Summary
As nucleosomes are widely replaced by protamine in mature human sperm, epigenetic contributions of sperm chromatin to embryo development have been considered highly limited. However, we find the retained nucleosomes significantly enriched at loci of developmental importance including imprinted gene clusters, miRNA clusters, HOX gene clusters, and the promoters of stand-alone developmental transcription and signaling factors. Importantly, histone modifications localize to particular developmental loci. H3K4me2 is enriched at certain developmental promoters, whereas large blocks of H3K4me3 localize to a subset of developmental promoters, regions in HOX clusters, certain non-coding RNAs, and generally to paternally-expressed imprinted loci, but not paternally-repressed loci. Notably, H3K27me3 is significantly enriched at developmental promoters that are repressed in early embryos, including many bivalent (H3K4me3/H3K27me3) promoters in embryonic stem cells. Finally, developmental promoters are generally DNA hypomethylated in sperm, but acquire methylation during differentiation. Taken together, epigenetic marking in sperm is extensive, and correlated with developmental regulators.
During spermiogenesis canonical histones are largely exchanged for protamines1, 2, small basic proteins that form tightly-packed DNA structures important for normal sperm function3. We find about 4% of the genome retained in nucleosomes (Supplementary Fig. 1a). The rare retained nucleosomes in sperm consist of either canonical or histone variant proteins, including a testes-specific histone H2B (TH2B) with an unknown specialized function4,5. Their presence may simply be due to inefficient protamine replacement, leading to a low random distribution genome-wide with no impact in the embryo. Alternatively, these retained nucleosomes, along with attendant modifications, might be enriched at particular genes/loci. This latter possibility would raise the possibility for programmatic retention for an epigenetic function in the embryo. To address these questions, we localized the nucleosomes retained in mature sperm from fertile donors using high-resolution genomic approaches.
Developmental loci bear nucleosomes
To address donor variability, we examined nucleosome retention in a single donor (D1) and/or a pool of four donors (donor pool). Sperm chromatin was separated into protamine-bound and histone-bound fractions. Briefly, mononucleosomes were isolated (>95% yield) through sequential MNase digestion and sedimentation (Supplementary Fig. 1b-e). This mononucleosome pool was utilized for ChIP (to select modified nucleosomes), or the DNA was isolated from the mononucleosome pool to represent all nucleosomes. Purified DNA was subjected to high-throughput sequencing (Illumina GAII), or alternatively, labeled and hybridized to a high-density promoter-tiling array (9 kb tiled; schematic, Supplementary Fig. 2).
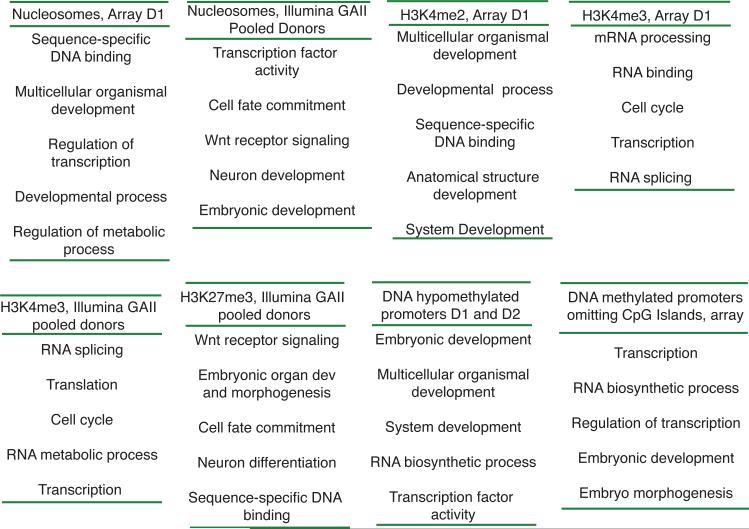
Our initial array approach examined three replicas of D1 (pairwise average R2 = 0.85). Notably, gene ontology (GO) analysis revealed nucleosomes significantly enriched at promoters that guide embryonic development, primarily developmental transcription factors and signaling molecules (GO-term false discovery rate (FDR) <0.01; Fig. 1, Supplementary Table 1: for all extended GO categories see Supplementary Tables and Supplementary Dataset 1). To conduct genome-wide profiling, we performed high-throughput sequencing of nucleosomes from D1 or the donor pool. Regions significantly enriched for histone relative to the input control (sheared total sperm DNA) were identified using a 300-bp window metric6. For display, we depict the normalized difference and FDR window scores (Fig. 2a, FDR transformation (−10 log10 (q-value FDR), 20 = 0.01, 25 = 0.003, 30 = 0.001, 40 = 0.0001). Histone-enriched loci for one individual (D1) were well correlated with a donor pool (r = 0.7). Globally, 76% of the top 9841 histone-enriched regions (FDR 40 cutoff) intersect genic regions, whereas the expected intersection given random distribution is 36% (p-value <0.001).
