Abstract
Various hypotheses have been proposed to explain the molecule processes of sarcomere assembly, partially due to the lack of systematic genetic studies of sarcomeric genes in an in vivo model. Towards the goal of developing zebrafish as a vertebrate model for this purpose, we characterized myofibrillogenesis in a developing zebrafish heart and went on to examine the functions of cardiac troponin T (tnnt2). We found that sarcomere assembly in zebrafish heart was initiated from a non-striated actin filament network at the perimembrane region, whereas sarcomeric myosin is independently assembled into thick filaments of variable length before integrating into the thin filament network. Compared to Z-discs that are initially aligned to form shorter periodic dots and expanded longitudinally at a later time, M-lines assemble later and have a constant length. Depletion of full-length tnnt2 disrupted the striation of thin filaments and Z-bodies, which sequentially affects the striation of thick filaments and M-lines. Conversely, truncation of a C-terminal troponin complex-binding domain did not affect the striation of these sarcomere sub-structures, but resulted in reduced cardiomyocyte size. In summary, our data indicates that zebrafish are a valuable in vivo model for studying both myofibrillogenesis and sarcomere-based cardiac diseases.
Keywords: Myofibrillogenesis, Sarcomere, Zebrafish, Heart, Tnnt2
Introduction
A sarcomere is the basic contractile unit in striated muscle. It is a highly organized structure that, through the interaction between actin and myosin, ensures the generation of coordinated force. The assembly of this elegant structure, which consists of Z-disc, M-line, I-band and A-band, is a complicated process that has fascinated cell biologists as a basic scientific question for more than 100 years. More recently, the question of myofibrillogenesis gained clinical significance due to the discovery that mutations in many sarcomeric genes are linked to human patients with cardiomyopathies and/or muscular dystrophies (Clark et al., 2002). Despite extensive studies, several models of myofibrillogenesis co-exist and have not been unified (Boateng and Goldspink, 2008; Sanger et al., 2005). The first model emphasizes on the existence of a stress fiber-like structure (SFLS), which functions as the initial backbone for sarcomere assembly (Dlugosz et al., 1984). The second model hypothesized that actin filaments and Z-discs form subunits independently from A-band subunits, with the two subsequently joined together to form a myofibril (Holtzer et al., 1997; Ojima et al., 1999; Schultheiss et al., 1990). The third model emphasizes on the existence of premyofibril along the cell membrane, a short mini-sarcomere structure that consists of Z-bodies composed of actin and α-actinin, and miniature A-bands. The binding of titin N-terminus to the Z-bodies expands the premyofibrils to longer nascent myofibrils (Dabiri et al.,1997; Du et al., 2003, 2008; Rhee et al., 1994). Although seemingly independent of each other, these three models are not mutually exclusive, because they emphasize on different assembly steps during myofibrillogenesis. In contrast to these three models, the fourth model argues that sarcomere assembly is a concurrent event without any intermediate structure (Ehler et al., 1999, 2004).
Early studies of myofibrillogenesis in the heart have been conducted using in vitro cell culture; however, the limitations of this system have been recognized. The proper 3-D cell–cell communication that might be essential for sarcomere assembly is disturbed and what has been studied is a reassembly process of the de-assembled sarcomere components, instead of de novo assembly process during the differentiation of cardiomyocytes (Gregorio and Antin, 2000; Holtzer et al., 1997; Rudy et al., 2001; Sanger et al., 1984; Wang et al., 1988; Wu et al., 1999). To address these concerns, in vivo animal models have recently been adopted, including chicken, quail and mouse (Du et al., 2008; Ehler et al., 1999; Hirschy et al., 2006; Tokuyasu and Maher, 1987). However, significant technical challenges in imaging sarcomere assembly in these animal models have arisen, since the heart progenitor cells are embedded deeply inside the embryos and are hard to access. Imaging is particularly tedious and expensive in mouse models, since the embryos develop in utero. Compared to the mouse, heart samples from stages of development where the heart is beating can be more easily harvested from chicken and quail, as their embryos develop ex utero. Thanks to improved imaging technology, the premyofibril structure, which cannot be detected in chicken (Ehler et al., 1999), was recently detected in quail embryos (Du et al., 2008). The disadvantage of the chicken/quail models, however, is the lack of knock-out and transgenic techniques, which prevents the systematic genetic analysis of myofibrillogenesis.
The zebrafish is a vertebrate model that possesses a desirable balance between embryology and genetics for the study of myofibrillogenesis. Zebrafish embryos are transparent and develop ex utero. Myofibrillogenesis happens in a single layer of cardiomyocytes, which is ideal for imaging and GFP technology. The zebrafish genome can be easily manipulated by either gain-of-function experiments using mRNA injection or transgenic technology, or by loss-of-function experiments using morpholino knockdown technology (Nasevicius and Ekker, 2000). Large-scale mutagenesis screens have already identified several mutants in sarcomere genes that affect myofibrillogenesis (Berdougo et al., 2003; Rottbauer et al., 2006; Sehnert et al., 2002; Xu et al., 2002; Zhao et al., 2008). In these reports, conclusions regarding gene function in myofibrillogenesis were mainly reached by transmission electron microscopy (TEM) technology, which has been used to characterize the sarcomere assembly process in the wild-type zebrafish heart (Wanga et al., 2001). Immunohistological studies have not been routinely carried out, mainly due to the lack of a description of the baseline process of myofibrillogenesis in a wild-type zebrafish heart. Since more antibodies have been tested and found to react with zebrafish proteins (Costa et al., 2003; Hinits and Hughes, 2007), we have used some of these antibodies to re-examine pickwick and tel mutants that have been previously characterized (Rottbauer et al., 2006; Xu et al., 2002). Interestingly, our antibody studies have revealed much detailed and novel information about the functions of titin and cmlc2 in myofibrillogenesis (Chen et al., 2008; Seeley et al., 2007). Therefore, a systematic immunohistological study of the sarcomere assembly process in a zebrafish heart is justified, which should greatly facilitate the use of this animal model for studying myofibrillogenesis.
Tnnt2 is a component of the troponin complex that regulates the interaction between myosin and actin in response to the Ca2+ wave (Parmacek and Solaro, 2004). The N-terminus of Tnnt2 binds tropomyosin (Tm), which sequentially anchors the troponin complex to the thin filament, while a C-terminal domain of Tnnt2 binds troponin I and troponin C, the other two components of the troponin complex (Pearlstone et al., 1986; Takeda et al., 2003; Zot and Potter, 1987). Mutations in TNNT2 have been found to be responsible for 15% of cardiomyopathies in humans (Watkins et al., 1995). In contrast to mutations in other sarcomeric genes such as myosin heavy chain that typically lead to a hypertrophic response, mutations in TNNT2 might result in mild hypertrophy but sudden cardiac death (Watkins et al., 1995). Transgenic Tnnt2 mouse models recapitulated these phenotypes and exhibited a small-heart phenotype (Tardiff et al., 1998, 1999). Depletion of tnnt2 in either zebrafish or mouse led to a silent heart (Nishii et al., 2008; Sehnert et al., 2002), suggesting that it has an important function in myofibrillogenesis. TEM studies also suggest a role for tnnt2 in thin filament assembly (Nishii et al., 2008; Sehnert et al., 2002).
In this report, we first conducted a detailed immunohistochemical study of the myofibrillogenesis process in the zebrafish heart. We then reveal in-depth mechanism regarding the function of tnnt2 in the assembly of each sarcomere sub-structure. Interestingly, we found that truncation of Tnnt2 at its C-terminus led to reduced cardiomyocyte cell size, recapitulating the small-heart phenotype in a transgenic mouse model. Our data underscore the value of zebrafish as a useful animal model for the genetic analysis of sarcomere assembly and pave the way for systematic studies of sarcomeric genes in this animal model.
Materials and methods
Zebrafish husbandry
The investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996). All zebrafish (Danio rerio) and embryos were maintained at 28 °C and staged as previously described (Westerfield, 1995). The sih mutant was kindly provided by Dr. Neil Chi and Dr. Didier Stainier (Sehnert et al., 2002), University of California at San Francisco.
Immunofluorescence microscopy and image analysis
Whole-mount immunofluorescence staining was performed as previously described (Chen et al., 2008; Seeley et al., 2007). To keep sarcomeres in a relaxed state, embryos were incubated in relaxation buffer (20 mM imadazole, 5 mM EGTA, 7 mM MgCl2, 5 mM creatine phosphate, 10 mM ATP, 100 mM KCl) for 1.5 h before flxation (Brixius et al., 2000; Li et al., 2006). The following antibodies were used at the indicated dilutions: anti-sarcomeric α-actinin (clone EA53, Sigma) at 1:1000, F59 (Developmental Studies Hybridoma Bank, DSHB) at 1:10, MEF2 (C-21, Santa Cruz Biotechnology) at 1:50, anti-myomesin (DSHB) at 1:100, anti-tropomyosin (CH1, Sigma) at 1:100, anti-troponin T (Sigma) at 1:200, anti-cardiac and skeletal troponin I (ABR-Affinity BioReagents) at 1:200, and Zn5 (ZIRC) at 1:500. Phalloidin was used at 1:10 (Molecular Probes). Alexa-conjugated secondary antibodies were used (Molecular Probes). Following staining, embryos were dissected to expose the heart region and imaged using an AxioplanII Zeiss microscope equipped with an ApoTome.
