Abstract
RNA 3’-phosphate cyclase (RtcA) synthesizes RNA 2’,3’ cyclic phosphate ends via three steps: reaction with ATP to form a covalent RtcA-AMP intermediate; transfer of adenylate to an RNA 3’-phosphate to form RNA(3’)pp(5’)A; and attack of the vicinal O2’ on the 3’-phosphorus to form a 2’,3’ cyclic phosphate. Here we report the 1.7 Å crystal structure of the RtcA-AMP intermediate, which reveals the mechanism of nucleotidyl transfer. Adenylate is linked via a phosphoamide bond to the His309 Nε atom. A network of hydrogen bonds to the ribose O2’ and O3’ accounts for the stringent ribonucleotide preference. Adenine is sandwiched in a hydrophobic pocket between Tyr284 and Pro131 and the preference for adenine is enforced by Phe135, which packs against the purine C2 edge. Two sulfates bound near the adenylate plausibly mimic the 3’-terminal and penultimate phosphates of RNA. The structure illuminates how the four α2/β4 domains contribute to substrate binding and catalysis.
Keywords: 2’,3’ cyclic phosphodiester; adenylyltransferase; covalent catalysis; RNA processing
INTRODUCTION
RNA 2’,3’ cyclic phosphate ends play important roles in RNA metabolism, especially as intermediates in tRNA splicing and repair (Konarska et al., 1982; Greer et al., 1983; Filipowicz and Shatkin, 1983; Laski et al., 1983; Amitsur et al., 1987; Zillman et al., 1991; Zofallova et al., 2000; Nandakumar et al., 2008). There are two enzymatic routes to generate RNA 2’,3’ cyclic phosphate termini. The first is via transesterification, wherein an internal ribose 2’-OH attacks the adjacent 3’-5’ phosphodiester and expels a 5’-OH terminated RNA leaving strand (Raines, 1998). The second pathway entails de novo cyclization of an RNA 3’-phosphomonoester in a three-step reaction that consumes ATP (Filipowicz et al., 1983).
The RNA 3’-terminal phosphate cyclase (Rtc) enzymes that catalyze de novo cyclization comprise an enzyme family with members distributed widely among bacterial, archaeal, and eukaryal taxa (Genschik et al., 1997). Cyclization occurs via a series of three nucleotidyl transfer reactions (Filipowicz et al., 1985; Reinberg et al., 1985). In the first step, Rtc reacts with ATP and a divalent cation to form a covalent Rtc-AMP intermediate and liberate PPi. The AMP is linked via a P–N bond to an invariant histidine side chain of Rtc. In the second step, the adenylate is transferred from Rtc-AMP to the RNA 3’-phosphate terminus to form an activated phosphoanhydride intermediate, RNA(3’)pp(5’)A. In the third step, the terminal ribose 2’-OH attacks the 3’-phosphate of RNA(3’)pp(5’)A to generate an RNA 2’,3’ cyclic phosphate product and release AMP.
The crystal structure of the apoenzyme of E. coli RtcA revealed a fold composed of four tandem modules, each comprising a four-stranded β sheet overlying two α helices (Palm et al., 2000). The essential His309 nucleophile is located within the C-terminal module (Billy et al., 1999). Though lacking any bound substrates or cofactors, the apoRtcA crystal structure contained a citrate anion docked in the N-terminal module via electrostatic and hydrogen bonding interactions with conserved amino acid side chains, including three arginines that are essential for the RNA cyclase and adenylyltransferase activities of the human homolog Rtc1 (Tanaka and Shuman, 2009).
The distinctive 3’ end-modification pathway of the Rtc enzymes and the prominence of cyclic ends in RNA repair reactions prompted our interest in extending the structural and mechanistic framework developed by Filipowicz and colleagues (Billy et al., 1999; Palm et al., 2000). Here we present the 1.7 Å crystal structure of the covalent RtcA-adenylate intermediate, and a structure-guided mutational analysis, which provide new insights to the mechanism of nucleotidyl transfer, the determinants of nucleotide substrate specificity, and the distinctive contributions of the four domain modules to formation of the active site.
RESULTS AND DISCUSSION
Crystallization and structure determination
Recombinant E. coli RtcA was produced as an N-terminal His10-tagged fusion protein and purified from a soluble bacterial extract by sequential Ni-agarose, anion exchange, and gel filtration chromatography steps. Adenylyltransferase assays, entailing incubation of 2 µM RtcA with 50 µM [α32P]ATP and magnesium, elicited scant label transfer to the RtcA polypeptide, to an extent that only 2% of the input RtcA was adenylylated in vitro. The low yield of RtcA-[32P]AMP in vitro could reflect a high extent of covalent adenylylation of RtcA during its production in vivo, in which case the activity might be unmasked by inclusion of pyrophosphate in the reaction mixtures, to promote deadenylylation by reversal of step1 of the cyclase pathway. Indeed, we observed that 100 µM PPi elicited a >10-fold gain of in vitro adenylylation, to an extent of 23% of the input RtcA protomers. These findings suggested that the recombinant RtcA was predominantly RtcA-AMP. To convert any residual active apoenzyme to RtcA-AMP prior to crystallization, we incubated a concentrated RtcA preparation with magnesium and ATP and then isolated the protein by gel filtration. RtcA crystals that grew in buffer containing ammonium sulfate and PEG(5000)-monomethyl ether were in space group P21 and diffracted to better than 1.7 Å resolution. Phases were obtained by molecular replacement with apoRtcA. The structure was ultimately refined at 1.68 Å resolution with R/Rfree values of 0.168/0.207 and excellent geometry (Table I). The crystals contained four RtcA protomers in the asymmetric unit.
Table I.
