Abstract
Aedes aegypti kynurenine aminotransferase (AeKAT) is a multifunctional aminotransferase. It catalyzes the transamination of a number of amino acids and uses many biologically relevant α-keto acids as amino group acceptors. AeKAT also is a cysteine S-conjugate β-lyase. The most important function of AeKAT is the biosynthesis of kynurenic acid, a natural antagonist of NMDA and α7-nicotinic acetylcholine receptors. Here, we report the crystal structures of AeKAT in complex with its best amino acid substrates, glutamine and cysteine. Glutamine is found in both subunits of the biological dimer, and cysteine is found in one of the two subunits. Both substrates form external aldemines with pyridoxal 5-phosphate in the structures. This is the first instance in which one pyridoxal 5-phosphate enzyme has been crystallized with cysteine or glutamine forming external aldimine complexes, cysteinyl aldimine and glutaminyl aldimine. All the units with substrate are in the closed conformation form, and the unit without substrate is in the open form, which suggests that the binding of substrate induces the conformation change of AeKAT. By comparing the active site residues of the AeKAT–cysteine structure with those of the human KAT I–phenylalanine structure, we determined that Tyr286 in AeKAT is changed to Phe278 in human KAT I, which may explain why AeKAT transaminates hydrophilic amino acids more efficiently than human KAT I does.
The sequence of Aedes aegypti kynurenine aminotransferase (AeKAT)1 is 47% identical with that of human KAT I and 51.9% identical with that of human KAT III (1). Human, rat, and mouse KAT I enzymes have been extensively studied (2, 3). Mammalian KAT I, also called glutamine transaminase K (GTK), is a multifunctional aminotransferase that acts on a number of amino acids (glutamine, cysteine, phenylalanine, kynurenine, methionine, etc.) and uses many biologically relevant keto acids as amino group acceptors (3, 4). AeKAT and a glutamine:phenylpyruvate aminotransferase from Thermus thermophilus, homologues of mammalian KAT I, have also been systematically characterized (5, 6). One of the enzymatic products of KAT I is kynurenic acid (KYNA), which is the only known endogenous antagonist of the N-methyl-d-aspartate (NMDA) subtype of glutamate receptors (7–9). KYNA is also the antagonist of the α7-nicotinic acetylcholine receptor (10–12). Very recently, KYNA has been identified as an endogenous ligand of an orphan G protein-coupled receptor (GPR35) that is predominantly expressed in immune cells (13). The tryptophan metabolic pathway is activated during inflammatory conditions, such as viral invasion, bacterial lipopolysaccharide, or interferon stimulation (14, 15). The activation of tryptophan metabolism causes a reduced plasma tryptophan level and an elevated KYNA concentration (16). Therefore, this receptor–ligand pair may also play a role in immunological regulation.
Many other functions have been proposed for mammalian KAT I enzymes, including maintaining low levels of phenylpyruvate, closing the methionine salvage pathway, regulating α-keto acid levels, sparing the essential amino acids, and maintaining a continual equilibrium among the amino acids (3, 17). However, quantitatively, the most important amine donor for KAT I enzymes in vivo is glutamine. The product of glutamine transamination (α-ketoglutaramate) is rapidly removed by cyclization and/or conversion to α-ketoglutarate. Transamination is therefore “pulled” in the direction of glutamine utilization (3). AeKAT showed high activity toward a number of amino acids, including glutamine, cysteine, tyrosine, methionine, phenylalanine, histidine, and kynurenine. Interestingly, cysteine has an intriguing effect on AeKAT activity toward kynurenine, enhancing its activity at relatively low concentrations and inhibiting its activity at higher concentrations (5, 18). Crystal structures of AeKAT, glutamine:phenylpyruvate aminotransferase from T. thermophilus, and human KAT I have been available for substrate recognition study (19–21). They are fold type I aminotransferases (22, 23), characterized by the presence of an N-terminal arm, a small domain, and a large domain. However, complex structures with their best natural amino acid substrates (such as glutamine and cysteine) were unavailable. The crystal structure of AeKAT indicated that Cys284 in AeKAT might be the cysteine regulation target of AeKAT. Cys284 is located in the large domain of the subunit and forms a thiol–thiolate hydrogen bond with Cys284 from the other subunit. The cysteine pair is close to the active center of AeKAT. Under oxidative conditions, these cysteine residues in AeKAT can reasonably form a disulfide bond because of the short distance between the sulfur atoms, requiring only a decrease of 1.5–1.6 Å (20). To understand the structural basis underlying substrate recognition and catalysis, we determined the three-dimensional crystal structures of AeKAT in complex with its best specific substrates, cysteine and glutamine. This is the first time that the crystal structures of the enzyme–glutamine and enzyme–cysteine complexes of KAT I enzymes have been reported.
MATERIALS AND METHODS
Expression and Purification of Recombinant AeKAT
AeKAT lacking the NH2-terminal mitochondrial leader sequence (amino acids 1–48) was expressed in a baculovirus/insect cell protein expression system and purified for crystallization (18, 20). The purified recombinant AeKAT was concentrated to 10 mg/mL protein in 5 mM phosphate buffer (pH 7.5) using a Centricon YM-30 concentrator (Millipore).
AeKAT Crystallization
The crystals were grown via hanging drop vapor diffusion methods with the volume of reservoir solution at 500 µL and the drop volume at 2 µL, containing 1 µL of protein sample and 1 µL of reservoir solution based on the previous reported method (20), except that the precipitant was changed from PEG 8000 to ammonium sulfate. The new crystallization buffer contained 2 M ammonium sulfate and 0.1 M Tris at pH 8.5. The AeKAT–cysteine complex was cocrystallized by adding 2.5 mM l-cysteine to the crystallization buffer described above. The crystals of the glutamine–enzyme complex were obtained by soaking the AeKAT crystals overnight in 2.5 mM l-glutamine in the crystallization buffer described above.
Data Collection and Processing
Individual AeKAT complex crystals were cryogenized in crystallization buffer containing 30% PEG 400 as a cryoprotectant. Diffraction data of AeKAT complex crystals were collected at Brookhaven National Synchrotron Light Source beamline X29A (λ = 1.1 Å). Data collection was carried out using an ADSC Q315 CCD detector. All data were indexed and integrated using HKL (24). Scaling and merging of diffraction data were performed using SCALEPACK (25). The parameters of the crystals and information regarding data collection are listed in Table 1.
Table 1.
