Summary
In mammals, ceramide, a key intermediate in sphingolipid metabolism and an important signaling molecule, is synthesized by a family of six ceramide synthases (CerS), each of which synthesizes ceramides with distinct acyl chain lengths. There are a number of common biochemical features between the CerS, such as their catalytic mechanism, and their stucture and intracellular localization. Different CerS also display remarkable differences in their biological properties, with each of them playing distinct roles in processes as diverse as cancer and tumor suppression, in the response to chemotherapeutic drugs, in apoptosis, and in neurodegenerative diseases.
Keywords: Ceramide, sphingolipid, metabolism, biosynthesis, intracellular signaling, endoplasmic reticulum
Ceramide Synthesis
Sphingolipids (SLs) are one of the three major classes of membrane lipids in eukaryotic cells. The building block of all SLs is ceramide, which has received wide attention over the past couple of decades due to its proposed role as an important intracellular signaling molecule involved in regulating differentiation, proliferation and apoptosis (1,2). Ceramide consists of a sphingoid long chain base to which a fatty acid is attached via an amide bond. The biochemical pathways of ceramide and SL synthesis are well-established and most of the enzymes have been identified and cloned (3). However, despite significant advances in understanding the pathways of biosynthesis, little is known about the mode of regulation of ceramide synthesis in different physiological or pathophysiological conditions.
Ceramide is generated de novo (Fig. 1) in the endoplasmic reticulum (ER) by a pathway that begins with the condensation of L-serine and palmitoyl-CoA by serine palmitoyl transferase (4) generating 3-ketosphinganine, which is then reduced to sphinganine via 3-ketosphinganine reductase (5). This is followed by acylation of sphinganine via sphinganine N-acyl transferase to dihydroceramide, and subsequent reduction of dihydroceramide to ceramide by dihydroceramide desaturase. While de novo ceramide synthesis occurs in the ER, further metabolism to sphingomyelin and glycosphingolipids occurs in the Golgi apparatus (3), with ceramide being transported from the ER to the Golgi apparatus by either vesicular trafficking or by the ceramide transfer protein, CERT (6).
Fig. 1. Ceramide metabolism.
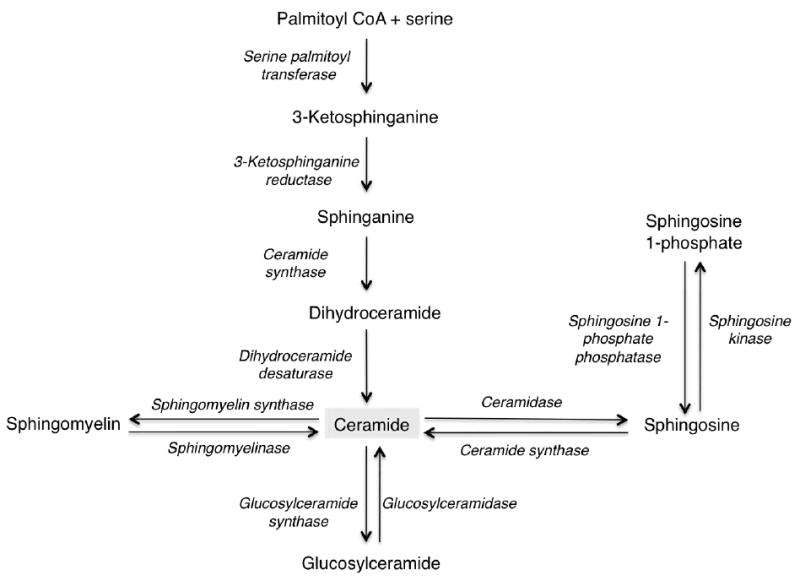
Overview of the major metabolic pathways of ceramide; enzymes are shown in italics.
This review focuses on the acylation of sphinganine by sphinganine N-acyl transferase, a reaction carried out in mammals by a family of six enzymes, the ceramide synthases (CerS). This reaction turns out to be unexpectedly complex and has been the focus of intense research over the past few years. The first comprehensive review of CerS biochemistry was published in 2006 (7), and the current review serves to update this earlier publication. We hope that readers will appreciate the dramatic progress made in this area, and the intense efforts being made to dissect the complexity of CerS biochemistry and biology.
