Summary
HER4/ErbB4 is a ubiquitously expressed member of the EGF/ErbB family of receptor tyrosine kinases that is essential for normal development of the heart, nervous system, and mammary gland. We report here crystal structures of the ErbB4 kinase domain in active and lapatinib-inhibited forms. Active ErbB4 kinase adopts an asymmetric dimer conformation essentially identical to that observed to be important for activation of the EGF receptor/ErbB1 kinase. Mutagenesis studies of intact ErbB4 in BaF3 cells confirm the importance of this asymmetric dimer for activation of intact ErbB4. Lapatinib binds to an inactive form of the ErbB4 kinase in a mode equivalent to its interaction with the EGF receptor. All ErbB4 residues contacted by lapatinib are conserved in the EGF receptor and HER2/ErbB2, which lapatinib also targets. These results demonstrate that key elements of kinase activation and inhibition are conserved among ErbB family members.
Introduction
HER4/ErbB4 is a member of the epidermal growth factor (EGF)/ErbB family of receptor tyrosine kinases (Carpenter, 2003; Plowman et al., 1993), which in humans also includes the epidermal growth factor (EGF) receptor (EGFR/ErbB1/HER1), ErbB2 (HER2/Neu), and ErbB3 (HER3) (Holbro and Hynes, 2004; Yarden and Sliwkowski, 2001). Each ErbB receptor is essential for normal animal development (Olayioye et al., 2000), and abnormal expression and activation of ErbBs is associated with many human cancers (Hynes and Lane, 2005). Loss of ErbB4 function in particular results in defects in the heart, nervous system, and mammary gland in mice (Gassmann et al., 1995; Tidcombe et al., 2003).
ErbBs mediate their biological effects through ligand-dependent elevation of their tyrosine kinase activities. ErbBs consist of an extracellular ligand-binding region, a single membrane-spanning region, a cytoplasmic kinase domain, and a 220-350 amino acid C-terminal ‘tail’ that becomes tyrosine phosphorylated following activation and mediates interactions between ErbBs and downstream effectors (Burgess et al., 2003). Ligand binding to the extracellular region results in formation of specific ErbB homo- and hetero-dimers, activation of the cytoplasmic kinase, and initiation of intracellular signaling cascades (Yarden and Sliwkowski, 2001). Despite 60-75% sequence identity, mechanistic differences beyond ligand and effector specificity have emerged among ErbB receptors. Both ErbB1 and ErbB4 behave in an archetypal manner and homodimerize and signal in response to ligand binding (Ferguson et al., 2000; Carpenter, 2003). ErbB2, however, lacks a known ligand and fails to homodimerize in normal circumstances. ErbB2 acts instead as a universal heterodimerization partner for each of the other ErbBs regardless of the stimulating ligand (Karunagaran et al., 1996; Klapper et al., 1999). An additional combination of properties is observed for ErbB3, which binds and responds to ligands but fails to homodimerize (Ferguson et al., 2002; Berger et al., 2004). The ErbB3 kinase domain appears to be inactive (Sierke et al., 1997), and ErbB3 must heterodimerize, most notably with ErbB2, to signal.
Structural studies of ErbBs have established a molecular context for understanding many features of ErbB signaling. Crystallographic studies of the extracellular regions of ErbB1, ErbB3, and ErbB4 show them to be made up of four discrete subdomains that adopt an autoinhibited conformation in the absence of ligand (Bouyain et al., 2005; Burgess et al., 2003; Cho and Leahy, 2002; Ferguson et al., 2003). Ligand binding promotes a domain rearrangement that exposes previously obscured surfaces and allows them to mediate dimerization (Burgess et al., 2003; Garrett et al., 2002; Ogiso et al., 2002). In contrast, crystal structures of the ErbB2 extracellular region show it to adopt a fixed, active-like structure in which the canonical ligand-binding surfaces are involved in an interdomain interaction. These unique interactions rationalize the absence of an ErbB2 ligand and the role of ErbB2 as a universal ErbB heterodimerization partner (Cho et al., 2003; Garrett et al., 2003).
Active kinases are characterized by several conformational features including the relative orientation of N- and C-terminal lobes, the disposition of the ‘activation’ loop, the orientation of the αC helix, and formation of a salt bridge involving a glutamate on the αC helix (Huse and Kuriyan, 2002). Recently, formation of a crystallographically-observed asymmetric dimer between EGFR kinase domains was shown to correlate with the presence of an activated kinase conformation and to be required for EGFR activation (Zhang et al., 2006). In this asymmetric kinase domain dimer, the C-lobe of a ‘donor’ contacts the N-lobe of an adjacent ‘acceptor’ and promotes conformational changes that activate the acceptor kinase. This type of activating interaction is reminiscent of the activating interaction between cyclins and the N-lobes of cyclin-dependent kinases and was suggested to be conserved in other ErbBs based on amino acid sequence conservation (Jeffrey et al., 1995; Zhang et al., 2006).
Understanding the molecular details of ErbB activation has direct clinical relevance. ErbB activation is associated with several human cancers, and drugs targeting ErbB1 and/or ErbB2 have been approved for treatment of breast, lung, and colon cancers (Johnston et al., 2006). These drugs include several small molecule kinase inhibitors, such as the 4-aniloquinazolines erlotinib (Tarceva), gefitinib (Iressa), and lapatinib (Tykerb), which all bind competitively at the ATP-binding site of the kinase (Denny, 2001; Petrov et al., 2006). Whereas erlotinib and gefitinib are relatively specific for ErbB1, lapatinib inhibits all active ErbB kinases (Wood et al., 2004). Activation of ErbB1 and ErbB2 clearly contributes to adverse tumor characteristics, but the converse may be true for ErbB4. Several observations suggest that ErbB4 activation inhibits cell proliferation and promotes apoptosis (Karamouzis et al., 2007). For this reason, development of ErbB inhibitors that specifically target ErbB1 and ErbB2 but not ErbB4 may prove beneficial.