Data Collection and Refinement Statistics
| AeKAT–glutamine | AeKAT–cysteine | |
|---|---|---|
| Crystal Data | ||
| Space group unit cell |
P212121 | |
| a (Å) | 56.373 | 56.050 |
| b (Å) | 95.590 | 95.678 |
| c (Å) | 165.901 | 166.752 |
| α = β = γ (deg) | 90.000 | 90.000 |
| Data Collection | ||
| X-ray source | BNLaX29 | |
| Wavelength | 1.1 | 1.1 |
| resolution (Å)b | 1.84 (1.91–1.84) | 1.96 (2.03–1.96) |
| total no. of reflections | 475345 | 455740 |
| no. of unique reflections | 78429 | 65484 |
| Rmergeb | 0.077 (0.275) | 0.072 (0.274) |
| redundancyb | 6.7 (4.0) | 7.4 (4.7) |
| completeness (%)b | 90.2 (45.3) | 94.4 (67.0) |
| Refinement Statistics | ||
| Rwork (%)b | 20.1 (24.4) | 21.8 (36.5) |
| Rfree (%)b | 24.3 (30.4) | 26.7 (40.2) |
| rmsd for bond lengths (Å) | 0.016 | 0.027 |
| Rmsd for bond angles (deg) | 1.713 | 1.926 |
| no. of ligand or cofactor molecules |
2 QLP | 1 LLP, 1 C6P |
| no. of water molecules | 584 | 469 |
| average overall B (Å2) | 32.8 | 42.5 |
| Ramachandran plot | ||
| allowed (%) | 99.7 | 99.7 |
| generously allowed (%) | 0 | 0.0 |
| disallowed (%) | 0.3 | 0.3 |
Brookhaven National Laboratory.
The values in parentheses are for the highest-resolution shell.
Structure Determination
The structures of AeKAT complexes were determined by the molecular replacement method using the published AeKAT structure without any ligands or waters (Protein Data Bank entry 1YIY) (20). Molrep (26) in the CCP4 suite was employed to calculate both the cross-rotation and translation function of the model. The initial model was subjected to iterative cycles of crystallographic refinement with Refmac, version 5.2 (27), and graphic sessions for model building used O (28). The substrate molecules were modeled when the R factor dropped to a value of 0.23 at full resolution for the AeKAT–glutamine structure and a value of 0.24 at full resolution for the AeKAT–cysteine structure based on both the 2Fo – Fc and Fo – Fc electron density maps. Solvent molecules were automatically added and refined with ARP/wARP (29) together with Refmac, version 5.2.
Structure Analysis
Superposition of structures was conducted using Lsqkab (30) from the CCP4 suite. Figures were generated using Pymol (31). Protein–substrate interaction was also analyzed using Pymol (31).
RESULTS
Overall Structure
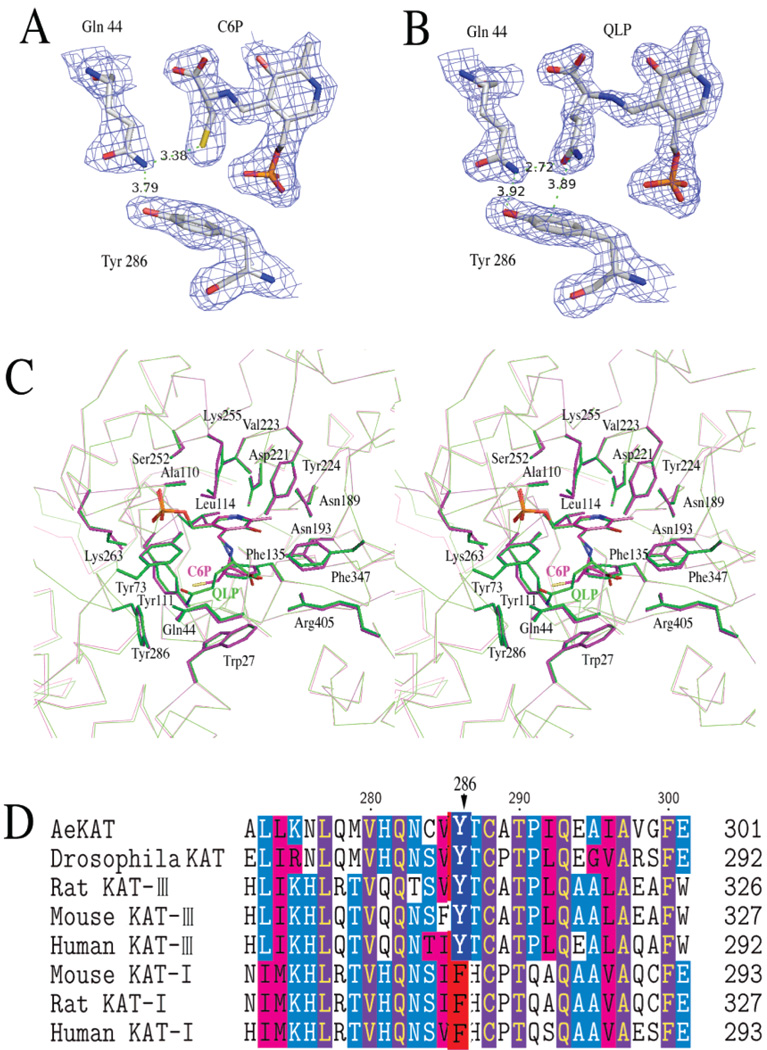
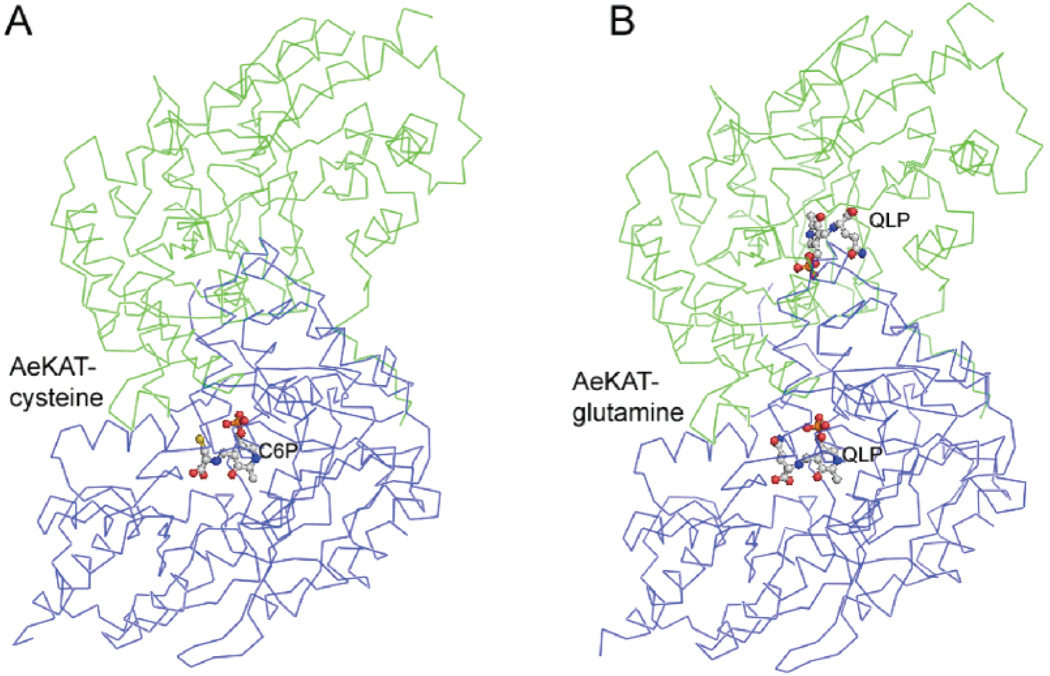
The structures of AeKAT–substrate complexes were determined through molecular replacement and refined to 1.96 Å resolution for the AeKAT–cysteine complex and 1.84 Å resolution for the AeKAT–glutamine complex. The final models contain 419 residues in each subunit and yield a crystallographic R value of 21.1% and an Rfree value of 24.3% for the AeKAT–glutamine complex and an R value of 21.8% and an Rfree value of 26.7% for the AeKAT–cysteine complex with ideal geometry evaluated with Molprobity (http://molprobity.biochem.duke.edu/) (Table 1). There are two protein molecules in an asymmetric unit that form one biological homodimer. The residues of the two subunits in AeKAT complexes are numbered 11 (A)–429 (A) for chain A and 11 (B)–429 (B) for chain B. The results of the refinement are summarized in Table 1. All residues except Tyr286 in all four chains are in favorable regions of the Ramachandran plot as defined with Procheck (32). Although Tyr286 (A) and (B) in all complex structures fall within a disallowed region of the Ramachandran plot of the determined structures, its excellent electron density allowed us to unambiguously assign the observed conformations (Figure 7A,B). Interestingly, Tyr286 (A) and (B) are in the favored region of the Ramachandran plot evaluated with Molprobity. The first 10 residues (1–10) were not defined in both the 2Fo – Fc and Fo – Fc electron