The length of myosin filaments and actin filaments was measured from the start to the end of each filament (or each actin dot in morphants) by using Axiovision software. Sarcomere length is defined as the distance between two neighbor α-actinin signals. Only α-actinin signals that appeared to be periodic along the same filament were measured. As no difference in sarcomere structure was detected between atrium and ventricle, we analyzed images of sarcomeres from the whole heart before 26 S, and sarcomeres from ventricle only at 48 hpf.
Bioinformatics
The sequence and exon–intron structure of zebrafish tnnt2 was determined using the Ensembl Database, while the sequence of human cardiac tnnt2 comes from Genbank. The nomenclature of human tnnt2 exons was based on(Farza et al., 1998). To confirm the in silico prediction, we designed the following primer pair to clone the cDNA of zebrafish tnnt2 by RT-PCR: Forward primer, 5′ ATGTCAGACAACGAAGAAGT 3′; Reverse primer, 5′ GCCTCAGAAAGTGAATGCTGTG 3′. The PCR product sequence was compared with zebrafish tnnt2 and human tnnt2 using DS Gene software (Accelrys Inc.), which confirmed all exons except exons 3, 16, and 17. Detailed information about zebrafish tnnt2 exon–intron structure is summarized in Supplemental Table S1.
Injection of morpholinos
MO-ATG and MO-E13 antisense morpholinos that target tnnt2 were purchased from Gene Tools LLC. Morpholinos were prepared and injected as previously described (Nasevicius and Ekker, 2000). The sequences are as follows: MO-ATG, same as the tnnt2 morpholino that was reported in Sehnert et al. (2002); and MO-E13, 5′ GAGACTTCATCTTACCTCATATTTC 3′.
Regular and real-time RT-PCR analysis of gene expression
Total RNA was extracted from 100 embryos at 48 hpf using the RNeasy Mini Kit (Qiagen) and reverse transcribed with SuperScript III Reverse Transcriptase (Invitrogen). The defective splicing event in MO-E13 morphants was detected using primer pair 1:
Forward primer: 5′ TAGAGAGACGGAGTGGAAAGAAACAGACTGAG 3′
Reverse primer: 5′ AATTTCTGATGGTCACTGACTCTGTTCCTC 3′
As a control, primer pair 2 targeting exon 8 was used to monitor the mRNA level of total tnnt2 transcripts:
Forward primer: 5′ CGTAAGCGCATGGAGAAGGACCTGAATG 3′
Reverse primer: 5′ TGGCATCATCTTCTGCTCTCTTTTTGGC 3′.
Gene expression was measured using SYBR Green-based real-time PCR analysis on an iCycler (Bio-Rad Laboratories). Primer sequences for measuring defective tnnt2 mRNA levels are:
E13RealF: 5′ TTCGGCAAACAGAAATATGAGATCAATGTC 3′
E18RealR: 5′ CACAGCATTCACTTTCTGAGGC 3′.
The expression of 18 S rRNA was used to normalize RNA content. The knockdown efficiency was calculated as previously described (Seeley et al., 2007).
Rescue experiments
Full-length tnnt2 cDNA was amplified from total RNA extracted from 72 hpf zebrafish embryos by RT-PCR using primers tnnt2-attB-F and tnnt2-attB-R1. The truncated tnnt2 cDNA fragment was amplified using primers tnnt2-attB-F and tnnt2-attB-R2. Their sequences are tnnt2-attB-F:
5′ GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGTCAGACAACGAAGAAGT 3′,
tnnt2-attB-R1:
5′ GGGGACCACTTTGTACAAGAAAGCTGGGTCCACAGCATTCACTTTCTGAGGC 3′
and tnnt2-attB-R2:
5′ GGGGACCACTTTGTACAAGAAAGCTGGGTCTCACTGAGAGCAGATTCATTG 3′.
The PCR products were cloned into the pDONR221 plasmid (Invitrogen). Together with the p5E-cmlc2 plasmid (a 5′ entry clone vector from the Tol2 Kit, (Kwan et al., 2007)), which contains a cmlc2 enhancer, and a p3E-IRES-EGFPpA plasmid (a 3′ entry clone vector from the Tol2 Kit, (Kwan et al., 2007)), which contains IRES EGFP, tnnt2 cDNAs were cloned into pDestTol2pA plasmid (a destination vector from the Tol2 Kit, Kwan et al. (2007)) by a recombination reaction. Linearized pCS2-TP plasmid (kindly provided by Koichi Kawakami) was used as template to generate capped mRNA encoding Tol2 transposase by in vitro transcription using SP6 polymerase (Message Machine, Ambion). Plasmids were co-injected with mRNA encoding Tol2 transposase into embryos at the single cell stage; the embryos were collected later at the proper stages for antibody staining. Since tnnt2 and egfp are encoded by one mRNA driven by the cmlc2 enhancer, it is predicted that both EGFP and tnnt2 are expressed in the same cardiomyocytes. Thus, fluorescent EGFP indicates the individual cardiomyocytes with ectopic tnnt2 expression.
Measurement of shortening fraction (SF) of the ventricular chambers
Movies of beating hearts from embryos at 48 hpf were recorded using a Zeiss microscope equipped with a Nikon camera. Images from movies were then used to measure the long axis length (a) and short axis length (b) between the myocardial borders of ventricles at diastole and systole, respectively. The percent shortening fraction (SF) was calculated using the formula: SF=(length at diastole−length at systole)/(length at diastole)×100. End-systolic or diastolic volumes of ventricle were calculated using the formula: volume=4/3πab2.
Quantification of ventricular cardiomyocyte cell size and cell number
The hearts of embryos at 48 hpf were double-stained to reveal the cell junctions of cardiomyocytes by using Zn5, an antibody that recognizes neurolin/DM-GRASP, a surface adhesin molecule (Fashena and Westerfield, 1999). Nuclei were stained by using a MEF2 antibody (Hinits and Hughes, 2007). The hearts were dissected out and images were acquired as Z-stacks for each heart sample using an AxioplanII Zeiss microscope. After processing the images using ‘extended focus,’ the surface area of individual cardiomyocytes was measured. Only cells with clearly visible outlines after being rendered in the XY plane were chosen for measurement. The cell number was considered to be the number of nuclei.
Statistics
Means and standard deviations of means (s.d.) were calculated from individual values, and a two-tailed t-test was used for comparison of two groups. Differences were considered significant when p<0.05.
Results
Assembly of thin and thick filaments during myofibrillogenesis in a zebrafish heart
In zebrafish, cardiac progenitor cells are localized in the anterior lateral plate mesoderm (ALPM) as two strips on both sides of zebrafish embryos at the 5 Somite (S) stage. These two groups of cardiac progenitor cells migrate towards the midline and meet at 18 S, fuse and reorganize to form a primitive heart tube that starts peristaltic contraction at 26 S, and finally form two cardiac chambers, namely the ventricle and atrium, that contract in a much stronger and coordinated fashion at 48 hpf. Therefore, it is predicted that functional contractile machinery should exist at 26 S and becomes more organized at 48 hpf.
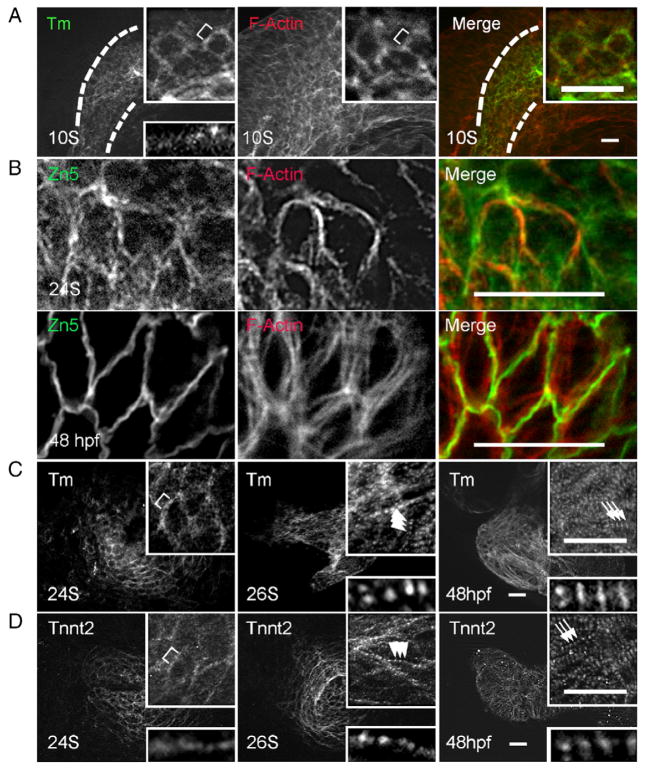
To test the above prediction and to characterize sarcomere assembly in a zebrafish heart, we studied the process of thin filament assembly by immunostaining. An antibody that recognizes tropomyosin, a thin filament component, reveals a filamentous network in the heart field as early as 10 S, when the cardiac progenitor is still within ALPM. This filamental network overlaps with and is part of the actin filamental network that also exists in neighboring tissues, as revealed by co-staining with phalloidin (Fig. 1A). The actin filamental network in cardiac progenitor cells is located in the cell membrane periphery region, as suggested by co-staining with Zn5 antibody and phalloidin (Fig. 1B), and remains non-striated until 26 S. The striation can first be detected at 26 S (Fig. 1C), when the heart tube forms and the heart first starts to beat. The event of thin filament striation formation can also be revealed by the anti-Tnnt2 antibody, which initially appears in the heart at 18 S (Fig. 1D, and Supplemental Fig. S2A). The thin filament continues the assembly process and becomes more organized at 48 hours post-fertilization (hpf), as indicated by the laterally broadened staining pattern of both tropomyosin and tnnt2 (Figs. 1C, D).
