Abstract
Background
Well characterized genes affecting warfarin metabolism (CYP2C9) and sensitivity (VKORC1) explain one-third of the variability in therapeutic dose before the International Normalized Ratio (INR) is measured.
Methods
To determine genotypic relevance after INR becomes available, we derived clinical and pharmacogenetic refinement algorithms using INR values on day 4 or 5 of therapy, clinical factors, and genotype.
Results
After adjusting for INR, CYP2C9 and VKORC1 genotypes remained significant predictors (P < 0.001) of warfarin dose. The clinical algorithm had an R2 of 48% (median absolute error [MAE]: 7.0 mg/week); the pharmacogenetic algorithm had an R2 of 63% (MAE: 5.5 mg/week) in the derivation set (N=969). In independent validation, the R2 was 26%-43% with the clinical algorithm, and 42%-58% adding genotype (P = 0.002).
Conclusion
After several days of therapy, a pharmacogenetic algorithm estimates the therapeutic warfarin dose more accurately than one using clinical factors and INR response, alone.
Keywords: pharmacogenetics, warfarin, dose-refinement
Introduction
Warfarin (Coumadin™, Marevan™, and others) is an ideal drug for testing the paradigm of personalized medicine. It is the most commonly prescribed oral anticoagulant in North America and many European and Asian countries (1) and is a leading cause of adverse drug reactions.(2-4) Warfarin has a narrow therapeutic index and large inter-individual variability in dose requirements, with some individuals requiring less than 1 and others more than 20 mg/day to maintain therapeutic International Normalized Ratio (INR) values.(5) Tailoring therapy based on individual INR response often takes weeks, during which the risk of adverse reactions is high.(6, 7)
To reduce this risk while maintaining effectiveness, pharmacogenetic algorithms have been developed to estimate the maintenance warfarin dose at the time of warfarin initiation.(8-16) Besides clinical factors, these initiation algorithms incorporate common single nucleotide polymorphisms (SNPs) in the cytochrome P450 (CYP) 2C9 system (CYP2C9*2 and CYP2C9*3) that are associated with impaired metabolism of warfarin(6, 17-19) and SNPs in the gene for vitamin K epoxide reductase complex 1 (VKORC1) that correlate with warfarin sensitivity.(8, 12, 16, 20-23) Together these SNPs explain one-third of the variability in therapeutic dose (R2 ~ 33%).(8, 12, 16, 20-23)
Pharmacogenetic initiation algorithms use these genes and clinical factors to estimate the therapeutic warfarin dose,(8, 10-13, 16, 23) but they have critical shortcomings. First, they offer no explicit guidance for warfarin dosing once the INR response to therapy is known. This limitation is compounded by the common delay of several days to get genotype results back from an outside laboratory. Some experts have argued that once VKORC1 and CYP2C9 genotype are available in practice, they are neither relevant (24) nor cost-effective.(25) Furthermore, with few exceptions,(8) prior initiation algorithms have been developed in small or single-centered studies and their predictive accuracy in broader populations is questionable. Although the US Food and Drug Administration (FDA) has included consideration of VKORC1 and CYP2C9 genotyping in the product label of Coumadin™/warfarin, several professional organizations do not endorse routine testing (American College of Chest Physicians, American College of Medical Genetics).(26-28)
One reason for such reluctance may be the lack of data on the ability to use VKORC1 and CYP2C9 genotype to further refine warfarin dose after INRs become available. The recent development of pharmacogenetic refinement algorithms looks promising,(29, 30) but, as these algorithms are tailored to the orthopedic population, they are not applicable to broad populations. In short, whether genotype can help refine an individual’s maintenance dose, after several days of warfarin therapy, remains unclear. (24, 31) Therefore, the international Warfarin Dose-Refinement (Warfarin DR) Collaboration, had two goals: (1) to develop and validate a pharmacogenetic refinement algorithm in an international cohort of patients receiving warfarin for varying indications, and (2) to determine if genotype is predictive of therapeutic dose even when an INR value is available on the 4th or 5th day of therapy.
Results
Derivation
In the derivation cohort (N = 969), therapeutic dose was inversely correlated with INR, VKORC1-1639 A, CYP2C9*2 and CYP2C9*3 alleles (P<0.001). Other significant, independent predictors of therapeutic dose were prior warfarin doses, age, BSA, stroke, diabetes, race, target INR, and use of amiodarone or fluvastatin. Other statins, individually and in combination, were not significant predictors of therapeutic dose in this dataset. Significant predictors of the therapeutic dose in the clinical refinement algorithm were similar (Tables 2 and 3) except that genotype was not offered into the model and race was not statistically significant.
Table 2.
Clinical Refinement Model in Derivation Cohort (N=969).
| Entry into model | variable | Effect on dose* (95% CI) | Cumulative Model R2 | P |
|---|---|---|---|---|
| 1 | ln(INR) † | -18% (-20% to -16%) | 22.2% | <0.001 |
| 2 | Dose-3, per mg‡ | +3% (2% to 4%) | 35.3% | <0.001 |
| 3 | Age, per year | -5% (-7% to -4%) | 39.4% | <0.001 |
| 4 | Dose-2, per mg | +4% (2% to 5%) | 42.4% | <0.001 |
| 5 | Dose-4, per mg | +2% (1% to 3%) | 44.5% | <0.001 |
| 6 | Stroke indication | -27% (-38% to -13%) | 45.4% | <0.001 |
| 7 | Target INR | +8% (4% to 11%) | 46.2% | <0.001 |
| 8 | Diabetes | -15% (-22% to -8%) | 46.9% | <0.001 |
| 9 | BSA║, per 0.25 m2 | +5% (2% to 7%) | 47.7% | <0.001 |
| 10 | Fluvastatin Use | -24% (-41% to -1%) | 48.0% | 0.040 |
| 11 | Amiodarone Use | -12% (-23% to 1%) | 48.2% | 0.060 |
Effect on the maintenance dose is calculated per 0.25 unit increase in ln(INR) or target INR.
ln is natural logarithm. INR is International normalized ratio.