density maps and are absent from the final models. The protein architecture revealed by AeKAT complex structures remains the same as that of the native AeKAT structure previously described (20). Electron density consistent with the presence of glutamine was observed in the active sites of both subunits of the AeKAT–glutamine complex, whereas only one of the monomers in the AeKAT–cysteine complex exhibited electron density consistent with the presence of cysteine (Figure 1). By binding the substrates, the protein chains changed their conformations to the closed forms in the AeKAT–glutamine complex. Similarly, chain B in the AeKAT–cysteine complex also assumed a closed form, but chain A without the ligand remained as the open form (chain A in the AeKAT–cysteine complex).
Figure 7.
(A) Tyr286 in the AeKAT–cysteine structure. The 2Fo – Fc electron density map covering C6P, Gln44, and Tyr286 is shown contoured at the 1.2σ level. (B) Tyr286 in the AeKAT–glutamine structure. The 2Fo – Fc electron density map covering QLP, Gln44, and Tyr286 is shown contoured at the 1.2σ level. (C) Stereo α-carbon representation of the AeKAT–cysteine complex (pink) superimposed onto the AeKAT–glutamine complex (green). The active site residues within 4 Å of the substrate are shown as sticks. (D) Sequence alignment of DrosophilaKAT and AeKAT with KAT I and KAT III sequences from mouse, rat, and human. Tyr286 in AeKAT and the aligned residues in other KATs are highlighted.
Figure 1.
α-Carbon representation of the structures of AeKAT complexes: (A) AeKAT–cysteine complex and (B) AeKAT–glutamine complex.
Substrate Recognition and Catalysis in the AeKAT–Cysteine Complex
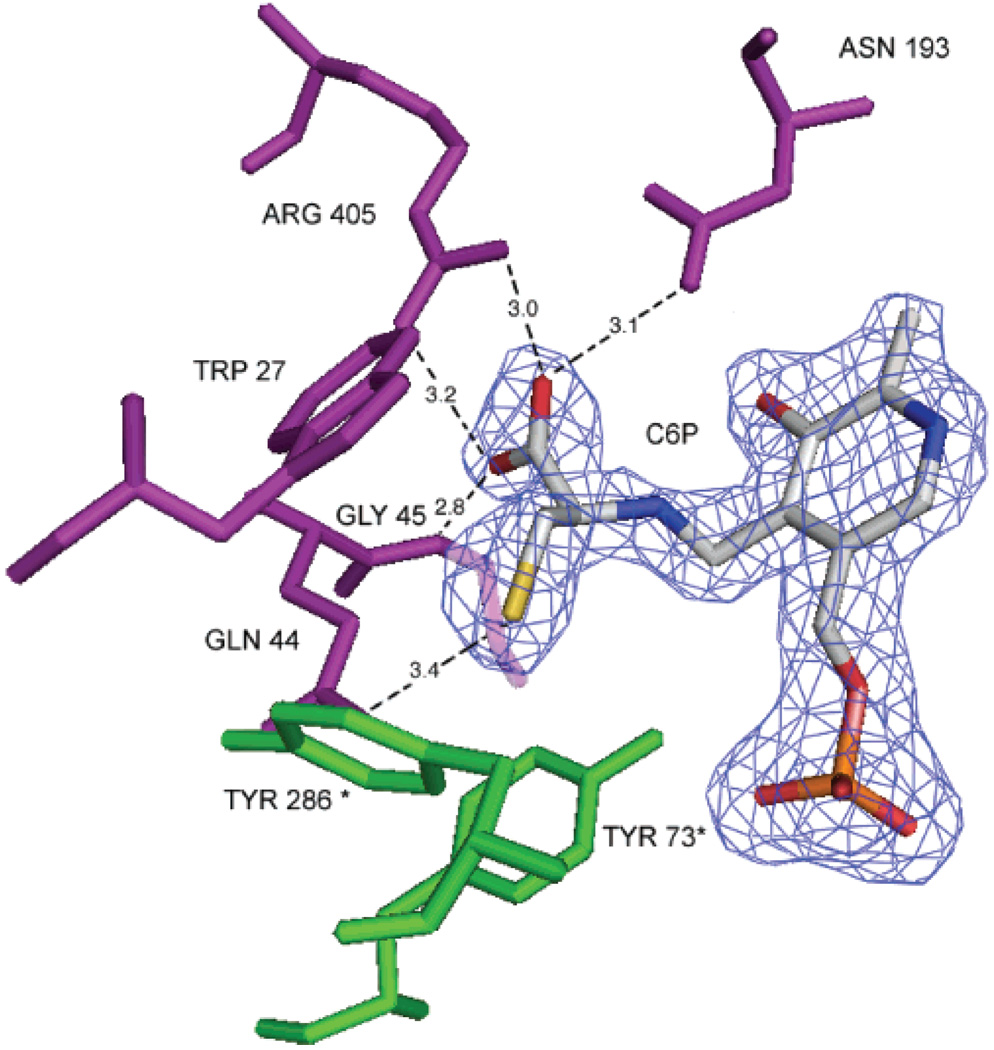
Inspection of the crystal structure of the AeKAT–cysteine complex revealed that the substrate lies near the PLP cofactor and forms an external aldimine with PLP. However, cysteine is seen in only one of the two subunits in a biological dimer. There are three water molecules at the active site of the subunit that do not contain a cysteine molecule and two waters in the subunit that contains a cysteine. A water molecule (W206) forms a hydrogen bond (3.6 Å) with the SG atom of C6P and also interacts with the NZ atom of Lys263 (2.7 Å), the OH atom of Tyr73 (A chain) (3.6 Å), and the OP2 atom of C6P (3.1 Å). Another water molecule (W368) forms a hydrogen bond (3.4 Å) with the O atom of C6P and also interacts with Gly45 (3.3 Å), Tyr340 (2.9 Å), and Tyr224 (2.9 Å). Several residues, including Arg405 (B), Tyr224 (B), Asn193 (B), Gln44 (B), Gly45 (B), Trp27 (B), Tyr73 (A), and Phe135 (B), define the substrate-binding site and contact the cysteine molecule. The carboxylate of cysteine forms a salt bridge with the guanidinium group of Arg405. The salt bridge is fixed by hydrogen bonding interactions with the NH atom of Gly45 (B) and the side chain of Asn193 (B) at both sides of the salt bridge. Gln44 (B) and Gly45 (B) are located at the turning point of the loop that dives into and partially plugs the enzyme active site, thus shielding the substrate-binding pocket from the bulk solvents (Figure 2). A thiol–thiolate hydrogen bond between Cys284 residues was observed in both AeKAT–cysteine and AeKAT–glutamine complexes as described in the previously reported unliganded structures (PLP form, 3.6 Å; PMP form, 3.5 Å) (20), but the thiol–thiolate hydrogen bond (3.7 Å) in the AeKAT–cysteine complex is slightly longer than that in the unliganded structures. However, the structure of the AeKAT–cysteine complex did not provide any clues regarding the potential interaction of free cysteine with the thiol–thiolate hydrogen bond. A mutation study seems necessary to determine if the thiol–thiolate hydorogen bond between Cys284 residues is the cysteine regulation target.