Fig. 1.
Assembly of thin filaments in the embryonic zebrafish heart. (A) Shown are images of 10 S embryos after two-color immunostaining to label Tm (green) and F-actin (red). Tm and F-actin networks are co-localized within the cardiac progenitor cells (outlined by dashed lines), which are labeled by Tm staining. The F-actin network also extends to the neighboring cells beyond the cardiac progenitor cells. (B) Shown are images of two-color immunostaining using the Zn5 antibody to label cell membrane (green) and phalloidin to label thin filaments (red). The thin filament network in the ventricle appears to associate with the cell membrane at both 24 S and 48 hpf. (C and D) Shown are images of 24 S, 26 S, and 48 hpf embryos after immunostaining using either anti-Tm (C) or anti-tnnt2 antibody (D). Both anti-Tm and anti-tnnt2 antibodies revealed a continuous filament network at 24 S, which becomes striated at 26 S, and lateral growth forming mature thin filaments at 48 hpf. Insets in (A, C, D) are of the same image at higher magnification. Insets at top are of networks, and insets at bottom are of myofibrils. Brackets, continuous thin filaments. Arrowheads, periodic dots of thin filaments after striation. Arrows, mature thin filaments after lateral growth. Scale bar=20 μm.
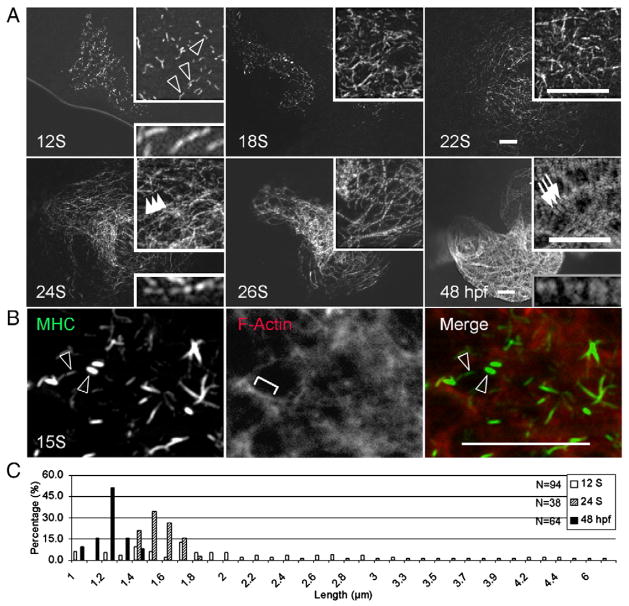
Compared to thin filament assembly, sarcomeric myosin filaments can be detected in the heart later, at 12 S, by immunostaining with F59, an antibody that recognizes the sarcomeric myosin heavy chain (MHC) (Fig. 2A). In contrast to a continuous thin filamental network, thick filaments exhibit rodlet structures with variable lengths, ranging from 1 μm to 6.1 μm (Fig. 2C). The pseudo H zone can be detected in shorter rodlets, suggesting their identity as bipolar polymer structures derived from the self-assembled myosin molecules in a head–tail/tail–head fashion. Similar rodlet structures of variable length were detected at the early stages of somite formation (data not shown). Initially, these myosin rodlets did not overlap with thin filaments, as indicated by two-color antibody staining at 15 S (Fig. 2B). These myosin rodlets gradually attach to the thin filament network at the perimembrane region and are organized into a periodic pattern at 24 S, just before the striation of thin filaments (Fig. 2A). Each preliminary sarcomeric unit consists of double rodlets separated by a pseudo H zone. The length of these myosin double rodlets becomes shorter and more uniform, ranging from 1.4 μm to 1.8 μm at 24 S, and from 1 μm to 1.4 μm at 48 hpf (Fig. 2C). The reduced length and increased width of each rodlet at 48 hpf suggests that myosin molecules continue to align with each other and register laterally to form mature A-bands.
Fig. 2.
Assembly of thick filaments in the embryonic zebrafish heart. (A) Thick filaments in embryos at different developmental stages as revealed by immunostaining for myosin heavy chain (MHC) using the F59 antibody. (B) Shown is the two-color immunostaining of 15 S embryos to reveal myosin filaments with the F59 antibody (green) and the F-actin filament network by phalloidin (red). Myosin rodlets and actin filaments do not overlap at this stage. (C) The distribution of lengths of myosin rodlets at 12 S and 24 S, and A-bands at 48 hpf. The myosin rodlets are of variable length until 24 S, when they turn uniform and become shorter at 48 hpf. The x-axis represents the length of myosin rodlets, while the y-axis represents the percentage of myosin rodlets with a particular length. N, total number of myosin rodlets quantified at each stage. Open arrowheads, myosin rodlets of variable length. Arrowheads, myosin rodlets of uniform length after assembling into the thin filament network. Arrows, mature A-bands. Brackets, continuous thin filaments. Insets are same images at higher magnification. Scale bar=20 μm.
Assembly of Z-discs and M-lines during myofibrillogenesis in a zebrafish heart
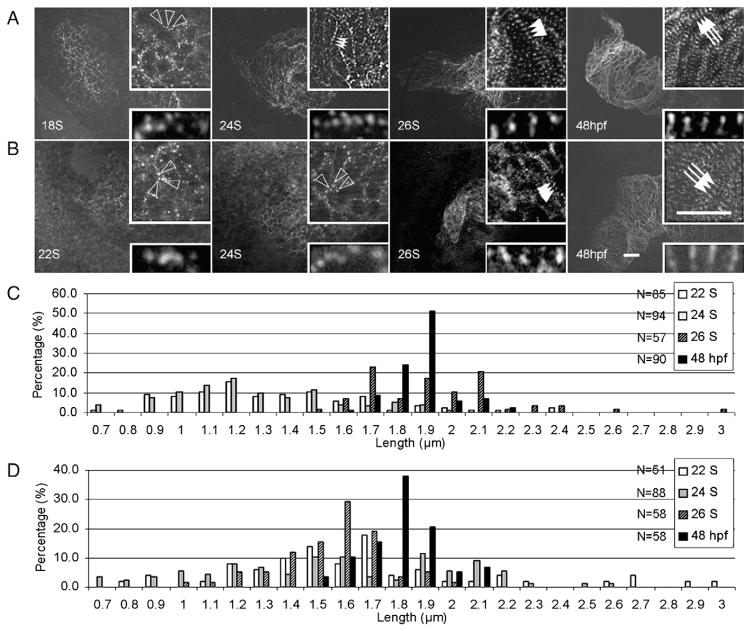
To investigate the assembly of the Z-disc, a sarcomeric structure that anchors thin filaments at their barbed ends and defines the border of each sarcomere, we performed immunostaining using an antibody that recognizes α-actinin (Fig. 3A). These α-actinin dots, which may represent Z-bodies, can be initially detected as irregular dots in the perimembrane region at 18 S, become periodic at 22–26 S, and undergo lateral growth between 26 S and 48 hpf to form mature Z-discs (Fig. 3A). The height of the Z-disc increases from 0.8±0.2 μm at 26 S to 1.6±0.3 μm at 48 hpf. Interestingly, the distance between the two neighboring Z-bodies expands from 1.4±0.4 μm at 22 S (Fig. 3C, and Supplemental Fig. S2B) to 1.9±0.3 μm at 26 S. The distance stays the same thereafter at least until 48 hpf (1.9±0.1 μm, Fig. 3C).
Fig. 3.
Assembly of α-actinin and myomesin in the embryonic zebrafish heart. Embryos at different stages were stained for α-actinin (A) and myomesin (B) to indicate assembly of the Z-disc and M-line. Before 22 S, α-actinin appears as irregular dot in the cell periphery region. The α-actinin dots become periodic at 24 S and the distance between them expands at 26 S. In contrast, myomesin dots are irregular before 24 S, and become periodic at 26 S. At 48 hpf, both α-actinin and myomesin dots undergo lateral growth to form the mature Z-disc and M-line. Open arrowheads, random dots of α-actinin (A) or myomesin (B). Indented arrowheads, α-actinin dots having shorter periodicity. Arrowheads, α-actinin dots (A) or myomesin (B) having longer periodicity. Arrows, mature Z-discs (A) or M-lines (B) after lateral growth. Insets are same images at higher magnification. Scale bar=20 μm. Distribution of the distance between two neighboring α-actinin or myomesin dots at different developmental stages is illustrated in C and D, respectively. The x-axis represents the length of sarcomere unit, while the y-axis shows the percentage of sarcomere units with a particular length. N, total number of sarcomere units quantified at each stage.
