Warren MJ. A story of chelatase evolution: identification and characterization of a small 13–15 kDa ‘ancestral’ cobaltochelatase (CbiXS) in the archaea. J. Biol. Chem. 2003;278:22388–22395. doi: 10.1074/jbc.M302468200. [DOI] [PubMed] [Google Scholar]
- 13.Hansen J, Muldbjerg M, Cherest H, Surdin-Kerjan Y. Siroheme biosynthesis in Saccharomyces cerevisiae requires the products of both the MET1 and MET8 genes. FEBS Lett. 1997;401:20–24. doi: 10.1016/s0014-5793(96)01423-8. [DOI] [PubMed] [Google Scholar]
- 14.Raux E, McVeigh T, Peters SE, Leustek T, Warren MJ. The role of Saccharomyces cerevisiae Met1p and Met8p in sirohaem and cobalamin biosynthesis. Biochem. J. 1999;338:701–708. [PMC free article] [PubMed] [Google Scholar]
- 15.Schubert HL, Raux E, Brindley AA, Leech HK, Wilson KS, Hill CP, Warren MJ. The structure of Saccharomyces cerevisiae Met8p, a bifunctional dehydrogenase and ferrochelatase. EMBO J. 2002;21:2068–2075. doi: 10.1093/emboj/21.9.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yin J, Xu LX, Cherney MM, Raux-Deery E, Bindley AA, Savchenko A, Walker JR, Cuff ME, Warren MJ, James MN. Crystal structure of the vitamin B12 biosynthetic cobaltochelatase, CbiXS, from Archaeoglobus fulgidus. J. Struct. Funct. Genomics. 2006;7:37–50. doi: 10.1007/s10969-006-9008-x. [DOI] [PubMed] [Google Scholar]
- 17.Al-Karadaghi S, Hansson M, Nikonov S, Jönsson B, Hederstedt L. Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis. Structure. 1997;5:1501–1510. doi: 10.1016/s0969-2126(97)00299-2. [DOI] [PubMed] [Google Scholar]
- 18.Schubert HL, Raux E, Matthews MA, Phillips JD, Wilson KS, Hill CP, Warren MJ. Structural diversity in metal ion chelation and the structure of uroporphyrinogen III synthase. Biochem. Soc. Trans. 2002;30:595–600. doi: 10.1042/bst0300595. [DOI] [PubMed] [Google Scholar]
- 19.Spencer JB, Stolowich NJ, Roessner CA, Scott AI. The Escherichia coli cysG gene encodes the multifunctional protein, siroheme synthase. FEBS Lett. 1993;335:57–60. doi: 10.1016/0014-5793(93)80438-z. [DOI] [PubMed] [Google Scholar]
- 20.Warren MJ, Roessner CA, Santander PJ, Scott AI. The Escherichia coli cysG gene encodes S-adenosylmethionine-dependent uroporphyrinogen III methylase. Biochem. J. 1990;265:725–729. doi: 10.1042/bj2650725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Warren MJ, Bolt EL, Roessner CA, Scott AI, Spencer JB, Woodcock SC. Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem. J. 1994;302:837–844. doi: 10.1042/bj3020837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stroupe ME, Leech HK, Daniels DS, Warren MJ, Getzoff ED. CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat. Struct. Biol. 2003;10:1064–1073. doi: 10.1038/nsb1007. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z. Oscillation data reduction program. In: Sawyer L, Isaacs N, Bailey S, editors. Data Collection and Processing. Warrington: SERC Daresbury; 1993. pp. 56–62. [Google Scholar]
- 24.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. Sect. D Biol. Crystallogr. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron-density maps and the location of errors in these models. Acta Crystallogr. Sect. A Found. Crystallogr. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. Sect. D Biol. Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 31.Lesk AM. NAD-binding domains of dehydrogenases. Curr. Opin. Struct. Biol. 1995;5:775–783. doi: 10.1016/0959-440x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 32.Mabbs FE, Collison D. Electron Paramagnetic Resonance of d Transition Metal Compounds. Amsterdam: Elsevier; 1992. [Google Scholar]
- 33.Peisach J, Blumberg WE. Structural implications derived from the analysis of electron paramagnetic resonance spectra of natural and artificial copper proteins. Arch. Biochem. Biophys. 1974;165:691–708. doi: 10.1016/0003-9861(74)90298-7. [DOI] [PubMed] [Google Scholar]