To investigate the molecular details of activation and inhibition of the ErbB4 kinase we have determined crystal structures of the ErbB4 kinase in the presence and absence of lapatinib. Without lapatinib the ErbB4 kinase adopts an active conformation and forms an asymmetric dimer virtually identical to the activating dimer observed for EGFR. A low-resolution structure of an inactive form of the ErbB4 kinase was also obtained in the absence of lapatinib. Mutagenesis studies confirm the importance of this dimerization mode for ErbB4 activation. With bound lapatinib the ErbB4 kinase adopts a conformation characteristic of inactive kinases and does not make the EGFR-like asymmetric dimer. Lapatinib binds to ErbB4 in a mode similar to its interaction with the EGFR kinase. All residues contacting lapatinib are conserved in both EGFR and ErbB4, which implicates kinase elements not directly involved in inhibitor contact in defining the different inhibitory parameters of lapatinib for EGFR and ErbB4.
Results
The HER4/ErbB4 kinase was expressed using a baculovirus expression system, purified, and crystallized in three different crystal forms (Table 1). The ErbB4 kinase structure in each crystal form was determined by molecular replacement using EGFR kinase structures as search models (Stamos et al., 2002; Wood et al., 2004; Zhang et al., 2006). Type I crystals were grown in the presence of the pan-ErbB inhibitor lapatinib and diffracted to 2.8 Å Bragg spacings. The ErbB4 kinase in these crystals adopts an inactive-like conformation (Figure 1B) (Huse and Kuriyan, 2002). Type II and type III crystals were grown in the presence of AMP-PNP and diffracted to 2.5 and 4.0 Å Bragg spacings, respectively. In type II crystals the ErbB4 kinase adopts an active-like conformation and forms the same asymmetric dimer interaction shown to be important for activation of the EGFR kinase (Zhang et al., 2006) (Figures 1A and 2). The ErbB4 kinase is a monomer in solution at 15 μM as judged by both analytical ultracentrifugation and size-exclusion chromatography indicating that, similar to the EGFR kinase, the kinase dimer interaction is relatively weak. A “symmetric dimer” equivalent to that observed for the EGFR kinase domain was not observed (Stamos et al., 2002). Type III crystals grew in the same space group with similar cell constants as type I crystals, and the ErbB4 kinase domain in these crystals also adopts an inactive-like conformation. These are the first crystals of an ErbB kinase domain in an inactive conformation not grown in the presence of either an inhibitor or a dimer-targeting mutation and are noted here for that reason despite their moderate resolution (Zhang et al., 2006). The existence of this crystal form suggests that high salt conditions may influence the conformational equilibria of this kinase.
Table 1. Crystallization, data collection and refinement statistics.
| Crystal | ErbB4 kinase complexed with lapatinib (Type I) | ErbB4 Kinase domain (Type II) | ErbB4 kinase domain (Type III) |
|---|---|---|---|
| Crystallization conditions | 4.3 M NaCl, 0.1 M Hepes, pH 7.0, 2% trifluoroethanol | 15%PEG3350, 0.1 M Tris, pH 8.5 | 2.2 M NaCl, 0.2 M MgCl2, 0.1 M Hepes, pH 7.0, 2% hexafluoroisopropanol |
| Spacegroup | P61 | P32 | P61 |
| Unit cell (Å) | a=b=102.6, c=185.1 | a=b=86.7, c=120.0 | a=b=102.7, c=181.3 |
| Resolution range (Å) | 30.0-2.80(3.00-2.80) | 30.0-2.50 (2.59-2.50) | 30.0-4.0 (4.14-4.00) |
| Molecules per ASU | 2 | 3 | 2 |
| Solvent content | 68% | 47.5% | 68% |
| Data completeness (%) | 99.9 (99.6) | 100 (100) | 98.6 (93.3) |
| redundancy | 4.8 | 2.6 | 7.5 |
| Rsym (%) | 6.2 (51.4) | 10.7 (45.7) | 25.3 (66.7) |
| I/σI | 24.5 (2.8) | 8.8 (1.9) | 7.6 (2.9) |
| Refinement | |||
| Phaser Search model | 1XKK | 1M14 | 3BBT without lapatinib |
| R/Rfree (%) | 25.0 / 28.9 | 20.2 / 25.5 | 30.2 / 35.4 |
| Number of protein atoms | 4357 | 6846 | 4354 |
| Number of non-protein/solvent atoms | 71 | 1 | 0 |
| Number of solvent atoms | 84 | 173 | 0 |
| Ramachandran plot | |||
| Most favored (%) | 89.2 | 89.6 | 84.8 |
| Additional allowed (%) | 9.7 | 9.9 | 13.9 |
| Generously allowed (%) | 1.1 | 0.5 | 1.3 |
| Rmsd bond length (Å) | 0.01 | 0.01 | 0.01 |
| Rmsd bond angle (°) | 1.3 | 1.3 | 1.6 |
| Mean B-value | 26.2 | 19.2 | 63.7 |
| PDB code | 3BBT | 3BCE | 3BBW |
Figure 1.