We then analyzed the assembly of the M-line, a structure that bundles the central part of thick filaments and defines the middle point of a sarcomere, by immunostaining using an antibody that recognizes myomesin. Myomesin immunoactivity can be first detected as irregular dots in the cell periphery region at 22 S, becomes organized into periodic dots at 26 S, and then undergoes lateral growth to form broader mature M-lines at 48 hpf with the height being increased from 0.7±0.3 μm to 1.9±0.3 μm (Fig. 3B). In contrast to the periodic α-actinin dots, the distance between the two neighboring myomesin dots remains constant (1.9±0.5 μm at 22 S; 1.8±0.5 μm at 24 S; 1.7±0.2 μm at 26 S; 1.8±0.1 μm at 48 hpf) (Fig. 3D). This finding suggests that M-line assembly is later than Z-disc assembly during cardiac myofibrillogenesis.
MO-ATG and MO-E13 knock down tnnt2 function and exhibit different cardiac phenotypes
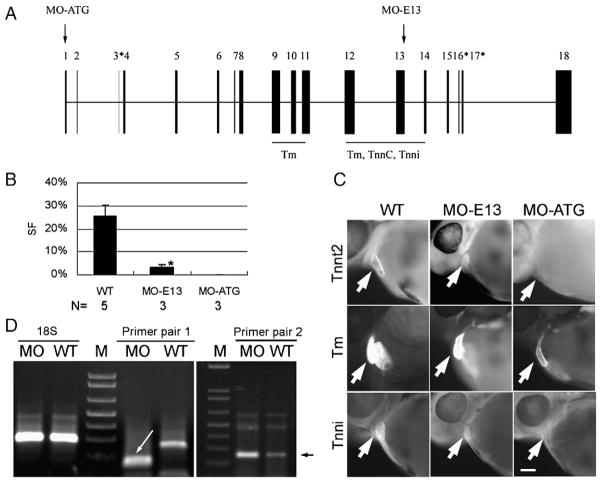
Having characterized the baseline of de novo sarcomere assembly in a zebrafish heart, we set out to conduct systematic genetic studies of sarcomeric genes in this vertebrate model in vivo. In this paper, we report our studies on cardiac troponin T (tnnt2) using the morpholino antisense technology. Based on the current annotation of the zebrafish tnnt2 genomic region and by comparing it to the peptide sequence encoded by human TNNT2 exons, we predict that zebrafish tnnt2 consists of at least 18 exons (Fig. 4A, and Supplemental Table S1). Previous biochemical and crystal structural studies suggested that exons 9–11 and 12–14 encode two domains that are needed to bind tropomyosin and thus attach the troponin complex to the thin filament, while exons 12–14 encode a domain that binds both troponin i (tnni) and troponin C (tnnc), the other two troponin complex components (Pearlstone et al.,1986; Takeda et al., 2003; Zot and Potter, 1987). To dissect how tnnt2 functions in sarcomere assembly, we designed MO-ATG, a morpholino that targets the translation start site of the tnnt2 transcript. We optimized the dosage of injection by following the silent heart phenotype, as suggested by sih, a null allele of tnnt2 in zebrafish (Sehnert et al., 2002). Injection of 12 pg of MO-ATG resulted in the total silencing of the heart (Fig. 4B). The expression of tnnt2 at the protein level was effectively reduced, as indicated by immunofluorescence staining using an anti-troponin T antibody (Fig. 4C). Consistent with the previous report with the sih mutant (Nishii et al., 2008; Sehnert et al., 2002), the expression level of tropomyosin was mildly reduced and tnni was severely reduced (Fig. 4C).
Fig. 4.
MO-ATG depletes full-length tnnt2, while MO-E13 generates truncated tnnt2. (A) Schematic illustration of the zebrafish tnnt2 gene. Exons are indicated by black boxes, with the alternatively spliced exons indicated by asterisks. The exons that encode an N-terminal Tm binding domain and a C-terminal Tm/tnnc/tnni binding domain are underlined (Watkins et al., 1995). The sites targeted by the MOs are indicated with arrows. For more detailed annotation, see Supplemental Table S1. (B) Quantification of cardiac contractility in WT and morphants. Shown are mean±s.d. of the shortening fraction of ventricular chambers. In contrast to a silent heart in MO-ATG morphants, weak contractility still exists in MO-E13 morphants. For video, see the Supplemental Movies. *p<0.01, if compared with WT. N, number of hearts quantified. (C) Shown are embryos of WT, morphants of MO-ATG or MO-E13 at 48 hpf after immunostaining to reveal tnnt2, Tm, and tnni, respectively. Both tnnt2 and tnni are severely reduced in MO-ATG morphants while Tm is weakly reduced. tnnt2 is weakly reduced in the MO-E13 morphant while Tm and Tnni remain unchanged. Arrows indicate the ventricular chambers. Scale bar=20 μm. (D) RT-PCR analysis of tnnt2 transcripts in MO-E13 morphants at 48 hpf. Primer pair 1 reveals a shorter product in the MO-E13 morphant (indicated by a white arrow), indicating an exon-skipping event that links exon 12 to exon 14. Primer pair 2 targeting at exon 8 detected an increased level of tnnt2 mRNA in MO-E13 morphants. The specific PCR product that is amplified by primer pair 2 is indicated by a black arrow. A primer pair targeting 18 S rDNA was used to optimize the starting amount of PCR template.
We also designed MO-E13, a morpholino that targets the splicing donor site after exon 13 of the tnnt2 gene. In contrast to the total silent heart phenotype observed in MO-ATG morphants, residual contractions can always be detected in MO-E13 morphants (Fig. 4B), even when injected at a dosage as high as 120 pg. The injection of 50 pg of MO-E13 resulted in 97% knockdown efficiency, as estimated by a real-time qRT-PCR-based assay. The major end product of this morphant is a defective tnnt2 transcript that lacks exon 13 due to an exon-skipping event, as suggested by RT-PCR followed by direct sequencing (Fig. 4D). At the protein level, it is predicted that a truncated Tnnt2 protein would be generated that deletes 54 amino acids at the C-terminus with the addition of three non-Tnnt2 amino acids. The truncated protein can be detected by immunostaining using our anti-tnnt2 antibody, although at a level somewhat lower than in wild-type (Fig. 4C). The reduced protein level is likely due to translational regulation such as protein stability, rather than a change at the transcriptional level, since the total level of tnnt2 transcripts is increased in the morphants (Fig. 4D). In contrast to MO-ATG morphants, neither the expression of tropomyosin nor troponin I protein is disturbed in the hearts of MO-E13 morphants (Fig. 4C). In summary, the above data indicate that, compared to the depletion of full-length tnnt2, the disruption of the C-terminal domain of tnnt2 resulted in different phenotypes in terms of both cardiac function and thin filament protein expression.
Full-length tnnt2 is required for the striation of thin filament
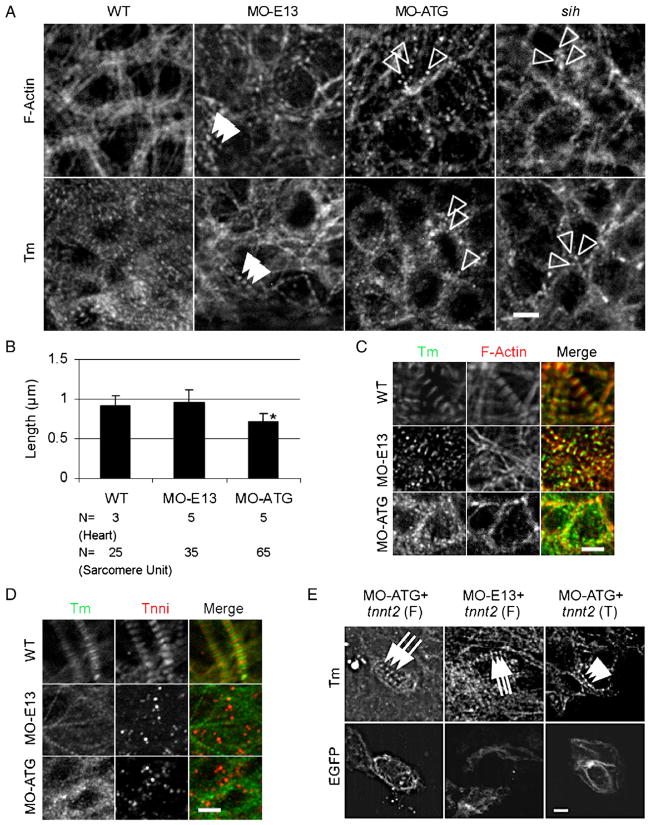
To understand how tnnt2 functions in sarcomere assembly in zebrafish heart, we first examined the effects of tnnt2 morpholinos on the assembly of the thin filaments. The continuous thin filament network can still be detected in both MO-ATG and MO-E13 morphants before 24 S (Supplemental Fig. S3), suggesting that tnnt2 is not required for the initial assembly of thin filaments. Later, the striation of the thin filament failed to happen in MO-ATG morphants, as suggested by non-striated staining pattern of both Tm and phalloidin at 30 S (Supplemental Fig. S3) and irregular dots at 48 hpf (Fig. 5A). In contrast, the striation of thin filament can still happen in MO-E13 morphants, as indicated by periodic dots of Tm and phalloidin (Fig. 5A, and Supplemental Fig. S3). The lateral growth of thin filaments was prevented in both morphants. The average length of phalloidin dots in MO-ATG morphants is around 0.71 μm, which is significantly shorter than those of the WT control and MO-E13 morphants, which are around 0.91 μm (Fig. 5B). We reasoned that more severely affected thin filament assembly in MO-ATG morphants might be due to disrupted tropomyosinactin filament interaction. Consistent with this hypothesis, tropomyosin immunoactivity appears partially disassociated from actin filaments in MO-ATG morphants, but not in MO-E13 morphants (Fig. 5C).