The Discovery of the Ceramide Synthases
CerS were originally known as Lass (Longevity Assurance) genes based on their homology to the yeast protein, longevity assurance gene 1 (LAG1p) (8-10), but were renamed upon their biochemical characterization (7). The most prominent feature of the CerS is that even though they all carry out the same chemical reaction (i.e. N-acylation of the sphingoid long chain base), each CerS has a high specificity towards the acyl CoA chain length used for N-acylation. Thus, the CerS are responsible for the fatty acid composition of ceramides (Fig. 2).
Fig. 2. The roles of CerS in synthesizing ceramides with different acyl chain lengths.
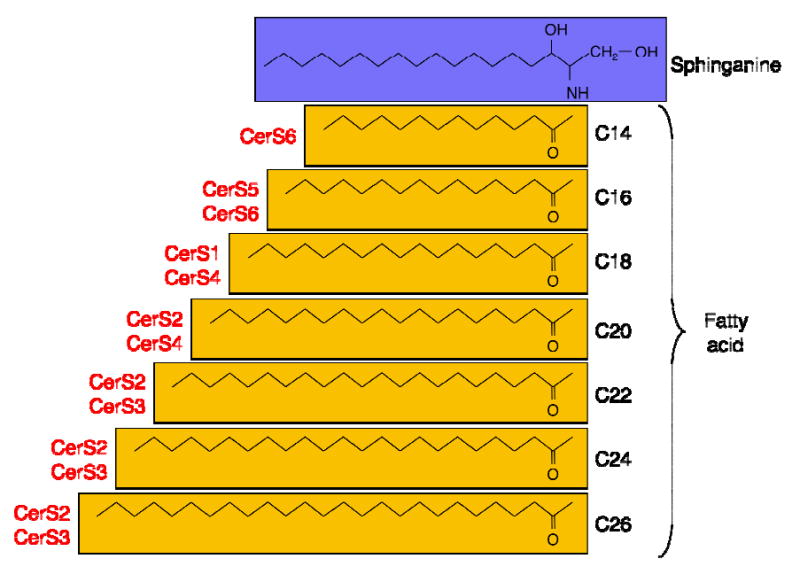
Ceramides can differ in their acyl chain length, as shown in the figure, as well as in their in their degree of saturation and α-hydroxylation (7). Sphinganine is show in blue, the acyl chain in yellow, and the CerS that synthesizes each ceramide is shown in red.
The founding member of the Lass gene family (Table 1) was discovered in yeast >15 years ago (11). Since LAG1 deletion prolonged the life span of S.cerevisiae by ∼50%, it was named LAG1. LAC1 was then identified as a close homolog of LAG1, and a double deletion of the two genes resulted in lethality (12) or poor-growth (13). LAG1 and LAC1 were subsequently shown to be required for the synthesis of the very long chain (C26) ceramides found in yeast (14). Once the human homolog of LAG1 was cloned, it was shown to be able to complement LAG1 in yeast longevity (11).
TABLE 1. Comparison of human CerS.