Dose −i is dose given i days before INR is measured.
BSA is Body Surface Area.
Table 3.
Pharmacogenetic Refinement Model in Derivation Cohort (N=969).
| Entry into model | variable | Effect on dose* (95% CI) | Cumulative Model R2 | P |
|---|---|---|---|---|
| 1 | VKORC1-1639 G>A | -20% (-23% to -17%) | 23.2% | <0.001 |
| 2 | ln(INR) † | -12% (-14% to -11%) | 35.5% | <0.001 |
| 3 | Dose-3, per mg‡ | +2% (1% to 3%) | 44.1% | <0.001 |
| 4 | Age, per year | -7% (-9% to -6%) | 49.9% | <0.001 |
| 5 | CYP2C9*3 | -28% (-32% to -23%) | 55.1% | <0.001 |
| 6 | CYP2C9*2 | -15% (-19% to -11%) | 57.0% | <0.001 |
| 7 | BSA║, per 0.25 m2 | +7% (4% to 9%) | 58.9% | <0.001 |
| 8 | Target INR | +7% (4% to 10%) | 59.7% | <0.001 |
| 9 | African Origin | -11% (-17% to -5%) | 60.4% | 0.001 |
| 10 | Stroke | -23% (-33% to -10%) | 61.0% | <0.001 |
| 11 | Dose-4, per mg | +1% (1% to 2%) | 61.5% | <0.001 |
| 12 | Dose-2, per mg | +2% (1% to 3%) | 62.0% | <0.001 |
| 13 | Diabetes | -9% (-16% to -3%) | 62.3% | 0.007 |
| 14 | Amiodarone Use | -12% (-21% to -1%) | 62.5% | 0.028 |
| 15 | Fluvastatin Use | -17% (-33% to 4%) | 62.6% | 0.106 |
Effect on the estimate of the maintenance dose is calculated per variant allele, and per 0.25 unit increase in ln(INR) or target INR.
ln is natural logarithm. INR is International normalized ratio.
Dose −i is dose given i days before INR is measured.
BSA is Body Surface Area.
This clinical refinement algorithm explained 48% of the variation in the derivation cohort and had a median absolute dosing error of 7.0 mg/week (1.0 mg/day). The pharmacogenetic refinement algorithm explained 63% of the variation in the derivation cohort and had a median absolute dosing error of 5.5 mg/week (0.78 mg/day).
Internal Validation
First, we assessed the performance of the pharmacogenetic refinement algorithm using the 204 patients in the internal validation cohort who had an INR available on the 4th day of therapy. Here, R2 was 58%, which was significantly (P=0.002) greater than the R2 of the clinical refinement algorithm (R2=43%, Table 4). The MAE of the pharmacogenetic refinement algorithm (4.9 mg/week) was less than that of the clinical refinement algorithm (6.1 mg/week) (P=0.020).
Table 4.
Accuracy of clinical and pharmacogenetic dose refinement algorithms in the internal validation cohort (N=244).
| Cohort Subset | Clinical | Pharmacogenetic | P | ||
|---|---|---|---|---|---|
| R2 | MAE* | R2 | MAE | ΔMAE† | |
| Subset with Day 4 INR (N=204) | 43% | 6.1 mg/week | 58% | 4.9 mg/week | 0.020 |
| Subset with Day 5 INR (N=105) | 44% | 7.4 mg/week | 60% | 6.3 mg/week | 0.160 |
Some participants (N = 65) had an INR (International Normalized Ratio) measured on both days 4 and 5 of therapy.
MAE is Median Absolute Error.
ΔMAE is difference in MAE.
When evaluating algorithms in the smaller internal validation set of patients who had an INR measured on the 5th day (N = 105), the results were similar. In this subset, R2 for the pharmacogenetic algorithm was 60% which was significantly (P=0.009) more accurate than the clinical refinement algorithm (R2=44%, Table 4). The MAE of the pharmacogenetic refinement algorithm (6.3 mg/week) was less than that of the clinical refinement algorithm (7.4 mg/week) but this difference was not significant (P=0.16).
Final Algorithms
After pooling the derivation and internal validation cohorts (N=1213) and re-deriving a final model using the same methods, we found that the pharmacogenetic refinement algorithm was: Maintenance dose (mg/week) =EXP [3.10894 - 0.00767 × Age, per year - 0.51611 × ln(INR) - 0.23032 × VKORC1-1639 G>A - 0.14745 × CYP2C9*2 - 0.3077 × CYP2C9*3 + 0.24597 × BSA + 0.26729 × Target INR -0.09644 × African Origin - 0.2059 × Stroke - 0.11216 × Diabetes - 0.1035 × Amiodarone Use - 0.19275 × Fluvastatin Use + 0.0169 × Dose-2 + 0.02018 × Dose-3 + 0.01065 × Dose-4].
The clinical refinement algorithm was: Maintenance dose (mg/week) = EXP [2.81602 - 0.76679 × ln(INR) - 0.0059 × Age, per year + 0.27815 × Target INR - 0.16759 × Diabetes + 0.17675 × BSA -0.22844 × Stroke - 0.25487 × Fluvastatin Use + 0.07123 × African Origin - 0.11137 × Amiodarone Use + 0.03471 × Dose-2 + 0.03047 × Dose-3 + 0.01929 × Dose-4].