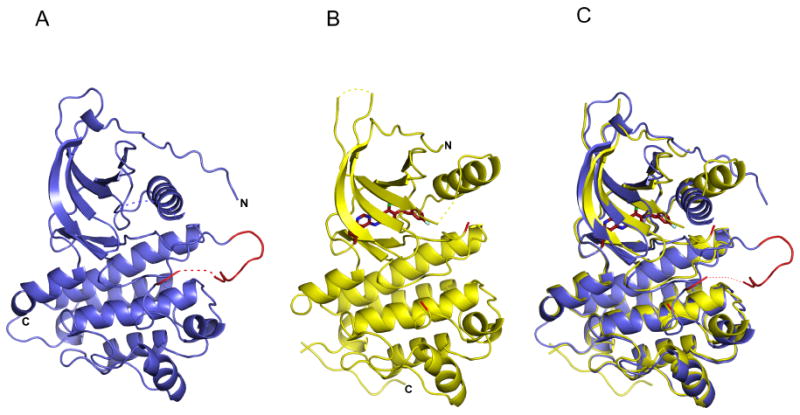
Ribbon diagrams of ErbB4 kinase structures. (A) The active conformation of the ErbB4 kinase. The activation loop is colored red. (B) An inactive conformation of the ErbB4 kinase in complex with lapatinib, which is shown as a red stick model. The activation loop is disordered, but the loop termini are colored red. (C) Superposition of the active and inactive conformations of the ErbB4 kinase.
Figure 2.
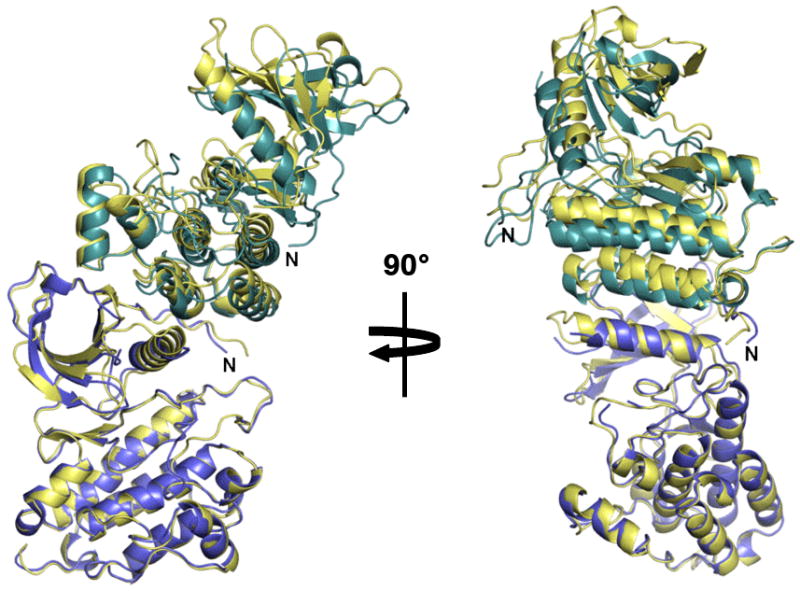
Superposition of the asymmetric dimers of EGFR and ErbB4 kinase. The ErbB4 kinase subunits are colored blue and cyan. An EGFR dimer is colored light yellow. Only the single blue colored ErbB4 kinase subunit and corresponding EGFR subunit were included in the superposition.
The kcat, Kmapp for ATP, Kmapp for a peptide substrate (GGMEDIYFEFMGGKKK), and IC50 and Kiapp for lapatinib were also measured for the ErbB4 kinase and compared with values determined for the ErbB2 kinase domain of kcat, Kmapp ATP, and Kmapp for the same peptide substrate (Table 2). These results are generally comparable to published values with the exception that the kcat for ErbB4 is ∼10-fold greater than previously measured (Brignola et al., 2002).
Table 2. Enzymatic parameters.
| Kmapp ATP (μM) | Kmapp peptide (μM) | kcat (min-1) | [Mn2+] (mM) | IC50 lapatinib (μM) | Kiapp lapatinib (nM) | |
|---|---|---|---|---|---|---|
| ErbB4 KD | 10 ± 2 | 169 ± 35 | 52 ± 10 | 2 | 1.4 ± 0.1 | 700 |
| ErbB2 KD | 24 ± 3 | 57 ± 5 | 6.1 ± 0.2 | 10 | -- | - |
| EGFR ICD* | 5 | 899 ± 200 | 1.32 | 0.62 | -- | 3 ± 0.2 |
| ErbB2 ICD* | 13 | 157 ± 40 | 3.60 | 1.60 | -- | 13 ± 1 |
| ErbB4 ICD* | 21 | 144 ± 22 | 5.58 | 0.20 | > 1 | 347 ± 16 |
These are previously reported values. (Brignola et al. 2002 and Wood et al. 2004).
All 17 C-lobe residues involved in the asymmetric dimer contact are conserved between ErbB4 and EGFR; 14/18 ErbB4 N-lobe residues involved in this contact are also conserved in EGFR. The ErbB4 asymmetric dimer buries 2252 Å2 with a shape complementarity value of .657 (Lawrence and Colman, 1993). Comparable values for the EGFR asymmetric dimer are 2077 Å2 and .614. As observed for the EGFR asymmetric kinase dimer, the ErbB4 kinase asymmetric dimer subunits are related by a crystallographic 3-fold screw axis (Zhang et al., 2006). If single subunits of the EGFR and ErbB4 kinase asymmetric dimer are superimposed, the positions of the other dimer subunits are related by a ∼7° rotation such that structural elements distant from the dimer interface superimpose progressively less well (Figure 2). This slight difference seems unlikely to have any functional consequences. To validate the importance of the asymmetric dimer for ErbB4 function, selected substitutions of both N- and C-lobe interface residues were introduced into full-length ErbB4. In each case, these substitutions diminish ligand-dependent autophosphorylation (Figure 3), similar to the effect of equivalent mutations in EGFR (Zhang et al., 2006). Curiously, no electron density for a bound nucleotide is observed in type II crystals despite the active kinase conformation and the presence of 1 mM AMP-PNP in the crystallization buffer. No electron density consistent with tyrosine phosphorylation is observed.
