Abstract
We investigated the association of 14 polymorphisms in the endothelial nitric oxide synthase gene (NOS3) with ankle brachial index (ABI) in non-Hispanic white hypertensives belonging to hypertensive sibships. Subjects (n = 659, mean age 61±9 y, 54% women) underwent measurement of ABI using a standard protocol, and the lowest of 4 ABI values was used in the analyses. Non-synonymous SNPs with a minor allele frequency > 0.02 and tag SNPs selected based on a measure of linkage disequilibrium (r2) were genotyped. We reduced the chance of false positives by testing for replication, randomly selecting 1 hypertensive sib from each sibship to create Subset 1 (n = 330) and Subset 2 (n = 329). Multivariable linear regression models were used to assess the associations of single NOS3 polymorphisms and haplotypes with ABI after adjustment for covariates (age, sex, body mass index, smoking, total cholesterol, HDL cholesterol, and diabetes). Two specific SNPs in significant LD with each other (rs891512 and rs1808593) were significantly associated with ABI in both subsets. Based on a sliding window approach with a window size of 2, estimated haplotypes from 2 SNP pairs (rs2070744–rs3918226 & rs1808593–rs7830) were also significantly associated with ABI in both subsets. In conclusion, specific NOS3 SNPs and haplotypes were associated with inter-individual variation in ABI, a non-invasive marker of peripheral arterial disease, in replicate subsets of hypertensive subjects. These findings motivate further investigation of the role of NOS3 variants in determining susceptibility to peripheral arterial disease.
Keywords: genetics, nitric oxide synthase, ankle-brachial index, peripheral arterial disease
1. Introduction
Peripheral arterial disease (PAD) affects 8–10 million people in the United States [1,2] and is associated with a marked impairment in quality of life and an increased risk of stroke, myocardial infarction, and cardiovascular death [3]. Noninvasive assessment of atherosclerotic PAD is performed by measuring the ankle-brachial index (ABI), the ratio of systolic blood pressure (SBP) at the ankle to the SBP in the arm. Normally ABI is ≥ 1.0, but with increasing degrees of severity of arterial disease in the lower extremities, there is a progressive fall in SBP at the ankle. An ABI of ≤0.90 or ≤0.95 is used to make the diagnosis of PAD in the clinical setting. Individuals with PAD may not have typical symptoms of exertional leg discomfort, and ABI is a useful diagnostic measure to ascertain the presence of PAD.
Coronary artery disease (CAD), cerebrovascular disease, and PAD are all influenced by the atherosclerotic disease process. As such, many of the well-established risk factors for atherosclerosis, such as increasing age, dyslipidemia, hypertension, cigarette smoking, and diabetes [4], are common for these diseases. The Cardiovascular Health Study showed increased relative risk for PAD among hypertensive individuals compared to normotensives and an association between total cholesterol and PAD [5]. Despite these similarities, PAD differs from CAD and cerebrovascular disease in its risk factor profile and clinical presentation. For example, while men are typically more prone to atherosclerosis and the related consequences (i.e.- CAD or cerebrovascular disease) than women, studies have demonstrated that PAD may be less gender specific [6,7]. Furthermore, while cigarette smoking and diabetes are also closely related to CAD and cerebrovascular disease, the associations between these two risk factors and PAD are even stronger [2]. It is therefore likely that both genetic and environmental factors contribute differently to PAD than to CAD or cerebrovascular disease. While several conventional risk factors have been identified for PAD, these explain <20% of inter-individual variation in ABI [8], and the contribution of other ‘novel’ biochemical and genetic risk factors is less well characterized. In particular, little is known regarding genetic factors influencing inter-individual variation in ABI.
Nitric oxide (NO) is an important endogenous anti-atherogenic molecule, and variations in the endothelial nitric oxide synthase gene (NOS3) may influence bioavailability of NO and reduce vascular reactivity, thereby predisposing to the development of atherosclerotic vascular diseases including PAD [9]. NOS3 knock out mice have been shown to develop endothelial dysfunction and hypertension [10], and expression of recombinant NOS3 enhances endothelial function in the arteries of experimental animals [11]. Variation in NOS3 has been associated with reduced blood pressure (BP) fall after exercise training [12], lower basal coronary blood flow and reduced coronary vasodilatation to adenosine [13], enhanced systemic pressor response to phenylephrine [14], and reduced flow-mediated dilatation of the brachial artery [15].
We investigated the association of 14 single nucleotide polymorphisms (SNPs) in NOS3 with ABI in a cohort of hypertensive subjects. We tested the hypothesis that genetic variation in NOS3 is associated with differences in ABI by using non-synonymous and tag SNPs that represented much of the common variation in this gene. Because of the phenomenon of linkage disequilibrium (LD), tag SNPs can be used to capture the underlying common genetic variation within regions of significant LD, thereby reducing genotyping costs [16,17]. While single SNP associations indicate that variation within a gene may be associated with particular phenotypes, the role of haplotype organization of variation is important to consider [18]. As such, we also investigated the association of estimated haplotypes within the selected region of NOS3 with ABI. Although association studies are favored over linkage studies for unraveling the genetic bases of complex disorders, lack of replication in a majority of such studies has been a concern. Therefore, we divided our sample into 2 subsets to test for replication of the associations across subsets of our sample.
