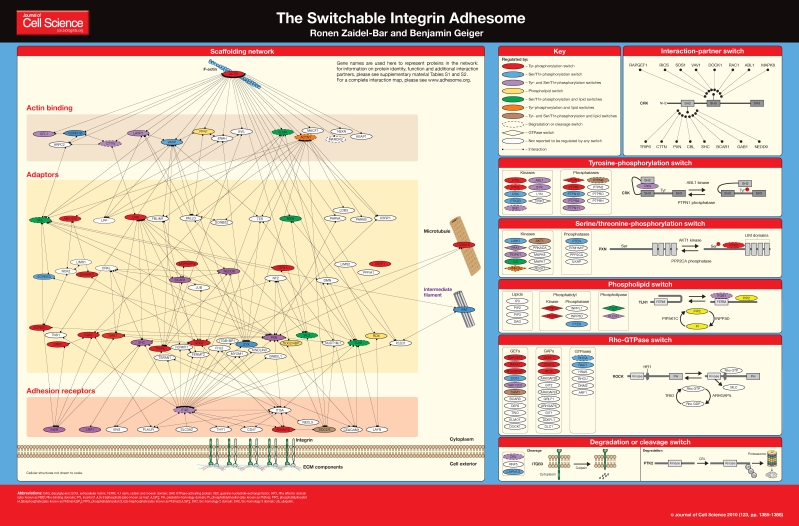
Adhesion sites, which interconnect cells with their neighbors or with the extracellular matrix (ECM), are large multiprotein complexes that provide mechanical coupling as well as a means for cells to sense the chemical and physical properties of their environment (Bershadsky et al., 2003; Chen et al., 2004; Geiger et al., 2001; Sastry and Burridge, 2000). The complex interplay between the mechanical role of cell adhesions and their ‘instructive role’, which is manifested by the activation of a wide variety of signaling networks, is mediated by a group of proteins collectively known as the ‘adhesome’. The concerted activity of adhesome components affects essentially all cellular functions, including morphogenesis, migration, proliferation, differentiation and viability (Berrier and Yamada, 2007; Streuli, 2009; Thiery, 2003; Vicente-Manzanares et al., 2009).
Adhesion to the ECM is mediated via heterodimeric transmembrane receptors, namely, α- and β-integrins. Combinations of among 18 α-chains and eight β-chains form different heterodimers to yield a rich diversity of ECM receptors, enabling different cell types to respond differentially to variations in the ECM (Arnaout et al., 2005). On the cytoplasmic side of the adhesion sites, integrins can interact, via their cytoplasmic tails, with at least 12 different adaptor proteins (Zaidel-Bar et al., 2007a). Among these molecules, tensin (encoded by the TNS1 gene) (hereafter, the gene encoding each protein will be shown in italics in parentheses), filamin (FLNA), talin (TLN1), plectin (PLEC1) and α-actinin (ACTN1) can provide a direct link to the actin cytoskeleton. Plectin can also directly interact with the intermediate-filament protein vimentin (VIM), and paxillin (PXN) can provide a direct link to microtubules. (Information concerning all interactions mentioned in the text and shown in the poster can be found in supplementary material Tables S1 and S2.) Links with the cytoskeleton are further reinforced by a second and third tier of adaptor molecules, which stabilize the adhesome network and connect to the various filament systems of the cell. For example, α-integrin (ITGA) can bind to paxillin, which can bind to actopaxin (PARVA), which binds to actin, and β-integrin (ITGB) can bind to kindlin-1 (FERMT1), which can bind to migfilin (FBLIM1), which can further bind to VASP, which binds to actin. Adaptors also bind to a variety of signaling proteins, some of which regulate the adhesion site itself (see below), and others feed into diverse cellular signaling pathways.
The integrin-actin connection is pivotal for the mechanosensory function of the adhesion: forces applied to integrins by contractile actomyosin bundles, for example, play an important role in recruitment to, and reinforcement of, the adhesion site, although the exact mechanism remains unclear (Bershadsky et al., 2006; Chen et al., 2004).