External validation
When evaluating the final algorithms in patients who had an INR measured on the 4th day (N = 517) the R2 was 40% for the final pharmacogenetic algorithm and 28% for the final clinical refinement algorithm. The MAE of both the final pharmacogenetic refinement algorithm (6.9 mg/week) and final clinical refinement algorithm (6.9 mg/week) was ~1 mg/day.
When evaluating algorithms in patients who had an INR measured on the 5th day (N = 438) the R2 was 42% for the final pharmacogenetic algorithm and 26% for the final clinical refinement algorithm. Again, the MAE of the final pharmacogenetic refinement algorithm (6.7 mg/week) and final clinical refinement algorithm (6.4 mg/week) was < 1 mg/day.
To account for the transient increase in the INR after valve replacement, the correction factor using the final pharmacogenetic or clinical algorithm was ~1.21 (i.e., the new predicted dose was 21% greater than predicted by the algorithm).
Discussion
The public is eager to see a return on its enormous investment in the Human Genome Project. The first payoffs are anticipated in the area of pharmacogenetics, where warfarin has been called the poster child.(32) Warfarin is a classic test-case because it has a narrow therapeutic index, is influenced by well-characterized genetic factors, and frequently causes adverse events. Now that pharmacogenetic dosing of warfarin is commercially available and several genotyping platforms are FDA approved, a logistical barrier has become apparent: most medical centers do not have same-day VKORC1 and CYP2C9 genotyping. Even in centers that do have access, the question of how to use genotype once an INR is available remained unanswered. In fact, skeptics have argued that genetic information may be irrelevant after several days of dosing, because INR response may capture warfarin sensitivity.(24, 31) In part, the skeptics were right. The clinical refinement algorithm (Table 2), which uses a single INR on day 4 or 5 of therapy, explained 26%-48% of variability in warfarin dose. For comparison, prior pharmacogenetic initiation algorithms,(8, 10-16, 23) explain only slightly more variability in dose (~50%). This similarity indirectly supports the claim that pharmacogenetics may add relatively little to predictive accuracy once INR data are available.
However, to compare how much genotype adds to predictive accuracy, one must compare pharmacogenetic accuracy with clinical accuracy on a particular day of therapy. We found that after 4 or 5 days of therapy, the addition of genetics improves the R2 by 12-17% (P < 0.002). These figures are averages; patients with uncommon genotypes likely benefit even more. Consider a 70-year-old patient, 5’9” (175 cm) and 200 lbs (91 kg), whose target INR is 2.5. If he presents with an INR of 1.4 after three 5-mg doses of warfarin, his predicted therapeutic dose (using the final pharmacogenetic algorithm) could be as low as 14.6 mg/week, or as high as 43.0 mg/week, depending on VKORC1-1639 and CYP2C9 genotype. For comparison, the clinical algorithm predicts 34.2 mg/week. Thus, genotype is a critical factor determining his therapeutic dose, even when INR is monitored after 4 or 5 doses. To assist the reader in performing similar calculations, we have made the final dose-refinement and initiation algorithms freely available on www.WarfarinDosing.org.
Several additional observations warrant discussion. First, VKORC1 was a more important predictor of therapeutic dose, than in our prior study of 92 orthopedic patients. Previously, we found that VKORC1 contributed modestly to dose variability once the INR after 3 doses was known.(29) However, all of the orthopedic patients had received pharmacogenetic therapy prospectively, so the initial warfarin doses already reflected VKORC1 genotype. Second, the effect of incorporating VKORC1 into the model causes the contribution of INR to R2 to be blunted from 22.2% in the clinical model to 12.3% in the pharmacogenetic one. Thus, the pharmacogenetic model should be more robust to errors in initial INR measurements. This robustness may be helpful in patients receiving therapeutic doses of unfractionated or low-molecular-weight heparin, anticoagulants that sometimes inflate initial INR values.(33)
While much of the variance can be explained by INR, prior doses, age, and (in the pharmacogenetic model) genotype, other variables also affect dose. The lower warfarin requirements in patients who have had a stroke is a new finding, and may reflect under-nutrition that is common post stroke.(34) Our observation that diabetes is a marker for lower warfarin requirements is consistent with prior literature.(35)
Several limitations also need to be discussed. As with any international collaboration, we are limited in the number of variables universally available for analysis. For example, some medications (e.g. fluconazole, rifampin, and barbiturates) interact with warfarin(36) but were too rare to be incorporated into the model and clinicians will have to account for them (the outliers in figure 1 demonstrate this necessity). The CYP4F2 V433M genotype was not collected at each site, and incorporation of this genotype might have improved the R2.(37) Estimated blood loss was not analyzed here, but can transiently inflate the INR after major surgery.(29, 30) Likewise, the algorithms do not account for decompensated heart failure or patient-specific environmental factors (e.g. dietary vitamin K intake), which may affect warfarin requirements.(38) Finally, although the population of participants of African ancestry is relatively large (N = 123), this analysis is still based on a predominantly Caucasian population.
Figure 1. Pharmacogenetic predicted versus actual therapeutic doses in the internal and external validation cohorts (using an INR on day 4 of therapy).