2. Materials and methods
2.1 Study Population
Subjects included non-Hispanic white participants in the Genetic Epidemiology Network of Arteriopathy (GENOA) study, a community-based study of hypertensive sibships that aims to identify genes influencing BP [19]. The study was approved by the Institutional Review Board of Mayo Clinic, Rochester MN. Written informed consent was obtained from each participant. In the initial phase of the GENOA study (9/1995 to 6/2001), sibships containing ≥2 individuals with essential hypertension diagnosed before age 60 years were selected for participation. At the Rochester, MN field center, 1583 non-Hispanic whites were enrolled. Participants returned in Phase II of GENOA for physical examination, laboratory tests, and measurement of the ABI. Through November of 2004, ABI had been measured in 866 participants. We attempted to reduce the chance of false positives in our SNP association study by testing for replication in 2 sample subsets. To create our replication subsets, we randomly selected 1 hypertensive sib from each sibship without replacement to create Subset 1 and then randomly selected another hypertensive sib from each sibship to create Subset 2. After excluding normotensive individuals and those without available genotype information, our sample contained a relatively small number of singletons (i.e.- no matching sibs) that were equally divided between the 2 samples. Division of an odd number of singletons led to a 1-subject difference between the 2 subsets. The total study sample combined 659 subjects in Subset 1 and Subset 2.
2.2 Clinical Assessments and Covariate Definitions
Height was measured by stadiometer, weight by electronic balance, and body mass index (BMI) was calculated as weight in kilograms divided by the square of height in meters. Resting SBP and diastolic blood pressure (DBP) were measured by a random zero sphygmomanometer. Blood was drawn by venipuncture after an overnight fast. Serum total cholesterol and high-density lipoprotein (HDL) cholesterol were measured by standard enzymatic methods. Low-density lipoprotein (LDL) cholesterol levels were estimated using the Friedewald formula [20]. The diagnosis of hypertension was established based on BP levels measured at the study visit (>140/90 mmHg) or a prior diagnosis of hypertension and current treatment with antihypertensive medications. Diabetes was considered present if the subject was being treated with insulin or oral agents or had a fasting glucose level ≥ 126 mg/dL. Current smoking was defined as having smoked within the year preceding examination.
2.3 Ankle-brachial index
ABI was measured in the supine position following a 5-min rest. Appropriately-sized BP cuffs were placed on each arm and ankle, and a Doppler ultrasonic instrument (Medisonics, Minneapolis MN) was used to detect each pulse. The cuff was inflated to 10 mm Hg above SBP and deflated at 2 mm Hg/s. The first reappearance of the pulse was taken as the SBP. To calculate ABI, the SBP at each ankle site was divided by the higher of the 2 brachial pressures. The lowest of the 4 ratios was designated as the ABI. The correlation of the lowest ABI with the average of the 2 ABIs from the same leg was 0.98, and inferences were similar using the lowest ABI or the average ABI.
2.4 SNP selection
Our algorithm for SNP selection first identified non-synonymous SNPs with a minor allele frequency (MAF) > 0.02 based on data from the Seattle SNPs database (http://pga.mbt.washington.edu). Second, we identified all SNPs with a MAF > 0.1 and unique sequence context that could potentially be typed in any of the 3 ethnic groups sampled in GENOA (non-Hispanic white, African-American, Hispanic). From the latter SNPs, tag SNPs were selected based on the r2 method described by Carlson et al. [21]. The final list of SNPs to be genotyped was established by selecting 1 SNP from each bin pair according to the following selection prioritization: 1st) a tag SNP in a conserved region (compared to mouse); 2nd) a tag SNP not in a conserved region; 3rd) a non-tag SNP in a conserved region; 4th) neither a tag SNP nor a SNP in a conserved region. We used this priority system because several bins had multiple tag SNPs, and some bins had no identified tag SNPs. Using the above approach, we identified 14 SNPs in NOS3 including the non-synonymous Asp298Glu polymorphism (rs1799983) (Table 2).
Table 2.