Figure 1.
Developmental genes are associated with particular chromatin attributes in human sperm. GoMiner was used to identify enriched categories, and all categories displayed have an FDR < 0.01. The top five general categories are listed, after omitting nearly identical/redundant classes. An expanded GO table with the unfiltered top 30-60 categories, the total genes, number of changed genes, enrichment, and FDR are provided in the Supplemental Material.
Figure 2.
Profiling of nucleosomes and their modifications at HOXD. For high-throughput sequencing, we display the mapped sequencing reads from D1 or a donor pool (red or orange bars; normalized difference score), and their significance (green or blue bars; FDR of 20 is <1% and FDR of 30 is <0.1%). a, The HOXD locus (black box) and an uncharacterized flanking locus (green oval). b, Profiling of nucleosome modifications at HOXD (in part a). The y-axis is signal intensity (log2, for ChIP-chip), or the normalized difference score for sequencing. The regions not tiled on the array are underlined in red.
Interestingly, sequencing revealed significant (FDR <0.001) histone retention at many loci important for embryo development including embryonic transcription factors and signaling pathway components (Fig. 1, Supplementary Tables 2 and 3). We display this enrichment at HOX loci (Fig. 2, Supplementary Fig. 3), but also observe this at stand-alone developmental transcription factors (Supplementary Fig. 4) and signaling factors (Supplementary Fig. 5). An FDR of 60 yields 4,556 genes, 1, 683 of which are grouped with developmental GO categories (2,848 total developmental genes). The magnitude of nucleosome enrichment at developmental loci is modest, with high significance provided by a moderate average increase at a large number of loci. Histones are also significantly enriched at the promoters of miRNAs (p-value <0.05) (Supplementary Fig. 6) and at the class of imprinted genes (p-value <0.0001) (Fig. 3), addressed in detail later. Selected loci were tested and confirmed by qPCR (Supplementary Fig. 7a-e). Outside of these enriched regions, we observe sequencing reads at low levels distributed genome-wide (e.g. Fig. 2a, Fig. 3a), an observation consistent with low levels of nucleosomes genome-wide, though contributions from non-nucleosomal contamination cannot be ruled out.
Figure 3.
Nucleosomes enrichment at imprinted gene clusters, with high H3K4me3 at paternally-expressed non-coding RNAs, and paternally-demethylated regions. a, Histone enrichment at the 11p15.5 imprinted cluster (ending near OSBPL5), but not in the adjacent region. b and c, An expanded view of the DMRs (yellow rectangles) of H19 (paternally-methylated) and MEST (paternally-demethylated). d, High H3K4me3 at the promoters of the paternally-expressed genes BEGAIN, DLK1, and RTL, and the lack of H3K4me3 at the methylated intergenic-differentially methylated region (IG-DMR) of GTL2 in sperm. Notably, both H3K4me3 and H3K27me3 reside at the promoter of GTL2, which later acquires methylation in the embryo.