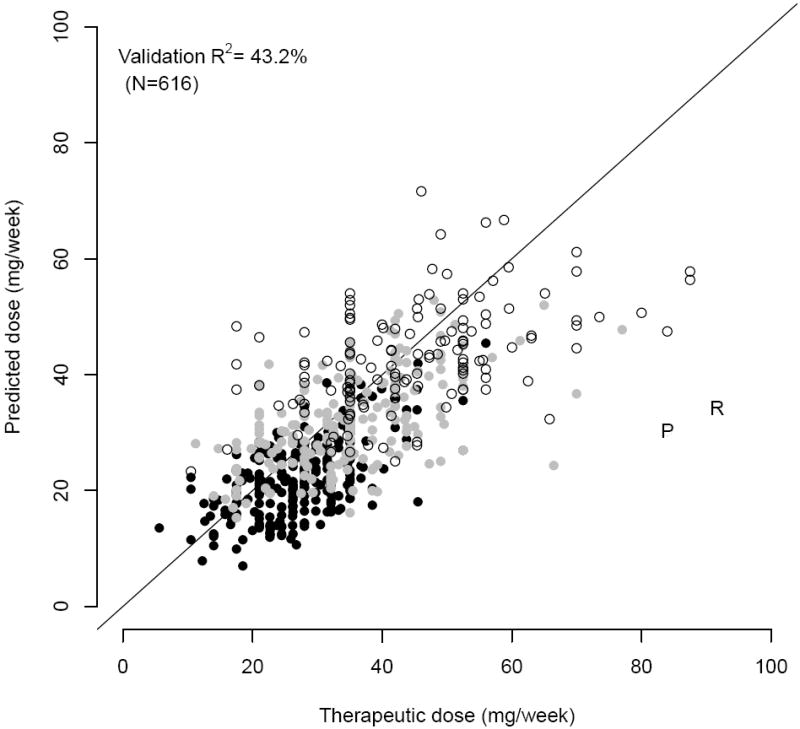
Open circles represent individuals for whom no variants in CYP2C9 and VKORC1 were detected. Grey circles represent individuals who are carrying one variant allele (either VKORC1-1639 A or a *2 or *3 allele. Black circles represent individuals with at least two variants. The ‘P’ was an individual taking phenytoin. The ‘R’ was an individual taking rifampin.
As a reminder of the importance of considering the limitations of any particular algorithm, we look to the external validation of the final algorithm. Many of these participants (N = 139; 20%) were receiving warfarin for valve replacement. Probably because of destruction and loss of functional clotting factors during cardiopulmonary bypass and because of decreased dietary intake around valve replacement surgery, this population has a transient increased sensitivity to warfarin post-operatively.(39-41) This indication, however, was rare in the internal datasets, so the algorithms had a tendency to under predict the therapeutic dosing requirements for all Inje University patients, resulting in a lower R2 and a greater MAE in the external validation cohort.
In contrast to traditional warfarin nomograms that rely on fixed initial doses and INR response alone,(42-45) the refinement algorithms developed here also accommodate demographics, warfarin indication, concurrent medications, flexible prior warfarin doses, comorbidities, and genotype. The pharmacogenetic refinement algorithm had a greater R2 and lower dosing error than previous pharmacogenetic algorithms,(8, 10-16, 23) with the exception of those tailored to specific, homogenous populations.(29, 30) Whether the high accuracy of the new genetic-based dosing algorithms will improve INR control or clinical outcomes is unknown, but is being addressed in three multi-centered, randomized trials in the US (Clarification of Optimal Anticoagulation through Genetics (COAG)), Genetics InFormatices Trial (GIFT) of Warfarin to Prevent DVT, and the Clinical and Economic Implications of Genetic Testing for Warfarin Management, and one in Europe (Pharmacogenetics of Anti-Coagulant Therapy (PACT)).
Personalized medicine will accomplish an important achievement if the pharmacogenetic algorithm developed here improves laboratory or clinical outcomes in ongoing trials. The hypothesized success would not belong to genetic testing, per se, but rather to a comprehensive approach whereby many patient-specific factors are accounted for explicitly. If this approach to warfarin management is any indication of what to expect from the investment in the Human Genome Project, genetics will add to, rather than replace, the list of factors that clinicians will need to consider when personalizing therapy.
Methods
Population
After Institutional Review Board approval at the participating sites, we obtained clinical and genetic data on 1213 patients in 3 continents: University of Alabama (N=62), Hospital for Special Surgery (N=11), Kaiser Permanente Colorado (N=30), University of Liverpool (N=149), Marshfield Clinic (N=147), Washington University in St. Louis School of Medicine (N=264), Ramathibodi Hospital, Mahidol University, Bangkok, Thailand (N=29), University of Pennsylvania (N=86), Uppsala University, Uppsala, Sweden (N=2), Intermountain Medical Center (N=155), Karolinska Institutet, Stockholm, Sweden (N=278). Patients were excluded if they did not achieve a therapeutic dose (defined below), if an INR on day 4 or 5 was not available, if their baseline (pre-warfarin) INR was above 1.4, if they were not genotyped for CYP2C9*2, CYP2C9*3 or VKORC1, or if they were prescribed fresh frozen plasma or vitamin K prior to their INR measurement. We randomly sampled 80% of the data for derivation, setting aside 20% for internal validation (Table 1). Dosing protocols varied among sites, with some participants (31%) being initiated on warfarin therapy using pharmacogenetic dosing algorithms.(29, 30, 46) However, stratifying by whether or not sites used a pharmacogenetic dosing protocol did not improve predictive accuracy. After development and internal validation, we studied 584 patients from 4 additional sites to validate the final algorithm (which was derived from combining the derivation and internal validation cohorts): Vanderbilt University (N=132), Inje University College of Medicine, South Korea (N=139), and University of Utah Hospital (N=117). The University of Liverpool also genotyped additional patients for external validation (N=196). Data from these 584 additional patients comprised the external validation cohort. Some of the data in the present analysis, were used for other pharmacogenetic analyses (8, 9, 16, 29, 30, 31, 51, 52, 53).