SNP annotation and allele frequencies for Subset 1 and Subset 2
| SNP | SNP Location on Chr.2* | Allele 1 | Allele 2 | Subset 1/Allele 1 N (%) | Subset 2/Allele 1 N(%) | Bin | tag | |
|---|---|---|---|---|---|---|---|---|
| 1 | rs1800783 | 150127045 | T | A | 381(61%) | 390(62%) | 6 | Yes |
| 2 | rs2070744 | 150127727 | T | C | 391(61%) | 386(61%) | 6 | Yes |
| 3 | rs3918226 | 150127824 | C | T | 582(89%) | 579(89%) | N/A | N/A |
| 4 | rs3793342 | 150132843 | G | A | 540(85%) | 530(85%) | 1 | No |
| 5 | rs1799983 | 150133759 | G | T | 418(66%) | 431(66%) | N/A | N/A |
| 6 | rs1800780 | 150136527 | A | G | 314(51%)† | 317(52%) | 7 | Yes |
| 7 | rs3918186 | 150140080 | A | T | 595(92%) | 583(91%) | 4 | Yes |
| 8 | rs3918188 | 150140429 | C | A | 397(63%) | 400(63%) | 3 | Yes |
| 9 | rs3730305 | 150142048 | C | A | 579(93%) | 567(91%)† | 4 | Yes |
| 10 | rs891511 | 150142491 | G | A | 428(68%) | 431(69%) | 1 | Yes |
| 11 | rs891512 | 150145737 | G | A | 485(77%) | 501(79%) | 2 | Yes |
| 12 | rs1808593 | 150145950 | T | G | 468(75%) | 466(75%) | 2 | Yes |
| 13 | rs7830 | 150147219 | G | T | 426(67%) | 419(67%) | 11 | Yes |
| 14 | rs3800787 | 150151284 | G | C | 405(64%) | 390(62%) | 11 | Yes |
Based on NCBI human reference sequence and dbSNP build 125;
Hardy-Weinberg Equilibrium P-value <0.05
Bold type SNP is non-synonymous
2.5 Genotyping
DNA was isolated using the PureGene DNA Isolation Kit from Gentra Systems (Minneapolis MN). Genotyping, based on polymerase chain reaction amplification techniques, was conducted at the University of Texas-Health Sciences Center at Houston using the TaqMan assay and ABI Prism® Sequence Detection System (Applied Biosystems, Foster City CA). Primers and probes are available from the authors upon request. Quality control measures for genotyping assays included robotic liquid handling; separate pre- and post-PCR areas; standard protocols and quality control analyses including 5% duplicates, positive and negative controls, computerized sample tracking, and data validity checks.
2.6 Statistical Analysis
Allele frequencies were estimated by gene counting. Agreement of NOS3 genotype frequencies with Hardy-Weinberg equilibrium expectations were tested using a χ2 goodness-of-fit test. The proportions, means, and standard deviations of the conventional cardiovascular risk factors were reported. Multivariable linear regression models were used to assess the association of NOS3 polymorphisms with ABI after adjustment for covariates (age, sex, BMI, smoking, total cholesterol, HDL cholesterol, and diabetes). Our decision to include these covariates in the adjustment models was directed by the published literature on PAD. Supporting this decision, univariate analyses of our data demonstrated many expected relationships between these established risk factors and ABI.
An analysis of variance (ANOVA) was performed in each subset to determine if significant interactions existed between the NOS3 polymorphisms and conventional risk factors. If statistically significant interactions were found based on the ANOVA results, tests of homogeneity were performed to determine if the interaction was homogeneous across subsets. Given that we were testing for SNP associations in 2 subsets, we used an alpha-level of 0.10 as our criterion for statistical significance in each set.
Using a sliding window approach with a window size of 2, haplotypes were estimated using Phase 2.0 [22]. For each individual, the estimated haplotype with the greatest probability above the assigned cut point of 0.75 was assumed. Within each window, individuals were excluded from the haplotype analyses if none of their respective estimated haplotype probabilities exceeded the assigned cut point. Haplotypes were constructed for each individual based on the estimation results of each window. Multivariable linear regression models were used to assess the association of each 2-marker NOS3 haplotype with ABI after adjustment for covariates (age, sex, BMI, smoking, total cholesterol, HDL cholesterol, and diabetes). We estimated the average effects of the identified haplotypes based on the methods described by Templeton [23].
3. Results
The descriptive statistics for the 2 subsets of GENOA hypertensive individuals investigated in this study are compared in Table 1. The mean age in both subsets was 61 years. The mean BMI was 31 kg/m2 in Subset 1 and 32 kg/m2 in Subset 2. The average ABI was 1.11 in Subset 1 and 1.09 in Subset 2. Participants in Subset 1 included 48% men, 15% diabetics, and 9% smokers. Subset 2 included 44% men, 22% diabetics, and 7% smokers. Only the proportion of diabetics in the 2 samples was significantly different (P = .02).
Table 1.
Subject characteristics.
| Subset 1 (n=330) | Subset 2 (n=329) | Subset Difference | |||
|---|---|---|---|---|---|
| Mean ± SD* | Range | Mean ± SD* | Range | P-value | |
| Age, years | 61 ± 9 | 32–82 | 61 ± 9 | 37–84 | 0.49 |
| BMI, kg/m2 | 31 ± 6 | 18–62 | 32 ± 6 | 20–59 | 0.14 |
| SBP, mm Hg | 135 ± 17 | 97–185 | 135 ± 16 | 85–194 | 0.95 |
| DBP, mm Hg | 75 ± 10 | 44–105 | 75 ± 9 | 41–103 | 0.63 |
| ABI | 1.11 ± 0.16 | 0.48–1.65 | 1.09 ± 0.15 | 0.38–1.49 | 0.11 |
| Pulse pressure, mm Hg | 60 ± 16 | 28–106 | 60 ± 16 | 30–126 | 0.82 |
| Total cholesterol, mg/dL | 193 ± 33 | 122–395 | 195 ± 33 | 105–307 | 0.41 |
| HDL cholesterol, mg/dL | 50 ± 14 | 26–108 | 51 ± 14 | 23–100 | 0.32 |
| LDL cholesterol**, mg/dL | 125 ± 37 | 46–331 | 126 ± 37 | 46–249 | 0.74 |
| Male, n (%) | 160 (48%) | 146 (44%) | 0.33 | ||
| Smoker, n (%) | 30 (9%) | 23 (7%) | 0.40 | ||
| Diabetes, n (%) | 48 (15%) | 71 (22%) | 0.02 | ||
Column entries are means ± standard deviation or numbers (percentages)
Calculated according to the Friedewald formula
SD, standard deviation; ABI, ankle-brachial index; BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein
We performed univariate analyses in each subset to determine which of the conventional risk factors for cardiovascular diseases were associated with ABI. Age, sex, BMI and smoking all demonstrated statistically significant associations with ABI in both subsets. The associations between diabetes status and HDL cholesterol with ABI were statistically significant in Subset 1 only. Total cholesterol and LDL cholesterol did not demonstrate statistically significant associations with ABI in either subset.