Figure 2.
Cysteine binding site. The cysteinyl aldimine (C6P) and the protein residues within 4 Å of cysteine substrate are shown. Residues colored pink are from chain B and residues colored green from chain A. The 2Fo – Fc electron density map covering C6P is shown contoured at the 1.3σ level. This figure was generated with PyMol.
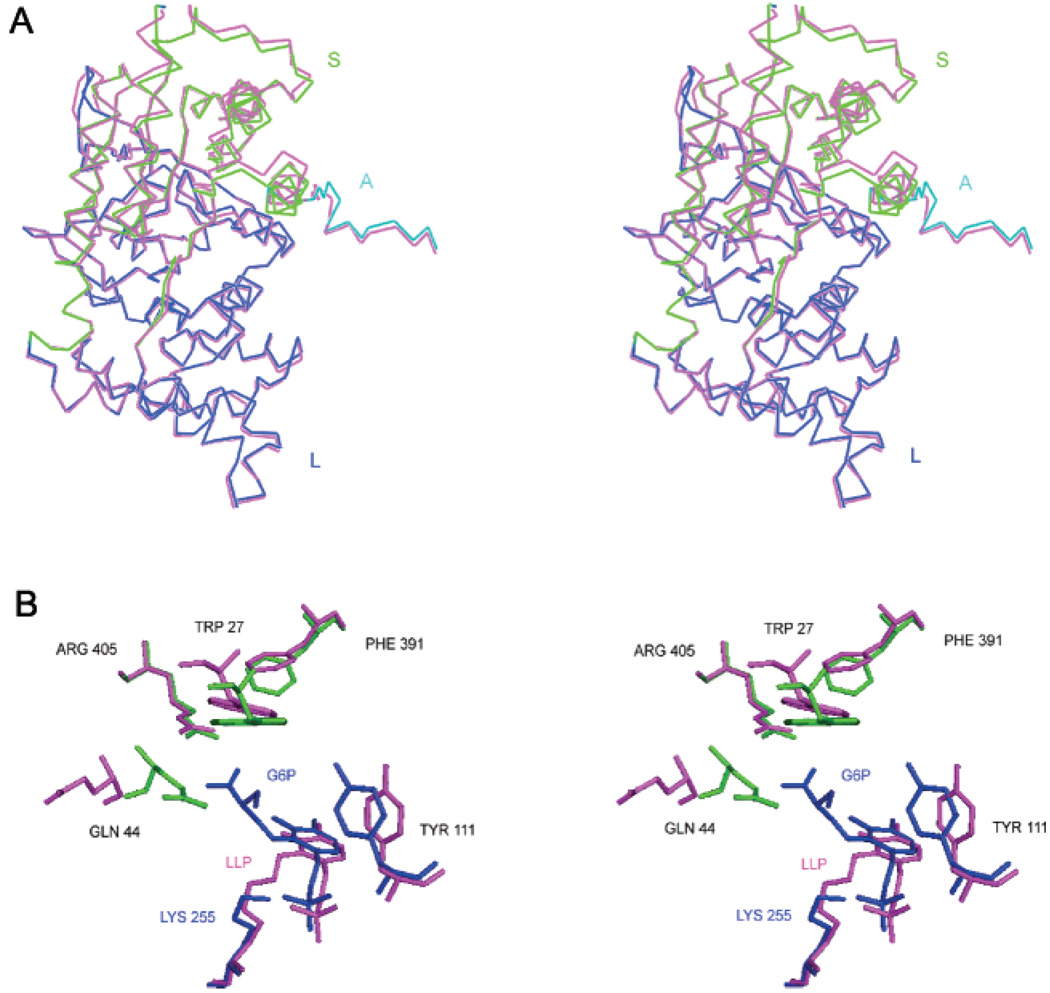
On superposition of chain A onto chain B of the AeKAT–cysteine complex (rmsd = 0.7666 Å) (Figure 3A), we identified the residues within 5 Å of cysteine and found that chain A and chain B differ in the side chain conformations of Trp27, Gln44, Tyr111, Phe391, and Arg405 that tilt or move closer to the substrate cysteine. In particular, the NE2 atom of Gln44 in chain B moved as much as 7.9 Å to form hydrogen bonds with the substrate (Figure 3B). By binding the substrate cysteine, the small domain of chain B rotates toward the cofactor (Figure 3A), which makes most of the residues around the active center closer to the cofactor and encapsulates the substrate within the center. Therefore, chain B has a closed conformation. The built-in cavity at the protein surface is clearly seen as the open form in chain A (Figure 4A), in which a tunnel-like unoccupied space in the cavity appears to serve as the entry path for the substrate. The PLP cofactor can be clearly seen through the tunnel-like structure. In contrast, the tunnel is closed in chain B that contains a cysteine (Figure 4B). The built-in cavity is constructed by the residues at the domain interface and the subunit interface of the enzyme. The main residues building the cavity wall are Leu69, Phe135, Asn193, and Tyr224 from the large domain at chain B, Trp27, Ile31, Ala35, Lys38, Asn41, Gln44, Gly45, Phe46, Phe347, and Arg405 from the small domain at chain B, and Tyr73 and Tyr286 from chain A.