The very high level of apparent connectivity that characterizes the adhesome network points to an intriguing aspect of the regulation of integrin-mediated adhesion. On one hand, the dense connectivity of adhesome components can contribute to the robustness of integrin adhesions, which remain intact even after the removal of many nodes (components) (Zaidel-Bar et al., 2007a); yet, on the other hand, dynamic analyses (e.g. fluorescence recovery after photobleaching and quantitative time-lapse microscopy) indicate that the receptors, adaptors and actin are in a constant state of flux (Hu et al., 2007; Worth and Parsons, 2008). Therefore, it is expected that most interactions between adhesome components are transient, and it is the dynamic nature of the adhesion site that makes it sensitive and responsive to external signaling cues and forces. So, how can the adhesome be both robust and highly dynamic at the same time?
The switchable adhesome
The large number of proteins identified over the years as components of integrin-mediated cell-ECM adhesions is daunting. The last published inventory included 156 components (Zaidel-Bar et al., 2007a), and this number continues to grow. The updated version of the adhesome network presented herein includes 180 nodes, with the addition of proteins discovered in RNA-interference and yeast two-hybrid screens and from in vitro localization studies that have been published in the interim.
Even more overwhelming is the number of direct interactions reported between these components, which, according to our update, is at least 742. As the adhesome network grows in complexity and connectivity, it is becoming increasingly apparent that, to accommodate the need for both robustness and dynamic plasticity at the adhesion site, most if not all of these interactions can be switched ‘on’ or ‘off’. Thus, viewing the entire network with its wealth of interactions as if it were in an ‘on’ state at all times is most likely misleading. It is somewhat analogous to starting a car and having all of the dashboard operation indicators and warning lights flashing when you turn the key in the ignition – a situation you don't expect to occur in any real driving scenario.
In this poster article, we present only the scaffolding network of the integrin adhesome (i.e. adhesion receptors, adaptors, actin regulators and the associated cytoskeleton) with its known interactions. Regulatory proteins are shown, but they are not connected by arrows to their targets. Instead, information about the regulation of each protein is color-coded to indicate whether it is, for example, phosphorylated at a tyrosine residue (red), a serine or threonine residue (blue), phosphorylated at both tyrosine and serine or threonine residues (purple) and so on. The purpose of this presentation is to highlight the fact that more than half of the adhesome proteins can be regulated. Detailed information about all known adhesome components and their interactions is available in table format in supplementary material Tables S1 and S2, and as an interactive map at www.adhesome.org.
As discussed above, many of the interactions within the adhesome are regulated such that the same protein might be engaged in different interactions under varying conditions. We refer to such changes in interaction as ‘switches’. To illustrate the ‘switchability’ of the adhesome network, in the sections below we define the basic switching mechanisms that are found in integrin adhesion networks. Throughout our discussion, we intentionally avoid classifying the switches as either activating or inhibiting, as a given modification might activate some molecular functions but inhibit others. For the sake of simplicity, we discuss the different switches in the adhesome separately; however, it is clear that these switches are often interconnected.
Interaction-partner switch
On average, each adhesome protein has approximately nine different potential partners (Zaidel-Bar et al., 2007a). Although some multidomain proteins, such as Vav and Trio, can bind to several partners simultaneously, most proteins have fewer interaction domains than they do potential molecular partners. Hence, most proteins will be bound at any given time to only a fraction of their potential binding partners. For example, Crk (CRK), which has three major interaction domains [a Src-homology 2 (SH2) domain and two SH3 domains], can interact with at least 16 different proteins, half of which can interact with the SH3 domains via their proline-rich sequences, and half of which interact with the SH2 domain via phosphorylated tyrosine residues. The SH2-domain partners are mostly other adaptor proteins, whereas the SH3-domain partners are mostly guanine-nucleotide-exchange factors (GEFs) for GTPases. Because only some of the potential binding partners are expressed in any given cell type, the actual partner ‘switchboard’ might be simpler than what is derived from the adhesome database. However, many of these switches are real, and the ‘choice’ of a particular interaction partner can affect the structure and function of the adhesion site. Furthermore, different subpopulations of a given protein might interact with different binding partners in parallel, in the same cell and possibly within the same adhesion site, compounding the complexity of the functional networking of adhesome components.