The genomic annotations and allele frequencies of the 14 SNPs genotyped in this study are listed in Table 2. There were 2 SNPs for which genotype frequencies deviated significantly from Hardy-Weinberg expectations in one subset or the other but not both. In Figure 1, we illustrate the LD pattern of these SNPs.
Figure 1.
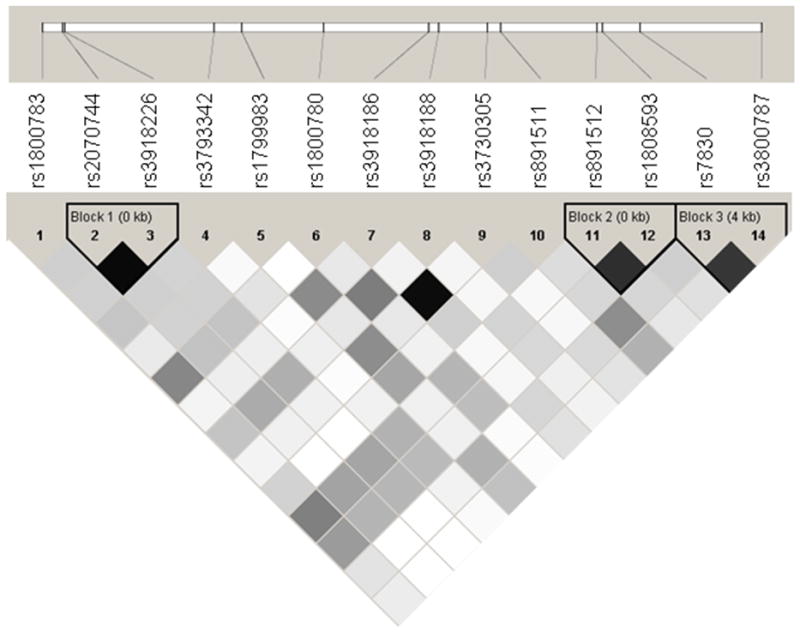
A linkage disequilibrium map of NOS3 (based on r2). Pooled analysis accounts for family structure; shading schemes of cells: r2 = 0 (white); 0 < r2 < 1 (shades of grey); r2 = 1 (black).
We adjusted ABI for independent predictors of PAD (age, sex, BMI, smoking, total cholesterol, HDL cholesterol, and diabetes), and the association of each SNP with ABI was assessed after adjustment for these covariates. Despite the fact that they were not statistically significant independent predictors of ABI in our study, we included diabetes, total cholesterol, and HDL cholesterol in our adjustment models, since the literature suggests that diabetes and lipid levels are risk factors for PAD [24].
The adjusted single SNP associations with ABI are summarized in Table 3. Overall, there were more NOS3 SNPs associated with ABI than would be expected by chance alone. Two specific SNPs (rs891512, and rs1808593) in LD (Figure 1) were significantly associated with ABI in both Subset 1 and Subset 2. One additional SNP (rs2070744) was highly significant in Subset 1 and marginally significant in Subset 2. The lowest frequency homozygote classes for the 2 replicate SNPs showed low relative frequencies in both subsets. Furthermore, no significant mean ABI differences between the lowest frequency homozygote classes and the corresponding heterozygote classes were detected. As such, these genotype classes were combined in order to determine the respective genotype-phenotype relationships. For both of the replicate SNPs, the combined genotype class had significantly lower mean ABI values compared to the respective highest frequency homozygote genotype classes, suggesting that carrying the minor allele for one of these polymorphisms could be associated with an increased risk for PAD. In Figure 2, we illustrate this trend by displaying the mean ABI values for the highest frequency homozygote and combined lowest frequency homozygote/heterozygote genotype classes for SNP rs1808593. The figure demonstrates not only the differences in the mean ABI values among genotype classes but also variation in ABI which may be indicative of underlying gene-environment or gene-gene interactions [25].
Table 3.