| Name | Chromosomal location | Gene size (base pairs) | Protein size (Da) | Tissue mRNA expression profilea | Acyl chain-length specificitya |
|---|---|---|---|---|---|
| CerS1 | 19p12 | 25,837 | 39,536 | Brain, skeletal muscle, testis <4 molecules/ng of total RNA |
C18 |
| CerS2 | 1q21.2 | 9,792 | 44,876 | Kidney, liver 30–40 molecules RNA/ng of total RNA |
C20-C26 |
| CerS3 | 15q26.3 | 144,326 | 46,217 | Testis, skin <20 molecules/ng of total RNA |
C22-C26 |
| CerS4 | 19p13.2 | 53,046 | 46,399 | Low expression levels in all tissues, more in skin, leukocytes, heart, liver <8 molecules/pg of total RNA |
C18-C20 |
| CerS5 | 12q13.3 | 37,565 | 45,752 | Low expression levels in all tissues <2 molecules/ng of total RNA |
C16 |
| CerS6 | 2q24.3 | 318,394 | 44,890 | Low expression levels in all tissues <3 molecules/ng of total RNA |
C14 and C16 |
In 1991 a mammalian gene, upstream of growth and differentiation factor-1 (UOG-1), was discovered while screening for transforming growth factor-β family members, and was found to be expressed as part of a bicistronic RNA together with growth/differentiation factor-1 (gdf1) (15). Replica plating demonstrated that it was able to functionally complement the LAG1 and LAC1 double deletion in yeast (12). However, it was not until 2002 that UOG-1 was defined as the first mammalian CerS, when its over-expression resulted in increased ceramide synthesis in mammalian cells; remarkably, the ceramides produced by UOG-1 only contained one kind of fatty acid, namely stearic acid (C18) (8). Subsequent bioinfomatics analyses (12,16,17) revealed additional mammalian Lag homologs, originally characterized as translocating chain-associating membrane protein homologs (TRH) (8,10), with the ceramides synthesized by TRH1 shown to contain stearic (C18) and arachidic (C20) acids, whereas ceramide synthesized by TRH4-overexpressing cells were preferentially enriched in palmitic acid (C16) (10). Six mammalian homologs are now known (Table 1) with each using a relatively restricted sub-set of acyl CoAs for ceramide synthesis. The CerS can be divided up into those with homology to fungal Lag1p homologs and to UOG1/CerS1, or to those with homology to a subfamily (CerS2-6) containing a homeobox-like domain (17).
Ceramide Synthases: Common Features
Identification of the active site
Little work has been done on structure-function characterization of the CerS. However, the importance of the Lag1p motif in CerS activity has been demonstrated by site-directed mutagenesis (18). Within this motif, two conserved histidine residues appear to play a key role in catalysis and/or substrate binding (16), since their mutation in mammalian CerS1 (18) or in yeast Lag1 adversly affects catalytic activity (19), although no direct proof of their role in catalysis has been provided.
Reaction mechanism
As discussed above, the most prominent features of the CerS is their use of a restricted sub-set of acyl CoAs for N-acylation. Thus, each CerS shows a similar Km towards sphinganine (20), even when different CoAs are used; for instance, CerS4, which can use either C18- or C20-CoA (10), has a similar Km towards sphinganine of ∼2 μM irrespective of the fatty acid (20), suggesting that the sphinganine binding site is similar between the different CerS. Nothing is known about the binding sites for acyl CoA or for sphinganine, but a recent study using the sphinganine analog, FTY720, gave some clues about the mode of substrate binding (21). Surprisingly, FTY720 inhibited CerS activity by non-competitive inhibition towards acyl CoA and uncompetitive inhibition towards sphinganine. Uncompetitive inhibitors bind to enzymes only after formation of an E-S complex, with this type of inhibition most commonly encountered in multi-substrate reactions where the inhibitor is competitive with respect to one substrate but uncompetitive with respect to another. Thus, sphinganine appears to first bind to CerS forming an E-S (CerS-sphinganine) complex, and only after formation of this complex, can FTY720 bind. Alternatively, there may be two sphinganine binding sites which act allosterically with respect to one another, or CerS themselves may form dimers which interact allosterically.
Structural features
CerS contain a unique C-terminal domain, the TLC domain, named after the three protein families in which it was originally found (Tram, Lag and CLN8) (16). Mammalian and yeast CerS have anywhere between 5-8 transmembrane domains, depending on the alogorithm used to identify putative transmembrane domains (19,22). Mammalian CerS also contain a putative glycosylation site at the N-terminus (at NX(S/T)) (22), but it is not known whether the topology of this residue is consistent with it being actively glycosylated. All of the mammalian CerS, apart from CerS1, have a Hox-like domain. The Hox domain is derived from homeobox proteins, sequence-specific transcription factors important in development (23). However, the first 15 amino acids of this domain are missing in CerS, as is the key residue (N51) involved in DNA binding; therefore it is unlikely that the Hox-like domain functions as a genuine transcription factor. The majority of the Hox-like domain is not required for catalytic activity (24), although a highly conserved region of 12 amino acids at the C-terminus of the Hox-like domain is essential for CerS5 and CerS6 activity, with two positively charged conserved residues (K134, K140) in this region critical for catalytic activity (24).