Conformational switch
Incidentally, Crk also serves as an excellent example of a conformational switch. A switch in protein conformation can be triggered by a change in temperature, pH or ion concentration; by the binding of another protein; or, as with Crk, by a post-translational modification. Some adhesome proteins, such as vinculin (VCL), talin, focal adhesion kinase (PTK2), Src and testis-derived transcript (TES), can exist in either a closed (inactive) or open (active) conformation (Critchley, 2004; Garvalov et al., 2003; Mitra and Schlaepfer, 2006). In the closed conformation, which is usually maintained by specific interactions between the N- and C-terminal domains, important activity modules are concealed within the folded protein. The closed-to-open conformational switch involves a dramatic change in protein folding. Other conformational switches invoke smaller, but no less crucial, changes in amino acid position. For example, the extracellular domains of integrins are activated to bind ligands following specific interactions between their cytoplasmic tails and talin (Shimaoka et al., 2002).
Notably, it has also been proposed that force-induced conformational switches might play a role in mechanosensing by the adhesome (Bershadsky et al., 2006; Lee et al., 2007; Sawada et al., 2006). Proof of this concept comes from recent work demonstrating that the application of physiologically relevant forces to single talin rods causes stretching that exposes cryptic binding sites for vinculin (del Rio et al., 2009), and from the finding that a tension-induced conformational switch in α5β1 integrin facilitates a higher-strength crosslinked bond with fibronectin (Friedland et al., 2009).
Tyrosine-phosphorylation switch
Rapidly assembling and disassembling integrin adhesion sites are highly tyrosine phosphorylated, and the phosphorylation state of proteins such as Src, focal adhesion kinase and paxillin is directly associated with their dynamics at these sites (Panetti, 2002; Zaidel-Bar et al., 2007b). Nine kinases and nine phosphatases regulate the tyrosine-phosphorylation status of adhesion receptors, adaptors and actin regulators, as well as many of the regulatory proteins of the adhesome. (Proteins regulated by tyrosine phosphorylation are colored in the poster in red, purple, orange or brown, depending on whether they are also regulated by another type of switch.) A phosphorylated tyrosine residue can affect a protein in several ways: it can, for example, create a docking site for an SH2 domain of a partner protein or even of the same protein, often leading to a conformational change. This modification, in turn, can modulate enzymatic activity or binding affinities.
In the example for a tyrosine-phosphorylation switch shown in the poster, Crk is locked into a folded state by an interaction between its N-terminal SH2 domain and phosphorylated tyrosine 221, which is located close to its C-terminus (Rosen et al., 1995). Tyrosine 221 is phosphorylated by Abelson murine leukemia viral oncogene homolog 1 (ABL1), which can also bind to Crk in the folded conformation. Conversely, dephosphorylation of Crk by protein tyrosine phosphatase, non-receptor type 1 (PTPN1) opens up the protein, rendering its SH2 domain available for binding to one of several phospho-proteins (e.g. paxillin).
Compared with the overall proteome, tyrosine-kinase and SH2 domains are significantly enriched among components of the adhesome, underscoring the importance of the tyrosine-phosphorylation switch in regulating adhesome dynamics (Zaidel-Bar et al., 2009). Importantly, tyrosine phosphorylation seems to be an important transducer of mechanical forces into biochemical signals. For example, protein tyrosine phosphatase receptor type α (PTPRA)-dependent activation of Src-family kinases is required for the force-dependent formation of focal complexes, which are an early form of integrin-mediated ECM contacts (von Wichert et al., 2003). Interestingly, dephosphorylation of paxillin and p130cas (BCAR1) seems to play a role in the polar response of endothelial cells to shear flow (Zaidel-Bar et al., 2005).