E. coli RtcA crystallographic data and refinement statistics
| Crystal Dimensions @ 130 K | P21 a=83.73 Å b=81.93 Å c=105.18 Å β=103.4 | ||||
| Radiation Source | NSLS-X29 λ=1.1 Å | ||||
| Crystallographic Data Quality (SCALEPACK): | |||||
| Resolution Å | 40.0 – 1.68 (1.70 – 1.68) | ||||
| Rsym (for I > -σI) | 7.3% (51.1%) | ||||
| Unique Reflections1 | 154840 (5035) | ||||
| Mean Redundancy | 6.07 (4.1) | ||||
| Completeness | 98.5% (96.7%) | ||||
| Mean I/σI | 26.8 (2.1) | ||||
| Molecular Replacement Statistics (PHASER): | |||||
| MR scoring2: | RFZ=8.0, 8.0, 5.5, 6.3 | ||||
| TFZ=4.8,11.7,14.6,17.0 | |||||
| LLG=93, 290, 461, 588 | |||||
| Refinement Statistics (PHENIX): | |||||
| Resolution Å | 35.13 – 1.68 (1.74 – 1.68) | ||||
| R-factors (free/work) | 0.207 / 0.168 (0.259 / 0.216) | ||||
| Estimated Coordinate Error | 0.22 Å | ||||
| Model Statistics: | |||||
| Model Contents | 1351 amino acids from 4 RtcA monomers | ||||
| 74 alternate conformations | |||||
| 8 sulfate, 10 glycerol, 5 sodium ions | |||||
| 1508 water molecules | |||||
| 5 residue peptide (chain E – His-tag) | |||||
| RMS Deviation for Bonds / Angles | 0.012 Å / 1.46° | ||||
| Ramachandran Outliers | None | ||||
| B-factors (Å2): | |||||
| Wilson / Overall | 23.3 / 32.1 | ||||
| By chain (A, B, C, D) | 27.5, 28.9, 31.6, 33.0 | ||||
| Heteroatoms / Water | 59.3 / 42.3 | ||||
| Mean Atomic Anisotropy3 | 0.4 Å | ||||
| NCS Deviations4 (Å) | B | C | D | ||
| (Cα residues 4–339) | A | 0.76 | 0.26 | 0.60 | |
| B | 0.72 | 0.65 | |||
| C | 0.65 | ||||
Standard definitions are used for all of the parameters. Figures in parentheses refer to data in the highest resolution bin.
F+ and F− were treated as equivalent observations throughout scaling.
MR statistics are the Z-scores for Rotation (RFZ) and Translation (TFZ) and the Log-Likelihood Gain (LLG) following optimization of each of the 4 monomers of the search model.
Anisotropy values are derived from TLS refinement.
No NCS restraints were used during refinement.
The omit density maps of all four protomers revealed the presence of AMP covalently linked via a P–N bond to His309. Fig. 1 shows the density map of the histidinyl-AMP adduct and several surrounding amino acids in protomer A. Thus, the structure captures the RtcA-AMP catalytic intermediate in the RNA cyclization pathway. There was little variability between the four RtcA polypeptides (Table I). A least-squares superposition of all four RtcA protomers showed close alignment of the histidinyl-AMP and vicinal amino acids side chains (Supplemental Fig. S1). Henceforth, our discussion of the RtcA-AMP structure will refer to the A protomer.
Figure 1. Covalent linkage of AMP to His309 Nε.
Stereo view of a finely sampled 1.7 Å composite omit density map of the RtcA active site contoured at 1.3 σ (blue mesh; grid spacing 0.2 Å). The red mesh is the Fo-Fc difference density for the bound adenylate contoured at 3.5 σ. See also Figure S1 for a superposition of the active sites of the four RtcA protomers in the asymmetric unit.
Comparision of RtcA-AMP and apoRtcA
The fold of the RtcA-AMP intermediate is depicted in Fig. 2A with the four component domain modules colored as follows: N-terminal domain 1 in magenta (aa 4–85), domain 2 in yellow (aa 86–177), domain 3 in blue (aa 186–277) and domain 4 in green (aa 178–185 plus 278–339). Domains 1, 2 and 4 comprise a single globular unit with pseudo-three-fold symmetry, into which domain 3 is inserted. Domain 3 packs against domain 2 and forms a “lid” over the purine base of AMP (Fig. 2A). Superposition of RtcA-AMP on apoRtcA (from pdb 1QMH, colored gray in Fig. 2A) highlighted key differences in the two crystal structures. For example, apoRtcA crystallized as a disulfide-linked homodimer (Palm et al. 2000). The covalent bond between the Cys308 side chains of adjacent apoRtcA protomers is denoted by the arrowhead in Fig. 2A. By contrast, RtcA-AMP crystallized as a monomer and the shortest distance between Cys308 side chains of neighboring protomers was 41 Å (not shown). Thus, E. coli RtcA is not an obligate homodimer.
Figure 2. Comparison of the RtcA-AMP and apoRtcA structures.
(A) The A protomer of RtcA-AMP is superimposed on the RtcA apoenzyme (the B protomer from the 2.1 Å structure in pdb 1QMH). The four domains of RtcA-AMP are colored as follows: N-terminal domain 1 in magenta (aa 4–85), domain 2 in yellow (aa 86–177), domain 3 in blue (aa 186–277) and domain 4 in green (aa 178–185 plus 278–339). The AMP bound covalently to His309 is depicted as a stick model with green carbons. Two sulfate anions bound to domain 1 are depicted as stick models. The apoRtcA is colored gray. A citrate anion bound to domain 1 of apoRtcA is shown as a stick model with gray carbons. ApoRtcA crystallized as a disulfide-linked homodimer; the disulfide crosslink (denoted by the arrowhead) is shown between Cys308 in the B protomer and Cys308 in the equivalent α-helix (in orange) of the apoRtcA A protomer. (B) Differences in main chain Cα positions (Å) and conformation (phi and psi angles) of the superimposed RtcA-AMP and apoRtcA structures are plotted for each amino acid in the primary structure. The margins of the four domain modules are indicated above the x-axis.
Whereas the β-sheet scaffolds of the four domains are virtually identical in apoRtcA and RtcA-AMP, there were local variations elsewhere in main chain conformation (reflected in phi/psi angle changes) and substantial movements of α-helices and connecting loops (reflected in Cα position changes) in the transition from apoenzyme to enzyme-adenylate (Fig. 2B). These movements were centered principally around the AMP-binding site – entailing shifts in several α-helices of domains 2 and 4 plus a large movement (3.8 Å) in a domain 2 loop (aa 129–131) – and served, together with side chain conformational changes, to open up an otherwise obstructed active site (discussed below). Domain 1 also experienced a significant structural change focused on an oxyanion binding pocket, occupied by a citrate anion in apoRtcA and by two sulfate anions in RtcA-AMP (Fig. 2A). The proximity of the sulfates to the covalent adenylate suggests that they plausibly mimic either the phosphates of the PPi leaving group during the RtcA adenylylation step or the terminal and penultimate RNA phosphates during the RNA adenylylation step.