Intracellular localization
The subcellular location of the CerS proteins is the ER (8,10,19,22,25-27), although it should be emphasized that most of these studies have been performed with ectopically-expressed proteins since specific antibodies are not readily available to most endogenous CerS proteins (with the exception of CerS6). CerS has been partially purified from a mitochondria-enriched fraction (28), the major cellular site of apoptosis regulation (29), but as with all subcellular fractionation, careful analysis of the extent of contaminating membranes must be performed. CerS have also shown to be associated with mitochondria upon mitochondrial injury in cerebral ischemia/reperfusion (30).
Regulation
Little is known about how and whether CerS are regulated post-translationally. There is some evidence supporting a role for phosphorylation of some of the CerS; thus, activation of protein kinase C (PKC) increases de novo ceramide synthesis (31), which was attributed to up-regulation of CerS5 activity (32). Data from high performance mass spectrometry support a suggestion that mouse liver CerS2 and CerS5 might be phosphorylated (33).
Ceramide Synthases: Distinctive Features
CerS1
CerS1, which synthesizes C18-ceramide (8), is structurally and functionally distinctive from the other CerS and is found on an entirely separate branch of the phylogenetic tree (7). Moreover, it is the only CerS that does not contain a Hox-like domain (17,22). Since CerS1 was the first mammalian CerS to be identified, the number of studies on CerS1 is relatively high compared to other CerS.
Non-catalytic subunits
The lack of a hox-like domain in CerS1 is perhaps not surprising since CerS1 is most closely related to yeast Lag1 (7), and yeast CerS do not contain a Hox-like domain. However, yeast Lag1p requires another protein, Lip1, for its activity (34). Although CerS1 has not been purified to homogeneity, there is currently no data implying that it requires an additional subunit for its activity. CerS5, in contrast, has been purified, but no additional subunits were co-purified after immunoprecipitation (35). These data suggest a unique role for Lip1p in yeast that is not required in mammalian cells.
Tissue distribution
Since specific antibodies for use in immunofluorescence studies are not available for most of the the endogenous CerS proteins, studies on the tissue distribution of these enzymes (Fig. 3) are limited to analyses of their mRNA levels. CerS1 was originally found to be mainly expressed in brain and at low levels in skeletal muscle and testis (22), which was later confirmed by real-time PCR (10,27,36). Analyses of CerS1 distribution in brain tissue by in situ-hybridization revealed that CerS1 was expressed in most neurons but only at low or undetectable amounts in cells of the white matter (37). CerS1 was slightly up-regulated postnatally, which may reflect the synthesis of neuronal plasma membranes. The high level of CerS1 in neurons is consistent with the acyl chain composition of neuronal SLs, which comprises mainly C18-fatty acids (37). Whether CerS1 is also involved in inherited metabolic disorders, such as ceroid lipofuscinoses (38), requires further study.
Fig. 3. CerS mRNA expression in human tissues.