Adjusted* single SNP associations with ABI by Subset
| Subset 1 | Subset 2 | ||
|---|---|---|---|
| SNP | P | P | |
| 1 | rs1800783 | 0.0014 | 0.2445 |
| 2 | rs2070744 | 0.0002 | 0.1017 |
| 3 | rs3918226 | 0.5690 | 0.0355 |
| 4 | rs3793342 | 0.2645 | 0.7778 |
| 5 | rs1799983 | 0.1382 | 0.1468 |
| 6 | rs1800780 | 0.3829 | 0.2374 |
| 7 | rs3918186 | 0.7644 | 0.7221 |
| 8 | rs3918188 | 0.3427 | 0.9407 |
| 9 | rs3730305 | 0.8102 | 0.5799 |
| 10 | rs891511 | 0.2371 | 0.5210 |
| 11 | rs891512† | 0.0016 | 0.0402 |
| 12 | rs1808593† | 0.0012 | 0.0302 |
| 13 | rs7830 | 0.1804 | 0.1008 |
| 14 | rs3800787 | 0.3557 | 0.4060 |
Adjusted for gender, age, BMI, smoking, total cholesterol, HDL, and diabetes; ABI, ankle brachial index; BMI, body mass index; SNP, single nucleotide polymorphism
Model P-value <0.10 in both Subsets
Figure 2.
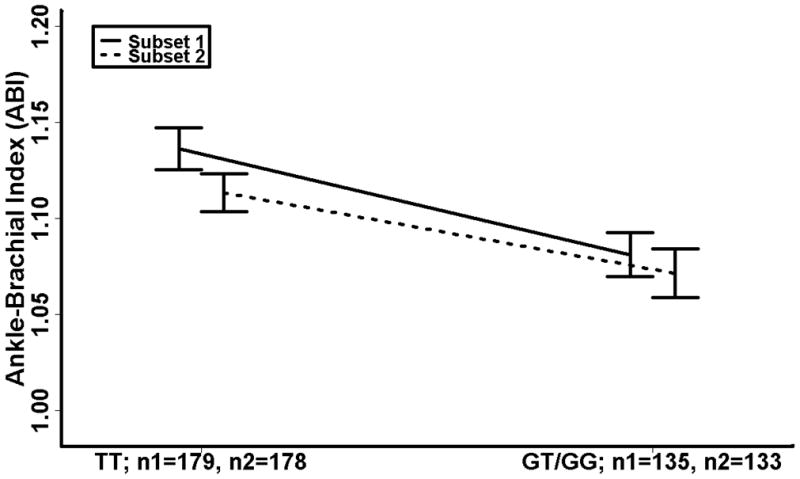
rs1808593 genotype specific mean ABI (highest frequency homozygote vs. combined lowest frequency homozygote/heterozygote) with standard error bars. ABI is adjusted for age, sex, BMI, smoking, total cholesterol, HDL cholesterol, and diabetes; ABI, ankle brachial index; BMI, body mass index.
Based on a sliding window approach with a window size of 2, we assessed the associations between 13 haplotypes and ABI. Haplotypes from 5 SNP pairs in Subset 1 and 3 SNP pairs in Subset 2 were significantly associated with ABI. Replication of significant associations between ABI and haplotypes from the rs2070744–rs3918226 SNP pair (Subset 1, P = 0.01; Subset 2, P = 0.02) and haplotypes from the rs1808593–rs7830 SNP pair (Subset 1, P = 0.02; Subset 2, P = 0.07) was detected. To determine whether the replication of P-values was associated with the same genotype-phenotype relationship, we performed tests of homogeneity according to subset. Both SNP pairs demonstrated homogeneous genotype-phenotype relationships across subsets.
In Figure 3, we display the average effects of the rs180593–rs7830 haplotypes which were observed in both subsets. Haplotypes that were not represented in both subsets were not included in the haplotype analyses as they would be non-informative with respect to our replication design. The TT haplotype increased mean ABI in both subsets. The TG haplotype also demonstrated an intermediate positive average effect in both subsets, increasing mean ABI in both Subset 1 and Subset 2. Conversely, the GG haplotype demonstrated a negative effect in both subsets (Subset 1 = −0.024, Subset 2 = −0.023), suggesting that the GG haplotype from this SNP pair may be associated with lower ABI values.
Figure 3.

Average effects of observed haplotypes (TT, TG & GG) on ABI from the rs1808593–rs7830 SNP pair. ABI is adjusted for age, sex, BMI, smoking, total cholesterol, HDL cholesterol, and diabetes; ABI, ankle brachial index; BMI, body mass index.
4. Discussion
Multiple studies have investigated the association of polymorphisms in candidate genes with the occurrence of essential hypertension and coronary heart disease, but relatively few studies have explored the relationship between specific candidate gene polymorphisms and PAD. We investigated the association of variants of NOS3 with ABI, a non-invasive measure of PAD, in hypertensive subjects. The motivating hypothesis was that genetic polymorphisms implicated in essential hypertension may influence PAD risk by means of a common pathophysiological pathway. Out of 14 NOS3 SNPs studied, allelic variants of 5 SNPs were significantly (P <0.10) related to inter-individual variation in ABI in at least 1 subset of the sample. Two SNPs were significant in both subsets.