Chemical linkage of AMP to histidine
Protein phosphohistidine intermediates and products are encountered in many instances of enzymatic phosphoryl transfer other than RNA cyclization. There appears to be no strict rule as to which histidine nitrogen atom serves as the nucleophile. For example, nucleoside diphosphate kinase forms a histidinyl-Nδ-phosphate intermediate during phosphoryl transfer to an NDP phosphoacceptor (Morera et al. 1995; Xu et al. 1997). By contrast, succinyl-CoA-synthetase forms a histidinyl-Nε-phosphate intermediate, also during phosphoryl transfer to an NDP phosphoacceptor (Fraser et al. 1999, 2000). Covalent catalysis via histidine also applies to diverse nucleotidyl transfer reactions, exemplified by galactose-1-phosphate uridylyltransferase (GalT), which acts via an enzyme-UMP intermediate wherein the UMP phosphorus is linked to histidine-Nε in the active site (Wedekind et al. 1996). GalT belongs to the histidine-triad (HIT) family of nucleotidyl hydrolases and transferases. Other members of the HIT family that utilize adenosine nucleotide substrates have been cocrystallized as transition state mimetics with adenosine and tungstate, wherein the tungsten atom is covalently linked to histidine-Nε in the active site (Lima et al. 1997).
In the case of RNA cyclase, Billy et al. (1999) employed differential proteolysis and mass spectrometry to assign His309 as the site of covalent adenylylation in RtcA. However, their analysis did not discriminate whether the P–N bond was to histidine Nδ or Nε. The RtcA-AMP structure resolves this issue, insofar as the omit density map shows unambiguously that AMP is linked via a P–N bond to the Nε atom of His309 (Fig. 1). Despite a shared use of histidine Nε as the attacking nucleophile, there is no structural similarity between RtcA and the HIT protein family; their active sites are entirely different.
Architecture of the adenylate-binding pocket
The adenosine nucleoside is bound to RtcA in an anti conformation with the adenine base buried in a hydrophobic pocket (Fig. 1). The adenine is sandwiched between Tyr284 on one side, with which it forms a π stack, and Pro131 and Phe251 on the other, with which it makes van der Waals contacts (Figs. 1 and 3). Phe135 forms the back end of the adenine pocket and the planar aromatic ring is oriented orthogonal to the edge of the purine ring. The adenine C2 atom is pointed directly at Phe135 at a distance of 3.6 Å from the plane of the phenyl ring (Fig. 1). The close van der Waals contacts of adenine C2 and Phe135 side chain (conserved as a tyrosine in hRtc1) explains the strong preference of RtcA and hRtc1 for ATP versus GTP as the substrate for enzyme-adenylylation and RNA cyclization (Genshik et al. 1997, 1998; Vicente and Filipowicz, 1988), i.e., because the exocyclic 2-amino group of GTP would clash sterically with Phe135.
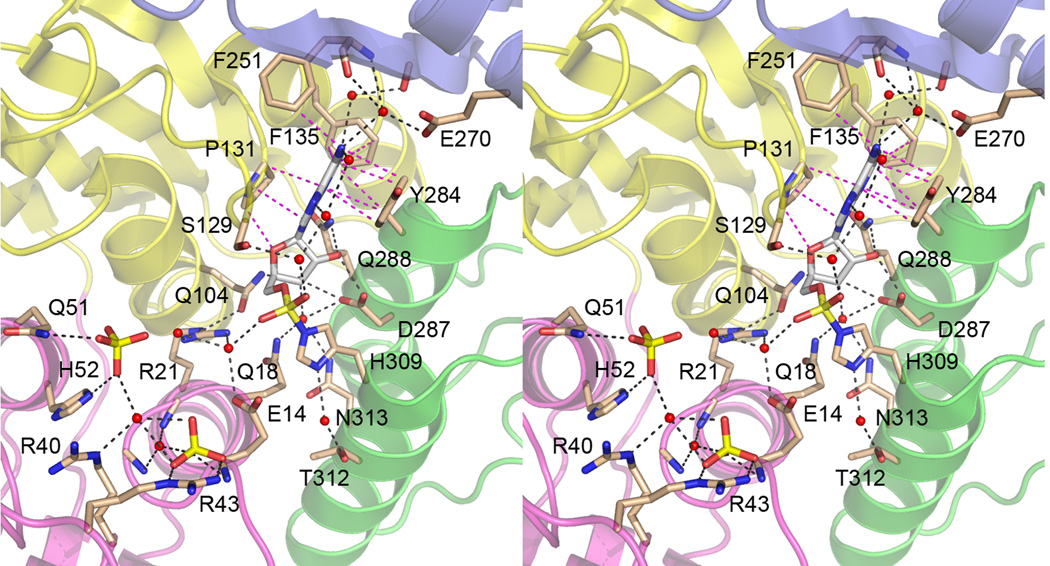
Figure 3. The RctA active site.
Stereo view of the active site in the A promoter of RtcA-AMP. The folds of the four domains are colored as in Fig. 2. Selected amino acid side chains and main chain atoms, the covalently bound adenylate, and the two sulfate anions bound to domain 1 are rendered as stick models. Waters are depicted as red spheres. Hydrogen bonds are indicated by black dashed lines; van der Waals contacts are denoted by magenta dashed lines.
There are no direct polar contacts between the adenine base and RtcA amino acids. Rather, the polar edge of the adenine is decorated with a network of interacting waters that bridge to main chain and side chain atoms of RtcA, especially those in domain 4 (Fig. 3). Water-mediated contacts between adenine N1 and N6 atoms and the Glu270 side chain, the Phe251 amide and carbonyl, and the Arg249 carbonyl – though not adenine-specific – could explain why RtcA is purine specific (Genshik et al. 1998).
