Abstract
Organelles are separate yet interdependent units of eukaryotic cells. They provide an appropriate milieu for the catalysis of many biochemical reactions, and they must establish physical links to communicate and exchange metabolites throughout the cell. Interorganelle communication is an important, yet still poorly understood, aspect of cell biology. We recently identified a protein complex that we refer to as ERMES [endoplasmic reticulum (ER)-mitochondria encounter structure], the main function of which is to provide a tethering force between the ER and the mitochondria. This complex, composed of both ER and mitochondrial transmembrane proteins, is located at the interface of the two organelles and serves to zipper them together. Previous work had implicated several ERMES components in many different physiological processes. The identification of ERMES as an interorganelle tether allows us to re-examine the amply documented phenotypic consequences of the loss of this complex in the light of this new function, thus providing a unique opportunity to assess the physiological relevance of ER-mitochondria junctions. These phenotypes hint at a broad role for ER-mitochondria connections in regulating mitochondrial and cell physiology. In this Hypothesis, we speculate on the potential role of ER-mitochondria connections as hubs in regulating several aspects of mitochondrial biology, including the regulation of mitochondrial membrane biosynthesis, genome replication, Ca2+ signaling and protein import. Finally, we discuss how cells might use ER-mitochondria communication to fine-tune these processes according to their metabolic needs.
Keywords: Ca2+ regulation, ERMES, Endoplasmic reticulum, Membrane biogenesis, Mitochondria, Mitochondrial DNA
Introduction
For decades, electron microscopists have observed that organelles such as the endoplasmic reticulum (ER) and mitochondria are physically connected (Robertson, 1960; Mannella et al., 1998), and electron-dense structures that bridge the two organelles have been observed (Csordás et al., 2006). We recently identified a protein complex that tethers the two organelles (Kornmann et al., 2009). To achieve this, we screened for yeast mutants that had a phenotype that could be suppressed by expressing a synthetic protein designed to artificially tether the ER and mitochondria. Using this approach, we identified a protein complex that we refer to as ERMES (ER-mitochondria encounter structure). Mutations in the protein components of ERMES cause diverse mitochondrial phenotypes, including mitochondrial morphology defects and the inability of cells to grow on non-fermentable media. These phenotypes can be reversed by the expression of the artificial tether, suggesting that the main function of the complex is to act as a mechanical link between the ER and mitochondria. ERMES is a complex composed of at least four proteins: Mdm10 and Mdm34, which are integral outer mitochondrial membrane (OMM) proteins, Mmm1, which is integral to the ER membrane, and Mdm12, which is a cytosolic protein. The association of these proteins into ERMES thus zippers the ER and mitochondria. However, previous studies indicate that ERMES plays roles in many different cellular processes, such as mitochondrial motility and inheritance, mitochondrial genome replication, protein import into mitochondria, and mitochondrial phospholipid homeostasis (Boldogh et al., 2003; Hobbs et al., 2001; Kornmann et al., 2009; Meeusen and Nunnari, 2003; Meisinger et al., 2007; Osman et al., 2009; Youngman et al., 2004). What do these diverse pathways have in common that might involve an ER-mitochondria connection? In this Hypothesis, we review the evidence implicating ERMES in these physiological processes and propose that ER-mitochondria connections are used as hubs for integrating cellular cues and transmitting such information to mitochondria.
ER-mitochondria phospholipid exchange
A large body of literature has implicated ER-mitochondria connections in membrane-lipid exchange between the two organelles. The main site of lipid synthesis in the cell is the ER, and mitochondria must import lipid components of both the inner mitochondrial membrane (IMM) and OMM from the ER. However, because mitochondria are not connected by vesicular trafficking pathways, they must obtain ER lipids by non-vesicular mechanisms (Daum and Vance, 1997). ER membranes have long been known to cosediment with mitochondria in cell-fractionation experiments, owing to the presence of ER-mitochondria tethers (Vance, 1990). Interestingly, these cosedimenting ER membranes have a different composition than the rest of the ER, giving rise to the concept of mitochondria-associated membranes (MAMs) (Rusiñol et al., 1994). MAMs define a laterally differentiated subcompartment of the ER that is dedicated to performing lipid biosynthesis and perhaps other functions necessary for the maintenance of mitochondrial membranes. MAMs are enriched in enzymes involved in phospholipid biosynthesis, and have been suggested to promote phospholipid exchange in vitro (Achleitner et al., 1999).