Figure 3.
(A) α-Carbon representation in stereo of chain B (closed form) superimposed onto chain A (open form, colored pink) of the AeKAT–cysteine complex. The small domain (S), arm (A), and large domain (L) are colored blue, green, and light blue, respectively. (B) Superposition of chain B onto chain A of the AeKAT–cysteine complex. Only protein residues having different conformations within 5 Å of the cysteine substrate are depicted as sticks. The residues and ligand from chain A are colored pink and those from chain B colored green or blue.
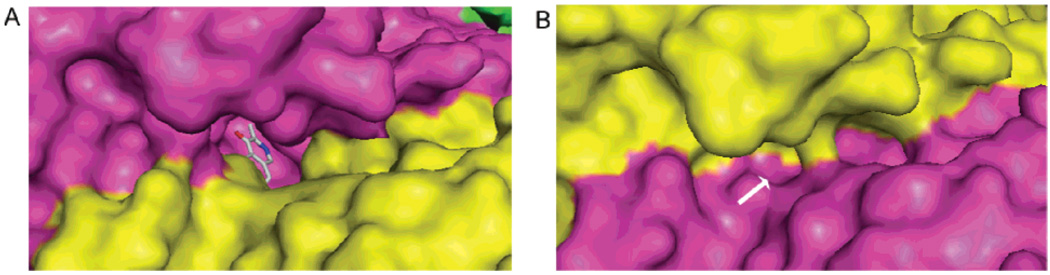
Figure 4.
Molecular surface of the AeKAT–cysteine complex. (A) Surface around the active center of the open form, chain A. The built-in cavity is clearly seen, and the cofactor is seen through the tunnel. (B) Surface around the active center of the closed form, chain B. The built-in cavity is much smaller than the cavity in the open form, and the tunnel to the cofactor is closed.
Substrate Recognition and Catalysis in the AeKAT–Glutamine Complex
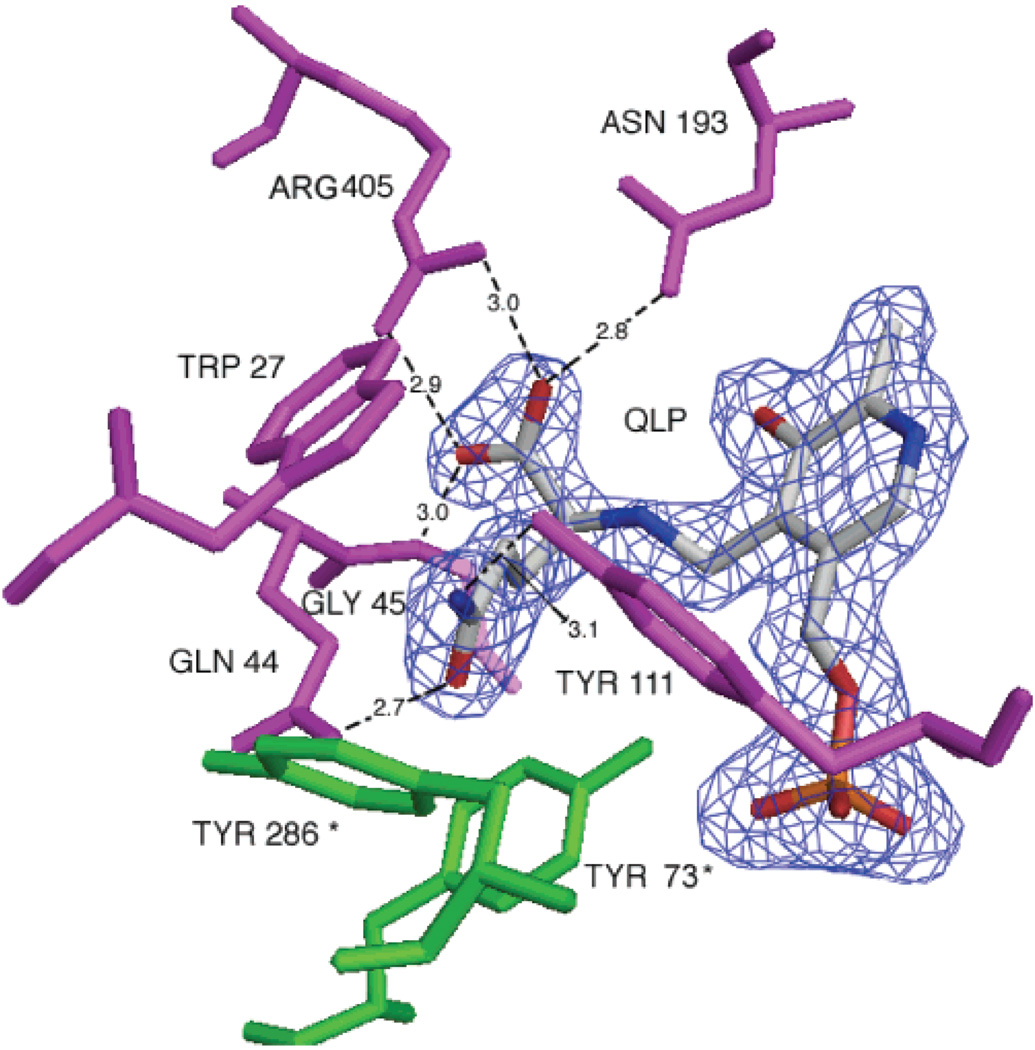
Similar to the crystal structure of AeKAT–cysteine complex, the substrate lies near the PLP cofactor and forms an external aldimine with PLP in each subunit of the AeKAT–glutamine complex (Figure 5). Several residues, including Arg405, Tyr224, Asn193, Gly45, Trp27, Gln44, Tyr111, and Phe135, define the substrate-binding site and contact the glutamine molecule in both subunits of the biological dimer (Figure 5). Upon superposition of chain A onto chain B of the AeKAT–glutamine complex (rmsd = 0.3263 Å), we determined that both chains have a closed conformation. The interactions between glutamine and protein are basically the same as the interactions between cysteine and protein, with the exception that glutamine has an increased number of interactions. For example, the glutamine R group of QLP is stabilized by hydrogen bonds with the OH atom of Tyr73 (A chain) (3.9 Å), the OH atom of Tyr111 (3.1 Å), and the NE2 atom of Gln44 (2.7 Å), and also through hydrophobic interactions with the ring of Tyr286 (A chain) (3.8 Å). In contrast, the cysteine R group of C6P interacts with only the NE2 atom of Gln44 (3.4 Å).
Figure 5.
Glutamine binding site. The glutaminyl aldimine (QLP) and the protein residues within 4 Å of the glutamine substrate are shown. The residues colored pink are from chain A and residues colored green from chain B. The 2Fo – Fc electron density map covering the QLP is shown contoured at the 1.2σ level. This figure was generated with PyMol.