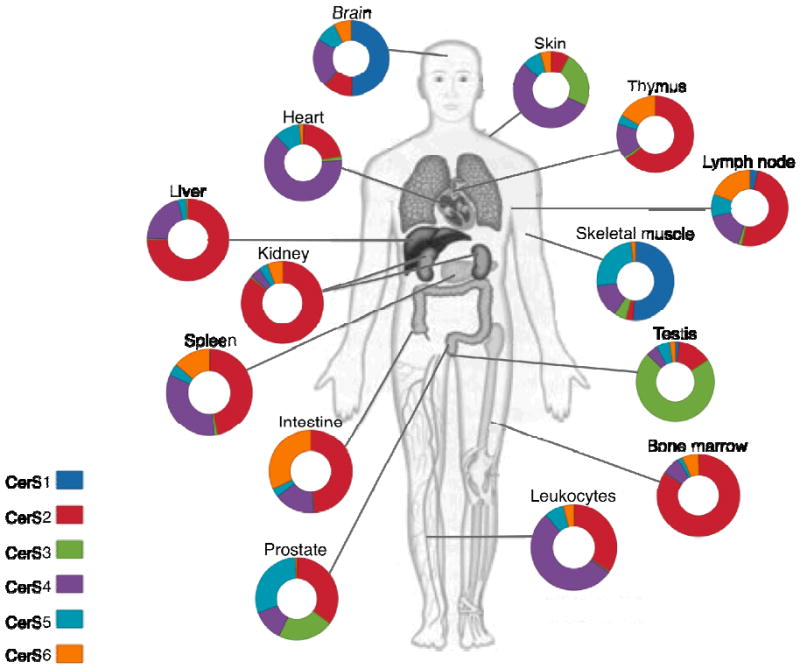
The distibution is shown as a pie chart for each organ, with each CerS color-coded as in the legend. The data is based Ref. (27).
Cancer and chemotherapeutic drugs
A role for CerS1 in the regulation of the growth of head and neck squamous cell carcinoma (HNSCC) has been suggested, based on data demonstrating the down-regulation of C18-ceramide in the majority of HNSCC tissues compared to adjacent normal tissue, which was not detected in nonsquamous tumors (39). Moreover, the balance between levels of C16-and C18-ceramide may be related to the development of HNSCC tumor tissue (39,40), which is also associated with the state of clinical progression of HNSCC (41).
CerS1 also has a unique role in regulating sensitivity to chemotherapeutic drugs. CerS1 sensitized human embryonic kidney cells to a wide range of drugs including cisplatin, carboplatin, doxorubicin and vincristine, whereas CerS5 only sensitized cells to doxorubicin and vincristine and CerS4 did not affect sensitivity to any of the drugs (42). Likewise, CerS1 plays an important role in the regulation of cell death in HNSCC cells via gemcitabine/doxorubicin-induced caspase activation (43). Treatment of cultured chronic myeloid leukemia cells with imatinib (a drug used to treat certain types of cancer) mediates the generation of C18-ceramide via CerS1, which is involved in imatinib-induced apoptosis in sensitive K562 cells; interestingly, overexpression of sphingosine kinase 1 (SK1) in resistant-cells prevented cell death in response to imatinib by altering the balance between C18-ceramide and sphingosine 1-phosphate (S1P) (44).
Degradation and translocation
Upon application of different stresses, including drugs used in cancer chemotherapy, and ultra violet radition, CerS1 turns-over rapidly by ubiquitination and proteasomal degradation, suggesting that it has a short half life (45). CerS1 is also phosphorylated in vivo, and activation of protein kinase C (PKC) increases its phosphorylation (45), with turnover regulated by the opposing functions of p38 MAP kinase and PKC, such that p38 MAP kinase acts as a positive regulator of turnover while PKC is a negative regulator. Moreover, some drugs or stresses induce a novel and specific translocation of CerS1 from the ER to the Golgi apparatus (46). These stresses cause a specific cleavage of CerS1; the cleavage is dependent on the action of the proteasome, as inhibition of proteasome function inhibits stress-induced CerS1 translocation, indicating that proteolytic cleavage precedes translocation. Modulation of PKC showed that it plays a central role in regulating CerS1 translocation. The study suggests that diverse stresses initiate responses through different signaling pathways, which ultimately converge to regulate CerS1 localization (46). Thus, CerS1 is uniquely regulated with respect to the other CerS enzymes.
CerS2
CerS2 [also known as LASS2, TRH3 (10,47), SP260 (48) or LAG1Hs-2 (12)] was first cloned in 2001 (49). It contains the conserved TLC domain and the Hox-like domain, and synthesizes very long acyl chain ceramides (Table 1).