Fig. 5.
Thin filament striation was disrupted in MO-ATG morphants, but not in MO-E13 morphants. (A) Shown are images of 48 hpf embryos after immunostaining using either Tm antibody or phalloidin staining to reveal the F-actin network. Irregular actin and Tm dots were detected in MO-ATG morphants and sih mutants, while periodic dots of actin and Tm dots were detected in MO-E13 morphants. Open arrowheads, random dots of F-actin or Tm. Arrowheads, periodic F-actin or Tm dots. (B) Quantification of the length of F-actin filaments in 48hpf embryos. Shown are mean±s.d. *p<0.01, if compared with WT. N, number of hearts or sarcomere units quantified. (C) Shown are images of 48 hpf embryos after two-color immunostaining to reveal Tm (green) and actin filament (red). Association of Tm and F-actin was disrupted in MO-ATG morphants, but not in MO-E13 morphants. (D) Shown are images of 48 hpf embryos after two-color immunostaining for Tm (green) and Tnni (red). Assembly of Tnni into thin filaments and sarcomeres was disrupted in both morphants. (E) Shown in the top panels are images of 48 hpf embryos after immunostaining with CH1 antibody to reveal sarcomere assembly. Shown in the lower panels are individual cardiomyocytes with EGFP signal, indicating the expression of ectopic tnnt2 transcripts. Injection of full-length tnnt2 in either MO-ATG or MO-E13 morphants completely rescued sarcomere assembly, while injection of C-terminal truncated tnnt2 in MO-ATG morphants partially rescued sarcomere assembly to achieve a striated pattern. Arrows, mature sarcomeres. Arrowheads, partially rescued sarcomeres with striated pattern. Scale bar=5 μm.
In wild-type embryos, tnni immunoactivity exhibits a striated pattern at 48 hpf (Fig. 5D), indicating the attachment of the troponin complex to the thin filaments. In contrast, in hearts of MO-ATG morphants, tnni exhibits random dots that are dispersed throughout the cytoplasm without any association to tropomyosin (Fig. 5D). A similarly dispersed tnni staining pattern was also detected in MO-E13 morphants. These data confirm that a major function of tnnt2 is to attach the troponin complex to the thin filament, as has been previously suggested by both genetic studies in the tnnt2 knock-out mouse (Nishii et al., 2008) and structural studies of the troponin complex (Takeda et al., 2003). Moreover, our data indicate that the C-terminal domain of Tnnt2, encoded by exons 13 to 18, is needed for this function.
To confirm the specificity of the thin filament phenotypes in MO-ATG and MO-E13 morphants, we performed rescue experiments by co-injection of a tnnt2 plasmid driven by a cmlc2 cardiac-specific enhancer. We used either full-length Tnnt2 or a truncated version of Tnnt2 that deletes the C-terminal 54 amino acids (Supplemental Fig. S5). Individual cardiomyocytes with ectopic tnnt2 expression can be distinguished from the neighboring cells by green fluorescence (see Materials and methods). The ectopic expression of a full-length tnnt2 is able to rescue sarcomeric phenotypes in either MO-ATG or MO-E13 morphants, as indicated by the presence of mature sarcomere structures (Fig. 5E). On the other hand, ectopic expression of a truncated version of tnnt2 is capable of rescuing thin filament assembly in MO-ATG morphants to the degree of having a striated periodic pattern (Fig. 5E), as was seen in MO-E13 morphants (Fig. 5A). The contractile function of these EGFP+ cardiomyocytes is fully recovered upon co-injection of full-length tnnt2 (Supplemental Movies 2 and 4) and is partially restored upon co-injection of truncated tnnt2 (Supplemental Movie 5).
Taken together, the above data suggest that depletion of full-length Tnnt2 disrupts the striation and reduces the length of thin filaments, probably by disrupting the expression level of tropomyosin and/or the interaction of tropomyosin with actin filaments. Since the C-terminal domain of Tnnt2 is dispensable for this function, the tropomyosin interaction domain located at the N-terminal part of Tnnt2 is likely responsible for this function.
Full-length tnnt2 is required for the striation of the Z-disc, M-line and thick filaments
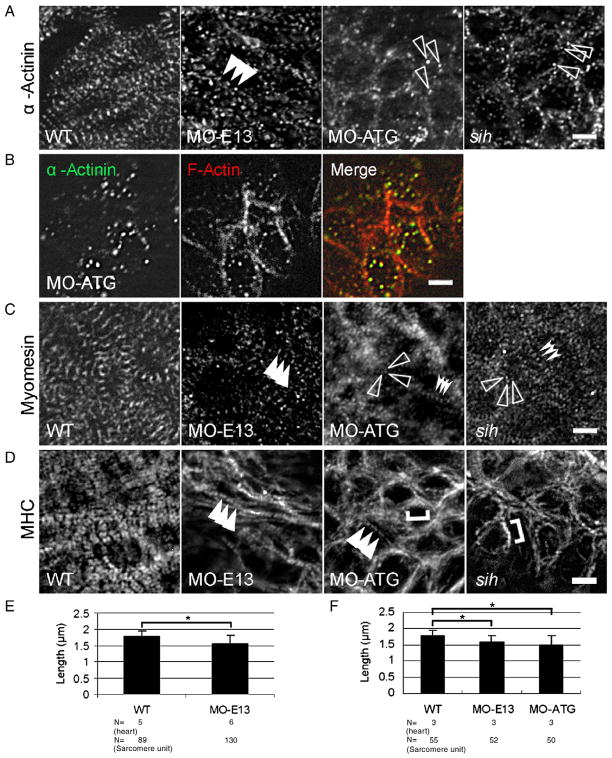
To understand how tnnt2 functions in the assembly of other sarcomere structures, we investigated the formation of Z-disc, M-line and thick filaments in tnnt2 morphants. Similar to the irregular dotted patterns of phalloidin and Tm staining seen in MO-ATG morphants (Fig. 5A), irregular α-actinin dots were detected in the region of the cell periphery at both 30 S and 48 hpf (Fig. 6A and Supplemental Fig. S4). Most of these random dots of α-actinin immunoactivity co-localize with F-actin (Fig. 6B), suggesting their identity as Z-bodies that fail to assemble any further. This irregular Z-body phenotype is a specific consequence of depleting full-length tnnt2 because of the following observations. First, irregular α-actinin dots were also noted in the sih mutant. However, in contrast, a regular periodic pattern can be detected in MO-E13 morphants (Fig. 6A). Of note, the distance between two neighboring α-actinin dots is shorter than that observed in the WT control (Fig. 6E).
Fig. 6.
Disrupted assembly of the Z-disc, thick filaments and the M-line in morphants. (A) Shown are images of 48 hpf embryos after immunostaining to reveal Z-disc assembly using α-actinin antibody. Irregular dots were detected in both MO-ATG morphants and sih mutants, whereas periodic dots were detected in MO-E13 morphants. Arrowheads, periodic α-actinin dots. Open arrowheads, irregular α-actinin dots. (B) Shown are images of MO-ATG morphant embryos at the 48 hpf stage after two-color immunostaining to reveal α-actinin (green) and actin filament (red). α-actinin dots and the F-actin network are still co-localized. (C) Shown are images of 48 hpf embryos after immunostaining to reveal M-line assembly using the anti-myomesin antibody. Arrowheads, striated M-lines. Open arrowheads, irregular myomesin dots. Indented arrowheads, periodic myomesin dots separated by a short distance. (D) Shown are images of 48 hpf embryos after immunostaining to reveal thick filaments using the F59 antibody. Arrowheads, striated thick filaments. Brackets, continuous thick filaments. (E) Quantification of the distance between two neighboring α-actinin dots in ventricles from WT and MO-E13 morphants at 48 hpf. Shown are mean±s. d. N, number of heart and sarcomere units quantified. (F) Quantification of the distance between two periodically neighboring myomesin dots in ventricles from WT and morphants at 48 hpf. Shown are mean±s.d. N, number of heart and sarcomere units quantified. *p<0.01. Scale bar=5 μm.
Likely due to a sequential event of a disrupted thin filaments and/or Z-discs, the assembly of M-lines is disturbed in tnnt2 morphants. Periodic dots of myomesin staining can be observed in MO-E13 morphants at 48 hpf, but they failed to undergo lateral growth to form mature M-lines (Fig. 6C). Most myomesin dots appear irregular in both MO-ATG morphants and sih mutants, although periodic dots can occasionally be detected. The distance between these regular dots in both morphants is shorter than that in wild-type embryos (Fig. 6F), suggesting defective M-line/thick filament assembly.