DISCUSSION
By cocrystallizing with the substrates cysteine, glutamine, kynurenine, and tyrosine, AeKAT was cocrystallized with only an excess of the substrate cysteine, yielding crystals of a true intermediate complex, cysteinyl aldimine. The AeKAT–cysteine complex represents an excellent model of the complex structure free from the bias caused by structural differences between substrate analogues (or inhibitors). Cysteine not only is one of the best substrates for AeKAT but also affects enzyme activity toward kynurenine, enhancing kynurenic acid production between 0.6 and 2.5 mM and inhibiting kynurenic acid formation at >5 mM (5). The inhibition can be explained by the mechanism of substrate competition, but it is unclear how cysteine increases enzyme activity toward kynurenine. The enzyme is crystallized at 2.5 mM cysteine in the crystallization buffer but is not crystallized at the same concentration of other substrates, which suggests that cysteine may help stabilize the enzyme. Cysteine is found in only one subunit of the biological dimer. In contrast, crystals of the AeKAT–glutamine complex are obtained by soaking the native crystals with glutamine, and glutamine is found in both subunits of the AeKAT–glutamine complex structure. Both cysteine and glutamine are good amino group donors for AeKAT with Km values of 1.3 mM with cysteine and 3.8 mM with glutamine (5). The concentrations of cysteine and glutamine are the same in the cocrystallization buffer and the soaking buffer. Theoretically, cysteine should be somewhat easier than glutamine to bind to the active site of AeKAT based on their affinity for the enzyme. The presence of cysteine in just one of the two subunits as determined by cocrystallization and the presence of glutamine in both subunits as determined by soaking suggest that the kinetic constants of the enzyme are different between crystal and solution forms. This should serve as an example illustrating the different behaviors between crystal and solution forms of an enzyme.
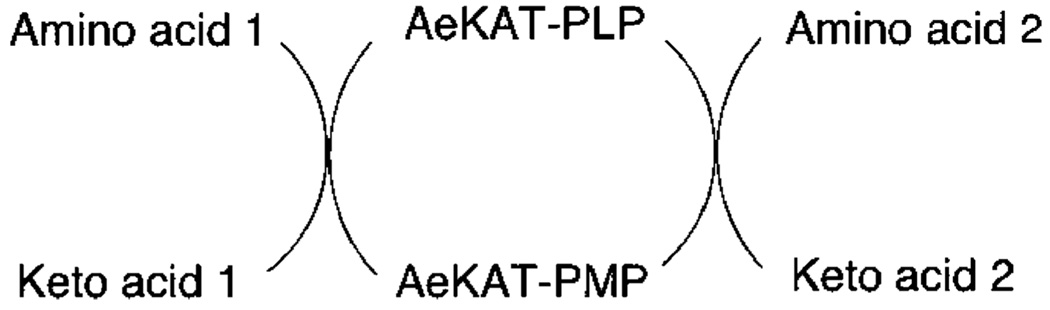
AeKAT conducts ping-pong bi-bi reactions to catalyze keto acid 1 and amino acid 2 to amino acid 1 and keto acid 2, respectively. As shown in Scheme 1, amino acid 1 adds to the AeKAT-PLP form of the enzyme, producing the AeKAT–amino acid 1 complex, but then the enzyme is covalently altered to form a complex between a stable enzyme form, AeKAT-PMP, and the first product of keto acid 1. Keto acid 1 is then ejected, and a second substrate, keto acid 2, adds to the enzyme. The amino group originally transferred to AeKAT-PLP to form AeKAT-PMP is then transferred to keto acid 2, forming amino acid 2 and regenerating the AeKAT-PLP form. Finally, amino acid 2 is ejected, and the cycle is complete (23). The fact that cysteine is found in only one subunit of the biological dimer in the cocrystallized crystal structure is interesting. In the complex of Paracoccus denitrificans aromatic amino acid aminotransferase with 3-phenylpropionate (33) and the complex of A. aegypti alanine glyoxylate aminotransferase with alanine (34), the substrate also was observed in only one subunit of their homodimers. On the basis of these results, it seems reasonable to suggest that the two subunits of AeKAT are not synchronized during the enzyme-catalyzed reaction through a ping-pong bi-bi mechanism and that the catalyzing mechanism of the aminotransferases might be more complicated than we thought.
Scheme 1.
AeKAT-Catalyzed Transamination Reaction
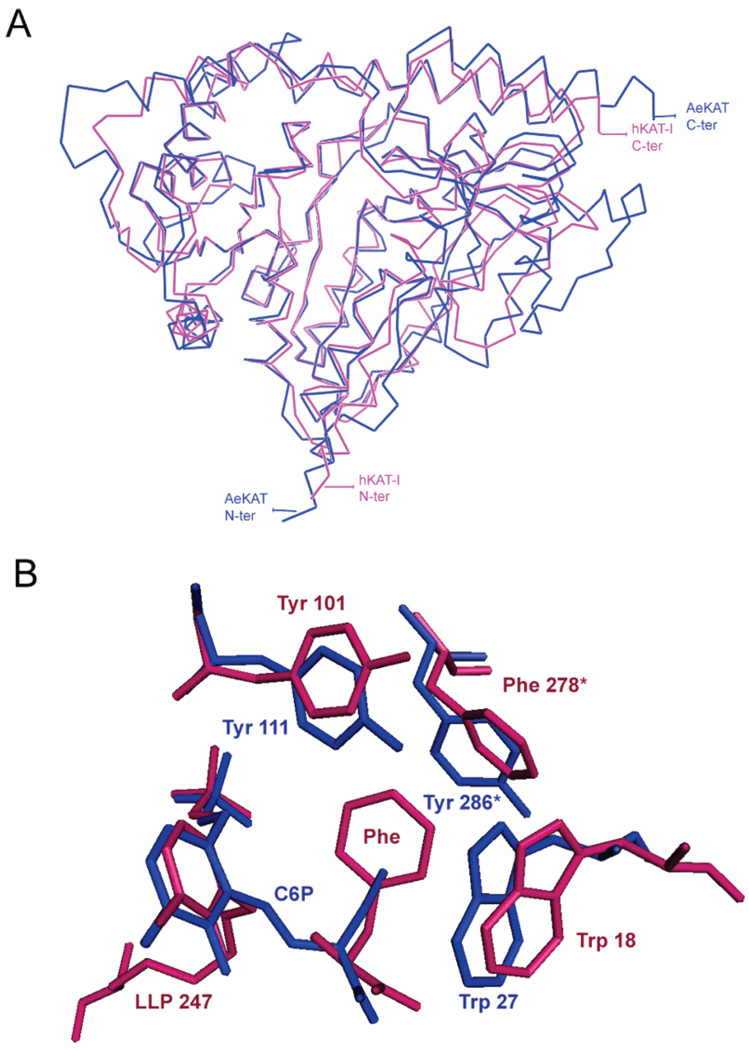
The substrate profiles of AeKAT and hKAT I are essentially the same, including aromatic amino acids (kynurenine, phenylalanine, tryptophan, and tyrosine), sulfurcontaining amino acids (methionine and cysteine), and other amino acids (glutamine, leucine, histidine, asparagine, and aminobutyrate). However, they exhibit catalytic efficiencies quite different from those of the amino acids mentioned above. hKAT I is more efficient in transaminating hydrophobic amino acids (such as phenylalanine, leucine, and tryptophan), while AeKAT is more efficient in transaminating relatively hydrophilic amino acids (such as cysteine, histidine, tyrosine, and methionine) (4, 5). Upon superposition of a dimer of the AeKAT–cysteine complex onto the hKAT I–phenylalanine structure (Figure 6A), we identified the residues within 5 Å of both ligands and found that most residues were the same. An exception was that Tyr286 of AeKAT was replaced with Phe278 in hKAT I (Figure 6B). This difference between the two enzymes might explain their difference in terms of catalytic efficiency with hydrophobic and hydrophilic substrates. A sequence alignment shows that KAT I sequences from different mammalian species have a Phe residue in this position, but insect KAT and mammalian KAT III sequences have Tyr residue at the same site (Figure 7D). Upon superposition of an AeKAT–cysteine complex ligand binding chain onto a chain of the AeKAT–glutamine complex (Figure 7C), we did not observe any significant conformational changes in the two structures. Inspection of the electron density map indicates that the OH atom of Tyr286 can interact with the NE2 atom of Gln44, which in turn may impact substrate binding (Figure 7A,B). Therefore, the Tyr286 at the active site of AeKAT might enhance the binding of relatively hydrophilic substrates, while the corresponding Phe residue at the same position might better interact with relatively hydrophobic substrates. However, the difference (tyrosine vs phenylalanine) does not seem to change the substrates of the KAT enzymes, because AeKAT and human KAT I possess the same substrate profiles.