Serine/threonine-phosphorylation switch
Fifteen kinases and phosphatases regulate the phosphorylation status of specific serine or threonine residues of proteins from all functional groups of the adhesome. (Proteins regulated by serine/threonine phosphorylation are colored in the poster in blue, purple, green or brown, depending on whether they are also regulated by another type of switch.) These phosphorylation events often activate a conformational switch, which can further affect enzymatic activity, receptor availability or the binding affinity for other proteins. Both activating and inhibiting effects of serine or threonine phosphorylation have been documented. In the example shown in the poster, paxillin (PXN) phosphorylation at serine 178 by Akt (AKT1) is required for the binding of focal adhesion kinase (PTK2) (Huang et al., 2008); this interaction is reversed upon dephosphorylation by PPP2CA.
Phospholipid switch
Phosphoinositides are minor phospholipids that originate in the plasma membrane and can bind to some receptors, adaptors and actin regulators. (In the poster, proteins regulated by phospholipids are colored yellow, green, orange or brown, depending on whether they are also regulated by another type of switch). The precursor of all phosphoinositides is phosphatidylinositol (PtdIns). Two phosphatidyl kinases and two phosphatases regulate the formation of phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] from PtdIns. The kinase phosphoinositide 3-kinase (PI3K) can further phosphorylate PtdIns(4,5)P2 into phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3], an action that can be reversed by the PtdIns(3,4,5)P3 phosphatase PTEN (PTEN); phospholipases can hydrolyze PtdIns(4,5)P2 into inositol (1,4,5)-trisphosphate [Ins(1,4,5)P3] and diacylglycerol (DAG) (van den Bout and Divecha, 2009). With the exception of Ins(1,4,5)P3, which is soluble, all of the phosphoinositides remain at the plasma membrane and can act as switches only on proteins that are close to the membrane. Levels of PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in the plasma membrane influence the recruitment of proteins harboring pleckstrin homology (PH) domains (Irvine, 1998). Notably, the PH domain is the second most prevalent motif found in the adhesome, being present in 23 components.
Many of the proteins regulated by PtdIns(4,5)P2 are actin-binding proteins, such as ezrin, radixin, moesin (ERM), filamin and α-actinin. These proteins regulate the architecture of the cytoskeleton at the adhesion site (Sechi and Wehland, 2000).
In the example illustrated in the phospholipid-switch panel of the poster, talin (TLN1) arrives at the membrane in a folded conformation in which it cannot bind β1 integrin (ITGB1), but it can recruit phosphatidylinositol 4-phosphate 5-kinase (PIP5K) to the membrane, where it catalyzes the formation of PtdIns(4,5)P2. Binding of PtdIns(4,5)P2 to talin unlocks this important protein, revealing binding sites for β1 integrin, as well as for actin and vinculin (Goksoy et al., 2008).
Rho-GTPase switch
Rho GTPases are found in either an active GTP-bound state or in an inactive GDP-bound state. They are switched ‘on’ by GEFs, and ‘off’ by GTPase-activating proteins (GAPs) (Hall, 2005). In their active state, Rho GTPases can bind to specific Rho-binding domains in some of the kinases and phosphatases of the adhesome (shown in the poster as diamonds) and switch them on. For example, the serine/threonine kinase activity of Rho-associated coiled-coil-containing protein kinase (ROCK) is turned on after binding to active Rho. Phosphorylation by ROCK activates the myosin light chain and inhibits the myosin light chain phosphatase, leading to an increase in actomyosin contractility (Totsukawa et al., 2000). Many GEFs and GAPs bind to adaptors within the adhesome, and their recruitment can locally activate or deactivate Rho GTPases in the vicinity of the adhesion site. For example, phosphorylated paxillin recruits p120RasGAP, which releases p190RhoGAP and leads to local inhibition of RhoA during early cell spreading (Tsubouchi et al., 2002). However, the effects of Rho GTPases on the adhesion site are mostly indirect and result from their effects on the actin cytoskeleton (Kaverina et al., 2002).