As expected, the early step of myosin rodlet assembly was not disturbed in both MO-ATG and MO-E13 morphants at 12 S (Supplemental Fig. S4). Although delayed slightly at 24 S, these myosin rodlets can still align to form periodic striated fiber structures at 48 hpf in MO-E13 morphants (Fig. 6D), although they fail to undergo further lateral growth to form mature A-bands. In contrast, thick filaments can still integrate into the perimembrane thin filament network in both MO-ATG morphants and sih mutants. However, these thick filaments predominantly appear in a non-striated pattern (Fig. 6D). The striated pattern can be occasionally detected in MO-ATG morphants.
Truncated tnnt2, but not full-length tnnt2, results in the reduced cell size of cardiomyocytes
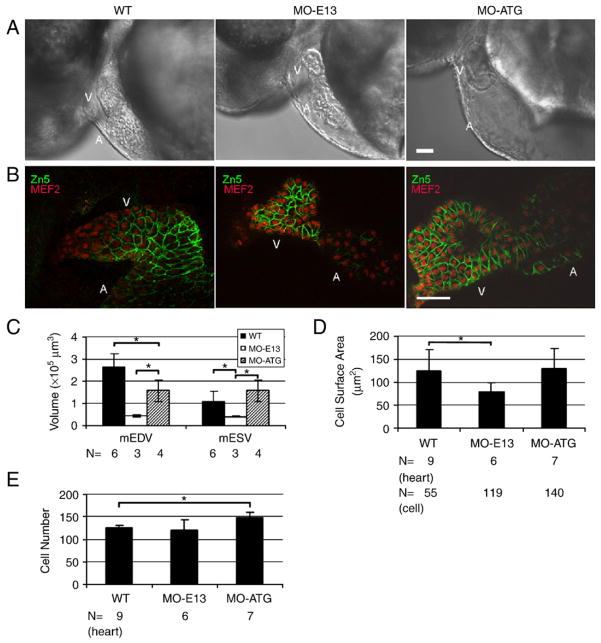
Visual inspection of tnnt2 morphants suggested that ventricular size in MO-E13 morphants is smaller than that in both WT and MO-ATG morphants (Fig. 7A). Indeed, both mean end-diastolic volume (mEDV) and the mean end-systolic volume (mESV) of ventricles in 48 hpf MO-E13 morphant embryos are significantly smaller than those indices in either wild-type or MO-ATG morphants (Fig. 7C). To reveal the mechanism of reduced heart size at the cellular level, we measured the cell size and number of cardiomyocytes by antibody staining (Fig. 7B, quantified in Figs. 7D and E). We found that the surface area of individual cardiomyocytes is significantly reduced in MO-E13 morphants (78.5±19.9 μm2 in MO-E13 morphants vs. 125.4±45.4 μm2 in WT, Figs. 7B, D), while the number of cardiomyocytes remains unchanged (122±21 in MO-E13 morphants vs. 125±7 in WT, Figs. 7B, E). As a control, the surface area of individual cardiomyocytes in MO-ATG morphants remains normal (130.5±42.8 μm2 in MO-ATG morphants vs. 125.4±45.4 μm2 in WT, Figs. 7B, D), while the number of cardiomyocytes increases (149±11 in MO-ATG morphants vs. 125±7 in WT, Figs. 7B, E). In summary, we conclude that the truncated tnnt2 results in the reduced size of individual cardiomyocytes without disturbing cardiomyocyte number.
Fig. 7.
Injection of MO-ATG and MO-E13 results in different changes in the size and number of ventricular cardiomyocytes. (A) Shown are lateral images of hearts in live WT and morphant embryos, 48 hpf. Ventricle in MO-E13 morphants appears smaller than WT. V, ventricle. A, atrium. (B) Shown are images of 48 hpf embryos after two-color immunostaining using Zn5 to reveal cell borders of cardiomyocytes (green) and mef2c to reveal nuclei of cardiomyocytes (red). V, ventricle. A, atrium. (C) Quantification of end-systolic and diastolic ventricular volume (mESV and mEDV, respectively) at 48 hpf. Shown are mean±s.d. N, number of hearts quantified. (D) Quantification of the surface area of individual cardiomyocytes from a ventricular chamber at 48 hpf. Shown are mean±s.d. N, number of hearts and cells quantified. (E) Quantification of ventricular cardiomyocyte number at 48 hpf. N, number of hearts quantified. *p<0.01. Scale bar=40 μm.
Discussion
Myofibrillogenesis in the development of zebrafish heart
In this paper, we have characterized the process of sarcomere assembly in a wild-type zebrafish heart. The accessible zebrafish embryology allows us to resolve at least five different stages of myofibrillogenesis, as detailed below (Supplemental Fig. S1). Our data support key perspectives of the first three models of myofibrillogenesis, while disagree with the fourth model of myofibrillogenesis.
First, between 10 and 12 S, thin filaments, consist of both actin and tropomyosin, and form a continuous non-striated network in the perimembrane region that correspond to the stress fiber-like structure (SFLS). This observation in an in vivo animal model supported the existence of SFLS, a key focus of the first model of myofibrillogenesis that was proposed based on studies in cultured cardiomyocytes (Dlugosz et al., 1984). Our data also suggested that the assembly of sarcomere in a zebrafish heart started from an actin filament network that exists in many cells, including cardiomyocytes. The first molecular event that distinguishes this filamental network in the heart from those in the neighboring tissues is the expression and binding of Tropomyosin at 10 S. In contrast to the SFLS model, we cannot detect the transition between striated and non-striated stretches between the center and the cellular periphery of the cell (Messerli et al., 1993), as all filaments are perimembranous. This situation is probably due to the limited volume of cytoplasm in cardiomyocytes at early embryonic stages, which is in contrast to the increased cytoplasm in mature cardiomyocytes after developmental hypertrophy.
At the same stage, we detected sarcomeric myosin rodlets assembled independently and did not overlap with the thin filament network, which is consistent with the second model of sarcomere assembly (Holtzer et al., 1997; Ojima et al., 1999; Schultheiss et al., 1990). This discovery is also supported by a recent report in quail heart, where myosin initially appears as short rodlets independently of thin filaments (Du et al., 2008). Together, however, these in vivo data argue against the statement that defined 1.6 μm thick filaments in a mature A-band are assembled before they are integrated into the sarcomere, an idea that was proposed mainly on the basis of cell culture studies (Allen and Pepe, 1965; Holtzer et al., 1997; Kelly, 1969). Instead, thick filament length must be determined during the assembly process between the thick and thin filaments. Interestingly, we found that the length of these myosin rodlets initially varies from 1 μm to 6.1 μm in zebrafish, which is different from the short rodlets detected in quail with a length of 0.75 μm (Du et al., 2008). The difference in rodlet length between fish and quail is likely a species-specific difference.
Second, between 18 S and 22 S, α-actinin and myomesin start to be expressed and appear as irregular dots in the periphery region of cardiomyocytes. Troponin T appears in thin filaments.
Third, from 22 S to 24 S, irregular α-actinin dots become periodic with a distance of 1.2 μm between the neighboring dots, which is shorter than that in mature sarcomere (1.9 μm). This structure corresponds to the premyofibril structure of the third model for myofibrillogenesis (Dabiri et al., 1997; Rhee et al., 1994). At the same time, thick filaments start to assemble into the thin filament network and become striated. The mechanism of how irregular α-actinin dots become periodic is still unclear. Our data showed that Z-bodies in tnnt2 MO-ATG morphants and the sih mutant turned to be irregular, suggested that the formation of the periodicity of the Z-bodies depends on properly assembled thin filaments. It also remains to be determined whether non-muscle myosin IIB interacts with thin filament to assemble premyofibrils before sarcomeric myosin is recruited to form thick filament, as predicted by the premyofibril model.
Fourth, at 26 S, the distance between the neighboring α-actinin dots expands to 1.9 μm and is accompanied by the striation of the thin filaments and the formation of myomesin periodicity. This stage of sarcomeric structure might correspond to the nascent striated myofibril in the SFLS model or to the nascent myofibril in the premyofibril model (Dabiri et al., 1997; Rhee et al., 1994). Of note, this is the time when the heart starts beating peristaltically. The underlying mechanism of the expansion of α-actinin dots from 1.2 μm at 22–24 S to 1.9 μm at 26 S remains to be determined. It has been proposed that Titin plays an important function in the expansion from premyofibril to nascent myofibril; however, the direct genetic evidence is still lacking. In the future, zebrafish titin mutants will be examined to test this hypothesis.
Fifth, from 26 S to 48 hpf, the primitive sarcomere structure further aligns and undergoes lateral growth to form a mature sarcomere that is capable of supporting the much more coordinated beating of a two-chambered heart.
The interaction between thin and thick filaments is important for the later stages of sarcomere assembly
Although thin and thick filaments initially assemble independently, the interaction between the two filament types is important for the later stages of sarcomere assembly, especially when the sarcomere undergoes lateral growth. In this study, we show that disturbing thin filament assembly by depleting tnnt2 affects the striation of thick filaments and sequentially the lateral growth of the sarcomere. Conversely, disturbance of thick filament assembly by depletion of either ELC or RLC affects the lateral growth of the sarcomere (Chen et al., 2008). Furthermore, disruption of the interaction between thin and thick filaments without disturbing thin or thick filament assembly per se, as seen in tnnt2 MO-E13 morphants, also affects the lateral growth required for mature sarcomere formation. In addition, our data raise an interesting possibility that the interaction between thin and thick filaments might participate in determining sarcomere length. This prediction is derived from our observation that the length of the sarcomere is shorter in tnnt2 MO-ATG morphants, MO-E13 morphants and cmlc2 morphants, but longer in cmlc1 morphants (Chen et al., 2008). The prediction needs to be further examined in the context of genetic studies of titin, a gigantic protein that has been proposed as the template of the sarcomere.