Figure 6.
(A) α-Carbon representation of the AeKAT–cysteine structure (blue) superimposed onto the hKAT I–phenylalanine structure (pink). (B) Superposition of the AeKAT–cysteine structure onto the hKAT I–phenylalanine structure. Only different protein residues within 5 Å of the cysteine substrate or phenylalanine substrate are depicted as sticks. The residues and ligand form of AeKAT are colored blue and those from hKAT I colored red.
It is well-known that by binding its substrates, aspartate aminotransferase changes its conformation from the open form to the closed form (35–40). This substrate-mediated conformation change is also observed in other aminotransferases (21, 33, 41). The AeKAT–cysteine and AeKAT–glutamine structures show that the protein subunits are crystallized as closed forms when they bind substrates. It is possible that the approach of cysteine or glutamine to the built-in cavity, which is exposed to the solvent region, induces movement of a subdomain (small domain here) toward the large domain. By doing so, the enzyme encapsulates the substrate within the cavity to produce a Michaelis complex. Therefore, all the residues on the cavity wall, instead of only the binding residues, might be collectively involved in the substrate-mediated conformation change and substrate binding.
ACKNOWLEDGMENT
This work was carried out in part at the National Synchrotron Light Source, Brookhaven National Laboratory.
Footnotes
This work was supported by Grant AI 44399 from the National Institutes of Health.
The atomic coordinates and structure factors (entries 2r5c and 2r5e) have been deposited in the Protein Data Bank.
Abbreviations: AeKAT, Aedes aegypti kynurenine aminotransferase; C6P, cysteinyl aldimine; KAT, kynurenine aminotransferase; KYNA, kynurenic acid; LLP, lysine pyridoxal 5′-phosphate; PLP, pyridoxal 5′-phosphate; QLP, glutaminyl aldimine; rmsd, root-mean-square deviation.
REFERENCES
- 1.Yu P, Li Z, Zhang L, Tagle DA, Cai T. Characterization of kynurenine aminotransferase III, a novel member of a phylogenetically conserved KAT family. Gene. 2006;365:111–118. doi: 10.1016/j.gene.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 2.Schwarcz R. The kynurenine pathway of tryptophan degradation as a drug target. Curr. Opin. Pharmacol. 2004;4:12–17. doi: 10.1016/j.coph.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Cooper AJ. The role of glutamine transaminase K (GTK) in sulfur and α-keto acid metabolism in the brain, and in the possible bioactivation of neurotoxicants. Neurochem. Int. 2004;44:557–577. doi: 10.1016/j.neuint.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Han Q, Li J, Li J. pH dependence, substrate specificity and inhibition of human kynurenine aminotransferase I. Eur. J. Biochem. 2004;271:4804–4814. doi: 10.1111/j.1432-1033.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 5.Han Q, Li J. Cysteine and keto acids modulate mosquito kynurenine aminotransferase catalyzed kynurenic acid production. FEBS Lett. 2004;577:381–385. doi: 10.1016/j.febslet.2004.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosono A, Mizuguchi H, Hayashi H, Goto M, Miyahara I, Hirotsu K, Kagamiyama H. Glutamine:phenylpyru-vate Aminotransferase from an Extremely Thermophilic Bacterium, Thermus thermophilus HB8. J. Biochem. (Tokyo) 2003;134:843–851. doi: 10.1093/jb/mvg210. [DOI] [PubMed] [Google Scholar]
- 7.Leeson PD, Iversen LL. The glycine site on the NMDA receptor: structure-activity relationships and therapeutic potential. J. Med. Chem. 1994;37:4053–4067. doi: 10.1021/jm00050a001. [DOI] [PubMed] [Google Scholar]
- 8.Perkins MN, Stone TW. An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res. 1982;247:184–187. doi: 10.1016/0006-8993(82)91048-4. [DOI] [PubMed] [Google Scholar]
- 9.Birch PJ, Grossman CJ, Hayes AG. Kynurenic acid antagonises responses to NMDA via an action at the strychnine-insensitive glycine receptor. Eur. J. Pharmacol. 1988;154:85–87. doi: 10.1016/0014-2999(88)90367-6. [DOI] [PubMed] [Google Scholar]
- 10.Pereira EF, Hilmas C, Santos MD, Alkondon M, Maelicke A, Albuquerque EX. Unconventional ligands and modulators of nicotinic receptors. J. Neurobiol. 2002;53:479–500. doi: 10.1002/neu.10146. [DOI] [PubMed] [Google Scholar]
- 11.Hilmas C, Pereira EF, Alkondon M, Rassoulpour A, Schwarcz R, Albuquerque EX. The brain metabolite kynurenic acid inhibits α7 nicotinic receptor activity and increases non-α7 nicotinic receptor expression: Physiopathological implications. J. Neurosci. 2001;21:7463–7473. doi: 10.1523/JNEUROSCI.21-19-07463.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkondon M, Pereira EF, Yu P, Arruda EZ, Almeida LE, Guidetti P, Fawcett WP, Sapko MT, Randall WR, Schwarcz R, Tagle DA, Albuquerque EX. Targeted deletion of the kynurenine aminotransferase ii gene reveals a critical role of endogenous kynurenic acid in the regulation of synaptic transmission via α7 nicotinic receptors in the hippocampus. J. Neurosci. 2004;24:4635–4648. doi: 10.1523/JNEUROSCI.5631-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Simonavicius N, Wu X, Swaminath G, Reagan J, Tian H, Ling L. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006;281:22021–22028. doi: 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 14.Mellor AL, Munn DH. IDO expression by dendritic cells: Tolerance and tryptophan catabolism. Nat. Rev. Immunol. 2004;4:762–774. doi: 10.1038/nri1457. [DOI] [PubMed] [Google Scholar]