Table 1.
Demographic and clinical information in the derivation and internal validation cohorts
| Derivation | Internal Validation | |
|---|---|---|
| Demographic Variables | (N=969) | (N=244) |
| Male, N (%) | 545 (56.2) | 144 (59) |
| African Origin, N (%) | 95 (9.8) | 28 (11.5) |
| Caucasian Race, N (%) | 818 (84.4) | 207 (84.8) |
| Asian Race, N (%) | 28 (2.9) | 6 (2.5) |
| Other/Unknown Race, N (%) | 28 (2.9) | 3 (1.2) |
| Allele Frequencies | ||
| VKORC1-1639 G> A | 35.2% | 39.2% |
| CYP2C9*2 | 10.4% | 11.5% |
| CYP2C9*3 | 5.9% | 5.5% |
| Indication | ||
| Atrial Fibrillation/Flutter, N (%) | 306 (31.6) | 81 (33.2) |
| Orthopedic Surgery, N (%) | 264 (27.2) | 61 (25.0) |
| DVT or PE, N (%)* | 251 (25.9) | 69 (28.3) |
| Valve Replacement, N (%) | 32 (3.3) | 5 (2.0) |
| Stroke, N (%) | 17 (1.8) | 6 (2.5) |
| Other or Missing Indication, N (%) | 99 (10.2) | 22 (9.0) |
| Clinical Variables | ||
| Age, mean (SD), years† | 62 (14.2) | 61 (14.5) |
| Height, mean (SD), cm‡ | 170 (10.4) | 173 (10.9) |
| Weight, mean (SD), kg§ | 87.5 (22.4) | 86.7 (23.3) |
| Therapeutic Warfarin Dose, geometric mean (SD), mg/week | 32.1 (1.6) | 31.7 (1.5) |
| INR on day 4, geometric mean (SD) ‡ | 1.8 (1.4) | 1.9 (1.4) |
| INR on day 5, geometric mean (SD) | 1.9 (1.4) | 1.9 (1.3) |
| Target INR, mean (SD) | 2.5 (0.2) | 2.5 (0.2) |
| 1st Warfarin Dose, mean (SD), mg | 7.7 (3) | 7.6 (3) |
| 2nd Warfarin Dose, mean (SD), mg | 6.6 (2.8) | 6.7 (2.9) |
| 3rd Warfarin Dose, mean (SD), mg | 5.1 (2.6) | 5.0 (2.4) |
| Fluvastatin Use, N (%) | 7 (0.7) | 1 (0.4) |
| Amiodarone Use, N (%) | 31 (3.2) | 8 (3.3) |
| Inducer Use, N (%)║ | 7 (0.7) | 1 (0.4) |
| Current Smoker, N (%) | 101 (10.4) | 27 (11.1) |
| Liver Disease, N (%)¶ | 15 (1.5) | 4 (1.6) |
| Diabetes, N (%) | 79 (8.2) | 21 (8.6) |
DVT is Deep Venous Thrombosis; PE is Pulmonary Embolism.
SD is Standard Deviation.
INR is the International Normalized Ratio.
Rifampin or carbamazepine.
Liver Disease is hepatic cirrhosis, a two-fold elevation of any liver transaminase, or an albumin < 3.6.
Study Outcomes
The outcome variable was the therapeutic (maintenance) warfarin dose, defined as the dose that led to stable therapeutic anticoagulation levels: all sites required therapeutic INR values on at least two consecutive visits. Thus, data from studies that originally required only a single INR to define therapeutic dose (8, 29, 30), were re-analyzed using the more stringent definition in this analysis.
Statistical Analysis of the Derivation Cohort
Using stepwise selection, we quantified the relationship between therapeutic doses and genetic and clinical information available on the fourth or fifth day of warfarin therapy (Tables 2 and 3). Variables were allowed to remain in the multivariable linear regression model if they achieved statistical significance (P ≤ 0.05), or were marginally significant (0.05 < P ≤ 0.20) with strong biological plausibility. Because some patients had INRs available on both the fourth and the fifth day of therapy (N=355), we repeated random selection of half of them for whom we would use their 4th-day INR and half of them for whom we would use their 5th-day INR when deriving the model using a bootstrap procedure with 1000 resamples. Height and weight were combined into body surface area (BSA) using the classic fomula.(47)
We assessed the predictive ability of demographics (gender, race, and ethnicity), warfarin indication (atrial fibrillation, orthopedic surgery, venous thromboembolism, cardiac valve, and stroke), current medications (amiodarone, CYP inducers and statins), comorbidities (diabetes or liver disease), genotype, and INR values. Categorical variables were coded ‘1’ if present and ‘0’ if absent. To preserve linearity we log transformed therapeutic doses and INR. CYP2C9 inducers included rifampin/rifamycin, or carbamazepine. Fluvastatin, simvastatin, lovastatin, rosuvastatin, and atorvastatin were tested individually and in combination. Information on other interacting medications was not consistently available from sites. If diabetes status, smoking status, statin use, amiodarone use, or inducer use was not recorded at a particular site (n=232, n=148, n=232, n=135, and n=485 respectively), their probabilities were estimated using a likelihood method. Probabilities were then used in the regression equation instead of missing dummy variables. Missing BSA (n=38) was imputed from height or weight (if available), sex, and presence or absence of diabetes.