The ribose moiety of histidinyl-AMP adopts a C(2')-endo conformation (Fig. 1), which is stabilized by five hydrogen bonds to the ribose O2’ and O3’ atoms (Fig. 3). Asp287 accepts bidentate hydrogen bonds to its carboxylate oxygens from the ribose 2’-OH and 3’-OH groups (Fig. 1). The 3’-OH receives additional H-bonds from Gln104 Nε and from a water coordinated by Gln18, Asn313 and Asp287 (Fig. 3). The ribose 2’-OH receives a hydrogen bond from the Gln288 Nε atom (Fig. 3). We speculate that these ribose contacts fix the position of the nucleoside and thereby the orientation of the ATP α-phosphate with respect to the His309 nucleophile. The existence of two dedicated hydrogen bonds to the ribose 2’-OH provides a rationale for the inability of RtcA (and hRtc1) to use dATP as a substrate (Reinberg et al. 1985; Vicente and Filipowicz, 1988; Genshik et al. 1997, 1998).
A striking feature of the RtcA-AMP structure is the absence of direct contacts between protein functional groups and the AMP phosphate oxygens (Fig. 3). Assuming that histidine adenylylation is catalyzed via an associative mechanism, we expect an extra negative charge to develop on the ATP α-phosphate in the transition state. Most phosphoryl transferases promote catalysis, at least in part, by neutralizing this negative charge, typically via coordination of the nonbridging phosphate oxygens by arginines and lysines and/or reliance on a divalent cation cofactor that interacts with the reactive phosphate. In RtcA-AMP the nonbridging oxygens are each coordinated by a water molecule that bridges to an amino acid side chain (Fig. 3). Whereas the Ser129-bridged water contact to the AMP phosphate is not observed in all four RtcA protomers; the Glu14-bridged water contact is conserved. RNA cyclase is strictly dependent on a divalent cation for RNA cyclization and for enzyme-adenylylation (Genschik et al. 1998; Reinberg et al., 1985; Vicente and Filipowicz, 1998). We propose that Glu14 comprises part of the metal-coordination complex, and that the metal is situated near the position of the water shown in Fig. 3, where it can coordinate the ATP α-phosphate (and, perhaps, the PPi leaving group as well).
Another notable aspect of the RtcA active site is the absence of direct protein interactions with the His309 Nδ atom. Other histidine-based covalent phosphoryl/nucleotidyl transferases typically rely on hydrogen bond donation from the protonated “non-nucleophile” histidine nitrogen atom to a main chain carbonyl (in the case of GalT and other HIT proteins) or a side chain carboxylate (in nucleoside diphosphate kinase and succinyl-CoA-synthetase) (Wedekind et al. 1996; Lima et al. 1997; Morera et al. 1995; Fraser et al, 1999). These contacts are thought to assist in orienting the histidine for its attack on the phosphorus center and in stabilizing the imidazolium group in the covalent intermediate (Wedekind et al. 1996). In RtcA-AMP, the His309 Nδ interacts with a water molecule that is in turn coordinated by Thr312 Oγ (Fig. 3A), suggesting that hydrogen bonding to the bridging water serves in lieu of the orienting histidine-protein contacts seen in other transferases. Human Rtc1 also has a threonine in the position equivalent to RtcA Thr312.
Active site remodeling precedes adenylylation
The positions and orientations of many of the amino acid side chains that bind AMP in the RtcA-AMP intermediate differ substantially from their conformations in the apoRtcA structure. In particular, Tyr284, which stacks on the AMP adenine, adopts a different rotamer in apoRtcA so that the phenol ring overlaps the AMP adenosine in the RtcA-AMP intermediate (Fig. 2A). Pro131 in apoRtcA also intrudes on the space occupied by adenine in the RtcA-AMP intermediate. Consequently, the AMP binding site is effectively occluded in the apoenzyme. The superposition of the two structures indicates that the “closed” active site seen in apoRtcA transits to an “open” conformation by: (i) reorientation of Tyr284; (ii) a 3.8 Å movement of the domain 2 loop that includes Pro131; and (iii) unkinking of the domain 2 helix that includes Phe135 (Fig. 2A). Key residues in domain 4 also undergo conformation switches. Asp287, which coordinates the ribose hydroxyls in RtcA-AMP, is pointed in the opposite direction in the apoRtcA structure (Fig. 2A). The His309 nucleophile is out of position by 2.5 Å in the apoenzyme and is not properly oriented with respect to the AMP phosphate (Fig. 2A). AMP binding elicits an inward movement of the proximal end of the His309-containing domain 4 helix and the preceding loop, which is necessarily coupled to reorientation of Asp287 (which would sterically prevent the helix movement if it remained in its apoenzyme rotamer). Finally, the domain 1 loop between β1 and α1 undergoes a significant rearrangement in RtcA-AMP versus apoRtcA in which the Glu14 Cα shifts by 5.4 Å (Fig. 2A and B). In apoRtcA, both of the Glu14 carboxylate oxygens are hydrogen-bonded to the Gly16 main chain amide. In RtcA-AMP, these contacts are severed, the loop moves, the proximal end of the α1 helix is unwound (Fig. 2A), and Glu14 makes new hydrogen bonds to the Gln18 main chain amide and to the phosphate-bridging water discussed above.
Structure-guided mutagenesis of the adenylate-binding pocket
Nine residues identified presently as constituents of the AMP binding site were targeted for mutational analysis. Gln104, Ser129, Phe135, Phe251, Glu270, Tyr284, Asp297, and Gln288 were replaced individually by alanine and Pro131 was changed to glycine. To sample the extreme case of complete loss of function, we also mutated the His309 nucleophile to alanine and glycine. The RtcA mutants were produced in E. coli as His10 fusions and purified from soluble lysates by Ni-agarose and anion exchange chromatography steps in parallel with wild-type RtcA (Fig. 4A).
Figure 4. Effects of mutations in the AMP-binding pocket on Rtc1 cyclase activity.