Degradation or cleavage switch
The intracellular-Ca2+-dependent protease calpain has been shown to cleave several key scaffolding components of the adhesome, including integrin, talin, tensin, vinculin, paxillin and filamin, as well as kinases and other regulators. In contrast to modifications mediated by all other switches, calpain-mediated cleavage is irreversible, and most probably has a role in the disassembly of the adhesion site (Perrin and Huttenlocher, 2002). The example shown in the poster for a calpain-mediated cleavage switch depicts cleavage of the cytoplasmic domain of β-integrin. Cleavage at one or more of four sites (Thr741, Tyr747, Phe754 and Tyr759) was shown to occur during platelet aggregation (Du et al., 1995).
The E3 ubiquitin ligase CBL has long been known to associate with integrin adhesions, where it binds and ubiquitylates numerous proteins (Schmidt and Dikic, 2005). Poly-ubiquitylation is a signal that targets proteins to the proteasome for degradation. However, at least in the case of paxillin, it was shown that ubiquitylation by another E3 ubiquitin ligase, RNF5, affects the localization of this protein without affecting its degradation (Didier et al., 2003). Adhesome proteins that are regulated by cleavage or degradation are indicated in the poster by dashed outlines.
Concluding remarks
In the first poster that illustrated the integrin adhesion network, published almost a decade ago, there were just over 50 components and fewer than 200 interactions, which allowed for the entire network to be drawn on one page (Zamir and Geiger, 2001). The sheer magnitude of today's adhesome makes such a presentation essentially impossible to follow. Therefore, we chose to highlight in this poster the high degree of switchability in the adhesome. The fact that only a subset of the documented interactions between adhesome components occurs at a given time and place makes the ‘actual’ network substantially simpler. However, teasing apart the different switches and determining which are biologically important are challenges that still lie ahead.
As mentioned above, adhesion switches can be interconnected, as in the case of Crk, for which a phosphorylation switch leads to a conformational switch and finally to a binding-partner switch. Beyond sequential switching within the same protein, many adhesome proteins are involved in a series of switches that resemble signaling cascades in other cellular systems. Moreover, in some instances, more than one signal might have to converge on a protein to elicit a switch. For example, the actin-related protein Arp2/3 complex transiently binds to vinculin following PtdIns(4,5)P2 and Rac1 activation (DeMali et al., 2002). Although it might be easier for us to picture linear cascades, the highly connected nature of the adhesome network suggests that a single switch in a ‘hub protein’, such as the tyrosine kinase Src, might lead to multiple switching cascades that have the potential to rapidly change the composition of the adhesion and its dynamics.
To understand adhesome regulation at a systems level, major challenges involve the dynamic modeling of such switching cascades and the elucidation of the mechanisms by which an environmental signal, be it chemical or mechanical, propagates through the network to trigger its reorganization and transduce a coherent cellular response. A combination of proteomics, molecular cell biology and in silico simulations should bring us closer to that goal.
Supplementary Material
Acknowledgments
Studies related to the contents of this article were supported by the following: a Weizmann Institute of Science – Mario Negri Institute grant; the Nanotechnology Center for Mechanics in Regenerative Medicine (part of the NIH Nanomedicine Development Center Network; NIH grant no. PN2 EY016586); and the Cell Migration Consortium (NIH grant no. U54 GM064346). B.G. holds the Erwin Neter Professorial Chair in Cell and Tumor Biology. R.Z.-B. was supported by NIH postdoctoral training grant GM078747 and by a fellowship from the Machiah Foundation. Deposited in PMC for release after 12 months.