We coded CYP2C9*2 and CYP2C9*3 SNPs as 0 if absent, 1 if heterozygous, and 2 if homozygous. Likewise, VKORC1–1639 G>A (rs9923231) was coded 0 (homozygous GG), 1 (heterozygous), or 2 (homozygous AA). If VKORC1–1639 G>A genotype (also called VKORC1 3673) was missing (N = 241, derivation; N=57, internal validation) we inferred it from VKORC1 1173/6484 C>T (rs9934438) or VKORC1 1542/6853 G>C (rs8050894), which are in high linkage disequilibrium.(20, 48, 49)
To accommodate INRs taken on either the fourth or the fifth day of therapy, we defined doses in terms of the number of days they were given before the INR was drawn. For example, for patients with an INR on the 5th day of therapy, dose-2 was the dose given 2 days prior (on the 3rd day of therapy); dose-3 was the dose given on the 2nd day of therapy, etc. In this manner, only one INR (taken on either day 4 or day 5) was required for any individual’s dose prediction.
We used 1000 bootstraps(50) to compare accuracy (R2 and median absolute error, MAE) between the pharmacogenetic and clinical refinement models as well as between the refinement algorithms and previously validated pharmacogenetic and clinical initiation algorithms.(8) Because warfarin use after valve replacement was rare in the derivation and internal validation cohorts (N = 37), we calculated a correction factor for these individuals in a post-hoc analysis to quantify the transient warfarin sensitivity after valve replacement (observed previously:(39-41)) after external validation, we regressed the residual from the final model onto a dummy variable indicating whether or not the patient (in any cohort) had this indication, using all data available.
Supplementary Material
Open circles represent individuals for whom no variants in CYP2C9 and VKORC1 were detected. Grey circles represent individuals who are carrying one variant allele (either VKORC1-1639 A or a *2 or *3 allele. Black circles represent individuals with at least two variants. The ‘P’ was an individual taking phenytoin. The ‘R’ was an individual taking rifampin.
Acknowledgments
We thank the NIH (R01s HL074724 and HL097036), Wellcome Trust, the Swedish Science Council (Medicine 04496), the Swedish Heart and Lung foundation, the Clinical Research Support (ALF) at Uppsala University, Nycomed Comp (Stockholm, Sweden), the UK Department of Health, the Thailand Senior Researcher Fund, Korea Health 21 R&D Project, Ministry for Health, Welfare and Family Affairs (A030001), and Susan Gatchel and Elena Deych from Washington University in St. Louis. Petra Lenzini had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Conflict of Interest Disclosures The authors have no conflicts of interest.
Authorship Contributions Drs. Gage and Eby were investigators on a grant from Osmetech that focused on a gene (CYP4F2) not included in this analysis. Dr. Caldwell has applied for a patent for the same (CYP4F2) gene. P. Lenzini and B. Gage designed the research, collected, analyzed, and interpreted data, and wrote the manuscript. M. Wadelius, S. Kimmel, J.L. Anderson, and A.L Jorgensen collected data, and wrote the manuscript. M. Pirmohamed, M.D. Caldwell, N. Limdi, J.K. Burmester, M.B. Dowd, P. Angchaisuksiri, A.R. Bass, J. Chen, N. Eriksson, A. Rane, J.D. Lindh, J.F. Carlquist, B.D. Horne, G. Grice, P.E. Milligan, C. Eby, J. Shin, H. Kim, D. Kurnik, C.M. Stein, G. McMillin, R.C. Pendleton, R.L. Berg, and P. Deloukas, collected data and edited the manuscript.
References
- 1.Pirmohamed M. Warfarin: almost 60 years old and still causing problems. Br J Clin Pharmacol. 2006 Nov;62:509–11. doi: 10.1111/j.1365-2125.2006.02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budnitz DS, Pollock DA, Weidenbach KN, et al. National surveillance of emergency department visits for outpatient adverse drug events. Jama. 2006 Oct 18;296:1858–66. doi: 10.1001/jama.296.15.1858. [DOI] [PubMed] [Google Scholar]
- 3.Wysowski DK, Nourjah P, Swartz L. Bleeding complications with warfarin use: a prevalent adverse effect resulting in regulatory action. Arch Intern Med. 2007 Jul 9;167:1414–9. doi: 10.1001/archinte.167.13.1414. [DOI] [PubMed] [Google Scholar]
- 4.Wester K, Jonsson AK, Spigset O, et al. Incidence of fatal adverse drug reactions: a population based study. Br J Clin Pharmacol. 2008 Apr;65:573–9. doi: 10.1111/j.1365-2125.2007.03064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindh JD, Holm L, Dahl ML, et al. Incidence and predictors of severe bleeding during warfarin treatment. J Thromb Thrombolysis. 2008 Apr;25:151–9. doi: 10.1007/s11239-007-0048-2. [DOI] [PubMed] [Google Scholar]
- 6.Higashi MK, Veenstra DL, Kondo LM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287:1690–8. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 7.Limdi NA, McGwin G, Goldstein JA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008 Feb;83:312–21. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008 Sep;84:326–31. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009 Jan 22;113:784–92. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlquist JF, Horne BD, Muhlestein JB, et al. Genotypes of the cytochrome p450 isoform, CYP2C9, and the vitamin K epoxide reductase complex subunit 1 conjointly determine stable warfarin dose: a prospective study. J Thromb Thrombolysis. 2006 Dec;22:191–7. doi: 10.1007/s11239-006-9030-7. [DOI] [PubMed] [Google Scholar]