(A) Rtc1 purification. Aliquots (4 µg) of recombinant wild-type (WT) RtcA and the indicated mutants were analyzed by SDS-PAGE. The Coomassie-Blue stained gel is shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left. (B) Cyclase activity. Reaction mixtures (10 µl) containing 50 mM Tris-HCl (pH 7.4), 2 mM DTT, 10 mM MgCl2, 100 µM ATP, 20 nM 5'-AAAAUAAAAGC*pCp (a 12-mer RNA 3'-phosphate substrate labeled with 32P at the penultimate phosphate, prepared according to Tanaka and Shuman, 2009), and 5 nM wild-type or mutant RtcA were incubated for 10 min at 37°C. The mixtures were adjusted to 50 mM sodium acetate (pH 5.2), 1 mM ZnCl2 and then digested with nuclease P1 (1 U) at 50°C for 10 min. The digests were analyzed by ascending polyethyleneimine cellulose TLC with a buffer consisting of 4 M ammonium sulfate, 3 M sodium acetate, isopropanol (80:18:2, v:v:v). The 32P-labeled nucleotides *pC (derived form the input RNA substrate) and *pC>p (derived from the cyclized RNA product) were visualized and quantified by scanning the TLC plate with a Fuji Film BAS-2500 imager. The extents of RNA cyclization by the WT and mutant RtcA proteins are plotted. Each datum is the average of three experiments ± SEM. (C) RtcA adenylylation. Reaction mixtures (10 µl) containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, 1 mM DTT, 50 µM [α32P]ATP, 100 µM pyrophosphate and 2 µM RtcA as specified were incubated at 37°C for 10 min. The reactions were quenched by adding 10 µl of 100 mM Tris-HCl (pH 6.8), 200 mM DTT, 4% SDS, 20% glycerol, 40 mM EDTA, 0.2% bromophenol blue. The samples were analyzed by electrophoresis through a 12% polyacrylamide gel containing 0.1% SDS. The RctA-[32P]AMP adducts were visualized by autoradiography of the dried gels and quantified by scanning the gels with a Fuji BAS-2500 imager. The extents of adenylylation by the WT and mutant RtcA proteins are plotted. Each datum is the average of three experiments ± SEM.
To assay RNA cyclization, we used a 12-mer RNA 3’-phosphate substrate labeled with 32P at the penultimate phosphate (5’-AAAAUAAAAGC*pCp). Wild-type RtcA and the RtcA mutants (5 nM each) were reacted with 20 nM RNA, 100 µM ATP, and 10 mM MgCl2. Note that the ATP concentration used was 5-fold higher than the Km value of 20 µM ATP for the cyclase reaction of E. coli RtcA (Genschik et al. 1998). The products were digested with nuclease P1 and the labeled *pC (derived from substrate) and the cyclized *pC>p product were resolved by PEI-cellulose TLC. Whereas wild-type RtcA cyclized 95% of the input RNA, the H309A and H309G mutants were inert, as expected (Fig.4B). Q104A, S129A and E270A were as active or nearly as active as wild-type RtcA in RNA cyclization (Fig. 4B). We surmise that the water-mediated contact of Ser129 with the AMP phosphate, the hydrogen bond of Gln104 to the ribose O3’, and the water mediated contact of Glu270 to adenine N6 are not essential for RtcA catalysis. These inferences were underscored by assays of RtcA adenylylation in the presence of PPi (Fig. 4C), wherein the Q104A and S129A proteins retained wild-type activity and E270A yielded half as much enzyme-adenylate as wild-type RtcA. In the case of Gln104 and Glu270, it is likely that their contacts with the AMP ribose and adenine, respectively, are functionally redundant with the other RtcA main chain or side chain contacts to the same adenylate atoms (Fig. 3).
The instructive findings were that Asp287 and Gln288, which coordinate the ribose hydroxyls, were both essential for RtcA activity. D287A and Q288A cyclized just 1% and 2% of the input RNA (Fig. 4B) and were virtually inert in RtcA adenylylation (Fig. 4C). There was a hierarchy of mutational effects at the four hydrophobic residues that comprise the adenine pocket. F135A was the most deleterious (3% cyclization), followed by Y284A (12% cyclization), F251A (28% cyclization), and P131G (33% cyclization) (Fig. 4B). The yields of RtcA-[32P]AMP in vitro by F135A, Y284A, F251A and P131G were 2%, 2%, 38% and 13% of the wild-type value, respectively (Fig. 4C). These results implicate Tyr284 and Phe135, which respectively stack parallel and orthogonal to the purine ring, as the most important constituents of the adenine pocket. Our mutational results here for E. coli RtcA agrees with the more limited mutational analysis of two of the adenosine-binding residues in human Rtc1, in which alanine changes at Tyr294 (equivalent to RtcA Tyr284) and Asp297 (equivalent to RtcA Asp287) abolished the cyclization and adenylyltransferase activities (Tanaka and Shuman, 2009).
An oxyanion-binding pocket in RtcA domain 1
The A and C protomers of RtcA-AMP both contained two sulfate anions bound to domain 1, at sites closely flanking the citrate anion that was bound to the RtcA apoenzyme (Fig. 2A). The B and D protomers contained only one sulfate bound to domain 1. The “invariant” sulfate occupies an oxyanion hole at the proximal end of the α1 helix that was unwound in the transition from apoenzyme to enzyme-adenylate, wherein the sulfate receives one direct and two water-mediated hydrogen bonds from the main chain amides of Glu14, Gly16, and Gly17, respectively (Fig. 3). The sulfate also forms a bidentate salt bridge to Arg43 (Fig. 3). Arg43 is conserved in human Rtc1 and essential for its RNA cyclase and adenylyltransferase activities (Tanaka and Shuman, 2009). The sulfate anion is situated 7 Å from the AMP phosphate. We envision that this sulfate mimics the 3’-phosphate of the RNA substrate and/or the phosphate in the PPi leaving group derived from the ATP γ-phosphate. The distance between this sulfate/phosphate site and the AMP phosphate would presumably shorten in the presence of a bridging divalent cation that counteracts the anionic charge repulsion. Arg21 is poised to assist in coordinating the RNA 3’-phosphate during this approach to the histidinyl-adenylate and/or in coordinating the ATP β phosphate during the adenylylation step. Arg21 is conserved in human Rtc1 and is essential for cyclase and adenylyltransferase activities (Tanaka and Shuman, 2009).