Tissue distribution
Early data obtained by Northen blot analysis demonstrated that CerS2 is much more widely expressed than CerS1 and is found in at least 12 human tissues, with high expression in kidney and liver, moderate expression in brain, heart, skeletal muscle, placenta and lung, and low expression in colon, thymus, spleen, small intestine and peripheral blood leukocytes (47). More recently, real time PCR analysis provided similar but quantitative data, demonstrating that CerS2 mRNA is ubiquitously expressed and is highly abundant in many tissues, especially in liver and kidney (27). In mouse brain, CerS2 is predominantly expressed in oligodendrocytes and Schwann cells and is up-regulated during the period of active myelination, suggesting that CerS2 might be responsible for the synthesis of myelin SLs (37).
Genomic organization
CerS2 has a compact gene size, a low number of introns, short 5′- and 3′-UTRs, with a high percentage of surrounding chromosomal sequences containing CpG and Alu elements, and contains a low percentage of LINE-1s. Further, CerS2 is located within chromosomal regions that are replicated early within the cell cycle (50). CerS2 consists of features typical of a ‘housekeeping’ gene, although no other CerS genes exhibit these characteristics (27).
Tumor Metastasis Suppressor Gene-1 (TMSG-1)
A truncated form of CerS2, which lacks 150 residues at the N-terminus, is also highly expressed in kidney and in liver, and was shown to inhibit colony formation of human hepatoma cells in vitro (49). Using two-hybrid screens, a number of interactions were detected between the truncated form of CerS2 and several membrane-associated receptors or transporters, but the relevance of these interactions for full length CerS2 is not clear. Interestingly, mRNA levels of the truncated form of CerS2 were extremely low in a metastatic cell line compared to a non-metastatic cell line, prompting the naming of this protein as Tumor Metastasis Suppressor Gene-1 (TMSG-1) (51). The relationship between TMSG-1 and full length CerS2 is not known, but it should also be noted that most of the CerS exists as splice variants which code different isoforms. Understanding the role of these isoforms and their roles in regulating expression of the full-length proteins (7) remains a challenge for the future. However, in one (9) of two studies (9,52), CerS2 was able to restore SL biosynthesis in a ΔLAG1ΔLAC1 yeast strain whereas TMSG-1 could not, suggesting that the biological role of TMSG-1 is unrelated to ceramide synthesis.
Regulation
The activity of CerS2 can be regulated by S1P, via interaction of S1P with two residues that are part of an S1P receptor-like motif found only in CerS2 (27). Mutation of each of these residues alone had no effect on S1P inhibition of CerS2 activity, but mutation of both residues completely abolished the inhibitory effect of S1P on CerS2; interestingly, lysophosphatidic acid, a lipid which has similar structural properties to S1P, had no effect on CerS2 activity (21). The opposing functions that ceramide and S1P play in signaling pathways suggests that this mode of regulation might be of significance in cell physiology, and may provide a molecular explanaton for the so-called ceramide/S1P rheostat (53), which proposes that the balance between S1P and ceramide determines whether a cell survives or dies. Another possible role of CerS2 is in regulation of ER stress. CerS2 knock-down resulted in the induction of autophagy and activation of the unfolded protein response (UPR) (54). Interestingly, when CerS2 was knocked-down using siRNA, other CerS showed increases in RNA but not protein levels (54).
Cancer and disease
Two recent studies have implicated CerS2 in breast cancer. In the first (55), total ceramide levels were increased in malignant tumor tissue, with a noticeable increase in levels of C16-, C24:1- and C24:0-ceramide, which correlated with an increase in mRNA levels for CerS2, CerS4 and CerS6. In the second study (56), CerS2 and CerS6 mRNA levels were significantly elevated in cancer tissue compared to paired normal tissue, with approximately half of the individuals showing elevated CerS2 and CerS6 mRNA. A significant correlation was found between CerS2 and CerS6 expression, and between CerS4 and CerS2/CerS6 expression. Together these results suggest an important role for the CerS2 gene in breast cancer etiology or diagnosis.