Protamine occupancy (3 replicas, R2 = 0.89, arrays only) yielded 7,151 enriched regions (>2.5-fold), but failed to identify any enriched GO term categories, though a few segments of the Y chromosome were notably enriched (including the testis-specific TSPY genes, data not shown). Regions of histone enrichment did not exclude protamine, consistent with a nucleosomeprotamine mixture existing even at histone-enriched loci. However, as protamine fragments averaged ~750 bp, protamine depletion would have to be extensive (regions >2 kb) to be apparent. Taken together, nucleosomes are significantly enriched in sperm at genes important for embryonic development, with transcription factors the most enriched class.
Localization of modified nucleosomes
As histones replace protamines genome-wide at fertilization7,8, unmodified histones retained in sperm would seem insufficient to influence gene regulation in embryos. Therefore, we examined three additional chromatin properties: 1) histone variants, 2) histone modifications, and 3) DNA methylation. ChIP-chip analysis of TH2B (two replicas, R2 = 0.93) reveals 0.3% of gene promoters with relatively high levels of TH2B (>2-fold enrichment). GO analysis revealed significant (FDR <0.06) enrichment at genes important for sperm biology, capacitation and fertilization (Supplementary Table 4), but not at developmental categories. ChIP-Seq analysis with H2A.Z nucleosomes (at standard conditions, 150 mM-250 mM salt) did not reveal significant enriched GO categories, with high enrichment limited to pericentric heterochromatin (Supplementary Fig. 8), consistent with prior immunostaining9.
Modified nucleosomes were localized by performing ChIP on mononucleosomes, followed by either array analysis or sequencing (schematic, Supplementary Fig. 2). We normalized the dataset for each modification to the dataset derived from input mononucleosomes, determined enriched regions (array, >2-fold; sequencing FDR 40), found the nearest neighboring gene, and performed GO analysis. In somatic cells, H3K4me2 is correlated with euchromatic regions. In sperm, ChIP-chip H3K4me2 was enriched at many promoters, and at significant levels at promoters for developmental transcription factors (two replicas R2 = 0.94; GO term FDR <0.06, Fig. 1, Supplementary Table 5). In somatic cells, H3K4me3 is localized to: 1) the TSS of active genes, 2) genes bearing ‘poised’ RNA polymerase II (Pol II), and 3) the proximal promoter of inactive developmental regulators in ES cells - promoters that also bear the silencing mark H3K27me310,11, and thus termed bivalent. Mature sperm are transcriptionally inert, and Pol II protein levels are barely detectable (data not shown), so the high H3K4me3 levels we observed in sperm chromatin (Supplementary Fig. 1f) seemed surprising. H3K4me3 was localized by both ChIP-chip (3 replicas, R2 = 0.96) and ChIP-Seq. The raw datasets were similar (r = 0.7) and the thresholded datasets were very similar (array 2-fold; Seq, FDR 40; 96% intersection, p-value <0.001). With both datasets, simple inspection revealed small peaks at many gene 5’ ends, with high levels and broader blocks at a subset of genes (ie. HOX loci. Fig. 2, Supplementary Fig. 3). GO term analyses with either dataset yielded genes important for changing nuclear architecture, RNA metabolism, spermatogenesis, and also selected transcription factors important for embryonic development (FDR <0.01, Fig. 1, Supplementary Tables 6 and 7, Supplementary Fig. 9). H3K4me3 at genes related to nuclear architecture and spermatogenesis can presumably be attributed to their prior activation during gametogenesis. RNA metabolism occurs both in gametogenesis and the early embryo, so attribution to a prior program as opposed to a potential poising for a future program cannot be unambiguously attributed. However, several transcription and signaling factors of importance in embryo development exhibited high levels and a broad distribution of H3K4me3, including EVX1/2, ID1, STAT3, KLF5, FGF9, SOX7/9, certain HOX genes, and certain non-coding RNAs (Fig. 2, Supplementary Fig. 3, 6).
