Abstract
Aging in the brain is characterized by increased susceptibility to neuronal loss and functional decline, and mitochondrial DNA (mtDNA) mutations are thought to play an important role in these processes. Due to the proximity of mtDNA to the main sites of mitochondrial free radical generation, oxidative stress is a major source of DNA mutations in mitochondria. The base excision repair (BER) pathway removes oxidative lesions from mtDNA, thereby constituting an important mechanism to avoid accumulation of mtDNA mutations. The complexity of the brain implies that exposure and defence against oxidative stress varies among brain regions and hence some regions may be particularly prone to accumulation of mtDNA damages. In the current study we investigated the efficiency of the BER pathway throughout the murine lifespan in mitochondria from cortex and hippocampus, regions that are central in mammalian cognition, and which are severely affected during aging and in neurodegenerative diseases. A regional specific regulation of mitochondrial DNA repair activities was observed with aging. In cortical mitochondria, DNA glycosylase activities peaked at middle-age followed by a significant drop at old age. However, only minor changes were observed in hippocampal mitochondria during the whole lifespan of the animals. Furthermore, DNA glycosylase activities were lower in hippocampal than in cortical mitochondria. Mitochondrial AP endonuclease activity increased in old animals in both brain regions. Our data suggest an important regional specific regulation of mitochondrial BER during aging.
Keywords: DNA repair, Base excision repair, Mitochondria, Aging, Brain
1. Introduction
The mitochondrial free radical theory of aging (Harman, 1972) continues to receive support from experimental investigations, and accumulation of mitochondrial DNA mutations has been suggested to play a determinant role in the aging process (Barja, 1999; Beckman and Ames, 1999; Kujoth et al., 2007; Sastre et al., 2000; Trifunovic et al., 2004). As a result of its proximity to the main site of mitochondrial reactive oxygen species (ROS) generation, mtDNA is subjected to continuous oxidation by free radicals. Hence, the steady-state levels of oxidative lesions in mtDNA are several-fold higher than those found in nuclear DNA (Barja and Herrero, 2000; Richter et al., 1988). Since many of the oxidative lesions are mutagenic, oxidative attack by ROS is an important source of mtDNA mutations. Several major pathways of DNA repair have been described, and the particular pathway used depends, in part, upon the type of DNA damage that is being repaired. Separate DNA repair mechanisms exist for the nuclear and mitochondrial compartments (Bohr and Anson, 1999), and not all the DNA repair pathways that are present in the nucleus have been found in the mitochondria. The main DNA repair pathway common to the two compartments is the base excision repair (BER). However, mitochondria possess an independent BER machinery, the components of which are coded by nuclear genes (Bohr, 2002).
BER is the main pathway for repair of small DNA modifications caused by alkylation, deamination or oxidation and includes four distinct steps (Bohr, 2002). First, DNA glycosylases, which have distinct substrate specificities, are responsible for recognition and removal of the modified bases, rendering an abasic site, which is then processed by AP endonuclease. Repair can then proceed through one of two subpathways: short- or long-patch BER. The short-patch BER involves the incorporation of a single nucleotide into the gap by DNA polymerase followed by strand ligation by DNA ligase. Long-patch BER involves incorporation of several nucleotides, typically two to seven, followed by cleavage of the resulting 5′ flap and ligation. Both subpathways of BER have been reported to occur in nuclei, and it is well established that short-patch BER takes place in mitochondria (Bohr, 2002). Interestingly, recent investigations suggest that long-patch BER is also involved in repairing mtDNA lesions (Akbari et al., 2008; Liu et al., 2008).
Recently, Nei-like homologues (NEILs) have been described to be present in mammalian cells (Hazra et al., 2002a). Due to their unique preference for recognizing and cleaving oxidatively damaged bases in bubble DNA substrates (Dou et al., 2003), NEIL glycosylases have been proposed to play an important role in repair of oxidative damage in replicated and transcribed DNA. NEIL mRNA has been detected in different mammalian tissues, including brain (Hazra et al., 2002a, b), and only one previous investigation has reported the presence of NEIL in mitochondria in liver (Hu et al., 2005a). In contrast to the lack of obvious phenotypes when DNA glycosylases are knocked out in rodents, an important symptomology resembling the metabolic syndrome in humans has been reported as a consequence of deleting NEIL1 in mouse (Vartanian et al., 2006).
Mitochondrial BER capacity has been reported to be organ-specific, with the brain being one of the tissues with lowest capacity (Karahalil et al., 2002), despite the fact that brain is one of the tissues with the highest oxidative load. The relatively low DNA repair capacity in brain mitochondria in mammals is critical due to the main role of the brain in homeostasis of the organism. Oxidative modifications and mutations in mtDNA accumulate with aging in the brain and are considered to play an important role in neuronal loss associated with aging and neurodegenerative diseases, such as Parkinson’s and Alzheimer’s diseases and amyotrophic lateral sclerosis (Beal, 2005; Melov, 2004; Vermulst et al., 2007). Different studies have previously investigated the total or nuclear BER activities in relation to brain aging and age-related neurodegenerative diseases, but only a few have attempted to investigate how mitochondrial BER is modulated during lifespan and the importance of this process in the functional decline in brain with aging (reviewed in Weissman et al., 2007).
The brain possesses a highly organized and complex architecture. Metabolic activities and antioxidant levels as well as distribution of trace metals implicated in oxidative stress have been described to vary among brain regions (Cardozo-Pelaez et al., 2000; Benkovic and Connor, 1993; Frederickson, 1989). Importantly, it has also been reported that some regions are more prone to accumulate oxidative DNA modifications than others (Cardozo-Pelaez et al., 1999; Giovannelli et al., 2003). Together with ROS, reactive nitrogen species (RNS) are also an important source of DNA damage in the brain, due to the role that nitric oxide plays as an intercellular messenger. RNS are mainly implicated in the formation of DNA strand breaks and deamination of DNA bases, with deoxy-uracil representing the main product of hydroxylative deamination of cytosine (Kilinc and Kilinc, 2005; Reddy et al., 2007).