Figure 3.
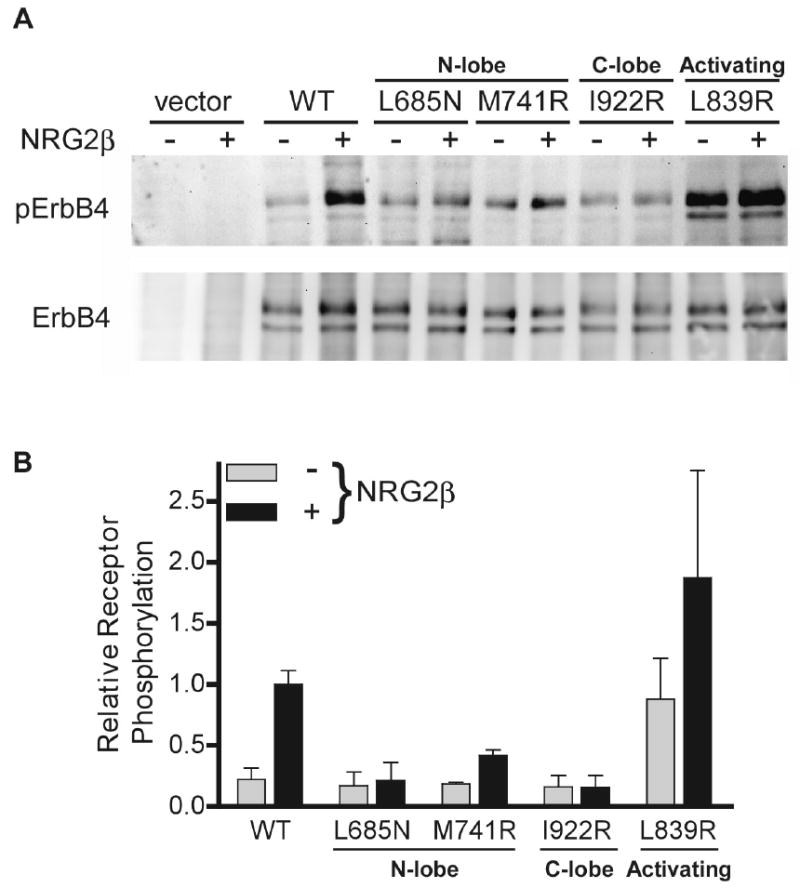
Receptor activation assay. (A) Western blot analysis of the indicated ErbB4 variants immunoprecipitated from cell lysates following treatment with or without Neuregulin-2β (NRG-2β). Blots were probed with an anti-phosphotyrosine antibody (top) or anti-ErbB4 antibody (bottom). (B) Quantitation of Western blot band intensities. The results are the mean and standard deviation from three independent experiments.
Electron density for lapatinib is present in type I crystals (Figure 4). The moderate resolution of type I crystals (2.8 Å) and the weak electron density for lapatinib limit clear interpretation of specific lapatinib conformations, but the lapatinib binding mode appears similar to its binding to the EGFR kinase (Figure 4) (Wood et al., 2004). No electron density is observed for the sulfone-furanyl moiety, and it was not included in the atomic model. Satisfactory density exists for the fluorophenyl and quinazolinamine moieties, and inspection of difference electron density maps strongly suggests that the phenyl group on the fluorophenyl is flipped 180° relative to its orientation in the lapatinib-EGFR complex, altering the relative position of the fluoro group (Figure 4). In addition, a slight movement of the αC helix, which contacts the fluoro group, is observed in ErbB4 complex relative to EGFR complex. Incomplete density exists for the internal chlorophenyl ring, with one carbon extending outside of clear density despite extensive modeling of different possible conformations (Figure 4A).
Figure 4.
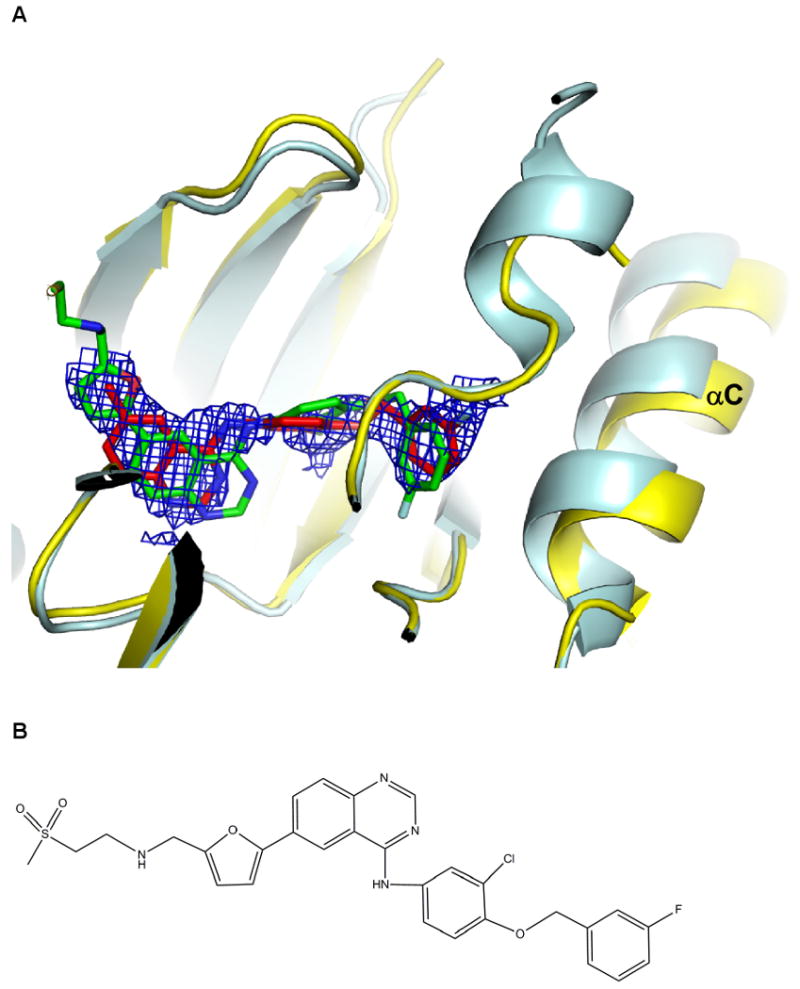
Bound lapatinib. (A) The 2Fo-Fc simulated annealing omit map for lapatinib contoured at 1σ. The structure of ErbB4 kinase is colored yellow, and lapatinib is shown as a red stick model. The structure of the EGFR kinase-lapatinib complex is superimposed with the EGFR kinase colored pale cyan and lapatinib green. (B) Chemical structure of lapatinib.