Footnotes
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/123/9/1385/DC1
References
- Arnaout M. A., Mahalingam B., Xiong J. P. (2005). Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell Dev. Biol. 21, 381-410 [DOI] [PubMed] [Google Scholar]
- Berrier A. L., Yamada K. M. (2007). Cell-matrix adhesion. J. Cell Physiol. 213, 565-573 [DOI] [PubMed] [Google Scholar]
- Bershadsky A. D., Balaban N. Q., Geiger B. (2003). Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 19, 677-695 [DOI] [PubMed] [Google Scholar]
- Bershadsky A., Kozlov M., Geiger B. (2006). Adhesion-mediated mechanosensitivity: a time to experiment, and a time to theorize. Curr. Opin. Cell Biol. 18, 472-481 [DOI] [PubMed] [Google Scholar]
- Chen C. S., Tan J., Tien J. (2004). Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 6, 275-302 [DOI] [PubMed] [Google Scholar]
- Critchley D. R. (2004). Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem. Soc. Trans. 32, 831-836 [DOI] [PubMed] [Google Scholar]
- del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., Sheetz M. P. (2009). Stretching single talin rod molecules activates vinculin binding. Science 323, 638-641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMali K. A., Barlow C. A., Burridge K. (2002). Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 159, 881-891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier C., Broday L., Bhoumik A., Israeli S., Takahashi S., Nakayama K., Thomas S. M., Turner C. E., Henderson S., Sabe H., et al. (2003). RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell. Biol. 23, 5331-5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X., Saido T. C., Tsubuki S., Indig F. E., Williams M. J., Ginsberg M. H. (1995). Calpain cleavage of the cytoplasmic domain of the integrin beta 3 subunit. J. Biol. Chem. 270, 26146-26151 [DOI] [PubMed] [Google Scholar]
- Friedland J. C., Lee M. H., Boettiger D. (2009). Mechanically activated integrin switch controls alpha5beta1 function. Science 323, 642-644 [DOI] [PubMed] [Google Scholar]
- Garvalov B. K., Higgins T. E., Sutherland J. D., Zettl M., Scaplehorn N., Kocher T., Piddini E., Griffiths G., Way M. (2003). The conformational state of Tes regulates its zyxin-dependent recruitment to focal adhesions. J. Cell Biol. 161, 33-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger B., Bershadsky A., Pankov R., Yamada K. M. (2001). Transmembrane crosstalk between the extracellular matrix-cytoskeleton. Nat. Rev. Mol. Cell Biol. 2, 793-805 [DOI] [PubMed] [Google Scholar]
- Goksoy E., Ma Y. Q., Wang X., Kong X., Perera D., Plow E. F., Qin J. (2008). Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell 31, 124-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A. (2005). Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33, 891-895 [DOI] [PubMed] [Google Scholar]
- Hu K., Ji L., Applegate K. T., Danuser G., Waterman-Storer C. M. (2007). Differential transmission of actin motion within focal adhesions. Science 315, 111-115 [DOI] [PubMed] [Google Scholar]
- Huang Z., Yan D. P., Ge B. X. (2008). JNK regulates cell migration through promotion of tyrosine phosphorylation of paxillin. Cell. Signal. 20, 2002-2012 [DOI] [PubMed] [Google Scholar]
- Irvine R. (1998). Inositol phospholipids: translocation, translocation, translocation. Curr. Biol. 8, R557-R559 [DOI] [PubMed] [Google Scholar]
- Kaverina I., Krylyshkina O., Small J. V. (2002). Regulation of substrate adhesion dynamics during cell motility. Int. J. Biochem. Cell Biol. 34, 746-761 [DOI] [PubMed] [Google Scholar]
- Lee S. E., Kamm R. D., Mofrad M. R. (2007). Force-induced activation of talin and its possible role in focal adhesion mechanotransduction. J. Biomech. 40, 2096-2106 [DOI] [PubMed] [Google Scholar]
- Mitra S. K., Schlaepfer D. D. (2006). Integrin-regulated FAK-Src signaling in normal and cancer cells. Curr. Opin. Cell Biol. 18, 516-523 [DOI] [PubMed] [Google Scholar]