In the present study, we have investigated the mitochondrial BER activity in cortex and hippocampus during the murine lifespan. These regions are central in mammalian cognition and have been reported to be severely affected during aging and in neurodegenerative diseases (Geinisman et al., 1995; Joelving et al., 2006; West et al., 1994). We have studied activities of individual enzymes catalyzing single steps in the mitochondrial BER pathway: DNA glycosylases and AP endonuclease. Oxoguanine DNA glycosylase (OGG1) and uracil DNA glycosylase (UNG1) recognize and remove 8-oxodG and deoxy-uracil (dU) from DNA, respectively, and endonuclease III homologue 1 (NTH1) is mainly responsible for removal of 5-hydroxy-deoxyuracil (5-OHdU). Moreover, we investigated the presence of NEIL activity in brain mitochondria, as well as how this activity is affected during the murine lifespan. Finally, changes in mitochondrial AP endonuclease (mtAPE) activity with aging in the two brain regions were studied. APE is responsible for repair of AP sites, and also single-strand DNA breaks with 3′ blocking groups generated under oxidative attack of DNA. Moreover, APE has been described to play a central role in neuronal survival (Vasko et al., 2005).
Our data indicate that a region specific regulation of mitochondrial BER activities takes place in the brain during aging. They also suggest that the capability of responding to mtDNA oxidative damage is lower in hippocampal than in cortical mitochondria, which might contribute to higher mtDNA instability in hippocampus and hence to the higher vulnerability reported in this region with aging.
2. Materials and methods
2.1. Animals
Male C57BL/6J mice were obtained from Taconic M&B (Denmark). Mice were fed ad libitum and kept in a 12 h light/12 h night cycle. Breeding of the animals started at different time points so all the animals reached the required age at the same time. All animals were sacrificed by decapitation at Taconic M&B facilities and brains were immediately harvested for dissection. Cortex, hippocampus and cerebellum were dissected and snap frozen. Four age groups were investigated: 1 month (young group); 5 months (adult group); 10 months (middle-aged group); and 15–20 months (old group).
2.2. Mitochondrial purification
Mitochondria were isolated as previously described (Hansford et al., 1999) with minor modifications. All steps were carried out at 4 °C. Briefly, brain regions were homogenized in MSHE buffer (210 mM mannitol, 70 mM sucrose, 10 mM HEPES, 1 mM EGTA, 1 mM EDTA; pH 7.4) with a glass–glass homogeniser. Homogenization was carried out in the presence of subtilisin (1.4 U/ml) in order to avoid contamination of nuclear proteins in the mitochondrial preparation. The protease was removed by centrifugation at 8500 ×g for 8 min. Supernatant was discarded and the pellet was resuspended in MSHE buffer. All subsequent steps were performed in the presence of protease inhibitors (0.15 mM spermine, 0.75 mM spermidine, 1 mM PMSF, 5 mM DTT, 5 μl protease inhibitor cocktail set III (Calbiochem)). After centrifugation at 500 × g for 12 min, mitochondria were spun down by centrifugation of the supernatant at 10,000 × g for 9 min. The mitochondrial pellet was resuspended in MSHE buffer and the final mitochondrial fraction was obtained after centrifugation at 8500 × g for 9 min. Mitochondria were resuspended in Mitobuffer (20 mM HEPES, 1 mM EDTA, 2 mM DTT, 5% glycerol; pH 7.4), aliquoted and stored at −80 °C until use. Due to the small size, hippocampi from two mice were pooled to obtain one preparation. Protein concentration of the mitochondrial preparations was determined by the Lowry method (Lowry et al., 1951).
2.3. Western blotting
Absence of nuclear contamination in the mitochondrial samples was confirmed by western blot analysis of the mitochondrial preparations from the different groups. Mitochondrial samples (40 μg protein) were separated on 4–12% NuPAGE Novex® Bis–Tris gels (Invitrogen) and transferred to PVDF membranes (Invitrogen). The absence of nuclear protein lamin B was assayed by using a polyclonal goat antibody (1:1000; Santa Cruz Biotechnology), and mitochondrial presence was assayed by detection of VDAC1 (polyclonal rabbit antibody 1:200; Abcam). AP endonuclease was detected by using a monoclonal mouse antibody (1:2000; Novus Biologicals). Secondary anti-goat, anti-rabbit or anti-mouse antibodies were applied at 1:3000–1:5000. Membranes were visualized using ECL plus® (GE Healthcare, Amersham).
2.4. Oligonucleotides
All oligonucleotides were purchased from DNA Technology (Aarhus, Denmark) and were 5′-end-labeled using T4 polynucleotide kinase (PNK) and γ-32P-ATP. Mixtures of 100 μg oligonucleotide containing the DNA lesion or an unmodified base (Table 1), 20 units of T4 PNK (Fermentas), PNK forward buffer A (Fermentas) and 333 μCi γ-32P-ATP were incubated for 90 min at 37 °C, and then stopped with 2 μl 0.5 M EDTA. The unincorporated γ-32P-ATP was removed by binding to G25 Microspin columns (GE Healthcare, Amersham). Duplexing of the 5′-end-labeled oligonucleotide with the corresponding complementary strand was carried out by heating to 90 °C followed by gradual cooling to room temperature.
Table 1.
Oligonucleotides used in assays for DNA repair activities
| Name | bp | Sequence |
|---|---|---|
| 8-oxodG | 30 | 5′-ATA TAC CGC G8C CGG CCG ATC AAG CTT ATT |
|
|
3′-TAT ATG GCG CCG GCC GGC TAG TTC GAA TAA | |
| dUracil | 30 | 5′-ATA TAC CGC GUC CGG CCG ATC AAG CTT ATT |
|
|
3′-TAT ATG GCG CGG GCC GGC TAG TTC GAA TAA | |
| 5-OHdU | 30 | 5′-ATA TAC CGC G5C CGG CCG ATC AAG CTT ATT |
|
|
3′-TAT ATG GCG CGG GCC GGC TAG TTC GAA TAA | |
| 5-OHdU bubble | 51 | 5′-GCT TAG CTT GGA ATC GTA TCA TGT A5A CTC GTG TGC CGT GTA GAC CGT GCC |

|
3′-CGA ATC GAA CCT TAG CAT AGG CAC CCG ACA AAC ACG GCA CAT CTG GCA CGG | |
| AP | 30 | 5′-ATA TAC CGC GGF CGG CCG ATC AAG CTT ATT |
|
|
3′-TAT ATG GCG CCG GCC GGC TAG TTC GAA TAA |
The modified bases are shown in bold letters. Underlined bases indicate unpaired area. bp: base pairs. (8) 8-oxodG; (U) dU; (5) 5-OHdU; (F) tetrahy-drofuran.