Interestingly, ChIP-Seq analysis revealed very significant levels of H3K27me3 at developmental promoters in sperm (Supplementary Table 8, Fig. 2, Supplementary Fig. 3,4), and overlapped significantly with H3K27me3-occupied genes in embryonic stem (ES) cells (p-value <0.01), which are silent prior to differentiation. Furthermore, bivalent genes (bearing H3K4me3 and H3K27me3) in ES cells had a very significant overlap with bivalent genes in sperm (FDR <0.001 for each mark). Of the 1999 genes identified as bivalent in ES cells, 861 were bivalent in sperm (p-value <0.01; Supplementary Table 9). Also notable but not explored further were many blocks of high H3K4me3 or H3K27me3 in regions lacking annotation (oval, Fig. 2a). Finally, H3K9me3 was not detected at the small set developmental promoters tested, but was high at pericentric regions (qPCR only, Supplementary Fig. 7d). Taken together, our results reveal extensive histone modification patterns, and significant similarities to patterns observed in ES cells.
DNA methylation profiles
DNA methylation profiles examined two fertile donors (D2 and D4) using a methylated DNA immunoprecipitation (MeDIP) procedure and promoter arrays (individual replicates average D2 R2 = 0.97 and D4 R2 = 0.89). Their methylation patterns were highly similar (pairwise R2 = 0.86), and extensive qPCR validated our array threshold (Supplementary Fig. 7e). GO analysis of genes with pronounced DNA hypomethylation yielded transcription and signaling factors that guide embryo development (FDR <0.05; Fig. 1, Supplementary Table 10) including HOX loci (Fig. 1, Fig. 4, blue and green bars, and Supplementary Fig. 4). Hypomethylation also overlapped very significantly with histone-enriched promoters (p-value <0.02, Supplementary Table 11). Bisulphite sequencing verified the MeDIP results, revealing extensive hypomethylation at developmental promoters in sperm (Supplementary Fig. 10b,c).
Figure 4.
Developmental promoters in sperm lack DNA methylation, but acquire methylation during development. a, DNA methylation of the HOXD locus in the mature sperm (blue bars) or primary fibroblasts (orange line overlay). The y-axis is the signal intensity (log2) and the x-axis is the annotated physical map (HG17). The regions not tiled on the array are underlined in red.
Interestingly, DNA hypomethylated promoters in mature sperm overlap greatly with developmental promoters bound by the self renewal network of transcription factors in human ES cells (e.g. OCT4, SOX2, NANOG, KLF4, and FOXD3 proteins12; intersection of OCT4 protein occupancy and DNA hypomethylation, p-value <0.01). In ES cells, these proteins promote self renewal and also work with repressive polycomb complexes (PRC2) to help repress a large set of developmental regulators (including HOX genes) to prevent differentiation10, 13-17 18-20. However, the hypomethylation of developmental genes in sperm is extensive (Fig. 4, Supplementary Fig. 4). In fact, when CpG islands are omitted from the datasets, GO term analysis of hypomethylated promoters still yields developmental genes (Fig. 1, Supplementary Table 12). Notably, many of these developmental genes become methylated following differentiation; differential analysis of sperm and primary human fibroblasts (MeDIP, two replicas R2 = 0.86) showed that the promoters occupied by PRC2/SUZ12 in human ES cells acquire methylation in fibroblasts (FDR <0.01, Supplementary Table 13 and 14; HOXD illustrated in Fig. 4, Supplementary Fig. 4 and 5). Finally, the promoters driving several key members of the self-renewal network are themselves markedly hypermethylated in sperm (OCT4, NANOG, FOXD3)(Fig. 10c), whereas their developmental target genes are hypomethylated (Fig. 10b), consistent with recent studies in mice21-24.