Relationship between M-line and Z-disc assembly
Compared to α-actinin, myomesin dots become periodic at 26 S, which is co-incident with the expansion of the α-actinin dots and the striation of the thin filaments. This striation happens later than the striation of the thick filaments, which interdigitate with the thin filaments and become periodic at 24 S. These observations in zebrafish heart support a previously proposed concept that M-line assembly is a later step of sarcomere assembly that happens after Z-disc assembly. Consistently, the disrupted M-line and thick filaments in MO-ATG morphants and the sih mutant suggest that a properly assembled thin filament/Z-disc is needed for the striation and periodicity of the thick filament/M-line. However, this statement is not supported by the existence of residue striated M-line in these morphants and the mutant. Instead, the latter observation might suggest that M-line assembly is regulated by a mechanism independent of Z-disc assembly, as has been proposed by a study of myofibrillogenesis in somites using the C2C12 cell culture system (Kontrogianni-Konstantopoulos et al., 2006). To further understand the relationship between M-line and Z-disc assembly, direct genetic studies of Z-disc and M-line proteins such as α-actinin and myomesin are needed.
Functions of tnnt2 in sarcomere assembly and cardiomyopathy
Based on our immunostaining results, it is clear that tnnt2 is not required for the initial assembly of the actin filament at early stages, as has been suggested by a previous study of tnnt2 knock-out mice (Nishii et al., 2008). This conclusion failed to be reached by the previous TEM analysis in zebrafish (Sehnert et al., 2002), underscoring our statement that immunostaining cannot be dismissed in analyzing sarcomere-related phenotypes. Partially due to the more accessible zebrafish embryos, we have uncovered additional information regarding the function of tnnt2 during the assembly of each substructure of the sarcomere. Our data suggest that tnnt2 is required for striation of the thin filaments, which in turn affects the periodicity of Z-bodies, M-lines and thick filaments. It is possible that tnnt2 regulates the assembly of thin filaments through a protein–protein interaction of its N-terminal domain with tropomyosin, as suggested by the reduced length of thin filaments and the disrupted co-localization between tropomyosin and actin filaments. In addition, depletion of tnnt2 may indirectly affect thin filament assembly by reducing the transcriptional levels of several thin filamental proteins such as tropomyosin and troponin I (this study and Sehnert et al., 2002). Of note, this latter mechanism might exist only in zebrafish, but not in mouse (Nishii et al., 2008; Sehnert et al., 2002).
In contrast to full-length Tnnt2, the C-terminal domain of Tnnt2 is not required for the striation of thin filaments and periodicity of Z-discs or for the expression level of tropomyosin transcripts. Periodic sarcomere-like structures still can assemble in tnnt2 MO-E13 morphants, although they are shorter and fail to undergo lateral growth. The shorter sarcomere length could be explained by the released inhibition of the troponin complex towards the thin filament-thick filament interaction, which results in a hypercontractile state that is irresponsive to either Calcium wave or relaxing solution treatment. Interestingly, truncation of the Tnnt2 C-terminus results in a smaller cell size in cardiomyocytes. This observation supports the statement that the reduced sarcomere length result in the reduced cardiomyocyte size, as has been previously proposed by genetic studies of ELC and RLC (Chen et al., 2008). Conversely, the disrupted interaction between thin and thick filaments is not sufficient to cause the small-cell phenotype, as normal cell size was observed in tnnt2 MO-ATG morphants. Of note, we cannot exclude the possibility that reduced number of sarcomeres per cardiomyocyte also contributes to the reduced cell size. A reliable method to quantify sarcomere number in each cardiomyocyte is yet to be established.
Alternatively, it is possible that the truncated Tnnt2 protein activates a signaling pathway that results in the reduction of cardiomyocyte size. This possibility is exciting since transgenic mouse models that contain a C-terminal truncated version of Tnnt2 exhibit a small-heart phenotype (Tardiff et al., 1998, 1999). To test this possibility, we have tried to inject mRNA or cDNA encoding truncated tnnt2 into wild-type embryos. Although we did not observe small-heart phenotypes, these negative results may simply due to the technical limitation of the transient assays. We are raising up stable transgenic fish lines to examine the potential dominant functions of the truncated tnnt2 in a zebrafish heart. Further studies are warranted to investigate this unique form of human cardiomyopathy using zebrafish models.
Our data highlight the value of zebrafish as a useful in vivo vertebrate model for the study of de novo sarcomere assembly. Its accessible embryology and unique genetic features complement the existing animal models such as chicken, quail and mouse. Our characterization of myofibrillogenesis in zebrafish heart supports different perspectives of the first three existing models of myofibrillogenesis, while disagrees with the fourth model. Clearly, sarcomere assembly is a progressive and closely regulated process, instead of just putting the sarcomeric proteins in the correct order. Our tnnt2 data further suggested that the assembly of the thin filament protein occurs at different stages and imposes an impact on the overall assembly of the sarcomere. More investigations in zebrafish are warranted to further reconcile different models of myofibrillogenesis. With the added possibility of generating a large number of zebrafish mutants that affect myofibrillogenesis, zebrafish possess great potential as a genomic model for systematic studies of both myofibrillogenesis and sarcomere-based cardiac diseases.
Supplementary Material
Acknowledgments
We are grateful to Beninio Jomok for maintaining our zebrafish facility; Drs. Neil Chi and Didier Stanier, UCSF, for sih fish; Dr. Chi-bing Chien, University of Utah, for Tol2 Kit vectors; and Dr. Koichi Kawakami, NIG, Japan, for the pCS2-TP plasmid. The F59 (developed by Dr. Frank E. Stockdale) and anti-myomesin (developed by Dr. J.C. Perriard) antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa, Department of Biological Sciences, Iowa City, IA 52242. This work was supported by a grant from the Muscular Dystrophy Association, NIH grant HL81753, and a startup fund from Mayo Clinic Foundation to X. Xu.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ydbio.2009.04.039.
Footnotes
Publisher's Disclaimer: This article appeared in a journal published by Elsevier. The attached copy is furnished to the author for internal non-commercial research and education use, including for instruction at the authors institution and sharing with colleagues.
Other uses, including reproduction and distribution, or selling or licensing copies, or posting to personal, institutional or third party websites are prohibited.