2.5. Mitochondrial DNA glycosylase activities
The activity of various DNA glycosylases in mitochondria from cortex and hippocampus was determined in vitro by incision assays essentially as described before (Souza-Pinto et al., 1999). After permeabilization in the presence of 0.05% Triton X-100 and 0.3 M KCl, mitochondria were incubated at 37 °C with 90 fmol of the corresponding duplexed 32P-labelled oligonucleotide (Table 1) in a 20 μl reaction containing 20 mM HEPES, 5 mM EDTA, 75 mM KCl, 1 mM MgCl2, 5% glycerol, 5 mM DTT and 0.1 mg/ml BSA. The amount of mitochondrial protein added in the reaction and the incubation time varied depending on the oligonucleotide used. Thus, 12 μg of mitochondrial extract and 1 h incubation time were chosen when removal of dU and 5-OHdU was investigated. Incubation time was 3 h and 24 μg of mitochondrial extract were added in the reactions with 8-oxodG substrate and 5-OHdU in bubble structure oligonucleotide. Moreover, 7.5 mM dNTPs were added when 5-OHdU in bubble structure was investigated in order to avoid unspecific cleavage of the substrate. Reactions were stopped by addition of 0.4% SDS and 0.2 μg/μl proteinase K, followed by 30 min incubation at 55 °C. Samples were mixed with 20 μl of formamide loading buffer (80% formamide, 10 mM EDTA, 1 mg/ml xylene cyanol FF, and 1 mg/ml Bromophenol Blue) heated to 90 °C for 5 min and loaded on a denaturing 20% polyacrylamide gel. The radioactively labeled DNA was visualized using a Personal Molecular Imager™ and quantified using Quantity One software (Bio-Rad).
2.6. Mitochondrial AP endonuclease activity
Mitochondrial APE activity was measured essentially as previously described (Stuart et al., 2004). 12 μg of mitochondrial protein were incubated for 1 h at 37 °C with a 30-mer oligonucleotide containing a tetrahydrofuran (THF, abasic site analog (Table 1)). Reaction conditions were identical to those described above for the DNA glycosylase assays. 20 μl of formamide loading buffer were added to each sample and substrate and product were resolved by electrophoresis on a denaturing 20% polyacrylamide gel.
2.7. Statistical analysis
All data are reported as mean ± standard error of the mean using three to five independent mitochondrial preparations. For each independent sample, all assays were performed in triplicate. Comparisons of groups were statistically analyzed by Student’s t-test.
3. Results
Brain function is modulated during the entire lifespan, with rapid cell proliferation and cell differentiation in developing brain during embryogenesis and the first post-natal weeks; after maturation, age-related functional decline occurs. In order to include the entire murine lifespan, four ages were studied in the present investigation: 1 month, when brain development processes are nearly completed; 5 months, mature adult animals; 10 months, middle-aged animals; and 15–20 months representing old animals.
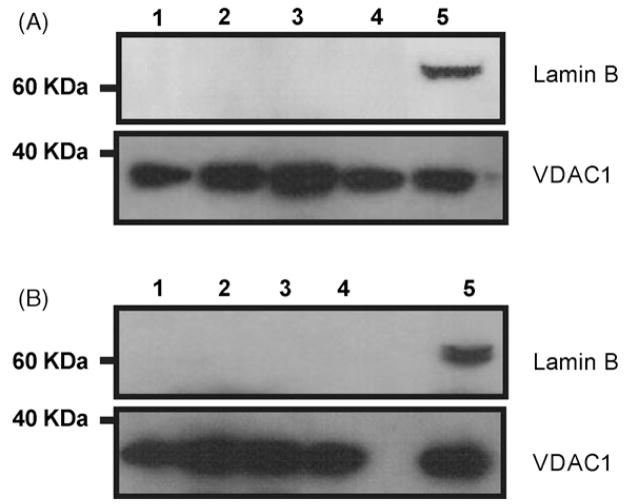
Mitochondrial preparations from cortex and hippocampus of the different age-groups were used to investigate the activities of the two first steps in the BER pathway during lifespan. DNA glycosylase activities were measured using double-stranded DNA oligonucleotides containing specific base damages and AP endonuclease activity was determined by using a tetrahydrofuran (THF) (an abasic site analogue) containing oligonucleotide (Table 1). Mitochondria from the brain regions were purified by differential centrifugation. The outer mitochondrial membrane protein VDAC1 was used to confirm mitochondrial presence (Fig. 1). Mitochondrial purifications were essentially devoid of nuclear contamination, as determined by western blotting for the nuclear protein Lamin B (Fig. 1). While lamin B was detected in whole brain extract, no signal was observed in mitochondrial fractions from any of the age groups.
Fig. 1.
Western blot analysis of Lamin B and VDAC1 in whole brain extract and mitochondrial fractions isolated from cortex and hippocampus. 40 μg of mitochondrial protein from hippocampus (A) and cortex (B) were loaded and resolved on a SDS-PAGE gel as described in Materials and Methods. Representative blot images are shown. Lane 1: young group; lane 2: adult group; lane 3: middle-aged group; lane 4: old group; lane 5: whole brain extract.