Attributes of HOX clusters and miRNAs
Nucleosome enrichment was clear across HOX loci and proximal flanking regions, but falls off precipitously outside (HOXD, Fig. 2a; HOXA Supplementary Fig. 3a). Histone-enriched HOXD regions with a single donor (D1) were largely shared with the donor pool (Fig. 2a) (D1 vs. donor pool, r = 0.7). Interestingly, retained nucleosomes have regional covalent modifications. For example, distinct and very large (5-20kb) blocks of H3K4me3 are clearly observed at all HOX loci, and also at imprinted genes (addressed below). At HOXD, high H3K4me3 extends for ~20 kb, encompassing all of EVX2 and extending to the 3’ region of HOXD13 (Fig. 2b). Remarkably, a similar profile is observed at the related HOXA locus (Supplementary Fig. 3a). At HOXD second block of H3K4me3 is observed in the region between HOXD4 and HOXD8 (Fig. 2b) a region that encodes multiple ncRNAs expressed during development. This region represents a marked difference from the chromatin status in ES cells; in ES cells HOXD8-D11 are all bivalent. The distribution of H3K4me2 (determined from two replicas of D1) is clearly different from H3K4me3 at HOX loci (Fig. 2b, Supplementary Fig. 3). For example, at HOXD, H3K4me2 is enriched in HOXD8-D11, the region most deficient in H3K4me3 (Fig. 2b). Interestingly, high H3K27me3 encompasses all HOX loci and their proximal flanking regions. In contrast, high levels of H3K9me (a mark of heterochromatin) (Supplementary Fig. 7d) or H2A.Z were not detected at the HOX loci.
Histones are enriched at many miRNAs, especially miRNA clusters (Supplementary Fig. 6). For example, 16 of the 29 miRNA clusters on autosomes were significantly enriched (P-value <0.05). Clusters include those bearing let7e, miR-17, miR-15a, miR-96, miR-135b, and miR-10a/b, as well as the stand-alone miRNAs miR-153-1, miR-488 and miR-760. Notably, many histone-occupied miRNAs are associated with embryonic development25 (p-value <0.01), and their promoters were largely hypomethylated (Supplementary Fig. 10d). Furthermore, 7 of the 12 miRNAs on autosomes that are occupied by OCT4, NANOG and SOX2 in human ES cells17 are significantly occupied by histone (from pooled sequencing data). However, we do not currently understand the logic for their modification status; certain miRNA clusters have high histone and bivalent status, while others lack either modification (Supplementary Fig. 6).
Attributes of primary and secondary imprinted genes
Nucleosomes are significantly enriched at most imprinted genes in sperm, but at both paternally- and maternally-expressed loci. However, we observe striking specificity of H3K4me3 localization, with high and broad levels present at genes and non-coding RNAs that are paternally expressed. Locus 11p15.5 (Fig. 3a) is a large imprinted cluster with IGF2, H19 and KCNQ1 and multiple miRNAs. Here, increased levels of histone are present throughout the imprinted region (up to OSBPL5), but not in the large adjacent region lacking imprinted genes (Fig. 3a). Notably, the paternally-silenced H19 locus upstream of KCNQ1 has a methylated DMR (Supplementary Fig. 10a) that lacks H3K4me3 (Fig. 3b). In contrast, MEST (a paternally-expressed gene) has high H3K4me3 that extends from its promoter and first exon (containing the demethylated differentially methylated region (DMR), Fig. 3c and 10a) through the second exon. The antisense non-coding RNA MESTIT (also paternally expressed) is transcribed from the first intron, which is also very high in H3K4me3 (Fig. 3c). Furthermore, the promoter region of the paternally-expressed antisense non-coding RNA KCNQ1OT1 displays H3K4me3 (Fig. 3a, and data not shown) and the DMR is DNA demethylated (Fig. 10a). Several additional examples of paternally-expressed loci with blocks of H3K4me3 are provided in Supplemental Figure 11, including PEG3, the non-coding RNAs AIR (antisense to IGF2R) and GNASAS (antisense to GNAS). In contrast, genes flanking KCNQ1 that are repressed by the non-coding RNA KCNQOT1 (such as OSBPL5, TSSC4 and CD81, Fig. 3a, expanded in Supplementary Fig. 11) contain histone, but lack H3K4me3. Notably, several paternally-silenced genes (bearing DNA methylation) bore moderate (2-3 fold) enrichment of H3K9me3, a mark absent at paternally-expressed genes (Supplementary Fig. 7d).