- 15.Taylor MW, Feng GS. Relationship between interferon-γ, indoleamine 2,3-dioxygenase, and tryptophan ca-tabolism. FASEB J. 1991;5:2516–2522. [PubMed] [Google Scholar]
- 16.Stone TW, Darlington LG. Endogenous kynurenines as targets for drug discovery and development. Nat. Rev. Drug Discovery. 2002;1:609–620. doi: 10.1038/nrd870. [DOI] [PubMed] [Google Scholar]
- 17.Han Q, Fang J, Li J. Kynurenine aminotransferase and glutamine transaminase K of Escherichia coli: Identity with aspartate aminotransferase. Biochem. J. 2001;360:617–623. doi: 10.1042/0264-6021:3600617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang J, Han Q, Li J. Isolation, characterization, and functional expression of kynurenine aminotransferase cDNA from the yellow fever mosquito, Aedes aegypti(1) Insect Biochem. Mol. Biol. 2002;32:943–950. doi: 10.1016/s0965-1748(02)00032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi F, Han Q, Li J, Li J, Rizzi M. Crystal structure of human kynurenine aminotransferase I. J. Biol. Chem. 2004;279:50214–50220. doi: 10.1074/jbc.M409291200. [DOI] [PubMed] [Google Scholar]
- 20.Han Q, Gao YG, Robinson H, Ding H, Wilson S, Li J. Crystal structures of Aedes aegypti kynurenine ami-notransferase. FEBS J. 2005;272:2198–2206. doi: 10.1111/j.1742-4658.2005.04643.x. [DOI] [PubMed] [Google Scholar]
- 21.Goto M, Omi R, Miyahara I, Hosono A, Mizuguchi H, Hayashi H, Kagamiyama H, Hirotsu K. Crystal Structures of Glutamine:Phenylpyruvate Aminotransferase from Thermus thermophilus HB8: Induced Fit and Substrate Recognition. J. Biol. Chem. 2004;279:16518–16525. doi: 10.1074/jbc.M311575200. [DOI] [PubMed] [Google Scholar]
- 22.Jansonius JN. Structure, evolution and action of vitamin B6-dependent enzymes. Curr. Opin. Struct. Biol. 1998;8:759–769. doi: 10.1016/s0959-440x(98)80096-1. [DOI] [PubMed] [Google Scholar]
- 23.Eliot AC, Kirsch JF. Pyridoxal phosphate enzymes: Mechanistic, structural, and evolutionary considerations. Annu. Rev. Biochem. 2004;73:383–415. doi: 10.1146/annurev.biochem.73.011303.074021. [DOI] [PubMed] [Google Scholar]
- 24.Otwinowski Z. In: HKL User’s Manual. Sawyer L, Isaacs N, Bailey S, editors. Warrington, U.K: Daresbury Laboratory; 1993. pp. 56–62. [Google Scholar]
- 25.Minor W. Ph.D. Dissertation. West Lafayette, IN: Purdue University; 1993. [Google Scholar]
- 26.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47(Part 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 29.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: Improvement and extension of crystallographic phases by weighted averaging of multiple refined dummy atomic models. Acta Crystallogr. 1997;D53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. Crystal Physics, Diffraction, Theoretical and General Crystallography. Acta Crystallogr. 1976;A32:922–923. [Google Scholar]
- 31.DeLano WL. PyMol. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 32.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 33.Okamoto A, Nakai Y, Hayashi H, Hirotsu K, Kagamiyama H. Crystal structures of Paracoccus denitrificans aromatic amino acid aminotransferase: A substrate recognition site constructed by rearrangement of hydrogen bond network. J. Mol. Biol. 1998;280:443–461. doi: 10.1006/jmbi.1998.1869. [DOI] [PubMed] [Google Scholar]
- 34.Han Q, Robinson H, Gao YG, Vogelaar N, Wilson SR, Rizzi M, Li J. Crystal Structures of Aedes aegypti Alanine Glyoxylate Aminotransferase. J. Biol. Chem. 2006;281:37175–37182. doi: 10.1074/jbc.M607032200. [DOI] [PubMed] [Google Scholar]
- 35.Rhee S, Silva MM, Hyde CC, Rogers PH, Metzler CM, Metzler DE, Arnone A. Refinement and comparisons of the crystal structures of pig cytosolic aspartate aminotransferase and its complex with 2-methylaspartate. J. Biol. Chem. 1997;272:17293–17302. [PubMed] [Google Scholar]
- 36.Miyahara I, Hirotsu K, Hayashi H, Kagamiyama H. X-ray crystallographic study of pyridoxamine 5′-phosphate-type aspartate aminotransferases from Escherichia coli in three forms. J. Biochem. (Tokyo) 1994;116:1001–1012. doi: 10.1093/oxfordjournals.jbchem.a124620. [DOI] [PubMed] [Google Scholar]
- 37.Malashkevich VN, Strokopytov BV, Borisov VV, Dauter Z, Wilson KS, Torchinsky YM. Crystal structure of the closed form of chicken cytosolic aspartate aminotransferase at 1.9 Å resolution. J. Mol. Biol. 1995;247:111–124. doi: 10.1006/jmbi.1994.0126. [DOI] [PubMed] [Google Scholar]
- 38.McPhalen CA, Vincent MG, Jansonius JN. X-ray structure refinement and comparison of three forms of mitochondrial aspartate aminotransferase. J. Mol. Biol. 1992;225:495–517. doi: 10.1016/0022-2836(92)90935-d. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto A, Higuchi T, Hirotsu K, Kuramitsu S, Kagamiyama H. X-ray crystallographic study of pyridoxal 5′-phosphate-type aspartate aminotransferases from Escherichia coli in open and closed form. J. Biochem. (Tokyo) 1994;116:95–107. doi: 10.1093/oxfordjournals.jbchem.a124509. [DOI] [PubMed] [Google Scholar]
- 40.Jager J, Moser M, Sauder U, Jansonius JN. Crystal structures of Escherichia coli aspartate aminotransferase in two conformations. Comparison of an unliganded open and two liganded closed forms. J. Mol. Biol. 1994;239:285–305. doi: 10.1006/jmbi.1994.1368. [DOI] [PubMed] [Google Scholar]
- 41.Hirotsu K, Goto M, Okamoto A, Miyahara I. Dual substrate recognition of aminotransferases. Chem. Rec. 2005;5:160–172. doi: 10.1002/tcr.20042. [DOI] [PubMed] [Google Scholar]