3.1. Mitochondrial DNA glycosylase activities are differentially regulated during aging in cortex and hippocampus
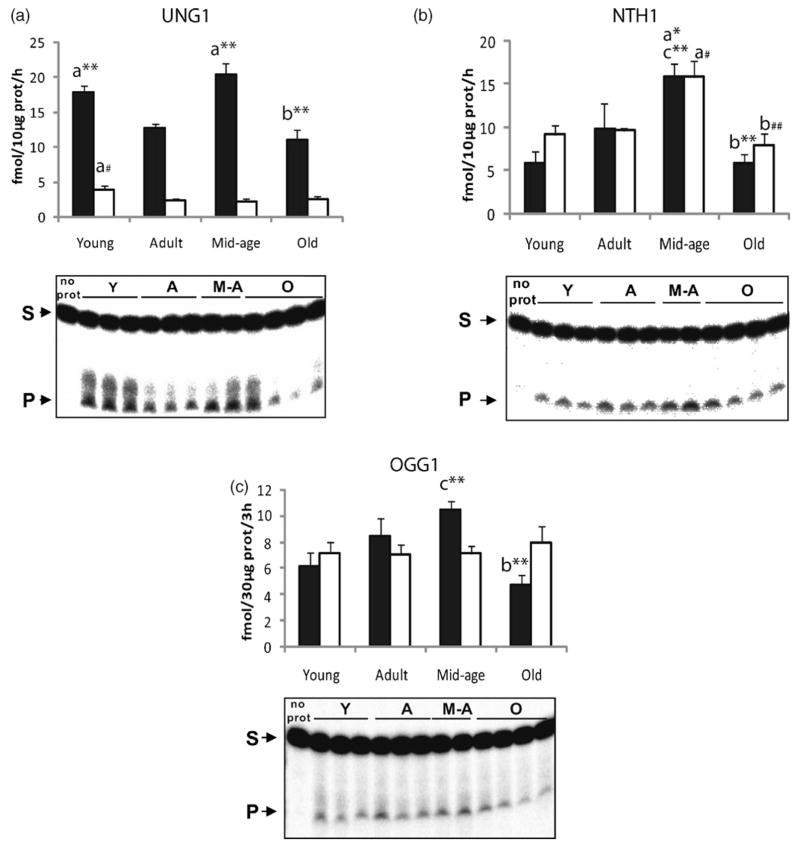
First, we investigated the activities of three main DNA glycosylases in mitochondria from hippocampus and cortex; UNG1, OGG1 and NTH1. Mitochondrial DNA glycosylase activities showed different patterns during lifespan depending on the brain region (Fig. 2). In cortical mitochondria, a significant increase in OGG1 and NTH1 activities occurred from young to middle-aged animals (p ≤ 0.01). Similarly, mitochondrial UNG1 activity was enhanced from adult to middle-aged mice (p ≤ 0.005). However, these increases were followed by a major drop at old age in all three activities (p ≤ 0.003), suggesting a failure of long-term up-regulation of DNA glycosylase activities in mitochondria from cortex. In contrast, no significant changes in the activity of DNA glycosylases were observed during the lifespan in hippocampal mitochondria, with the exception of NTH1 for which a significant peak at middle-age was observed (p ≤ 0.05). Moreover, a significant decrease in UNG1 activity from young to adult animals was observed in both regions (p ≤ 0.05). Notably, hippocampal mitochondria from mature animals generally showed lower incision activities than cortical mitochondria.
Fig. 2.
Changes in DNA glycosylase activities in mitochondria from cortex and hippocampus with aging. Top panels show quantification of DNA glycosylase activities and bottom panels show representative gels from cortical mitochondria. Incision activities were calculated from the amount of radioactivity in the products relative to the total radioactivity in the lane. Each lane in the gels represents one animal. Values presented are means ± S.E.M. of triplicate measurements from three to five mitochondrial preparations and are expressed as fmol per hour per 10 μg of mitochondrial protein for UNG1 and NTH1 and fmol per 3 h per 30 μg of mitochondrial protein for OGG1 activity. Filled bars represent cortical mitochondrial activities; Open bars represent hippocampal mitochondrial activities. (A) UNG1 activity; (B) NTH1 activity; (C) OGG1 activity. (a) Indicates significant difference versus adult group; (b) indicates significant difference versus middle-aged group; (c) indicates significant difference versus young group. Significant changes in cortical mitochondria: *p ≤ 0.06; **p ≤ 0.01; ***p ≤ 0.001; significant changes in hippocampal mitochondria: #p ≤ 0.06; ##p ≤ 0.01.
In order to examine whether the lack of modulation in the three DNA glycosylase activities in hippocampal mitochondria during lifespan was specific for this brain region, we further investigated the glycosylase activities in mitochondria from cerebellum. Cerebellar mitochondria were purified by the same procedure and checked for nuclear contamination by western blotting (data not shown). UNG1, OGG1 and NTH1 incision activities were then investigated. The activity of these DNA glycosylases in cerebellar mitochondria was higher in old than in adult animals (Fig. 3) suggesting that mitochondria from this particular region are capable of up-regulating DNA glycosylase activities to a higher extent than cortical mitochondria. Moreover, these results indicate that the lack of age-related regulation in mitochondrial DNA glycosylase activities was specific for hippocampus.
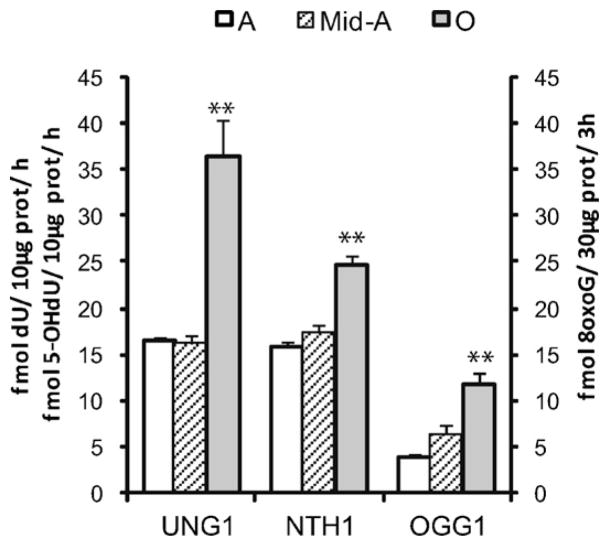
Fig. 3.
Changes in DNA glycosylase activities in mitochondria from cerebellum with aging. Quantification of DNA glycosylase activities as described for Fig. 2. Values presented are means ± S.E.M. of triplicate measurements from three to five mitochondrial preparations and are expressed as fmol per hour per 10 μg of mitochondrial protein for UNG1 and NTH1 and fmol per 3 h per 30 μg of mitochondrial protein for OGG1 activity. Asterisk (*) indicates significant difference versus middle-aged group; **p ≤ 0.01.
3.2. Presence and changes with aging in mitochondrial NEIL activity in cortex and hippocampus
Despite the postmitotic nature of neurons, replication processes do take place in their mitochondria. NEIL glycosylases are reported to be involved in replication, and NEIL mRNAs have been described to be present in brain tissue (Hazra et al., 2002a, b). We investigated NEIL activity in cortical and hippocampal mitochondria during lifespan using a 5-OHdU containing bubble substrate that is specifically recognized by these DNA glycosylases. Indeed, NEIL activity was observed in mitochondria from both regions (Fig. 4) at a rate similar to the DNA glycosylase OGG1. Mitochondrial NEIL activity showed the same age-related changes as the other DNA glycosylases, both in cortex and hippocampus, with no significant changes during lifespan in the latter, and with a significant peak at middle-age in the cortical region (p ≤ 0.05). Similar to UNG1 and OGG1, NEIL activity was lower in hippocampal than in cortical mitochondria.
Fig. 4.
NEIL activity in mitochondria from cortex and hippocampus in different age groups. Top panel shows quantification of NEIL activity and bottom panel shows representative gel. Quantification of NEIL activities as described for Fig. 2. Each lane in the gels represents one animal. Values presented are means ± S.E.M. of triplicate measurements from three to five mitochondrial preparations and are expressed as fmol per 3 h per 30 μg of mitochondrial protein. Filled bars represent cortical mitochondrial activities; Open bars represent hippocampal mitochondrial activities. (a) Indicates significant difference versus adult group; (b) indicates significant difference versus middle-aged group; *p ≤ 0.06.