The 14q32.33 region (DLK-DIO3) is complex and interesting; paternally-expressed genes such as DLK1 and RTL1 have moderate levels of H3K4me3 in their promoters, and the imprinting control locus (IG-DMR) lacks H3K4me3 (Fig. 3d) and is DNA methylated26-28. Notably, the promoter of MEG3/GTL2 (just downstream of the IG-DMR) lacks DNA methylation in sperm, but acquires DNA methylation in the embryo26-28, termed secondary imprinting. Remarkably, this MEG3/GTL2 promoter region that later acquires methylation initially bears both H3K4me3 and H3K27me3 in sperm; it is bivalent. One interpretation is that H3K4me3 may prevent DNA methylation in the sperm and early embryo, with H3K27me3 helping to ensure early silencing at this locus. Finally, our examination of the X chromosome inactivation center revealed an apparent bivalent status (and DNA hypomethylation) at the TSS of the XIST non-coding RNA, but not at TSIX, though future studies are required to determine whether these marks influence the regulation of this locus in the embryo (Supplementary Fig. 6, 10d; note, sequence reads on the X are half that on autosomes, as the X is present in only 50% of sperm).
Modifications and expression timing
Transcriptome analysis has been performed in 4-cell and 8-cell human embryos, with 29 or 65 mRNAs identified as enriched, respectively29. Interestingly, genes in sperm bearing H3K4me3 but not H3K27me3 correlated with genes expressed at the 4-cell stage (14/24, p-value = 0.059). Also, genes bearing high H3K4me2 were significantly enriched at genes expressed in the 4-8 cell stage (23 of 49, p-value <0.02; only 49 tiled on our array). In contrast, no significant correlation was observed with H3K27me3, which instead associates with TFs required for differentiation and organogenesis (discussed above). Finally, we verified by qPCR the presence of H3K4me2 or H3K4me3 at a subset of these stage-specific gene promoters (Supplementary Fig. 12). Thus, these findings reveal correlations of H3K4me2/3 enrichment, but not H3K27 enrichment, with early expression.
Conclusion
We provide several lines of evidence that the parental genome is packaged and covalently modified in a manner consistent with influencing embryo development. Previous analyses of DNA methylation in sperm identified hypomethylated promoters23,24,30,31, showed similarities to the pattern in ES cells24,31, and overlap between PRC2 and CpG islands15,17,21,22. We add that hypomethylated developmental promoters in human sperm overlap significantly with developmental promoters (in ES cells) occupied by the self-renewal network. Also, the promoters that acquire methylation in fibroblasts are primarily developmental transcription factors that are bound by PRC2 in human ES cells, consistent with recent work linking PRC2 to DNA methylation in development and neuronal differentiation in mice21,32,33. Thus, components of the self-renewal network emerge as candidates for helping to direct DNA hypomethylation in the germline, and also to guide DNA hypermethylation to particular loci during differentiation, possibly to help ‘lock in’ differentiation decisions, though this remains to be tested.