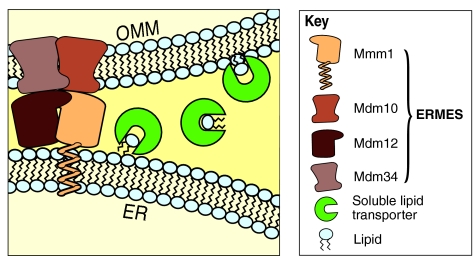
Consistent with a role of ERMES in phospholipid transfer, we found that cells in which protein components of this complex are mutated (hereafter referred to as ERMES mutants) partially phenocopied the loss of the mitochondrial phosphatidylserine decarboxylase Psd1 (Kornmann et al., 2009). Psd1 is the only mitochondrial enzyme in the aminoglycerophospholipid biosynthesis pathway. Therefore, its substrate and product must come from and return to the ER, respectively (Achleitner et al., 1999). We found that the metabolic flow through this pathway was slowed in ERMES mutants, consistent with the idea that ERMES-mediated ER-mitochondrial junctions are important for phospholipid exchange. However, the flow of phospholipids was not completely abrogated in ERMES mutants, and mitochondrial membranes in the mutant cells were not profoundly different compared with wild-type cells with respect to their lipid composition. Specifically, the mitochondrial membranes of ERMES mutants had a lower abundance of some phospholipid classes, such as cardiolipins and phosphatidylethanolamines, whereas the levels of other phospholipid classes, such as phosphatidylcholines or phosphatidylinositols, were mostly unaffected (Osman et al., 2009). This observation suggests that ERMES is not strictly necessary for providing lipids to mitochondria and that ERMES-independent pathways must exist. Such pathways might involve soluble lipid transporters such as members of the ceramide transporter (CERT) and the oxysterol-binding protein (OSBP) families that can mediate non-vesicular ER–plasma-membrane lipid exchange through an aqueous space (D'Angelo et al., 2008). It is possible that ERMES-induced ER-mitochondria apposition serves to enhance the efficiency of shuttling of such transporters between the two organelles (Fig. 1) and that different transporters with different specificities might be differentially affected by the loss of ERMES, explaining the lipid-specific phenotypes of ERMES mutants. Such a scenario would mean that a lack of tight ER-mitochondria connections would not abolish interorganelle lipid exchange but would reduce it to an extent that limited cell growth, without grossly affecting the steady-state phospholipid content.
Fig. 1.
Proposed role of ERMES in phospholipid exchange. ERMES-mediated ER-mitochondria apposition between the outer mitochondrial membrane (OMM) and the ER might facilitate the targeting and shuttling of soluble lipid-carrier proteins between the two organelles. Lipid insertion into the mitochondrial membrane is, in turn, crucial for the biogenesis of mitochondria.
In this view, controlling the tightness of ERMES-dependent association between the two organelles would provide a way to control the flow of lipid to and from mitochondrial membranes and fine-tune it according to the metabolic needs of the cell.
ER-mitochondrial Ca2+ crosstalk
Recent advances in Ca2+ imaging techniques have shown that Ca2+ is taken up by mitochondria upon its release from the ER (Rizzuto et al., 1993). This uptake is crucial for regulating mitochondrial activity and cell fate. Mitochondria can use their Ca2+-uptake capacity to buffer excessive Ca2+ in the cytosol, thereby preventing its possible cytotoxic effects. In turn, mitochondrial Ca2+ can tune mitochondrial activity through the activation of Ca2+-sensitive matrix dehydrogenases (Denton, 2009) and transporter proteins (Satrústegui et al., 2007). Conversely, a prolonged exposure to Ca2+ in the mitochondrial matrix can cause the opening of the permeabilization transition pore, leading to the release of cytochrome c from mitochondria into the cytosol and, ultimately, to apoptosis (Pinton et al., 2008). Thus, mitochondrial Ca2+ uptake has the potential to regulate essential aspects of cell physiology.