The second sulfate anion in protomers A and C is poised on the surface of domain 1, where it is coordinated by hydrogen bonds from Gln51 Nε and His52 Nε (Fig. 3). The first and second sulfates are 6.5 Å apart (S to S distance), which mimics the separation between adjacent phosphates (P to P) in a polynucleotide. Thus, we speculate that the second phosphate marks the position of the penultimate phosphate of the RNA substrate. The first and second sulfates are bridged by a water molecule coordinated by Arg40 (Fig. 3). Arg40 is conserved in human Rtc1 and essential for cyclase and adenylyltransferase activities (Tanaka and Shuman, 2009). We propose that it, along with Arg43, binds to the ATP γ phosphate and RNA 3’-phosphate during the first and second nucleotidyl transfer steps of the cyclase pathway. By contrast, although Gln51 and His52 are also conserved in human Rtc1, neither residue is essential. Changing Gln51 to alanine had no apparent effect on Rtc1 cyclization activity, whereas changing His52 to alanine elicited a 2-fold reduction (Tanaka and Shuman, 2009); these findings are consistent with a noncatalytic role for the contacts seen here with the second sulfate anion.
Mechanistic implications
The crystal structure of the enzyme-adenylate intermediate of the RNA cyclase reaction pathway is, to our knowledge, the second instance of an atomic structure of a nucleoside monophosphate covalently linked to histidine in an enzyme active site - the first being GalT linked to UMP (Wedekind et al. 1995). As discussed above, the active site of RtcA-AMP is sui generis and quite distinct from other histidine-based phosphoryl/nucleotidyl transferases. The present study extends initial structural insights from the apoenzyme, which focused on the novel arrangement of the four similarly folded domain modules. We can now invoke a fairly clear division of labor among the four structural modules, whereby: (i) essential side chains in domains 2 and 4 comprise the adenosine binding pocket; (ii) essential residues in domain 1 bind the PPi leaving group, the RNA 3’-terminal phosphates, and the metal cofactor; and (iii) domain 3 forms a lid over the adenine pocket and makes water-mediated purine-specific hydrogen bonds to N1 and N6. Comparison of the apoRtcA and RtcA-AMP structures reveals significant remodeling of the active site environment that must precede or accompany the enzyme-adenylylation step.
Although there is clearly more to learn by capturing RtcA structures in other states along the reaction pathway, the available structural and mutational data for E. coli RtcA and human Rtc1 prompt use to propose a catalytic mechanism for the nucleophilic attack of His309 Nε on the ATP α-phosphorus (Fig. 5) driven by the following enzymic contributions to orientation of the reactants and transition state stabilization: (i) orientation of the histidine by water-mediated hydrogen-bonding to the non-nucleophilic Nδ atom; (ii) positioning of the α-phosphorus by tight constraints on the ribose conformation imposed by Asp287, Gln288 and other contacts to the ribose hydroxyls; (iii) orientation of the PPi leaving group apical to attacking His309 achieved by ionic interactions of β and γ phosphates with the essential arginines of domain 1; and (iv) stabilization of the transition state of the α-phosphate by a metal ion coordinated by Glu14.
Figure 5. Proposed mechanism of RtcA adenylation.
The crystal structure and mutational effects suggest a model whereby nucleophilic attack of the His309 Nε on the ATP α-phosphorus is facilitated by: (i) proper orientation of His309 via a water mediated hydrogen bond of Nδ to Thr312 Oγ; (ii) orientation of the adenylate by hydrogen bonding of the ribose 2’ and 3’ hydroxyls to Asp287 and Gln288; (iii) stabilization of the transition state by a divalent cation (with imputed coordination of the metal by Glu14); and (iv) orientation of the pyrophosphate leaving group apical to the attacking His309 nucleophile via contacts of Arg21, Arg40 and Arg43 with the ATP β and γ phosphates.
MATERIALS AND METHODS
E. coli RtcA purification for crystallization experiments
The rtcA gene was PCR-amplified from E. coli genomic DNA with primers designed to introduce a NdeI site at the ATG codon for Met2 (amino acid sequence numbering according to Genschik et al. 1997) and a BamHI site immediately downstream of the stop codon. The PCR product was digested with NdeI and BamHI and then inserted between the NdeI and BamHI sites of pET16b. The plasmid insert was sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning. The pET-His10RtcA-(2–339) plasmid was transformed into Escherichia coli BL21-Codon Plus (DE3) (Novagen).
To prepare RtcA for crystallization experiments, a 1 L culture derived from a single transformant was grown at 37°C in LB medium containing 100 µg/ml ampicillin until the A600 reached 0.6 to 0.8. The culture was chilled on ice for 30 min and then adjusted to 0.1 mM isopropyl β-D-thiogalactoside and 2% (v/v) ethanol. Incubation was continued at 17°C for 16 h with constant shaking. Cells were harvested by centrifugation and stored at −80°C. All subsequent procedures were performed at 4°C. The cell pellet was suspended in 50 ml buffer A (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10% sucrose) and lysozyme was added to 0.2 mg/ml. After mixing gently for ~30 min, the suspensions were sonicated to reduce viscosity and insoluble material was removed by centrifugation at 20000g for 30 min. The soluble lysate was mixed for 1 h with 6 ml of a 50% slurry of Ni-NTA agarose (Qiagen) that had been equilibrated in buffer A. The agarose was recovered by centrifugation and resuspended in 10 ml of buffer B (50 mM Tris-HCl, pH 7.4, 250 mM NaCl, 10% glycerol) containing 25 mM imidazole. The cycle of centrifugation and resuspension of the agarose resin was repeated three times, after which the washed agarose beads (3 ml) were poured into a column. Bound proteins were eluted stepwise with 4-ml aliquots of 100, 200, 300 and 400 mM imidazole in buffer B. The elution profile was monitored by SDS-PAGE. His10RtcA was recovered in the 200, 300 and 400 mM imidazole eluates, which were pooled and diluted with buffer C (50 mM Tris-HCl, pH 7.4, 10% glycerol) to adjust the NaCl concentration to 100 mM. The protein solution was applied to a 1-ml column of DEAE-Sephacel equilibrated with 100 mM NaCl in buffer C. RtcA was recovered in the flow-through fraction (while residual nucleic acids were bound to the DEAE resin). The DEAE flow-through preparation was mixed with solid ammonium sulfate to achieve 60% saturation. The protein precipitate was collected by centrifugation and resuspended in 2 ml of buffer D (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM DTT, 1 mM EDTA, 5% glycerol) containing 2 mM ATP and 10 mM MgCl2. The protein sample was then gel-filtered through a 120-ml 16/60 HiPrep Sephacryl S-100 HR column (GE Healthcare) developed with buffer D at a flow rate of 1.0 ml/min. The peak RtcA-containing fractions were pooled and concentrated by centrifugal ultrafiltration.