3.3. AP endonuclease activity increases with aging in mitochondria from cortex and hippocampus
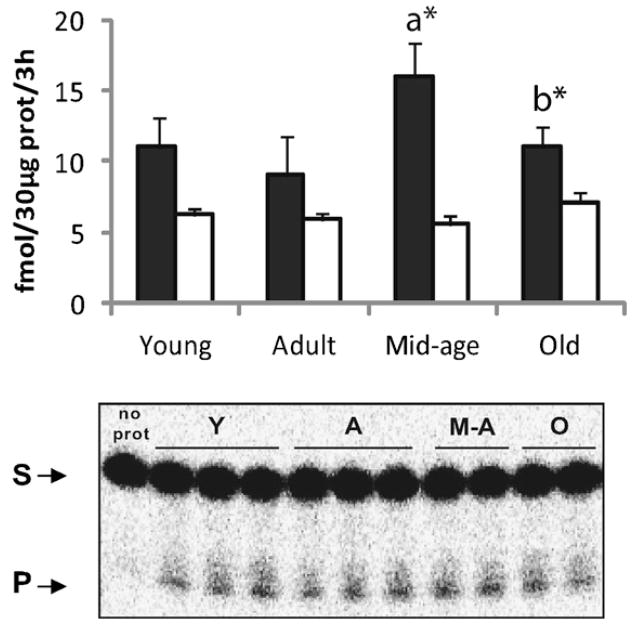
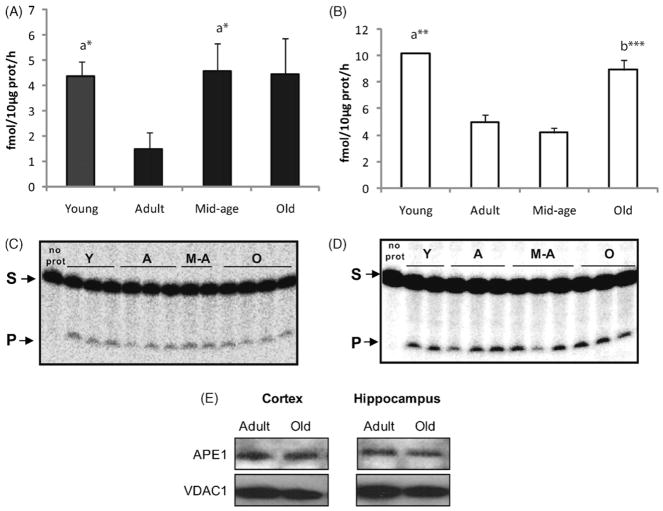
In order to investigate whether other single steps of the mitochondrial BER pathway were affected by aging in a similar manner as the DNA glycosylases, the AP endonuclease activity was determined in mitochondria from cortex (Fig. 5A) and hippocampus (Fig. 5B). Although mtAPE activity declined from immature animals to adult ones in both regions (p ≤ 0.033), a significant increase in mitochondrial AP activity was subsequently observed from adult to old animals. A regional difference was observed concerning the timing of enhancement in mtAPE activity. While hippocampal mtAPE activity was increased only in the oldest group (p ≤ 0.001), a significant increase was observed in mitochondria from cortex already at middle-age (p = 0.039). Moreover, and in contrast to mitochondrial DNA glycosylases, the elevated APE activity in cortical mitochondria was maintained in old mice. In order to determine whether the rise in mtAPE activity in old animals reflected an up-regulation in APE protein content, we investigated the protein amount of APE in mitochondrial extracts of the two regions. As shown in Fig. 5E, APE levels did not differ between mitochondrial fractions from adult and old animals.
Fig. 5.
MtAPE activity in cortical and hippocampal mitochondria with aging. Quantification of mtAPE activity in cortical (A) and hippocampal (B) mitochondria. Values presented are means ± S.E.M. of triplicate measurements from three to four mitochondrial preparations and are expressed as fmol per hour per 10 μg of mitochondrial protein. (C and D) Representative gels of AP incision in cortical and hippocampal mitochondria respectively. Each lane in the gels represents one animal. (E) Western blot analyses of APE in cortical and hippocampal mitochondria from adult and old groups. Representative blot image is shown. (a) Denotes significant difference versus adult group; (b) indicates significant difference versus middle-aged group; *p ≤ 0.06; **p ≤ 0.01; ***p ≤ 0.001.
4. Discussion
Brain aging and neurodegenerative diseases have often been related to DNA instability, particularly of mtDNA (Coskun et al., 2004; Howell et al., 2005; Smigrodzki et al., 2004) and to increasing levels of oxidative lesions (Markesbery and Lovell, 2006; Nakabeppu et al., 2007). Since oxidative lesions in DNA are mutagenic, mitochondrial DNA repair mechanisms play a central role in preventing accumulation of mtDNA mutations and in the maintenance of DNA stability. Cell survival in postmitotic tissues is critical in order to maintain tissue functionality. Such maintenance is especially important in the brain, where the postmitotic neurons play a central role in the control of most of the physiological functions of the organism.
The main aim of the current study was to investigate the mammalian DNA repair capacity throughout lifespan in mitochondria from two different brain regions, cortex and hippocampus. Brain function is modulated during the entire lifespan of mammals. During brain development, cell proliferation is accelerated and robust DNA repair is required in order to maintain DNA stability. In rodents, cell proliferation in developing brain gradually diminishes and generally ceases after the first month after birth. Previous studies on developing brain have reported higher activity of UNG1, OGG1 and APE in nuclear whole brain extracts during embryonic and neonatal stages compared to mature animals (Focher et al., 1990; Hildrestrand et al., 2007; Larsen et al., 2006). An important regulator in vertebrate development of the central nervous system is nitric oxide (Mantelas et al., 2007; Peunova et al., 2001), and hence the mitochondrial levels of RNS are likely to be high during brain maturation. As a consequence hereof, mtDNA is prone to deoxy-uracil base modifications due to deamination of cytosine induces by RNS. In agreement with that, our results showed that UNG1 and APE activities, involved in dU removal, were higher in mitochondria from cortex and hippocampus of young animals than in adult ones. Previous studies on nuclear extracts are hereby extended to mitochondria, suggesting that accurate mtDNA repair mechanisms are particularly important during the early stages of life in order to maintain neuronal viability.