In most cases authors are permitted to post their version of the article (e.g. in Word or Tex form) to their personal website or institutional repository. Authors requiring further information regarding Elsevier’s archiving and manuscript policies are encouraged to visit:
References
- Allen ER, Pepe FA. Ultrastructure of developing muscle cells in the chick embryo. Am J Anat. 1965;116:115–147. doi: 10.1002/aja.1001160107. [DOI] [PubMed] [Google Scholar]
- Berdougo E, Coleman H, Lee DH, Stainier DY, Yelon D. Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development. 2003;130:6121–6129. doi: 10.1242/dev.00838. [DOI] [PubMed] [Google Scholar]
- Boateng SY, Goldspink PH. Assembly and maintenance of the sarcomere night and day. Cardiovasc Res. 2008;77:667–675. doi: 10.1093/cvr/cvm048. [DOI] [PubMed] [Google Scholar]
- Brixius K, Mehlhorn U, Bloch W, Schwinger RH. Different effect of the Ca(2+) sensitizers EMD 57033 and CGP 48506 on cross-bridge cycling in human myocardium. J Pharmacol Exp Ther. 2000;295:1284–1290. [PubMed] [Google Scholar]
- Chen Z, Huang W, Dahme T, Rottbauer W, Ackerman MJ, Xu X. Depletion of zebrafish essential and regulatory myosin light chains reduces cardiac function through distinct mechanisms. Cardiovasc Res. 2008;79:97–108. doi: 10.1093/cvr/cvn073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- Costa ML, Escaleira R, Manasfi M, de Souza LF, Mermelstein CS. Cytoskeletal and cellular adhesion proteins in zebrafish (Danio rerio) myogenesis. Braz J Med Biol Res. 2003;36:1117–1120. doi: 10.1590/s0100-879x2003000800019. [DOI] [PubMed] [Google Scholar]
- Dabiri GA, Turnacioglu KK, Sanger JM, Sanger JW. Myofibrillogenesis visualized in living embryonic cardiomyocytes. Proc Natl Acad Sci U S A. 1997;94:9493–9498. doi: 10.1073/pnas.94.17.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlugosz AA, Antin PB, Nachmias VT, Holtzer H. The relationship between stress fiber-like structures and nascent myofibrils in cultured cardiac myocytes. J Cell Biol. 1984;99:2268–2278. doi: 10.1083/jcb.99.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du A, Sanger JM, Linask KK, Sanger JW. Myofibrillogenesis in the first cardiomyocytes formed from isolated quail precardiac mesoderm. Dev Biol. 2003;257:382–394. doi: 10.1016/s0012-1606(03)00104-0. [DOI] [PubMed] [Google Scholar]
- Du A, Sanger JM, Sanger JW. Cardiac myofibrillogenesis inside intact embryonic hearts. Dev Biol. 2008;318:236–246. doi: 10.1016/j.ydbio.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E, Rothen BM, Hammerle SP, Komiyama M, Perriard JC. Myofibrillogenesis in the developing chicken heart: assembly of Z-disk, M-line and the thick filaments. J Cell Sci. 1999;112 (Pt. 10):1529–1539. doi: 10.1242/jcs.112.10.1529. [DOI] [PubMed] [Google Scholar]
- Ehler E, Fowler VM, Perriard JC. Myofibrillogenesis in the developing chicken heart: role of actin isoforms and of the pointed end actin capping protein tropomodulin during thin filament assembly. Dev Dyn. 2004;229:745–755. doi: 10.1002/dvdy.10482. [DOI] [PubMed] [Google Scholar]
- Farza H, Townsend PJ, Carrier L, Barton PJ, Mesnard L, Bahrend E, Forissier JF, Fiszman M, Yacoub MH, Schwartz K. Genomic organisation, alternative splicing and polymorphisms of the human cardiac troponin T gene. J Mol Cell Cardiol. 1998;30:1247–1253. doi: 10.1006/jmcc.1998.0698. [DOI] [PubMed] [Google Scholar]
- Fashena D, Westerfield M. Secondary motoneuron axons localize DM-GRASP on their fasciculated segments. J Comp Neurol. 1999;406:415–424. doi: 10.1002/(sici)1096-9861(19990412)406:3<415::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Gregorio CC, Antin PB. To the heart of myofibril assembly. Trends Cell Biol. 2000;10:355–362. doi: 10.1016/s0962-8924(00)01793-1. [DOI] [PubMed] [Google Scholar]
- Hinits Y, Hughes SM. Mef2s are required for thick filament formation in nascent muscle fibres. Development. 2007;134:2511–2519. doi: 10.1242/dev.007088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschy A, Schatzmann F, Ehler E, Perriard JC. Establishment of cardiac cytoarchitecture in the developing mouse heart. Dev Biol. 2006;289:430–441. doi: 10.1016/j.ydbio.2005.10.046. [DOI] [PubMed] [Google Scholar]
- Holtzer H, Hijikata T, Lin ZX, Zhang ZQ, Holtzer S, Protasi F, Franzini-Armstrong C, Sweeney HL. Independent assembly of 1.6 microns long bipolar MHC filaments and I-Z-I bodies. Cell Struct Funct. 1997;22:83–93. doi: 10.1247/csf.22.83. [DOI] [PubMed] [Google Scholar]
- Kelly DE. Myofibrillogenesis and Z-band differentiation. Anat Rec. 1969;163:403–425. doi: 10.1002/ar.1091630305. [DOI] [PubMed] [Google Scholar]
- Kontrogianni-Konstantopoulos A, Catino DH, Strong JC, Bloch RJ. De novo myofibrillogenesis in C2C12 cells: evidence for the independent assembly of M bands and Z disks. Am J Physiol Cell Physiol. 2006;290:C626–C637. doi: 10.1152/ajpcell.00442.2005. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien CB. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- Li M, Lionikas A, Yu F, Tajsharghi H, Oldfors A, Larsson L. Muscle cell and motor protein function in patients with a IIa myosin missense mutation (Glu-706 to Lys) Neuromuscul Disord. 2006;16:782–791. doi: 10.1016/j.nmd.2006.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerli JM, Eppenberger-Eberhardt ME, Rutishauser BM, Schwarb P, von Arx P, Koch-Schneidemann S, Eppenberger HM, Perriard JC. Remodelling of cardiomyocyte cytoarchitecture visualized by three-dimensional (3D) confocal microscopy. Histochemistry. 1993;100:193–202. doi: 10.1007/BF00269092. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Nishii K, Morimoto S, Minakami R, Miyano Y, Hashizume K, Ohta M, Zhan DY, Lu QW, Shibata Y. Targeted disruption of the cardiac troponin T gene causes sarcomere disassembly and defects in heartbeat within the early mouse embryo. Dev Biol. 2008;322:65–73. doi: 10.1016/j.ydbio.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Ojima K, Lin ZX, Zhang ZQ, Hijikata T, Holtzer S, Labeit S, Sweeney HL, Holtzer H. Initiation and maturation of I-Z-I bodies in the growth tips of transfected myotubes. J Cell Sci. 1999;112 (Pt 22):4101–4112. doi: 10.1242/jcs.112.22.4101. [DOI] [PubMed] [Google Scholar]
- Parmacek MS, Solaro RJ. Biology of the troponin complex in cardiac myocytes. Prog Cardiovasc Dis. 2004;47:159–176. doi: 10.1016/j.pcad.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Pearlstone JR, Carpenter MR, Smillie LB. Amino acid sequence of rabbit cardiac troponin T. J Biol Chem. 1986;261:16795–16810. [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil Cytoskelet. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- Rottbauer W, Wessels G, Dahme T, Just S, Trano N, Hassel D, Burns CG, Katus HA, Fishman MC. Cardiac myosin light chain-2: a novel essential component of thick-myofilament assembly and contractility of the heart. Circ Res. 2006;99:323–331. doi: 10.1161/01.RES.0000234807.16034.fe. [DOI] [PubMed] [Google Scholar]
- Rudy DE, Yatskievych TA, Antin PB, Gregorio CC. Assembly of thick, thin, and titin filaments in chick precardiac explants. Dev Dyn. 2001;221:61–71. doi: 10.1002/dvdy.1125. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Mittal B, Sanger JM. Formation of myofibrils in spreading chick cardiac myocytes. Cell Motil. 1984;4:405–416. doi: 10.1002/cm.970040602. [DOI] [PubMed] [Google Scholar]
- Sanger JW, Kang S, Siebrands CC, Freeman N, Du A, Wang J, Stout AL, Sanger JM. How to build a myofibril. J Muscle Res Cell Motil. 2005;26:343–354. doi: 10.1007/s10974-005-9016-7. [DOI] [PubMed] [Google Scholar]
- Schultheiss T, Lin ZX, Lu MH, Murray J, Fischman DA, Weber K, Masaki T, Imamura M, Holtzer H. Differential distribution of subsets of myofibrillar proteins in cardiac nonstriated and striated myofibrils. J Cell Biol. 1990;110:1159–1172. doi: 10.1083/jcb.110.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley M, Huang W, Chen Z, Wolff WO, Lin X, Xu X. Depletion of zebrafish titin reduces cardiac contractility by disrupting the assembly of Z-discs and A-bands. Circ Res. 2007;100:238–245. doi: 10.1161/01.RES.0000255758.69821.b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, Schwartz S, Robbins J, Leinwand LA. A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J Clin Invest. 1998;101:2800–2811. doi: 10.1172/JCI2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardiff JC, Hewett TE, Palmer BM, Olsson C, Factor SM, Moore RL, Robbins J, Leinwand LA. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J Clin Invest. 1999;104:469–481. doi: 10.1172/JCI6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu KT, Maher PA. Immunocytochemical studies of cardiac myofibrillogenesis in early chick embryos. I. Presence of immunofluorescent titin spots in premyofibril stages. J Cell Biol. 1987;105:2781–2793. doi: 10.1083/jcb.105.6.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SM, Greaser ML, Schultz E, Bulinski JC, Lin JJ, Lessard JL. Studies on cardiac myofibrillogenesis with antibodies to titin, actin, tropomyosin, and myosin. J Cell Biol. 1988;107:1075–1083. doi: 10.1083/jcb.107.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanga J, Eckberg WR, Anderson WA. Ultrastructural differentiation of cardiomyocytes of the zebrafish during the 8–26-somite stages. J Submicrosc Cytol Pathol. 2001;33:275–287. [PubMed] [Google Scholar]
- Watkins H, McKenna WJ, Thierfelder L, Suk HJ, Anan R, O’Donoghue A, Spirito P, Matsumori A, Moravec CS, Seidman JG, et al. Mutations in the genes for cardiac troponin T and alpha-tropomyosin in hypertrophic cardiomyopathy. N Engl J Med. 1995;332:1058–1064. doi: 10.1056/NEJM199504203321603. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- Wu JC, Chung TH, Tseng YZ, Wang SM. N-cadherin/catenin-based costameres in cultured chicken cardiomyocytes. J Cell Biochem. 1999;75:93–104. [PubMed] [Google Scholar]
- Xu X, Meiler SE, Zhong TP, Mohideen M, Crossley DA, Burggren WW, Fishman MC. Cardiomyopathy in zebrafish due to mutation in an alternatively spliced exon of titin. Nat Genet. 2002;30:205–209. doi: 10.1038/ng816. [DOI] [PubMed] [Google Scholar]
- Zhao L, Zhao X, Tian T, Lu Q, Skrbo-Larssen N, Wu D, Kuang Z, Zheng X, Han Y, Yang S, Zhang C, Meng A. Heart-specific isoform of tropomyosin4 is essential for heartbeat in zebrafish embryos. Cardiovasc Res. 2008 doi: 10.1093/cvr/cvn177. [DOI] [PubMed] [Google Scholar]
- Zot AS, Potter JD. Structural aspects of troponin–tropomyosin regulation of skeletal muscle contraction. Annu Rev Biophys Biophys Chem. 1987;16:535–559. doi: 10.1146/annurev.bb.16.060187.002535. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.