Crystallization and data collection
Crystals of RtcA were grown by the hanging drop vapor diffusion method at room temperature. The RtcA protein sample (8 mg/ml, in buffer D) was mixed with an equal volume (2 µl) of the reservoir buffer containing 10 mM 2-(4-morpholino)-ethane sulfonic acid (MES), pH 6.5, 200 mM ammonium sulfate, and 20% (w/v) polyethyleneglycol-(5000)-monomethyl ether. Crystals grew to their full size (~400 × 150 × 50 µm) in 1 to 2 days. Single crystals were harvested, cryoprotected by suspension in 10 mM MES, pH 6.5, 200 mM ammonium sulfate, 25% polyethyleneglycol-(5000)-monomethyl ether, 7% glycerol and then flash frozen in liquid nitrogen. X-ray diffraction data were collected at the National Synchrotron Light Source (Brookhaven, NY) beamline X29 (tuned for maximum flux at 1.1 Å with a 100 µm beam aperture) equipped with an ADSC-q315 CCD detector. A single crystal was used to collect a total of 432 frames of rotation data in three separate sweeps. The first sweep consisted of 185 consecutive 1° oscillations and 1 s exposures. The second sweep was designed to measure low-resolution reflections without generating overloaded pixels and was collected with a longer crystal-to-detector distance, a ten-fold attenuated beam, and 62 consecutive 3° oscillations. The third pass was identical to the first but used 5 s exposures to capture weaker high-resolution data. All data were processed using HKL2000 and SCALEPACK. The diffraction statistics are compiled in Table I.
Structure determination and refinement
RtcA crystallized in monoclinic space group P21 (a = 83.73 Å, b = 81.93 Å, c = 105.18 Å; α = γ = 90.00° β = 103.44°) with four monomers in the asymmetric unit. Phases for RtcA were obtained via molecular replacement in PHASER (McCoy et al. 2007) using the previously solved apoRtcA structure (PDB 1QMH) as a search model. Following map calculation and initial model building in COOT (Emsley and Cowtan 2004), the side chains of the amino acids in the active site (and of those with unclear electron density) were truncated to alanine and the resulting model was refined against the crystallographic data in CNS (Brunger et al. 1998). Weighted difference density maps generated from this initial refinement were used to model the truncated side chains. A second round of refinement in CNS using a complete protein model, but no AMP or solvent, revealed difference density for all atoms in the covalent adenylate (as depicted in Fig. 1 by the red mesh). AMP was fit into the difference density map and refined using a combination of standard stereochemistry and phosphoramidate bond distance and angle target values derived from the geometry of the histidine-uridylate observed in the structure of GalT (PDB 1HXQ) and the small-molecule structure of monosodium phosphoramidate (Hobbs et al. 1953). The model was completed by adding heteroatoms and alternate amino acid conformations as warranted by the difference density maps and then processed further in PHENIX (Adams et al. 2002) via refinement of individual sites, simulated annealing, TLS (translation/libration/screw) refinement, individual atomic displacement parameters refinement, and occupancy refinement for alternate conformations. The assignment of protein regions to TLS groups was performed by the TLSMD server (Painter and Merritt, 2006). An additional round of refinement using no bond or angle restraints about the tetrahedral phosphate atom revealed deviations from the stereochemically restrained refinement of less than 0.05 Å for bonds and 2° for angles for all phosphorous bonded atoms.
The final refined model at 1.68 Å resolution (R/Rfree = 0.168/0.207) included 1351 amino acids from the 4 RtcA monomers, with Ramachandran values of 94.5% most favored, 5.5% allowed, and no residues with generous or disallowed conformations. The refinement statistics are compiled in Table I. The asymmetric unit of RtcA-AMP contains for monomers related by approximate two-fold rotations giving rise to pseudo-orthorhombic non-crystallographic symmetry. Molecules A and B are related by a 180.9° rotation about the (A × B) axis. Antiparallel main-chain hydrogen-bonding between residues 220 and 228 of each protomer resulted in an extended cross-protomer 8-strand β sheet. A similar cross-protomer interaction is seen between RtcA molecules C and D, which are themselves related by a 183.5° rotation about the X axis, when crystallographic symmetry is applied to the D protomer.
Mutational analysis
Missense mutations were introduced into the rtcA-(2–339) ORF via the two-stage PCR overlap extension method. The mutant rtcA genes were inserted into the pET16b. The plasmid inserts were sequenced completely to exclude the acquisition of unwanted changes during amplification and cloning. The pET-His10RtcA-(2–239) plasmids were transformed into Escherichia coli BL21-Codon Plus (DE3). Wild-type and mutant His10RtcA proteins were produced in parallel and purified from soluble bacterial lysates of 400-ml bacterial cultures by procedures similar to those described above, except for the following modifications: (i) 0.1% Triton X-100 was added to the cell suspension during the lysis procedure; (ii) the Niagarose RtcA preparations were dialyzed against buffer C containing 150 mM NaCl before passage over 0.5-ml columns of DEAE-Sephacel to remove nucleic acids; (iii) the ammonium sulfate precipitation and gel filtration steps were omitted. The DEAE flow-through fractions of wild-type and mutant RtcA were stored at −80°C. Protein concentrations were determined by using the Biorad dye reagent with bovine serum albumin as the standard. The RtcA yields from 400 ml cultures were between 8 and 30 mg of protein.