We have characterized the lapatinib-bound ErbB4 kinase conformation as inactive based on the location and orientation of the N- and C-lobes, the disposition of the αC helix, and the absence of a key salt bridge involving a glutamate on helix αC. Most of the activation loop in lapatinib-bound ErbB4 was disordered and not modeled (residues 844 to 857). It is therefore not clear whether there are direct inhibitory interactions between the activation loop and the αC helix in unactivated ErbB4. In EGFR, aliphatic side chains from a short helix in the activation loop interact directly with, and appear to buttress, a displaced helix αC (Zhang et al., 2006; Wood et al., 2004). Replacing these side-chains with polar groups disrupts the activation loop/helix αC interaction, and elevates the basal activity of the kinase. We are not able to determine whether similar autoinhibitory interactions occur in the inactive ErbB4 kinase structure. The relevant amino acids in the activation loop are conserved, however, and the L839R mutation enhances basal activity of ErbB4 just as the analogous L834R mutation does in EGFR (Figure 3).
Discussion
Discovery of the importance of an asymmetric EGFR kinase dimer for regulation of kinase activity provided a satisfying link between ligand-induced receptor dimerization and kinase activation (Burgess et al., 2003; Jeffrey et al., 1995; Zhang et al., 2006). Our observation of an essentially identical asymmetric dimer of the ErbB4 kinase and its importance for ErbB4 signaling provides strong support for its generality among ErbB family members. The asymmetric dimer mechanism introduces a new point of regulation of ErbBs that may be exploited by cellular factors or targeted therapies and is consistent with earlier observations of trans-phosphorylation among receptor tyrosine kinases (Ballotti et al., 1989; Lammers et al., 1990). In the asymmetric dimer one kinase is the ‘donor’ and the other the ‘acceptor’. The C-terminal tail of the donor is adjacent to the acceptor active site and readily phosphorylated, but the C-terminal tail of the acceptor is on the opposite side of the dimer >75 Å away from the donor active site, which is unable to phosphorylate the acceptor tail (Ballotti et al., 1989; Lammers et al., 1990).
Lapatinib inhibits ErbB4 kinase activity but less well than it inhibits EGFR and ErbB2, (Table 2, (Brignola et al., 2002)), but a comparison of the EGFR-lapatinib and ErbB4-lapatinib complex structures reveals no obvious explanation for this difference. Although the resolution of the the ErbB4-lapatinib structure is moderate, 2.8 Å, it is clear that lapatinib binds EGFR and ErbB4 in the same binding site, and all residues that come into van der Waals contact with lapatinib (atoms < 4 Å apart) are conserved between EGFR and ErbB4 (and ErbB2) (Supplementary Table 3). The only notable difference between the binding mode of lapatinib to EGFR and ErbB4 is a flip of the fluorophenyl ring, which alters the position of the fluorine but does not introduce or break contacts with residues not found in both EGFR and ErbB4 (Figure 4). This observation suggests that ErbB kinase elements not directly contacting lapatinib must contribute to the distinctive effects of lapatinib on EGFR and ErbB4 activity. Since lapatinib appears to bind preferentially to an inactive kinase conformation, one possibility is that ErbB elements that influence the interconversion between active and inactive states of the kinase, either by influencing the nature of intermediate states or the rates of their interconversion, are important components of inhibitor effectiveness (Levinson et al., 2006).
The different kinetic parameters obtained for ErbB kinases may reflect such differences in the balancing of factors that influence equilibria between active and inactive kinase conformations. As noted by Zhang et al (2006), activity of the EGFR kinase increases at either higher concentrations or when the L834R substitution is introduced into the activation loop. These authors note that these observations indicate some level of autoinhibition in the isolated EGFR kinase, which either increases in concentration or mutation are able to overcome. The kcat we measure for ErbB4 with a specific peptide substrate is ∼10-fold faster than previously reported for other ErbBs including ErbB4, suggesting our ErbB4 kinase may be intrinsically more active than other ErbBs. For comparison, the kcat we measure for ErbB2, which is ∼10-fold less than measured for ErbB4 (Table 2), is in line with previously reported values (Table 2 and (Brignola et al., 2002)). A possible explanation for this difference is that earlier authors worked with a larger fragment of the ErbB4 intracellular domain compared to our fragment, and the additional ErbB4 residues may have an autoinhibitory effect. A similar effect is not observed for longer versions of ErbB2, however, possibly indicating a different distribution of kinase control mechanisms (Table 2 and (Brignola et al., 2002)). The reduced sensitivity to lapatinib and the increased kcat for the ErbB4 kinase domain suggests that its equilibrium state may be biased toward the active form relative to EGFR. It is clear that even in the presence of shared regulatory mechanisms, fine tuning the details of ErbB kinase activity can have important effects on the response of ErbB receptors to various natural and artificial stimuli.