In order to reduce the probability of Type I error, 14 NOS3 SNPs were analyzed in two replicate samples. Even after adjustment for conventional risk factors, 2 of the NOS3 SNPs shared significant associations with ABI, suggesting that alterations in NOS3 may indeed influence inter-individual variation in ABI. We did genotype the well-known NOS3 non-synonymous SNP Asp298Glu (rs1799983), which has been postulated to alter function of NOS3 [26], but did not find the SNP to be associated with ABI in either subset. We identified modest nominal associations, and the single SNP associations detected in this analysis require confirmation.
Fowkes et al. [27] did not find an association between a 27 bp repeat in intron 4 of NOS3 and ABI in a community-based study of 940 men and women aged 60–79 years. However, since testing single variants is unlikely to capture all the variation in a gene, all or most of the common variants should be tested when investigating the association between a candidate gene and a disease phenotype [28]. By testing a set of tag SNPs, potential causal variants may be evaluated for an association with a particular phenotype, either directly or indirectly, via LD [29]. Although the optimal methodology to select tag SNPs at a locus is still evolving [16,30,31], the tag SNPs we selected captured most of the nucleotide variation at the NOS3 locus.
To further investigate the effect of variation in NOS3 on ABI, we conducted a series of haplotype analyses. Clark [18] has indicated that considering variation organized in haplotypes is important for three main reasons: 1) protein coding genes produce sequences that correspond to haplotypes, 2) population variation is structured into haplotypes, and 3) haplotypes reduce dimensionality of statistical tests and may lead to an increase of statistical power. We demonstrated significant associations between NOS3 haplotypes and inter-individual variation in ABI in our replicate hypertensive subsets. Several other studies have investigated the potential influence of NOS3 haplotypes on BP variation. Persu et al. [32] investigated the associations between several measures of BP and haplotypes based on SNPs with proposed functional effects (Asp298Glu, intron 4 variable number tandem repeat, and T-786C) and informativeness (intron 13 CA-repeat) and found clear relationships between constructed haplotypes and ambulatory SBP. Sandrim et al. [33] investigated the association between hypertension and haplotypes based on the same 3 SNPs with proposed functional effects. Their findings suggest that 2 NOS3 haplotypes may confer a protective effective against hypertension among blacks and whites, while 1 NOS3 haplotype may confer susceptibility to hypertension among whites. While we did not analyze the specific haplotypes in these studies, we did investigate the associations between 2 haplotypes involving the Asp298Glu polymorphism (rs1799983) and ABI. These associations were not statistically significant in either subset. We also investigated 2 haplotypes involving the T-786C polymorphism (rs2070744) and identified a replicated effect on ABI for 1 of the haplotypes that included this putatively functional variant. As such, our results lend support to other studies that have investigated the role of NOS3 haplotypes, and suggest that combinations of NOS3 variants may play a role in the etiology of complex common diseases in hypertensives.
Atherosclerotic vascular disease is complex, polygenic, and likely influenced by gene-gene and gene-environment interactions. A previous study reported an association between NOS3 polymorphisms and coronary artery disease in smokers but not in non-smokers [34]. In the present study, we detected an interaction between the NOS3 rs3918226 polymorphism and smoking (Subset 1, P = 0.08; Subset 2, P = 0.01) in the prediction of ABI. However, a test of homogeneity revealed that this interaction was heterogeneous across subsets (P<0.01) and, as a result, the replication of this finding could not be confirmed. Further investigation of SNP-covariate, SNP-SNP, and haplotype-covariate interactions in larger datasets may help to elucidate the multifactorial determinants of PAD risk.
In the clinical setting, ABI is used as a dichotomous variable, and a cut off of ≤0.90 or ≤0.95 is employed to confirm the presence of PAD. We did not analyze ABI as a dichotomous variable as this entailed loss of statistical power, particularly since the prevalence of an abnormal ABI (defined as < 0.90) was low in our study sample. Despite this, our analyses with ABI as a continuous outcome were warranted as recent studies suggest that, even in the range of 1.0–1.3, lower ABI may be related to PAD risk factors [35].
Some limitations of the present study need to be considered. Our approach was based on the premise that susceptibility alleles for common diseases (and related subclinical disease measures such as ABI) are not under strong negative selection, and common variants contribute to common disease traits (i.e. the ‘common disease/common variant’ hypothesis) [36]. However, the allelic spectrum for complex quantitative traits such as ABI is not fully delineated, and it is possible that multiple rare NOS3 polymorphisms influence ABI. Due to a lack of power, identifying such alleles would not be possible using the approach employed in this study. Given the large number of bins (i.e. - haplotype blocks) identified in this gene, we did not have the resources to genotype all tagSNPs. Consequently, not all of the genetic variation within NOS3 may have been captured. Our assigned probability cut point for estimated haplotype inclusion, while conservative, led to a moderate loss in power as a number of individuals were excluded from each window. Our inferences may not be generalizable to individuals who are younger, normotensive, or of other ethnicities. Although a priori power calculations indicated that we were adequately powered to detect relatively small SNP effects, insufficient sample sizes in each subset or random measurement error may have limited our power to detect genotype-phenotype associations.