Despite the lack of general changes in DNA glycosylase activities in cortical mitochondria from young to adult mice, we found a 1.6-fold increase in the mitochondrial activity of UNG1 and NTH1, and a 1.7-fold increase in NEIL1 activity in this brain region from adult to middle-aged animals. Similarly, mitochondrial OGG1 activity increased linearly 1.7-fold from young to middle-aged animals. These results suggest the existence of a general compensatory mechanism that leads to an up-regulation of mtDNA glycosylase activities in order to remove the accumulating mtDNA lesions with aging in cortex. Similarly, Souza-Pinto et al. (2001) previously reported a linear increase in OGG1 activity with aging in mitochondria from mouse liver. However, the augmentation of DNA glycosylase activities in cortical mitochondria at middle age was followed by a significant decrease in old mice, suggesting that the up-regulation cannot be sustained after middle age.
In contrast to the general changes found in cortical mitochondria, we observed only minor changes in mitochondrial DNA glycosylase activities in hippocampus during the lifespan. Furthermore, the capacity to remove DNA lesions was generally lower in hippocampal than in cortical mitochondria. The lack of regulation of DNA glycosylase activities during aging was specific to hippocampal mitochondria, since investigation of cerebellum revealed that an age-associated increase in DNA glycosylase activities also takes place in mitochondria from that region. Thus, a cellular adaptation to increasing mtDNA damage occurs with aging both in cortex and in cerebellum, although regional differences are observed, since in cortical mitochondria the DNA glycosylase activities drop in the late stage of life.
In accordance with the higher mtDNA repair capacity observed in cerebellum at old age, it has been reported that this region accumulates less mtDNA lesions with aging than other brain regions in humans and rodents (Corral-Debrinski et al., 1992; Filburn et al., 1996). Hippocampus has been described to be one of the brain regions most affected by aging (Geinisman et al., 1995), and at the same time, lipofuscin and mtDNA deletions have been described to accumulate to a higher extent in hippocampus than in cortex and cerebellum during aging (Abd El Mohsen et al., 2005; Filburn et al., 1996). Furthermore, hippocampus has been reported to be more vulnerable than cortex and cerebellum to oxidative stress conditions (Brown et al., 2004; Hota et al., 2007) and it is a main target in Alzheimer’s disease (West et al., 2000). Our results suggest that the lack of up-regulation of mitochondrial DNA glycosylase activities during aging in hippocampus, together with its lower general repair activity may play a major role in the age-related accumulation of mtDNA lesions in this region, and hence in the higher susceptibility to functional decline when compared to other mammalian brain regions, like cortex or cerebellum.
Although mRNAs of NEIL1 and NEIL2 have been detected in brain (Hazra et al., 2002a, b), NEIL activities have not previously been described in brain mitochondria. In the current investigation we report a significant activity of NEILs in mitochondria from cortex and hippocampus at a similar rate as other major DNA glycosylases, such as OGG1, described in the present study and in previous reports on brain mitochondria (Imam et al., 2006). NEIL1 has previously been described to be present in liver mitochondria (Hu et al., 2005a), and our results extend the location of NEILs to mitochondria of neuronal tissue. Together, these results suggest a new role of NEILs in the removal of DNA lesions in mitochondria during replication and transcription. Interestingly, the observed peak at middle-age in mitochondrial NEIL activity in the cortex was comparable with previous results on mRNA expression profiles of mitochondrial encoded genes in the same region (Manczak et al., 2005). This provides an interesting putative link between the capacity of mtDNA repair and mtDNA transcription in the cortex.
In order to analyze whether the efficiency of other steps of the BER pathway were also affected during aging, we investigated mtAPE activity in cortex and hippocampus. The results underscored the regional differences in mitochondrial BER modulation with aging. Although both regions showed a higher mtAPE activity in old animals than in younger ones, the increase occurred later in life in hippocampal than in cortical mitochondria, where a significant increase in the mtAPE activity was already observed in middle-aged animals. These results support the idea that mtDNA repair mechanisms are not as efficient in hippocampal mitochondria as in cortex during aging, leading to a higher instability of mtDNA in hippocampus (Filburn et al., 1996).
Despite the higher APE activity observed in mitochondria from old animals, no changes in APE protein levels were detected between adult and old groups in any of the regions. This result is in agreement with an earlier study on mtAPE levels during aging in brain regions (Imam et al., 2006). Our results point towards post-translational modification as the potential mechanism responsible for the increased mitochondrial activity with aging. Post-translational modifications have been described to modulate various aspects of the DNA damage response, including the activity of various DNA repair proteins (Hu et al., 2005b). APE has been described to be acetylated as well as phosphorylated (Bhakat et al., 2003; Luo et al., 2007). While acetylation of APE has been related to the action of the protein in transcriptional regulation (Bhakat et al., 2003), the phosphorylation of APE has been shown to modulate its DNA repair function (Luo et al., 2007). AP endonuclease has previously been reported to be essential for neuronal survival in part via apoptosis signalling (Vasko et al., 2005). The observed elevated mtAPE activity with aging in the current investigation might be a response mechanism to maintain cellular viability in both cortex and hippocampus, suggesting an important role of APE in mitochondria during aging.
In summary, our study denotes an important heterogeneity in the modulation of mtDNA repair enzymes with aging in the brain. The age-related mtDNA instability observed in hippocampus and the elevated susceptibility to neurodegenerative diseases in comparison to other brain regions might partly be due to a generally lower capacity to remove modified DNA bases and the incapacity of enhancing DNA glycosylase activities during aging. Up-regulation of APE activity in mitochondria from cortex and hippocampus occurs with aging and is maintained in old animals, supporting the essential role of this enzyme in mtDNA stability and cellular survival with aging through its role in the mitochondrial BER pathway. Finally, we have reported for the first time that brain mitochondria contain NEIL activity, supporting the role of NEIL glycosylases in the stability of mtDNA in brain mitochondria.
Acknowledgments
This research was supported by grants from the European Commission (LSHM-CT-2004-512020) and Lundbeck Foundation (4-55951-95094019) to TS. CG and RH were supported by the Danish Cancer Association. This research was also partially supported by funds from the intramural program of the National Institute on Aging, NIH. We thank Birija S. Patro and Rikke F. Frøhlich for critical reading of the manuscript.
Footnotes
Conflict of interest
The authors have no actual or potential conflict of interest associated with this research.