Experimental Procedures
Expression and purification of the ErbB4 kinase domain
A DNA fragment encoding residues 677-1004 of human ErbB4 was cloned into a modified pFASTBac vector. Amino-acid numbering refers to the Jma isoform and begins with the mature protein. This vector directs expression of the ErbB4 kinase region with an N-terminal strep II tag followed by a Tobacco Etch Virus (TEV) protease recognition site. Recombinant bacmid (Bac-to-Bac expression system, Invitrogen) was then transfected into Sf9 cells to produce recombinant baculovirus. Sf9 cells were grown in suspension, infected by the virus, and harvested 3 days after infection. Cell pellets were lysed by sonication in 50 mM Tris, pH 8.0, 150 mM NaCl, 10% glycerol, 1 mM DTT, and 1 mM PMSF. The lysate was centrifuged at 30,000 × g for 30 minutes. The supernatant was loaded onto a 5-ml Strep-Tactin Superflow column (IBA). Attached proteins were eluted using the lysis buffer plus 2.5 mM desthiobiotin. ErbB4 kinase containing fractions were treated with TEV protease overnight at 4°C to remove the N-terminal strep II tag, and the samples applied to a Ni-NTA column to remove the His-tagged TEV protease. The Ni-NTA column flow-through was collected and concentrated before loading onto a Superdex 75 size-exclusion column (Amersham). Fractions containing the kinase were pooled and concentrated to ∼4 mg/ml in the column buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM DTT).
Kinase Assays
To determine ErbB kinetic parameters, radiometric kinase assays were carried out in 50 mM HEPES (pH 7.5), 2 mM MnCl2, 37.5 mM NaCl, 1 mM DTT, 5% Glycerol, and 125 μg/mL BSA using Biotin-GGMEDIYFEFMGGKKK as the peptide substrate in a 25 μL reaction volume. The peptide substrate was prepared using standard Fmoc solid phase peptide synthesis followed by coupling biotin to the N-terminus using NHS-activated biotin (NovaBiochem) and purification by reverse phase HPLC. Reactions were initiated by the addition of 25-50 nM kinase, carried out for 3-6 min at 30°C, and stopped by the addition of 10 μL of 100 mM EDTA. To each sample, 10 μL of 10 mg/mL avidin was added and all samples were transferred to centrifugal filtration units with 30,000 NMWL membranes (Millipore) and washed three times with 100 μL wash solution (0.5 M Phosphate, 0.5 M NaCl, pH 8.5). The Kmapp for peptide substrate was determined with varying peptide concentrations (870, 435, 218, 109, 54.4, 27.2, and 13.6 μM) and fixed ATP concentration (100 μM) and the Kmapp for ATP was determined using varying ATP concentrations (120, 60, 30, 15, 7.5, 3.8, and 1.9 μM) and fixed peptide substrate concentration (435 μM). For the IC50 determination, the lapatinib (160, 40, 10, 2.5, 0.625, 0.156, and 0.039 μM) was pre-incubated with the enzyme on ice for 30 min, and the reactions were run with 10 μM ATP, 150 μM peptide, and 0.15% DMSO. Reactions for the HER2 kinase domain were performed under the same conditions with the exception that 10 mM MnCl2 and 320 nM kinase were used. Activities proved to be linear with time and kinase concentration in the ranges used, and the limiting substrate turnover was less than 10% for all rate measurements. Duplicate measurements were generally within 20%. Apparent Km and kcat values were obtained from non-linear curve fits to the Michaelis-Menten equation and the apparent Ki calculated assuming linear competitive inhibition versus ATP. Lapatinib was synthesized following published methods (Petrov et al., 2006).
Crystallization and structure determination
The ErbB4 kinase domain was incubated with either 0.2 mM lapatinib or 1 mM AMP-PNP and 2 mM MgCl2 on ice for an hour before setting up crystallization trials. All crystals were obtained by hanging drop method with a 1:1 mixture of the stock protein solution and the crystallization buffer. Crystals of the ErbB4 kinase complexed with lapatinib were grown in 4.3 M NaCl, 0.1 M Hepes, pH 7.0, and 2% trifluoroethanol. Crystals were transferred to 2.7 M sodium malonate, pH 7.0 as a cryoprotectant immediately prior to freezing in liquid nitrogen. Crystals of the kinase with AMP-PNP were obtained in two different conditions. Type I crystals were grown in 15% PEG3350, 0.1 M Tris, pH 8.5 and cryoprotected by addition of 10% glycerol to the crystallization solution immediately prior to freezing. Type III crystal were grown in 2.2 M NaCl, 0.2 M MgCl2, 0.1 M Hepes, pH 7.0, 2% hexafluoroisopropanol and cryoprotected by addition of 15% glycerol to the crystallization solution immediately prior to freezing. Diffraction data were collected at the Brookhaven NSLS X4C beamline. The data were processed with HKL2000 (Otwinowski and Minor, 1997). The structures were determined by molecular replacement using the program PHASER (Storoni et al., 2004) and structures of the EGFR kinase domain with lapatinib (1XKK) and apo-EGFR kinase (1M14) as the search models, respectively. The program COOT (Emsley and Cowtan, 2004) was used for model building, and CNS (Brunger et al., 1998) and REFMAC (Murshudov et al., 1997) were used for refinement. TLS groups were generated by TLSMD and used for refinement in REFMAC (Winn et al., 2001).