The Ca2+ transporters at the surface of mitochondria have a low affinity for Ca2+. This is surprising because the Ca2+ concentrations that are reached in the cytosol during Ca2+ discharge from the ER are much too low to allow these transporters to mediate efficient Ca2+ uptake into mitochondria. However, high Ca2+ concentrations
Box 1. ER-mitochondria connections in mammalian cells
Orthologs of ERMES components have, to date, only been identified in fungi, using standard homology searches. However, more refined analyses reveal that Mdm12 and Mmm1 contain an SMP domain (for synaptotagmin-like, mitochondrial and lipid-binding proteins) that is found in a plethora of membrane-bound proteins across the whole eukaryotic domain, often in conjunction with transmembrane, lipid-binding and Ca2+-binding domains (Lee and Hong, 2006). Because SMP domains are present in more than one subunit of ERMES, this domain might constitute a signature motif for ERMES function. The investigation of mammalian SMP-containing proteins thus promises to uncover functional orthologs of ERMES. Furthermore, the great variety of these SMP-containing proteins in metazoans might reveal additional levels of complexity in the ER-mitochondria connections in these organisms. Indeed, compared with fungi, mammalian cells seem to have more complex ER-mitochondria interfaces that are mediated by various protein complexes. The chaperone-mediated interaction between the mitochondrial porin voltage-dependent anion-selective channel protein 1 (VDAC1) and the ER Ca2+-channel inositol (1,4,5)-trisphosphate receptor [Ins(1,4,5)P3R; also known as IP3R] (Szabadkai et al., 2006), as well as the homotypic interaction of mitofusin 2 (Mfn2) partitioned between ER and mitochondrial membranes (de Brito and Scorrano, 2008), contribute to the physical link between the two organelles. Additional components, such as the cargo-sorting protein PACS2 (Simmen et al., 2005) and dynamin-related protein 1 (Drp1) (Szabadkai et al., 2004), might regulate this link indirectly. Although these proteins have been shown to influence Ca2+ exchange between the two organelles, it will be interesting to examine whether they also play roles in phospholipid exchange, mtDNA replication and/or mitochondrial protein import.
might occur locally at sites where mitochondria are in close proximity to sites of Ca2+ discharge from the ER (Rizzuto et al., 2004). Indeed, artificially increasing the linkage between the two organelles by expressing a synthetic ER-mitochondria tether increases the coupling between ER Ca2+ release and mitochondrial Ca2+ uptake in vivo. Conversely, decreasing this association by limited proteolysis decreases the coupling in vitro (Csordás et al., 2006).
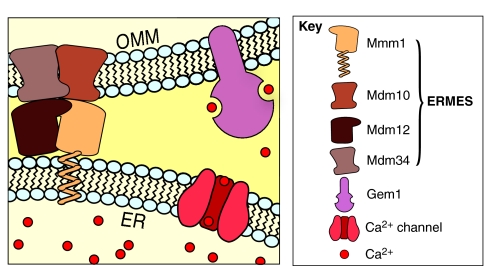
Using genome-wide genetic-interaction maps, we identified GEM1 [which encodes mitochondrial Rho GTPase 1 (Gem1)] as the gene whose mutation shows the most similar phenotype to ERMES mutants. Gem1 is a tail-anchored protein of the OMM and contains two Rho-like GTPase domains and two Ca2+-binding EF-hands. Gem1 is required for proper morphology and inheritance of mitochondria (Frederick et al., 2004). Mitochondria from GEM1-deleted strains harbor aberrant morphologies that resemble the aberrant mitochondria observed in ERMES mutants, but the molecular causes of this phenotype are unknown.