Accession number
The coordinates for the refined model of RtcA-AMP have been deposited in the RCSB protein structure database (PDB ID code 3KGD).
Supplementary Material
Acknowledgments
The work was supported by NIH grant GM46330. SS is an American Cancer Society Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billy E, Hess D, Hofsteenge J, Filipowicz W. Characterization of the adenylation site in the RNA 3’-terminal phosphate cyclase from Escherichia coli. J. Biol. Chem. 1999;274:34955–34960. doi: 10.1074/jbc.274.49.34955. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Strugala K, Konarska M, Shatkin AJ. Cyclization of RNA 3’-terminal phosphate by cyclase from HeLa cells proceeds via formation of N(3’)pp(5’)A activated intermediate. Proc. Natl. Acad. Sci. USA. 1985;8:1316–1320. doi: 10.1073/pnas.82.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Konarska M, Gross HJ, Shatkin AJ. RNA 3’-terminal phosphate cyclase activity and RNA ligation in HeLa cell extract. Nucleic Acids Res. 1983;11:1405–1418. doi: 10.1093/nar/11.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Vicente P. RNA 3’-terminal cyclase from Hela cells. Meth. Enzymol. 1990;181:499–510. doi: 10.1016/0076-6879(90)81147-m. [DOI] [PubMed] [Google Scholar]
- Fraser ME, James MNG, Bridger WA, Wolodko WT. A detailed structural description of Escherichia coli succinyl-CoA-synthetase. J. Mol. Biol. 1999;285:1633–1653. doi: 10.1006/jmbi.1998.2324. [DOI] [PubMed] [Google Scholar]
- Fraser ME, James MNG, Bridger WA, Wolodko WT. Phosphorylated and dephosphorylated structures of pig heart, GTP-specific succinyl-CoA-synthetase. J. Mol. Biol. 2000;299:1325–1339. doi: 10.1006/jmbi.2000.3807. [DOI] [PubMed] [Google Scholar]
- Genschik P, Billy E, Swianiewicz, Filipowicz W. The human RNA 3’-terminal phosphate cyclase is a member of a new family of protein conserved in eukarya, bacteria and archaea. EMBO J. 1997;10:2955–2967. doi: 10.1093/emboj/16.10.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Drabikowski K, Filipowicz W. Characterization of the Escherichia coli RNA 3’-terminal phosphate cyclase and its σ54-regulated operon. J. Biol. Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- Greer CL, Peebles CL, Gegenheimer P, Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- Hobbs E, Corbridge DEC, Raistrick B. The crystal structure of monosodium phosphoramidate, NaHPO3NH2. Acta Crystallogr. 1953;6:627–631. [Google Scholar]
- Konarska M, Filipowicz W, Gross HJ. RNA ligation via 2’-phosphomonoester, 3’,5’-phosphodiester linkage: requirement of 2’,3’-cyclic phosphate termini and involvement of a 5’-hydroxyl polynucleotide kinase. Proc. Natl. Acad. Sci. USA. 1982;79:1474–1478. doi: 10.1073/pnas.79.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laski FA, Fire AZ, RajBhandary UL, Sharp PA. Characterization of tRNA precursor splicing in mammalian extracts. J. Biol. Chem. 1983;258:11974–11980. [PubMed] [Google Scholar]
- Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moréra S, Chiadmi M, LeBras G, Lascu I, Janin J. Mechanism of phosphate transfer by nucleoside diphosphatekinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium. Biochemistry. 1995;34:11062–11070. [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Molecular Cell. 2008;31:278–286. doi: 10.1016/j.molcel.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter J, Merritt EA. TLSMD web server for the generation of multi-group TLS models. J. Applied Crystallogr. 2006;39:109–111. [Google Scholar]
- Palm GJ, Billy E, Filipowicz W, Wlodawer A. Crystal structure of RNA 3’-terminal phosphate cyclase, a ubiquitous enzyme with unusual topology. Structure. 2000;8:12–23. doi: 10.1016/s0969-2126(00)00076-9. [DOI] [PubMed] [Google Scholar]
- Raines RT. Ribonuclease A. Chem Rev. 1998;98:1045–1065. doi: 10.1021/cr960427h. [DOI] [PubMed] [Google Scholar]
- Reinberg D, Arenas J, Hurwitz J. The enzymatic conversion of 3’-phosphate terminated RNA chains to 2’,3’-cyclic phosphate derivatives. J. Biol. Chem. 1985;260:6068–6097. [PubMed] [Google Scholar]
- Tanaka N, Shuman S. Structure-activity relationships in human RNA 3’-phosphate cyclase. RNA. 2009;15:1865–1874. doi: 10.1261/rna.1771509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente O, Filipowicz W. Purification of RNA 3’-terminal phosphate cyclase from HeLa cells: covalent modification of the enzyme with different nucleotides. Eur. J. Biochem. 1988;176:431–439. doi: 10.1111/j.1432-1033.1988.tb14300.x. [DOI] [PubMed] [Google Scholar]
- Wedekind JE, Frey PA, Rayment I. The structure of nucleotidylated histidine-166 of galactose-1-uridylyltransferase provides insight into phosphoryl group transfer. Biochemistry. 1996;35:11560–11569. doi: 10.1021/bi9612677. [DOI] [PubMed] [Google Scholar]
- Xu YW, Moréra S, Janin J, Cherfils J. AlF3 mimics the transition state of protein phosphorylation in the crystal structure of nucleoside diphosphate kinase and MgADP. Proc. Natl. Acad. Sci. USA. 1997;94:3579–3583. doi: 10.1073/pnas.94.8.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillman M, Gorovsky MA, Phizicky EM. Conserved mechanism of tRNA splicing in eukaryotes. Mol. Cell. Biol. 1991;11:5410–5416. doi: 10.1128/mcb.11.11.5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofallova L, Guo Y, Gupta R. Junction phosphate is derived from the precursor in the tRNA spliced by the archaeon Haloferax volcanii cell extract. RNA. 2000;6:1019–1030. doi: 10.1017/s1355838200000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.