References
- Abd El Mohsen MM, Iravani MM, Spencer JP, Rose S, Fahim AT, Motawi TM, Ismail NA, Jenner P. Age-associated changes in protein oxidation and proteasome activities in rat brain: modulation by antioxidants. Biochem Biophys Res Commun. 2005;336:386–391. doi: 10.1016/j.bbrc.2005.07.201. [DOI] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 2008;7:605–616. doi: 10.1016/j.dnarep.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Barja G. Mitochondrial oxygen radical generation and leak: sites of production in states 4 and 3, organ specificity, and relation to aging and longevity. J Bioenerg Biomembr. 1999;31:347–366. doi: 10.1023/a:1005427919188. [DOI] [PubMed] [Google Scholar]
- Barja G, Herrero A. Oxidative damage to mitochondrial DNA is inversely related to maximum life span in the heart and brain of mammals. FASEB J. 2000;14:312–318. doi: 10.1096/fasebj.14.2.312. [DOI] [PubMed] [Google Scholar]
- Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- Beckman KB, Ames BN. Endogenous oxidative damage of mtDNA. Mutat Res. 1999;424:51–58. doi: 10.1016/s0027-5107(99)00007-x. [DOI] [PubMed] [Google Scholar]
- Benkovic SA, Connor JR. Ferritin, transferrin, and iron in selected regions of the adult and aged rat brain. J Comp Neurol. 1993;338:97–113. doi: 10.1002/cne.903380108. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Izumi T, Yang S, Hazra TK, Mitra S. Role of acetylated human AP-endonuclease (APE1/Ref-1) in regulation of the parathyroid hormone gene. EMBO J. 2003;22:6299–6309. doi: 10.1093/emboj/cdg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohr VA, Anson RM. Mitochondrial DNA repair pathways. J Bioenerg Biomembr. 1999;31:391–398. doi: 10.1023/a:1005484004167. [DOI] [PubMed] [Google Scholar]
- Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. doi: 10.1016/s0891-5849(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Brown MR, Geddes JW, Sullivan PG. Brain region-specific, age-related, alterations in mitochondrial responses to elevated calcium. J Bioenerg Biomembr. 2004;36:401–406. doi: 10.1023/B:JOBB.0000041775.10388.23. [DOI] [PubMed] [Google Scholar]
- Cardozo-Pelaez F, Song S, Parthasarathy A, Hazzi C, Naidu K, Sanchez-Ramos J. Oxidative DNA damage in the aging mouse brain. Mov Disord. 1999;14:972–980. doi: 10.1002/1531-8257(199911)14:6<972::aid-mds1010>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Cardozo-Pelaez F, Brooks PJ, Stedeford T, Song S, Sanchez-Ramos J. DNA damage, repair, and antioxidant systems in brain regions: a correlative study. Free Radic Biol Med. 2000;28:779–785. doi: 10.1016/s0891-5849(00)00172-6. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M, Horton T, Loft MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advance age. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci USA. 2004;101:10726–10731. doi: 10.1073/pnas.0403649101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- Filburn CR, Edris W, Tamatani M, Hogue B, Kudryashova I, Hansford RG. Mitochondrial electron transport chain activities and DNA deletions in regions of the rat brain. Mech Ageing Dev. 1996;87:35–46. doi: 10.1016/0047-6374(96)01696-x. [DOI] [PubMed] [Google Scholar]
- Focher F, Mazzarello P, Verri A, Hubscher U, Spadari S. Activity profiles of enzymes that control the uracil incorporation into DNA during neuronal development. Mutat Res. 1990;237:65–73. doi: 10.1016/0921-8734(90)90012-g. [DOI] [PubMed] [Google Scholar]
- Frederickson CJ. Neurobiology of zinc and zinc-containing neurons. Int Rev Neurobiol. 1989;31:145–238. doi: 10.1016/s0074-7742(08)60279-2. [DOI] [PubMed] [Google Scholar]
- Geinisman Y, Detoledo-Morrell L, Morrell F, Heller RE. Hippocampal markers of age-related memory dysfunction: behavioral, electrophysiological and morphological perspectives. Prog Neurobiol. 1995;45:223–252. doi: 10.1016/0301-0082(94)00047-l. [DOI] [PubMed] [Google Scholar]
- Giovannelli L, Decorosi F, Dolara P, Pulvirenti L. Vulnerability to DNA damage in the aging rat Substantia nigra: a study with the comet assay. Brain Res. 2003;969:244–247. doi: 10.1016/s0006-8993(03)02275-3. [DOI] [PubMed] [Google Scholar]
- Hansford RG, Tsuchiya N, Pepe S. Mitochondria in heart ischaemia and aging. Biochem Soc Symp. 1999;66:141–147. doi: 10.1042/bss0660141. [DOI] [PubMed] [Google Scholar]
- Harman D. The biologic clock: the mitochondria? J Am Geriatr Soc. 1972;20:145–147. doi: 10.1111/j.1532-5415.1972.tb00787.x. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra M. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA. 2002a;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra M, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002b;277:30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- Hildrestrand GA, Diep DB, Kunke D, Bolstad N, Bjoras M, Krauss S, Luna L. The capacity to remove 8-oxoG is enhanced in newborn neural stem/progenitor cells and decreases in juvenile mice and upon cell differentiation. DNA Repair (Amst) 2007;6:723–732. doi: 10.1016/j.dnarep.2006.12.008. [DOI] [PubMed] [Google Scholar]
- Hota SK, Barhwal K, Singh SB, Ilavazhagan G. Differential temporal response of hippocampus, cortex and cerebellum to hypobaric hypoxia: A biochemical approach. Neurochem Int. 2007;51:384–390. doi: 10.1016/j.neuint.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Howell N, Kubacka I, Keers SM, Turnbull DM, Chinnery PF. Co-segregation and heteroplasmy of two coding-region mtDNA mutations within a matrilineal pedigree. Hum Genet. 2005;116:28–32. doi: 10.1007/s00439-004-1203-x. [DOI] [PubMed] [Google Scholar]
- Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005a;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- Hu J, Imam S, Hashiguchi K, de Souza-Pinto N, Bohr VA. Phosphorylation of human oxoguanine DNA glycosylase (alpha-OGG1) modulates its function. Nucleic Acids Res. 2005b;33:3271–3282. doi: 10.1093/nar/gki636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam S, Karahalil B, Hogue B, Souza-Pinto N, Bohr V. Mitochondrial and nuclear DNA-repair capacity of various brain regions in mouse is altered in an age-dependent manner. Neurobiol Aging. 2006;27:1129–1136. doi: 10.1016/j.neurobiolaging.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Joelving FC, Billeskov R, Christensen JR, West M, Pakkenberg B. Hippocampal neuron and glial cell numbers in Parkinson’s disease—a stereological study. Hippocampus. 2006;16:826–833. doi: 10.1002/hipo.20212. [DOI] [PubMed] [Google Scholar]