The ErbB4-lapatinib complex crystals belong to space group P61 and contain two molecules in an asymmetric unit. Residues from 683-843 and 858-973 are visible in the electron density and included in the model. The activation loop (residues 844 to 857), 6 residues at the N-terminus, and 31 residues at the C-terminus were not visible in the electron density map and not included in the model. The ErbB4 kinase domain crystals grown in the presence of AMP-PNP belong to space group P32, contain three molecules in an asymmetric unit, and diffract to 2.5 Å Bragg spacings. Residues from 678 to 965 are visibile in the electron density and included in the model. 39 residues at the C-terminus are not visible and not included in the model. No electron density for a bound nucleotide is observed or included in the model. Diffraction data and refinement statistics are summarized in Table 1.
Receptor Activation Assays
Wild-type human ErbB4 JmbCyt2 isoform in pcDNA3.1(+) was kindly provided by Dr. Graham F. Carpenter (Vanderbilt University), and a Neuregulin-2 expression plasmid kindly provided by Dr. David Riese (Purdue). Point mutations were generated by site-directed mutagenesis using the Quikchange method (Stratagene, La Jolla, CA). Ba/F3 cells were maintained in RPMI-1640 media supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 2 mM sodium pyruvate and IL-3 (1 ng/ml). Plasmids directing expression of wild-type or mutated ErbB4 were electroporated into Ba/F3 cells using an Electrocell manipulator (ECM600, BTX Genetronics, San Diego, CA). Transfected cells were selected in medium containing 1 mg/ml G418 for 2 weeks, and viable cells were sorted by fluorescence-activated cell sorting on a FACSCalibur machine (BD Biosciences, San Jose, CA) for ErbB4 surface expression level using anti-ErbB4 antibody (Ab1) (Neomarkers, Fremont, CA) and R-phycoerythrin conjugated goat anti-mouse antibody (Invitrogen, Carlsbad, CA). Cells were serum starved for 24 hours in supplemented RPMI-1640 media (described above) lacking FBS. Starved cells were pre-chilled on ice, and incubated for 10 minutes in ice-cold starvation medium containing 200 ng/ml NRG2β, produced as described (Wilson et al., 2007). Cells were pelleted, washed extensively in phosphate-buffered saline (PBS), lysed in RIPA buffer (1% Na deoxycholate, 1% NP-40, 0.1% SDS, 150 mM NaCl, 10 mM Na phosphate buffer pH 7.2, 1 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 25 mM NaF, 5 mM Na2MoO4, 0.2 mM Na3VO4). ErbB4 receptors were immuno-precipitated using anti-ErbB4 (C18) (Santa Cruz Biotechnology, Santa Cruz, Ca) antibody and subjected to Western blot analysis using anti-ErbB4 antibody and phosphotyrosine antibody (PY20) (Zymed Laboratories Inc., San Francisco, CA). Receptor activation in Figure 3 was normalized for receptor expression level by dividing band intensities in the anti-phosphotyrosine Western blots by the intensities of the corresponding bands in anti-ErbB4 Western blots. The ratio of this value to the value obtained for wild-type ErbB4 with added NRG2β (arbitrarily set to 1 in each experiment) was then calculated and is displayed in Figure 3B. Intensities were quantified using Kodak Molecular Imaging software (v. 4.0.3). Mean ± standard deviation for three independent experiments are plotted.
Supplementary Material
Acknowledgments
We thank Johns Schwanof and Randy Abramowitz for assistance at NSLS beamline X4 and Jason McLellan and Craig Vander Kooi for assistance with X-ray data collection and comments on the manuscript. Supported by the Susan G Komen for the Cure Foundation (R.B.), the DOD CDMRP Award W81XWH-04-1-0499 (to DJL), and NIH grants R01-CA079992 (to M.A.L.), R01-CA90466 (to D.J.L), and R01-CA74305 (to P.A.C). S.H.C. is supported by a Pre-doctoral Fellowship from the Great Rivers Affiliate of the American Heart Association (Award 0715334U), and R.B is supported by the Susan G. Komen for the Cure Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballotti R, Lammers R, Scimeca JC, Dull T, Schlessinger J, Ullrich A, Van Obberghen E. Intermolecular transphosphorylation between insulin receptors and EGF-insulin receptor chimerae. Embo J. 1989;8:3303–3309. doi: 10.1002/j.1460-2075.1989.tb08491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MB, Mendrola JM, Lemmon MA. ErbB3/HER3 does not homodimerize upon neuregulin binding at the cell surface. FEBS Lett. 2004;569:332–336. doi: 10.1016/j.febslet.2004.06.014. [DOI] [PubMed] [Google Scholar]
- Bouyain S, Longo PA, Li S, Ferguson KM, Leahy DJ. The extracellular region of ErbB4 adopts a tethered conformation in the absence of ligand. Proc Natl Acad Sci U S A. 2005;102:15024–15029. doi: 10.1073/pnas.0507591102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignola PS, Lackey K, Kadwell SH, Hoffman C, Horne E, Carter HL, Stuart JD, Blackburn K, Moyer MB, Alligood KJ, et al. Comparison of the biochemical and kinetic properties of the type 1 receptor tyrosine kinase intracellular domains. Demonstration of differential sensitivity to kinase inhibitors. J Biol Chem. 2002;277:1576–1585. doi: 10.1074/jbc.M105907200. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Burgess AW, Cho HS, Eigenbrot C, Ferguson KM, Garrett TP, Leahy DJ, Lemmon MA, Sliwkowski MX, Ward CW, Yokoyama S. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol Cell. 2003;12:541–552. doi: 10.1016/s1097-2765(03)00350-2. [DOI] [PubMed] [Google Scholar]