- Panetti T. S. (2002). Tyrosine phosphorylation of paxillin, FAK, and p130CAS: effects on cell spreading and migration. Front Biosci. 7, d143-d150 [DOI] [PubMed] [Google Scholar]
- Perrin B. J., Huttenlocher A. (2002). Calpain. Int. J. Biochem. Cell Biol. 34, 722-725 [DOI] [PubMed] [Google Scholar]
- Rosen M. K., Yamazaki T., Gish G. D., Kay C. M., Pawson T., Kay L. E. (1995). Direct demonstration of an intramolecular SH2-phosphotyrosine interaction in the Crk protein. Nature 374, 477-479 [DOI] [PubMed] [Google Scholar]
- Sastry S. K., Burridge K. (2000). Focal adhesions: a nexus for intracellular signaling and cytoskeletal dynamics. Exp. Cell Res. 261, 25-36 [DOI] [PubMed] [Google Scholar]
- Sawada Y., Tamada M., Dubin-Thaler B. J., Cherniavskaya O., Sakai R., Tanaka S., Sheetz M. P. (2006). Force sensing by mechanical extension of the Src family kinase substrate p130Cas. Cell 127, 1015-1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M. H., Dikic I. (2005). The Cbl interactome and its functions. Nat. Rev. Mol. Cell Biol. 6, 907-918 [DOI] [PubMed] [Google Scholar]
- Sechi A. S., Wehland J. (2000). The actin cytoskeleton and plasma membrane connection: PtdIns(4,5)P(2) influences cytoskeletal protein activity at the plasma membrane. J. Cell Sci. 113Pt 21, 3685-3695 [DOI] [PubMed] [Google Scholar]
- Shimaoka M., Takagi J., Springer T. A. (2002). Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31, 485-516 [DOI] [PubMed] [Google Scholar]
- Streuli C. H. (2009). Integrins and cell-fate determination. J. Cell Sci. 122, 171-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery J. P. (2003). Cell adhesion in development: a complex signaling network. Curr. Opin. Genet. Dev. 13, 365-371 [DOI] [PubMed] [Google Scholar]
- Totsukawa G., Yamakita Y., Yamashiro S., Hartshorne D. J., Sasaki Y., Matsumura F. (2000). Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell Biol. 150, 797-806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi A., Sakakura J., Yagi R., Mazaki Y., Schaefer E., Yano H., Sabe H. (2002). Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 159, 673-683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bout I., Divecha N. (2009). PIP5K-driven PtdIns(4,5)P2 synthesis: regulation and cellular functions. J. Cell Sci. 122, 3837-3850 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Choi C. K., Horwitz A. R. (2009). Integrins in cell migration-the actin connection. J. Cell Sci. 122, 199-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wichert G., Jiang G., Kostic A., De Vos K., Sap J., Sheetz M. P. (2003). RPTP-alpha acts as a transducer of mechanical force on alphav/beta3-integrin-cytoskeleton linkages. J. Cell Biol. 161, 143-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worth D. C., Parsons M. (2008). Adhesion dynamics: mechanisms and measurements. Int. J. Biochem. Cell Biol. 40, 2397-2409 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Kam Z., Geiger B. (2005). Polarized downregulation of the paxillin-p130CAS-Rac1 pathway induced by shear flow. J. Cell Sci. 118, 3997-4007 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., Geiger B. (2007a). Functional atlas of the integrin adhesome. Nat. Cell Biol. 9, 858-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidel-Bar R., Milo R., Kam Z., Geiger B. (2007b). A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120, 137-148 [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R., Itzkovitz S., Geiger B. (2009). Protein networks in integrin-mediated adhesions. In Systems Biomedicine: Concepts and Perspectives (ed. Liu E. T., Lauffenburger D. A.), pp. 139-152 Oxford: Academic Press; [Google Scholar]
- Zamir E., Geiger B. (2001). Components of cell-matrix adhesions. J. Cell Sci. 114, 3577-3579 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.