- Karahalil B, Hogue BA, de Souza-Pinto NC, Bohr VA. Base excision repair capacity in mitochondria and nuclei: tissue-specific variations. FASEB J. 2002;16:1895–1902. doi: 10.1096/fj.02-0463com. [DOI] [PubMed] [Google Scholar]
- Kilinc K, Kilinc A. Mutagenic actions of nitrogen oxides. Indoor Built Environ. 2005;14:503–512. [Google Scholar]
- Kujoth GC, Bradshaw PC, Haroon S, Prolla TA. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:e24. doi: 10.1371/journal.pgen.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen E, Reite K, Nesse G, Gran C, Seeberg E, Klungland A. Repair and mutagenesis at oxidized DNA lesions in the developing brain of wild-type and Ogg1−/− mice. Oncogene. 2006;25:2425–2432. doi: 10.1038/sj.onc.1209284. [DOI] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung J, de Souza-Pinto N, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM, III, Shen B, Demple1 B. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol. 2008 doi: 10.1128/MCB.00457-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Luo Y, Ji X, Ling F, Li W, Zhang F, Cao G, Chen J. Impaired DNA repair via the base-excision repair pathway after focal ischemic brain injury: a protein phosphorylation-dependent mechanism reversed by hypothermic neuroprotection. Front Biosci. 2007;12:1852–1862. doi: 10.2741/2193. [DOI] [PubMed] [Google Scholar]
- Manczak M, Jung Y, Park BS, Partovi D, Reddy PH. Time-course of mitochondrial gene expressions in mice brains: implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J Neurochem. 2005;92:494–504. doi: 10.1111/j.1471-4159.2004.02884.x. [DOI] [PubMed] [Google Scholar]
- Mantelas A, Stamatakis A, Fameli M, Stylianopoulou F. Sex differences in the control of neuronal nitric oxide synthase by GABA-A receptors in the developing rat diencephalon. Brain Res Dev Brain Res. 2007;1149:38–49. doi: 10.1016/j.brainres.2007.02.075. [DOI] [PubMed] [Google Scholar]
- Markesbery WR, Lovell MA. DNA oxidation in Alzheimer’s disease. Antioxid Redox Signal. 2006;8:2039–2045. doi: 10.1089/ars.2006.8.2039. [DOI] [PubMed] [Google Scholar]
- Melov S. Modeling mitochondrial function in aging neurons. Trends Neurosci. 2004;27:601–606. doi: 10.1016/j.tins.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Tsuchimoto D, Yamaguchi H, Sakumi K. Oxidative damage in nucleic acids and Parkinson’s disease. J Neurosci Res. 2007;85:919–934. doi: 10.1002/jnr.21191. [DOI] [PubMed] [Google Scholar]
- Peunova N, Scheinker V, Cline H, Enikolopov G. Nitric oxide is an essential negative regulator of cell proliferation in Xenopus brain. J Neurosci. 2001;21:8809–8818. doi: 10.1523/JNEUROSCI.21-22-08809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy VP, Beyaz A, Perry G, Cooke MS, Sayre LM, Smith MA. The role of oxidative damage to nucleic acids in the pathogenesis of neurological disease. In: Evans MD, Cooke MS, editors. Oxidative Damage to Nucleic Acids. 2007. [Google Scholar]
- Richter C, Park JW, Ames BN. Normal oxidative damage to mitochondrial and nuclear DNA is extensive. Proc Natl Acad Sci USA. 1988;85:6465–6467. doi: 10.1073/pnas.85.17.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre J, Pallardo FV, Garcia de la Asuncion J, Vina J. Mitochondria, oxidative stress and aging. Free Radic Res. 2000;32:189–198. doi: 10.1080/10715760000300201. [DOI] [PubMed] [Google Scholar]
- Smigrodzki R, Parks J, Parker WD. High frequency of mitochondrial complex I mutations in Parkinson’s disease and aging. Neurobiol Aging. 2004;25:1273–1281. doi: 10.1016/j.neurobiolaging.2004.02.020. [DOI] [PubMed] [Google Scholar]
- Souza-Pinto NC, Croteau DL, Hudson EK, Hansford RG, Bohr VA. Age-associated increase in 8-oxo-deoxyguanosine glyco-sylase/AP lyase activity in rat mitochondria. Nucleic Acids Res. 1999;27:1935–1942. doi: 10.1093/nar/27.8.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza-Pinto N, Hogue BA, Bohr VA. DNA repair and aging in mouse liver: 8-oxodG glycosylase activity increases in mitochondrial but not in nuclear extracts. Free Radic Biol Med. 2001;30:916–923. doi: 10.1016/s0891-5849(01)00483-x. [DOI] [PubMed] [Google Scholar]
- Stuart JA, Hashiguchi K, Wilson DM, III, Copeland WC, Souza-Pinto NC, Bohr VA. DNA base excision repair activities and pathway function in mitochondrial and cellular lysates from cells lacking mitochondrial DNA. Nucleic Acids Res. 2004;32:2181–2192. doi: 10.1093/nar/gkh533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Törnell J, Jacobs JT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. PNAS. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasko MR, Guo C, Kelley MR. The multifunctional DNA repair/redox enzyme Ape1/Ref-1 promotes survival of neurons after oxidative stress. DNA Repair. 2005;4:367–379. doi: 10.1016/j.dnarep.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Vermulst M, Bielas JH, Kujoth GC, Ladiges WC, Rabinovitch PS, Prolla TA, Loeb LA. Mitochondrial point mutations do not limit the natural lifespan of mice. Nat Genet. 2007;39:540–543. doi: 10.1038/ng1988. [DOI] [PubMed] [Google Scholar]
- Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145:1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- West MJ, Coleman PD, Flood DG, Troncoso JC. Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer’s disease. Lancet. 1994;344:769–772. doi: 10.1016/s0140-6736(94)92338-8. [DOI] [PubMed] [Google Scholar]
- West MJ, Kawas CH, Martin LJ, Troncoso JC. The CA1 region of the human hippocampus is a hot spot in Alzheimer’s disease. Ann NY Acad Sci. 2000;908:255–259. doi: 10.1111/j.1749-6632.2000.tb06652.x. [DOI] [PubMed] [Google Scholar]