- Carpenter G. ErbB-4: mechanism of action and biology. Exp Cell Res. 2003;284:66–77. doi: 10.1016/s0014-4827(02)00100-3. [DOI] [PubMed] [Google Scholar]
- Cho HS, Leahy DJ. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- Cho HS, Mason K, Ramyar KX, Stanley AM, Gabelli SB, Denney DW, Jr, Leahy DJ. Structure of the extracellular region of HER2 alone and in complex with the Herceptin Fab. Nature. 2003;421:756–760. doi: 10.1038/nature01392. [DOI] [PubMed] [Google Scholar]
- Denny WA. The 4-anilinoquinazoline class of inhibitors of the erbB family of receptor tyrosine kinases. Farmaco. 2001;56:51–56. doi: 10.1016/s0014-827x(01)01026-6. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Berger MB, Mendrola JM, Cho HS, Leahy DJ, Lemmon MA. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol Cell. 2003;11:507–517. doi: 10.1016/s1097-2765(03)00047-9. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Darling PJ, Mohan MJ, Macatee TL, Lemmon MA. Extracellular domains drive homo- but not hetero-dimerization of erbB receptors. Embo J. 2000;19:4632–4643. doi: 10.1093/emboj/19.17.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Kofler M, Jorissen RN, Nice EC, Burgess AW, Ward CW. The crystal structure of a truncated ErbB2 ectodomain reveals an active conformation, poised to interact with other ErbB receptors. Mol Cell. 2003;11:495–505. doi: 10.1016/s1097-2765(03)00048-0. [DOI] [PubMed] [Google Scholar]
- Garrett TP, McKern NM, Lou M, Elleman TC, Adams TE, Lovrecz GO, Zhu HJ, Walker F, Frenkel MJ, Hoyne PA, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- Gassmann M, Casagranda F, Orioli D, Simon H, Lai C, Klein R, Lemke G. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378:390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu Rev Pharmacol Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- Johnston JB, Navaratnam S, Pitz MW, Maniate JM, Wiechec E, Baust H, Gingerich J, Skliris GP, Murphy LC, Los M. Targeting the EGFR pathway for cancer therapy. Curr Med Chem. 2006;13:3483–3492. doi: 10.2174/092986706779026174. [DOI] [PubMed] [Google Scholar]
- Karamouzis MV, Badra FA, Papavassiliou AG. Breast cancer: the upgraded role of HER-3 and HER-4. Int J Biochem Cell Biol. 2007;39:851–856. doi: 10.1016/j.biocel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Karunagaran D, Tzahar E, Beerli RR, Chen X, Graus-Porta D, Ratzkin BJ, Seger R, Hynes NE, Yarden Y. ErbB-2 is a common auxiliary subunit of NDF and EGF receptors: implications for breast cancer. Embo J. 1996;15:254–264. [PMC free article] [PubMed] [Google Scholar]
- Klapper LN, Glathe S, Vaisman N, Hynes NE, Andrews GC, Sela M, Yarden Y. The ErbB-2/HER2 oncoprotein of human carcinomas may function solely as a shared coreceptor for multiple stroma-derived growth factors. Proc Natl Acad Sci U S A. 1999;96:4995–5000. doi: 10.1073/pnas.96.9.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers R, Van Obberghen E, Ballotti R, Schlessinger J, Ullrich A. Transphosphorylation as a possible mechanism for insulin and epidermal growth factor receptor activation. J Biol Chem. 1990;265:16886–16890. [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Levinson NM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, Cole PA, Kuriyan J. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O, Fukai S, Yamanaka M, Kim JH, Saito K, Sakamoto A, Inoue M, Shirouzu M, Yokoyama S. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. Embo J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Petrov KG, Zhang YM, Carter M, Cockerill GS, Dickerson S, Gauthier CA, Guo Y, Mook RA, Jr, Rusnak DW, Walker AL, et al. Optimization and SAR for dual ErbB-1/ErbB-2 tyrosine kinase inhibition in the 6-furanylquinazoline series. Bioorg Med Chem Lett. 2006;16:4686–4691. doi: 10.1016/j.bmcl.2006.05.090. [DOI] [PubMed] [Google Scholar]
- Plowman GD, Culouscou JM, Whitney GS, Green JM, Carlton GW, Foy L, Neubauer MG, Shoyab M. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc Natl Acad Sci U S A. 1993;90:1746–1750. doi: 10.1073/pnas.90.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierke SL, Cheng K, Kim HH, Koland JG. Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J. 1997;322(Pt 3):757–763. doi: 10.1042/bj3220757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Tidcombe H, Jackson-Fisher A, Mathers K, Stern DF, Gassmann M, Golding JP. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci U S A. 2003;100:8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson KJ, Mill CP, Cameron EM, Hobbs SS, Hammer RP, Riese DJ., 2nd Inter-conversion of neuregulin2 full and partial agonists for ErbB4. Biochem Biophys Res Commun. 2007;364:351–357. doi: 10.1016/j.bbrc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Isupov MN, Murshudov GN. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr D Biol Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, et al. A unique structure for epidermal growth factor receptor bound to GW572016 (Lapatinib): relationships among protein conformation, inhibitor off-rate, and receptor activity in tumor cells. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