Conclusion
Using a parsimonious set of 14 SNPs that represented most of the polymorphic variation in NOS3, we found two of the SNPs to be consistently associated with inter-individual variation in ABI, a non-invasive marker of PAD, in replicate subsets of hypertensive patients. Two sets of haplotypes were also significantly associated with ABI in both subsets. These associations of select NOS3 variants with ABI provide rationale for further testing in other samples or in different experimental designs such as a case-control study. We regard our putative associations as potential hypotheses for independent replication studies. Further characterization of whether the NOS3 SNPs contribute to increased PAD and CHD risk will be useful for the potential early identification of hypertensive individuals at increased risk for these complications and for developing a better understanding of the etiology and pathophysiology of these diseases.
Acknowledgments
This work was supported by grant HL75794, HL68737, HL054481, HL54457 and the General Clinical Research Center Grant M01 RR00585 from National Institutes of Health.
References
- 1.Hiatt WR, Hoag S, Hamman RF. Effect of diagnostic criteria on the prevalence of peripheral arterial disease. The San Luis Valley Diabetes Study. Circulation. 1995;91:1472–1479. doi: 10.1161/01.cir.91.5.1472. [DOI] [PubMed] [Google Scholar]
- 2.Criqui MH. Peripheral arterial disease--epidemiological aspects. Vasc Med. 2001;6:3–7. doi: 10.1177/1358836X0100600i102. [DOI] [PubMed] [Google Scholar]
- 3.McDermott MM, Fried L, Simonsick E, Ling S, Guralnik JM. Asymptomatic peripheral arterial disease is independently associated with impaired lower extremity functioning: The Women’s Health and Aging Study. Circulation. 2000;101:1007–1012. doi: 10.1161/01.cir.101.9.1007. [DOI] [PubMed] [Google Scholar]
- 4.Cotran RS, Kumar V, Collins T. Pathologic Basis of Disease. 6. Philadelphia: WB Saunders Co; 1999. [Google Scholar]
- 5.Newman AB, Siscovick DS, Manolio TA, et al. Ankle-arm index as a marker of atherosclerosis in the Cardiovascular Health Study. Cardiovascular Heart Study (CHS) Collaborative Research Group. Circulation. 1993;88:837–45. doi: 10.1161/01.cir.88.3.837. [DOI] [PubMed] [Google Scholar]
- 6.Hirsch AT, Criqui MH, Treat-Jacobson D, et al. Peripheral arterial disease detection, awareness, and treatment in primary care. JAMA. 2001;286(11):1317–24. doi: 10.1001/jama.286.11.1317. [DOI] [PubMed] [Google Scholar]
- 7.Meijer WT, Hoes AW, Rutgers D, et al. Peripheral arterial disease in the elderly: The Rotterdam Study. Arterioscler Thromb Vasc Biol. 1998;18:185–92. doi: 10.1161/01.atv.18.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Kullo IJ, Bailey KR, Kardia SL, Mosley TH, Jr, Boerwinkle E, Turner ST. Ethnic differences in peripheral arterial disease in the NHLBI Genetic Epidemiology Network of Arteriopathy (GENOA) study. Vasc Med. 2003;8:237–242. doi: 10.1191/1358863x03vm511oa. [DOI] [PubMed] [Google Scholar]
- 9.Casas JP, Cavalleri GL, Bautista LE, Smeeth L, Humphries SE, Hingorani AD. Endothelial Nitric Oxide Synthase Gene Polymorphisms and Cardiovasculr Disease: A HuGE Review. Am J Epidemiol. 2006;164:921–935. doi: 10.1093/aje/kwj302. [DOI] [PubMed] [Google Scholar]
- 10.Huang PL, Huang Z, Mashimo H, Bloch KD, Moskowitz MA, Bevan JA, Fishman MC. Hypertension in mice lacking the gene for endothelial nitric oxide synthase. Nature. 1995;377:239–242. doi: 10.1038/377239a0. [DOI] [PubMed] [Google Scholar]
- 11.Kullo IJ, Mozes G, Schwartz RS, Gloviczki P, Tsutsui M, Katusic ZS, O’Brien T. Enhanced endothelium-dependent relaxations after gene transfer of recombinant endothelial nitric oxide synthase to rabbit carotid arteries. Hypertension. 1997;30:314–320. doi: 10.1161/01.hyp.30.3.314. [DOI] [PubMed] [Google Scholar]
- 12.Rankinen T, Rice T, Perusse L, Chagnon YC, Gagnon J, Leon AS, Skinner JS, Wilmore JH, Rao DC, Bouchard C. NOS3 Glu298Asp genotype and blood pressure response to endurance training: the HERITAGE family study. Hypertension. 2000;36:885–889. doi: 10.1161/01.hyp.36.5.885. [DOI] [PubMed] [Google Scholar]
- 13.Naber CK, Baumgart D, Altmann C, Siffert W, Erbel R, Heusch G. eNOS 894T allele and coronary blood flow at rest and during adenosine-induced hyperemia. Am J Physiol Heart Circ Physiol. 2001;281:H1908–1912. doi: 10.1152/ajpheart.2001.281.5.H1908. [DOI] [PubMed] [Google Scholar]
- 14.Philip I, Plantefeve G, Vuillaumier-Barrot S, Vicaut E, LeMarie C, Henrion D, Poirier O, Levy BI, Desmonts JM, Durand G, Benessiano J. G894T polymorphism in the endothelial nitric oxide synthase gene is associated with an enhanced vascular responsiveness to phenylephrine. Circulation. 1999;99:3096–3098. doi: 10.1161/01.cir.99.24.3096. [DOI] [PubMed] [Google Scholar]