The central findings of our work involve the significant enrichment of modified nucleosomes in the sperm genome at genes for embryo development, and a specificity to their modification patterns that might be instructive for the regulation of developmental genes, non-coding RNAs and imprinted loci. For example, histone retention and modification were clear at HOX loci and most of the targets of the self-renewal network in ES cells. One key concept in ES cell chromatin is the prevalence of developmental promoters with a bivalent status – bearing both H3K27me3 and H3K4me310. Interestingly, many promoters bivalent in ES cells are likewise bivalent in sperm, although some bear only H3K27me3 in sperm. Notably, H3K27me3 covers essentially all of the four HOX loci in sperm, whereas H3K4me3 is present in large blocks at only a subset of locations in HOX loci. Our work also provides correlations between H3K4me, but not H3K27me, and early expression in the embryo. In contrast, protamine-enriched loci did not reveal any significant GO categories. However, there were certain segments of the Y chromosome with protamine enrichment, including the testis-specific TSPY genes, though the significance is not known.
We also find histones enriched at imprinted gene clusters, and a striking correlation between H3K4me3 and paternally-expressed non-coding RNAs and genes; loci that lack DNA methylation in sperm. In contrast, maternally-expressed non-coding RNAs/genes, and especially paternally-methylated regions, lack H3K4me3 and (for the selected genes tested) contain moderate H3K9me3. Consistent with these observations, recent structural and in vitro data show that H3K4 methylation deters DNA methylation by DNMT3A2 and DNMT3L in mice36. However, experiments in model organisms are needed to address whether the modification patterns we report influence imprinting patterns in vivo. Taken together, we reveal chromatin features in sperm that may contribute to totipotency, developmental decisions, and imprinting patterns, and open new questions about whether aging and lifestyle affects chromatin in a manner that impacts fertility or embryo development.
Methods Summary
Biological Samples
Sperm samples were obtained from four men of known fertility attending the University of Utah Andrology laboratory, consented for research. Samples were collected after 2-5 days abstinence and subjected to a density gradient (to purify viable, motile, mature sperm) and treated with somatic cell lysis buffer (0.1% SDS, 0.5% Triton X in DEPC H2O) for 20 min on ice to eliminate white blood cell contamination. Samples were centrifuged at 10,000 G for 3 min and the sperm pellet was resuspended in 1X PBS and used immediately for chromatin preparation. Clontech human fibroblast cells (Lonza cc-2251) were cultured (37°C and 5% CO2) in DMEM containing 10% FBS and supplemented with penicillin and streptomyocin.
Chromatin IP
Standard ChIP methods were used35, but we omitted crosslinking and utilized the following salt concentrations in the numbered buffers (ref. 35): 1) 150 mM NaCl, 2) 250 mM NaCl, 3) 200 mM LiCl, and 4) 150 mM NaCl (the PBS wash). Antibodies: anti-H3K27me3 (Upstate 07-449), H3K4me3 (Abcam 8580), H3K4me2 (Abcam 32356), or TH2B (Upstate 07-680), H2A.Z (Abcam 4174), H3K9me3 (Abcam 8898). For each, 4 μL of antibody was coupled to 100 μL of Dynabeads (Invitrogen). Following ChIP, samples for sequencing were not amplified, whereas for arrays the DNA was amplified (WGA, Sigma) prior to hybridization.
Methylation Profiling Using MeDIP
MeDIP procedures for sperm and primary human fibroblasts (Clonetech) were performed as described previously30.
Sequencing
Sequencing utilized the Illumina GAII (Illumina Inc.) with standard protocols. Read numbers are final mapped microsatellite filtered reads (26-36 bases). Nucleosomes from D1: 19,658,110, D2-D4:18,842,467, D1-4: 25,933,196 with equal contribution from each donor (random subsampling). Input, human sperm DNA: 17,991,622, H3K4me3: 13,337,105, H3K27me3:10,344,413, and H2A.Z: 5,449,000. All genomics datasets have been deposited in GEO under the SuperSeries GSE15594.
Supplementary Material
Acknowledgements
We thank B. Dalley for microarray and sequencing expertise, B. Schackmann for oligos, Ken Boucher for statistical analysis, Jacqui Wittmeyer for yeast nucleosomes and helpful comments, and Tim Parnell for helpful comments. Financial support from the Department of Urology (genomics and support of S.S.H.), the Howard Hughes Medical Institute (genomics, biologicals and support of J.P. and H.Z.), CA24014 and CA16056 for core facilities, and the Huntsman Cancer Institute (bioinformatics and support of D.A.N.). B.R.C. is an investigator with HHMI.