The metazoan homolog of Gem1, known as Miro (mitochondrial Rho GTPase), is better characterized at the molecular level. Miro serves as an adaptor between mitochondria and the cytoskeleton by recruiting kinesin heavy chain (Wang and Schwarz, 2009). Upon Ca2+ binding to the two EF-hands of this protein, Miro undergoes a conformational change that allows it to sequester the motor head of kinesin heavy chain and, consequently, prevent it from binding to microtubules. As a result, treating neurons with calcimycin (a drug that permeates Ca2+ into the cell) causes mitochondria to stop their microtubule-driven movements. Interestingly, Miro responds to Ca2+ over a concentration range that is never attained in the cytosol of live cells (10-100 μM) (Wang and Schwarz, 2009). These values, however, are comparable to the affinity of the mitochondrial Ca2+ transporters that are thought to mediate mitochondrial Ca2+ uptake at ER-mitochondria contact sites. The high Ca2+ concentrations that are necessary to occupy the EF-hands of Miro might be attained at such sites.
Yeast strains expressing mutated versions of Gem1 that are unable to bind Ca2+ display the same phenotypes as GEM1-null mutants, suggesting that Gem1 is active in the Ca2+-bound configuration (Frederick et al., 2004). This observation, together with our genetic-interaction map, makes it tempting to speculate that Ca2+ originating from the ER activates Gem1 (Miro in metazoans) via binding to its EF-hands (Fig. 2), thereby regulating mitochondrial motility. This view must be balanced by the fact that the yeast ER contains much less Ca2+ than its mammalian counterpart (Strayle et al., 1999) and that Ca2+ discharge from the ER has not yet been described in yeast. The idea is consistent, however, with the observation that the mitochondria of ERMES mutants have defects in motility and inheritance (Boldogh et al., 2003).
Fig. 2.
Proposed role of ERMES in Ca2+ exchange. ERMES-mediated ER-mitochondria apposition might create cytosolic microdomains in which the Ca2+ concentration is higher than in the rest of the cytosol. The GTPase Gem1 (Miro in metazoans) might require these high levels for the binding of Ca2+ to its two EF-hands. Gem1, in turn, regulates mitochondrial morphology, motility and inheritance.
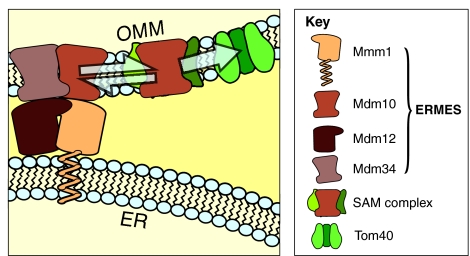
Mitochondrial protein import
Mdm10 is a constituent both of ERMES and of the sorting and assembly machinery (SAM) complex, which assembles β-barrel proteins in the OMM (Meisinger et al., 2004). Although these two complexes are physically and spatially separable, the fact that Mdm10 is part of both suggests a functional or regulatory connection between the two. The function of Mdm10 in the SAM complex is probably regulatory, rather than structural, in nature. Deletion of MDM10 differentially affects the assembly of various β-barrel proteins. It inhibits the assembly of the translocase of outer membrane (TOM)-complex component Tom40 but speeds up that of porins, showing that Mdm10 action within the SAM complex is not required for β-barrel assembly per se, but that it regulates it in a substrate-dependent manner. Interestingly, the deletion of a small component of the TOM complex, Tom7, has the opposite effect on the assembly of Tom40 and porins (Meisinger et al., 2006). Furthermore, Tom7 can titrate Mdm10 out of the SAM complex, providing a plausible explanation for the antagonistic effects of these two deletions on the assembly of β-barrel proteins. These data suggest that a limiting pool of Mdm10 might partition between the SAM complex and ERMES. Therefore, ERMES-mediated sequestering of Mdm10 might affect the assembly of β-barrel proteins, possibly generally regulating import of mitochondrial proteins, because Mdm10 action within the SAM complex seems to be specific for the assembly of the OMM translocase Tom40. Indeed, deletion of MMM1 or MDM12, which encode two ERMES components that are not part of the SAM complex, also causes defects in β-barrel protein assembly (Meisinger et al., 2007). Although alternative explanations for the effect of MMM1 and MDM12 on the assembly of β-barrel proteins are possible, we speculate that the link provided by Mdm10 between ERMES and the SAM complex might serve to regulate the protein-import activity of the mitochondria (Fig. 3), possibly following cues of the metabolic needs of the cell.