- 15.Savvidou MD, Vallance PJ, Nicolaides KH, Hingorani AD. Endothelial nitric oxide synthase gene polymorphism and maternal vascular adaptation to pregnancy. Hypertension. 2001;38:1289–1293. doi: 10.1161/hy1201.097305. [DOI] [PubMed] [Google Scholar]
- 16.Stram DO, Haiman CA, Hirschhorn JN, Altshuler D, Kolonel LN, Henderson BE, Pike MC. Choosing haplotype-tagging SNPS based on unphased genotype data using a preliminary sample of unrelated subjects with an example from the Multiethnic Cohort Study. Hum Hered. 2003;55:27–36. doi: 10.1159/000071807. [DOI] [PubMed] [Google Scholar]
- 17.Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA. Haplotype tagging for the identification of common disease genes. Nat Genet. 2001;29:233–237. doi: 10.1038/ng1001-233. [DOI] [PubMed] [Google Scholar]
- 18.Clark AG. The Role of Haplotypes in Candidate Gene Studies. Genet Epidemiol. 2004;27:321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- 19.Boerwinkle E. Multi-Center Genetic Study of Hypertension: The Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 20.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the Concentration of Low-Density Lipoprotein Cholesterol in Plasma, Without Use of the Preparative Ultrcentrifuge. Clinical Chemistry. 1972;18(6):499–502. [PubMed] [Google Scholar]
- 21.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens M, Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Templeton AR. The general relationship between average effect and average excess. Genetical Research. 1987;49:69–70. doi: 10.1017/s0016672300026756. [DOI] [PubMed] [Google Scholar]
- 24.Criqui MH, Denenberg JO, Langer RD, Fronek A. The epidemiology of peripheral arterial disease: importance of identifying the population at risk. Vascular Medicine. 1997;2(3):221–6. doi: 10.1177/1358863X9700200310. [DOI] [PubMed] [Google Scholar]
- 25.Reilly SL, Ferrell RE, Kottke BA, Sing CF. The gender-specific apolipoprotein E genotype influence on the distribution of plasma lipids and apolipoproteins in the population of Rochester, Minnesota. II. Regression relationships with concomitants. Am J Hum Genet. 1992;51:1311–1324. [PMC free article] [PubMed] [Google Scholar]
- 26.Wattanapitayakul SK, Mihm MJ, Young AP, Bauer JA. Therapeutic implications of human endothelial nitric oxide synthase gene polymorphism. Trends Pharmacol Sci. 2001;22:361–368. doi: 10.1016/s0165-6147(00)01692-8. [DOI] [PubMed] [Google Scholar]
- 27.Fowkes FG, Lee AJ, Hau CM, Cooke A, Connor JM, Lowe GD. Methylene tetrahydrofolate reductase (MTHFR) and nitric oxide synthase (ecNOS) genes and risks of peripheral arterial disease and coronary heart disease: Edinburgh Artery Study. Atherosclerosis. 2000;150:179–185. doi: 10.1016/s0021-9150(99)00366-4. [DOI] [PubMed] [Google Scholar]
- 28.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 30.Byng MC, Whittaker JC, Cuthbert AP, Mathew CG, Lewis CM. SNP subset selection for genetic association studies. Ann Hum Genet. 2003;67:543–556. doi: 10.1046/j.1529-8817.2003.00055.x. [DOI] [PubMed] [Google Scholar]
- 31.Chapman JM, Cooper JD, Todd JA, Clayton DG. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 32.Persu A, Vinck WJ, El Khattabi O, Janssen RGJH, Paulussen ADC, Devuyst O, Vlietinck R, Fagard RH. Influence of the endothelial nitirc oxide synthase gene on conventional and ambulatory blood pressure: sib-pair analysis and haplotype study. Journal of Hypertension. 2005;23:759–765. doi: 10.1097/01.hjh.0000163144.74588.ad. [DOI] [PubMed] [Google Scholar]
- 33.Sandrim VC, Coelho EB, Nobre F, Arado GM, Lanchote VL, Tanus-Santos JE. Suceptible and protective eNOS haplotypes in hypertensive black and white subjects. Atherosclerosis. 2006;186:428–432. doi: 10.1016/j.atherosclerosis.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Wang XL, Sim AS, Badenhop RF, McCredie RM, Wilcken DE. A smoking-dependent risk of coronary artery disease associated with a polymorphism of the endothelial nitric oxide synthase gene. Nat Med. 1996;2:41–45. doi: 10.1038/nm0196-41. [DOI] [PubMed] [Google Scholar]
- 35.McDermott MM, Liu K, Criqui MH, Ruth K, Goff D, Saad MF, Wu C, Homma S, Sharrett AR. Ankle-brachial index and subclinical cardiac and carotid disease: the multi-ethnic study of atherosclerosis. Am J Epidemiol. 2005;162:33–41. doi: 10.1093/aje/kwi167. [DOI] [PubMed] [Google Scholar]
- 36.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]