Footnotes
References
- 1.Ward WS, Coffey DS. DNA packaging and organization in mammalian spermatozoa: comparison with somatic cells. Biol Reprod. 1991;44:569–74. doi: 10.1095/biolreprod44.4.569. [DOI] [PubMed] [Google Scholar]
- 2.Wykes SM, Krawetz SA. The structural organization of sperm chromatin. J Biol Chem. 2003;278:29471–7. doi: 10.1074/jbc.M304545200. [DOI] [PubMed] [Google Scholar]
- 3.Balhorn R, Brewer L, Corzett M. DNA condensation by protamine and arginine-rich peptides: analysis of toroid stability using single DNA molecules. Mol Reprod Dev. 2000;56:230–4. doi: 10.1002/(SICI)1098-2795(200006)56:2+<230::AID-MRD3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 4.Gatewood JM, Cook GR, Balhorn R, Schmid CW, Bradbury EM. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. J Biol Chem. 1990;265:20662–6. [PubMed] [Google Scholar]
- 5.Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–9. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 6.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reik W, Santos F, Dean W. Mammalian epigenomics: reprogramming the genome for development and therapy. Theriogenology. 2003;59:21–32. doi: 10.1016/s0093-691x(02)01269-4. [DOI] [PubMed] [Google Scholar]
- 8.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 9.Rangasamy D, Berven L, Ridgway P, Tremethick DJ. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 2003;22:1599–607. doi: 10.1093/emboj/cdg160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein BE, et al. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–56. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–84. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–5. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 15.Tanay A, O'Donnell AH, Damelin M, Bestor TH. Hyperconserved CpG domains underlie Polycomb-binding sites. Proc Natl Acad Sci U S A. 2007;104:5521–6. doi: 10.1073/pnas.0609746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 17.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–13. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 19.Kopp JL, Ormsbee BD, Desler M, Rizzino A. Small Increases in the Level of Sox2 Trigger the Differentiation of Mouse Embryonic Stem Cells. Stem Cells. 2008 doi: 10.1634/stemcells.2007-0951. [DOI] [PubMed] [Google Scholar]
- 20.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 21.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–66. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 22.Illingworth R, et al. A novel CpG island set identifies tissue-specific methylation at developmental gene loci. PLoS Biol. 2008;6:e22. doi: 10.1371/journal.pbio.0060022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–85. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farthing CR, et al. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glazov EA, McWilliam S, Barris WC, Dalrymple BP. Origin, evolution, and biological role of miRNA cluster in DLK-DIO3 genomic region in placental mammals. Mol Biol Evol. 2008;25:939–48. doi: 10.1093/molbev/msn045. [DOI] [PubMed] [Google Scholar]
- 27.Takada S, et al. Delta-like and gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10:1135–8. doi: 10.1016/s0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- 28.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–16. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Li SS, Liu YH, Tseng CN, Singh S. Analysis of gene expression in single human oocytes and preimplantation embryos. Biochem Biophys Res Commun. 2006;340:48–53. doi: 10.1016/j.bbrc.2005.11.149. [DOI] [PubMed] [Google Scholar]
- 30.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 31.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–9. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vire E, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 33.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–6. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 34.Zalenskaya IA, Bradbury EM, Zalensky AO. Chromatin structure of telomere domain in human sperm. Biochem Biophys Res Commun. 2000;279:213–8. doi: 10.1006/bbrc.2000.3917. [DOI] [PubMed] [Google Scholar]
- 35.Gordon M, et al. Genome-Wide Dynamics of SAPHIRE, an Essential Complex for Gene Activation and Chromatin Boundaries. Mol Cell Biol. 2007;27:4058–69. doi: 10.1128/MCB.02044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 448:714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.