Fig. 3.
Proposed role of ERMES in mitochondrial protein import. ERMES and the sorting and assembly machinery (SAM) complex compete for a limited mitochondrial pool of Mdm10. This implies a common regulatory mechanism for the two complexes. Mdm10 in the SAM complex is required for the proper assembly of a subset of OMM β-barrel proteins. The main substrate of Mdm10 in the SAM complex is Tom40, the protein translocase of the OMM. Assembly and disassembly of ERMES might shift the equilibrium of Mdm10 incorporated into ERMES or SAM complexes, thereby regulating the assembly of Tom40 and general protein import in mitochondria.
Mitochondrial genome replication
Unlike most cellular organelles, mitochondria harbor a DNA molecule that is probably the vestigial genome of the endosymbiotic bacterium that gave rise to mitochondria. Although the mitochondrial genome only encodes a few dozen proteins, it is nevertheless essential for proper mitochondrial function. For example, yeast strains that have lost their mitochondrial DNA (mtDNA) are unable to grow on non-fermentable carbon sources (Goldring et al., 1971). mtDNA is compacted into structures called nucleoids. Replication of mtDNA is uncoupled from the cell cycle and happens continuously, but only in a fraction of nucleoids at a time (Meeusen and Nunnari, 2003). How mtDNA replication is coupled to cell growth is unknown.
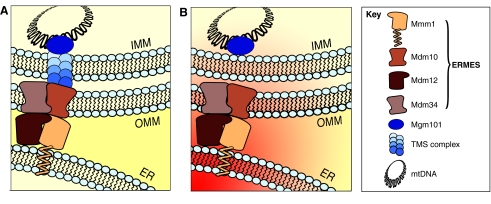
ERMES can be found in one to five foci along the mitochondrial surface per yeast cell. These foci are found in close proximity to mtDNA nucleoids in the mitochondrial matrix (Hobbs et al., 2001). Strikingly, ERMES foci almost exclusively associate with nucleoids that are undergoing active replication (Meeusen and Nunnari, 2003). ERMES mutants show defects in mitochondrial genome maintenance and frequently lose mtDNA. This suggests that ERMES might be directly involved in regulating mtDNA replication and/or genome maintenance. The spatial proximity of ERMES and actively replicating nucleoids, however, is not absolute. Even conventional light microscopy can detect an offset between the localization of nucleoids and ERMES foci, indicating that ERMES might regulate mtDNA replication over distances as great as 200 nm.
How can a complex situated at the ER-mitochondria interface regulate the replication of mtDNA in the mitochondrial matrix at a distance equal to that of many ribosomal diameters? One possibility is that ERMES is part of a larger protein assembly that spans the ER membrane as well as the IMM and OMM and that physically connects the ER lumen to the mitochondrial genome (Fig. 4A). The possibility that such a complex exists is suggested by the finding that the ER-resident protein Mmm1 coimmunoprecipitates with Mgm101, a DNA-binding protein of the nucleoid, in chemically crosslinked mitochondrial extracts (Meeusen and Nunnari, 2003). This proposed complex was called TMS (two-membrane-spanning complex). Our discovery that Mmm1 is an ER transmembrane protein indicates instead that such a complex actually spans three membranes. [Intriguingly, this alters the definition but not the acronym TMS (three-membrane-spanning complex).] An alternative possibility is that the distance between ERMES and mitochondrial nucleoids is spanned by a gradient of molecule(s) originating from the ER and diffusing into mitochondria from ERMES foci. This gradient might thus signal the mtDNA replication machinery and regulate its activity (Fig. 4B).