- 11.Sconce EA, Khan TI, Wynne HA, et al. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005 October;106:2329–33. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 12.Wadelius M, Chen LY, Downes K, et al. Common VKORC1 and GGCX polymorphisms associated with warfarin dose. Pharmacogenomics J. 2005 May 10;5:262–70. doi: 10.1038/sj.tpj.6500313. [DOI] [PubMed] [Google Scholar]
- 13.Herman D, Peternel P, Stegnar M, et al. The influence of sequence variations in factor VII, gamma-glutamyl carboxylase and vitamin K epoxide reductase complex genes on warfarin dose requirement. Thromb Haemost. 2006 May;95:782–7. [PubMed] [Google Scholar]
- 14.Schelleman H, Chen J, Chen Z, et al. Dosing algorithms to predict warfarin maintenance dose in Caucasians and African Americans. Clin Pharmacol Ther. 2008 Sep;84:332–9. doi: 10.1038/clpt.2008.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu AHB. Use of genetic and nongenetic factors in warfarin dosing algorithms. Pharmacogenomics. 2007;8:851–61. doi: 10.2217/14622416.8.7.851. [DOI] [PubMed] [Google Scholar]
- 16.Klein TE, Altman RB, Eriksson N, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009 Feb 19;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gage BF, Eby C, Milligan PE, et al. Use of pharmacogenetics and clinical factors to predict the maintenance dose of warfarin. Thromb Haemost. 2004 Jan;91:87–94. doi: 10.1160/TH03-06-0379. [DOI] [PubMed] [Google Scholar]
- 18.Margaglione M, Colaizzo D, D’Andrea G, et al. Genetic modulation of oral anticoagulation with warfarin. Thromb Haemost. 2000;84:775–8. [PubMed] [Google Scholar]
- 19.Wadelius M, Sorlin K, Wallerman O, et al. Warfarin sensitivity related to CYP2C9, CYP3A5, ABCB1 (MDR1) and other factors. Pharmacogenomics J. 2004 Dec 16;4:40–8. doi: 10.1038/sj.tpj.6500220. [DOI] [PubMed] [Google Scholar]
- 20.Rieder MJ, Reiner AP, Gage BF, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005 Jun 2;352:2285–93. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 21.D’Andrea G, D’Ambrosio RL, Di Perna P, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005 Jan 15;105:645–9. doi: 10.1182/blood-2004-06-2111. [DOI] [PubMed] [Google Scholar]
- 22.Yuan HY, Chen JJ, Lee MT, et al. A novel functional VKORC1 promoter polymorphism is associated with inter-individual and inter-ethnic differences in warfarin sensitivity. Hum Mol Genet. 2005 Jul 1;14:1745–51. doi: 10.1093/hmg/ddi180. [DOI] [PubMed] [Google Scholar]
- 23.Wadelius M, Chen LY, Lindh JD, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2008 Jun 23; doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bussey HI, Wittkowsky AK, Hylek EM, et al. Genetic testing for warfarin dosing? Not yet ready for prime time. Pharmacotherapy. 2008 Feb;28:141–3. doi: 10.1592/phco.28.2.141. [DOI] [PubMed] [Google Scholar]
- 25.Eckman MH, Rosand J, Greenberg SM, et al. Cost-effectiveness of using pharmacogenetic information in warfarin dosing for patients with nonvalvular atrial fibrillation. Annals of Internal Medicine. 2008;150:73–83. doi: 10.7326/0003-4819-150-2-200901200-00005. [DOI] [PubMed] [Google Scholar]
- 26.Ansell J, Hirsch J, Hylek E, et al. CHEST. 8. 2008. Pharmacology and management of the Vitamin K Antagonists. [DOI] [PubMed] [Google Scholar]
- 27.Flockhart DA, O’Kane D, Williams MS, et al. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med. 2008 Feb;10:139–50. doi: 10.1097/GIM.0b013e318163c35f. [DOI] [PubMed] [Google Scholar]
- 28.McClain MR, Palomaki GE, Piper M, et al. A rapid-ACCE review of CYP2C9 and VKORC1 alleles testing to inform warfarin dosing in adults at elevated risk for thrombotic events to avoid serious bleeding. Genet Med. 2008 Feb;10:89–98. doi: 10.1097/GIM.0b013e31815bf924. [DOI] [PubMed] [Google Scholar]
- 29.Millican E, Lenzini P, Milligan P, et al. Genetic-based dosing in orthopaedic patients beginning warfarin therapy. Blood. 2007;110:1511–5. doi: 10.1182/blood-2007-01-069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenzini PA, Grice GR, Milligan PE, et al. Laboratory and clinical outcomes of pharmacogenetic vs. clinical protocols for warfarin initiation in orthopedic patients. J Thromb Haemost. 2008 Oct;6:1655–62. doi: 10.1111/j.1538-7836.2008.03095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, Schwarz UI, Ritchie MD, et al. Relative contribution of CYP2C9 and VKORC1 genotypes and early INR response to the prediction of warfarin sensitivity during initiation of therapy. Blood. 2009 Apr 23;113:3925–30. doi: 10.1182/blood-2008-09-176859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carlquist J, Anderson J. Warfarin pharmacogenetics: up close and personalized. Future Medicine. 2008;5:125–8. [Google Scholar]
- 33.Soloman H, Randall J, Simmons V. Heparin-induced increase in the internatinal normalized ratio. Responses of 10 commercial thromboplastin reagents. AM J Clin Pathol. 1995;103:735–9. doi: 10.1093/ajcp/103.6.735. [DOI] [PubMed] [Google Scholar]