Fig. 4.
Proposed role of ERMES in mtDNA maintenance. ERMES complexes are found in close proximity to actively replicating nucleoids, which can be explained by two models. (A) ERMES might be part of a large protein assembly that spans the ER, the OMM and the IMM (three-membrane-spanning complex; TMS) and directly connects the ER lumen to the mitochondrial genome. (B)A gradient of signaling molecule(s) that originate from the ER (red gradient) might cue the mitochondrial genome for replication.
ERMES as a central hub for regulating mitochondrial physiology
Thus, ERMES lies at the crossroads of many crucial pathways of mitochondrial biology: membrane biosynthesis, protein import, motility and genome replication. We propose that ERMES plays a broad role in the regulation of mitochondrial biology by integrating cellular cues to instruct mitochondria to perform according to the metabolic needs of the cell.
The cell must exert tight control over these metabolic processes to ensure that they are tuned according to its physiological state. This control can be accomplished by transcription and translation, as most genes encoding mitochondrial proteins are found in the nucleus (Dyall et al., 2004) and are therefore under direct control of the nuclear transcription machinery. But, in order to gain control over mitochondrial behavior without requiring de novo transcription and translation, cells might use contact sites that allow direct interorganelle communication. Such sites might allow cells to regulate many mitochondrial processes in a localized and timely fashion, such as motility, protein import, initiation of replication of individual nucleoids, activity of the respiratory chain and/or opening of the permeability transition pore.
The membrane domains that are defined and perhaps corralled at ERMES-dependent contact sites might serve as platforms to recruit effectors responsible for the exchange of metabolites, such as lipid transporters or Ca2+ channels. Such metabolites might in turn diffuse further inside mitochondria and cue the respiratory apparatus, the protein-import system or the mtDNA replication machinery. We surmise that ERMES activity is regulated and that this regulation matches mitochondrial activity to cell growth.
Our understanding of how ERMES is regulated promises to further uncover a new language with which mitochondria communicate to the rest of the cell.
Acknowledgments
This work was supported by the NIH. B.K. is a fellow of the Swiss National Science Foundation. P.W. is an investigator at the Howard Hughes Medical Institute. Deposited in PMC for release after 12 months.
References
- Achleitner G., Gaigg B., Krasser A., Kainersdorfer E., Kohlwein S. D., Perktold A., Zellnig G., Daum G. (1999). Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur. J. Biochem. 264, 545-553 [DOI] [PubMed] [Google Scholar]
- Boldogh I. R., Nowakowski D. W., Yang H., Chung H., Karmon S., Royes P., Pon L. A. (2003). A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell 14, 4618-4627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K. F., Balla T., Mannella C. A., Hajnóczky G. (2006). Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915-921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo G., Vicinanza M., De Matteis M. A. (2008). Lipid-transfer proteins in biosynthetic pathways. Curr. Opin. Cell Biol. 20, 360-370 [DOI] [PubMed] [Google Scholar]
- Daum G., Vance J. (1997). Import of lipids into mitochondria. Prog. Lipid Res. 36, 103-130 [DOI] [PubMed] [Google Scholar]
- de Brito O. M., Scorrano L. (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605-610 [DOI] [PubMed] [Google Scholar]
- Denton R. M. (2009). Regulation of mitochondrial dehydrogenases by calcium ions. Biochim. Biophys. Acta 1787, 1309-1316 [DOI] [PubMed] [Google Scholar]
- Dyall S. D., Brown M. T., Johnson P. J. (2004). Ancient invasions: from endosymbionts to organelles. Science 304, 253-257 [DOI] [PubMed] [Google Scholar]
- Frederick R. L., McCaffery J. M., Cunningham K. W., Okamoto K., Shaw J. M. (2004). Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167, 87-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldring E., Grossman L., Marmur J. (1971). Petite mutation in yeast. J. Bacteriol. 107, 377-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs A. E., Srinivasan M., Jensen R. E., McCaffery J. M. (2001). Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152, 401-410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S. R., Schuldiner M., Nunnari J., Weissman J. S., Walter P. (2009). An ER-Mitochondria tethering complex revealed by a synthetic biology screen. Science 325, 477-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Hong W. (2006). Diverse membrane-associated proteins contain a novel SMP domain. FASEB J. 20, 202-206 [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Buttle K., Rath B. K., Marko M. (1998). Electron microscopic tomography of rat-liver mitochondria and their interactions with the endoplasmic reticulum. BioFactors 8, 225-228 [DOI] [PubMed] [Google Scholar]
- Meeusen S., Nunnari J. (2003). Evidence for a two membrane-spanning autonomous mitochondrial DNA replisome. J. Cell Biol. 163, 503-510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisinger C., Rissler M., Chacinska A., Szklarz L. K., Milenkovic D., Kozjak V., Schönfisch B., Lohaus C., Meyer H. E., Yaffe M. P., et al. (2004). The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 7, 61-71 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Wiedemann N., Rissler M., Strub A., Milenkovic D., Schönfisch B., Müller H., Kozjak V., Pfanner N. (2006). Mitochondrial protein sorting: differentiation of beta-barrel assembly by Tom7-mediated segregation of Mdm10. J. Biol. Chem. 281, 22819-22826 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfannschmidt S., Rissler M., Milenkovic D., Becker T., Stojanovski D., Youngman M. J., Jensen R. E., Chacinska A., Guiard B., et al. (2007). The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J. 26, 2229-2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Haag M., Potting C., Rodenfels J., Dip P. V., Wieland F. T., Brügger B., Westermann B., Langer T. (2009). The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184, 583-596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinton P., Giorgi C., Siviero R., Zecchini E., Rizzuto R. (2008). Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27, 6407-6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R., Brini M., Murgia M., Pozzan T. (1993). Microdomains with high Ca2+ close to IP3-sensitive channels that are sensed by neighboring mitochondria. Science 262, 744-747 [DOI] [PubMed] [Google Scholar]
- Rizzuto R., Duchen M. R., Pozzan T. (2004). Flirting in little space: the ER/mitochondria Ca2+ liaison. Sci. STKE 13, re1 [DOI] [PubMed] [Google Scholar]
- Robertson J. D., (1960). The molecular structure and contact relationships of cell membranes. Prog. Biophys. Mol. Biol. 10, 343-418 [PubMed] [Google Scholar]
- Rusiñol A. E., Cui Z., Chen M. H., Vance J. E. (1994). A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269, 27494-27502 [PubMed] [Google Scholar]
- Satrústegui J., Pardo B., Del Arco A. (2007). Mitochondrial transporters as novel targets for intracellular calcium signaling. Physiol. Rev. 87, 29-67 [DOI] [PubMed] [Google Scholar]
- Simmen T., Aslan J. E., Blagoveshchenskaya A. D., Thomas L., Wan L., Xiang Y., Feliciangeli S. F., Hung C., Crump C. M., Thomas G. (2005). PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717-729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayle J., Pozzan T., Rudolph H. (1999). Steady-state free Ca (2+) in the yeast endoplasmic reticulum reaches only 10 microM and is mainly controlled by the secretory pathway pump pmr1. EMBO J. 18, 4733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabadkai G., Simoni A. M., Chami M., Wieckowski M. R., Youle R. J., Rizzuto R. (2004). Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol. Cell 16, 59-68 [DOI] [PubMed] [Google Scholar]
- Szabadkai G., Bianchi K., Várnai P., De Stefani D., Wieckowski M. R., Cavagna D., Nagy A. I., Balla T., Rizzuto R. (2006). Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901-911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance J. (1990). Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248 [PubMed] [Google Scholar]
- Wang X., Schwarz T. L. (2009). The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman M. J., Hobbs A. E., Burgess S. M., Srinivasan M., Jensen R. E. (2004). Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 164, 677-688 [DOI] [PMC free article] [PubMed] [Google Scholar]