- 34.Andersson J, Gustafsson K, Fjellstrom C, et al. Five-day food intake in elderly female outpatients with Parkinson’s disease, rheumatoid arthritis or stroke. J Nutr Health Aging. 2004;8:414–21. [PubMed] [Google Scholar]
- 35.Hillman MA, Wilke RA, Caldwell MD, et al. Relative impact of covariates in prescribing warfarin according to CYP2C9 genotype. Pharmacogenetics. 2004 Aug;14:539–47. doi: 10.1097/01.fpc.0000114760.08559.dc. [DOI] [PubMed] [Google Scholar]
- 36.Holbrook AM, Pereira JA, Labiris R, et al. Systematic Overview of Warfarin and Its Drug and Food Interactions. Arch Intern Med. 2005 May 23;165:1095–106. doi: 10.1001/archinte.165.10.1095. [DOI] [PubMed] [Google Scholar]
- 37.Caldwell MD, Awad T, Johnson JA, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008 Apr 15;111:4106–12. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aquilante CL, Langaee TY, Lopez LM, et al. Influence of coagulation factor, vitamin K epoxide reductase complex subunit 1, and cytochrome P450 2C9 gene polymorphisms on warfarin dose requirements. Clin Pharmacol Ther. 2006 Apr;79:291–302. doi: 10.1016/j.clpt.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 39.Ageno W, Turpie AG. Exaggerated initial response to warfarin following heart valve replacement. Am J Cardiol. 1999 Oct 15;84:905–8. doi: 10.1016/s0002-9149(99)00463-4. [DOI] [PubMed] [Google Scholar]
- 40.Lee J, Lee B, Kim K, et al. Factors affecting warfarin therapy following cardiac valve surgery. The Annals of Pharmacotherapy. 2002 December;36:1845–50. doi: 10.1345/aph.1A431. [DOI] [PubMed] [Google Scholar]
- 41.Rahman M, Binesmael TM, Payne N, et al. Increased sensitivity to warfarin after heart valve replacement. Ann Pharmacother. 2006 Mar;40:397–401. doi: 10.1345/aph.1G407. [DOI] [PubMed] [Google Scholar]
- 42.Kovacs MJ, Rodger M, Anderson DR, et al. Comparison of 10-mg and 5-mg warfarin initiation nomograms together with low-molecular-weight heparin for outpatient treatment of acute venous thromboembolism. A randomized, double-blind, controlled trial. Ann Intern Med. 2003 May 6;138:714–9. doi: 10.7326/0003-4819-138-9-200305060-00007. [DOI] [PubMed] [Google Scholar]
- 43.Crowther MA, Ginsberg JB, Kearon C, et al. A randomized trial comparing 5-mg and 10-mg warfarin loading doses. Arch Intern Med. 1999;159:46–8. doi: 10.1001/archinte.159.1.46. [DOI] [PubMed] [Google Scholar]
- 44.Harrison L, Johnston M, Massicotte MP, et al. Comparison of 5-mg and 10-mg loading doses in initiation of warfarin therapy. Ann Intern Med. 1997;126:133–6. doi: 10.7326/0003-4819-126-2-199701150-00006. [DOI] [PubMed] [Google Scholar]
- 45.Siguret V, Gouin I, Debray M, et al. Initiation of warfarin therapy in elderly medical inpatients: a safe and accurate regimen. Am J Med. 2005 Feb;118:137–42. doi: 10.1016/j.amjmed.2004.07.053. [DOI] [PubMed] [Google Scholar]
- 46.Anderson JL, Horne BD, Stevens SM, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007 Nov 27;116:2563–70. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 47.DuBois D, DuBois E. Clinical Calorimetry; a formula to estimate the approximate surface area if height and weight be known. Arch Int med. 1916;17:863–71. [Google Scholar]
- 48.Wang D, Chen H, Momary KM, et al. Regulatory polymorphism in vitamin K epoxide reductase complex subunit 1 (VKORC1) affects gene expression and warfarin dose requirement. Blood. 2008 Aug 15;112:1013–21. doi: 10.1182/blood-2008-03-144899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Limdi NA, Beasley TM, Crowley MR, et al. VKORC1 polymorphisms, haplotypes and haplotype groups on warfarin dose among African-Americans and European-Americans. Pharmacogenomics. 2008 Oct;9:1445–58. doi: 10.2217/14622416.9.10.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Efron B, Tibshirani R. An introduction to the bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 51.Kim HS, Lee SS, Oh M, et al. Effect of CYP2C9 and VKORC1 genotypes on early-phase and steady-state warfarin dosing in Korean patients with mechanical heart valve replacement. Pharmacogenet Genomics. 2009 Feb;19:103–112. doi: 10.1097/FPC.0b013e32831a9ae3. [DOI] [PubMed] [Google Scholar]
- 52.Zhang JE, Jorgensen AL, Alfirevic A, et al. Effects of CYP4F2 genetic polymorphisms and haplotypes on clinical outcomes in patients initiated on warfarin therapy. Pharmacogenet Genomics. 2009 Oct;19:781–9. doi: 10.1097/FPC.0b013e3283311347. [DOI] [PubMed] [Google Scholar]
- 53.Jorgensen AL, Al-Zubiedi S, Zhang JE, et al. Genetic and environmental factors determining clinical outcomes and cost of warfarin therapy: a prospective study. Pharmacogenet Genomics. 2009 Oct;19:800–12. doi: 10.1097/FPC.0b013e3283317ab5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Open circles represent individuals for whom no variants in CYP2C9 and VKORC1 were detected. Grey circles represent individuals who are carrying one variant allele (either VKORC1-1639 A or a *2 or *3 allele. Black circles represent individuals with at least two variants. The ‘P’ was an individual taking phenytoin. The ‘R’ was an individual taking rifampin.