CerS2 may also be involved in the control of body weight and food intake. Upon administration of leptin (an adipocyte-derived hormone), a decrease in ceramide levels was observed in rat white adipose tissue, as were expression levels of a number of genes in the SL metabolic pathway, including CerS2 and CerS4 (57). In chickens, CerS2 is a T3- (thyroid hormone) depressed gene (58). T3 inhibits secretion of growth hormone (GH) from the pituitary gland (59) thereby inhibiting growth, suggesting a link between CerS2 and liver growth and development via the T3 hormone.
CerS2 KO mice
Recently, a CerS2 null mouse was generated (60). In this mouse, ceramide and downstream sphingolipids were devoid of very long (C22-C24) acyl chains, consistent with the substrate specificity of CerS2. Unexpectedly, C16-ceramide levels were elevated and as a result, total ceramide levels were unaltered. Levels of sphinganine were also significantly elevated, by up to 50-fold. Differences were also observed in the biophysical properties of lipid extracts isolated from liver microsomes, with membranes from CerS2 null mice displaying higher membrane fluidity and showing morphological changes (60). As a result of these changes, CerS2 null mice developed severe non-zonal hepatopathy from about 30 days of age and displayed increased rates of hepatocyte apoptosis and proliferation (61). In older mice, extensive and pronounced hepatocellular anisocytosis was observed with widespread formation of nodules of regenerative hepatocellular hyperplasia. Interestingly, even though CerS2 is found at equally high mRNA levels in kidney and liver, there were no changes in renal function and no pathological changes in the kidney. High throughput analysis of RNA expression in liver revealed up-regulation of genes associated with cell cycle regulation, protein transport, cell-cell interactions and apoptosis, and down-regulation of genes associated with intermediary metabolism, such as lipid and steroid metabolism, adipocyte signaling and amino acid metabolism (61). Thus, inhibition of very long acyl chain ceramide synthesis has a profound effect on hepatocyte physiology and pathophysiology. Pathological changes were also observed in the brain of these mice (62).
In summary, CerS2 and perhaps its splice variants seems to be somewhat unique among the CerS inasmuch as its expression levels changes in a number of disease states. This may be related to the lack of redundancy of CerS enzymes to synthesize very long acyl chain (C22-24) ceramides, which can only be synthesized by CerS2.
CerS3
CerS3 (also known as LASS3 or T3I), which synthesizes C24-ceramides and ceramides with longer acyl chains, is found mainly in skin and testis (36,63). CerS3 is highly expressed in keratinocytes (Table 1) and its expression increases during keratinocyte differentiation (64). CerS3 is able to synthesize ceramides containing α-hydroxy (2-hydroxy) fatty acids, which are abundant in the skin (65,66) where they are involved in maintaining the water permeability barrier function (67). CerS3 is also found at high levels in testis (27) where it is probably involved in sperm formation and androgen production (36). Cers3 mRNA is limited to germ cells, where its levels are up-regulated >700-fold during juvenile testicular maturation. Indeed, the increase in levels of glycosphingolipids containing very long chain (C26-C32) polyunsaturated fatty acid (LC-PUFA), which are essential for spermatogenesis, coincides with a strong elevation of CerS3 mRNA (63). In contrast, CerS3 was undetectable by in situ hybridization in the brain or in sciatic nerve (37). Interestingly, CerS3 is the only CerS for which splice variants have not been reported (7).
CerS4
CerS4 (also known as LASS4 (22) or TRH1 (10)) is perhaps the least studied of the CerS. CerS4 synthesizes ceramide containing C18-22 fatty acids (10), and is expressed at highest levels in skin, leukocytes, heart and liver (27); however, its absolute mRNA expression levels in these tissues are significantly lower than those of other CerS, such as CerS2. Also, unlike CerS1 and CerS5, CerS4 did not affect the sensitivity of cells to chemotherapeutic drugs (42). Interestingly, CerS4 (and CerS2) expression was elevated in the brain of an Alzheimer's disease mouse model (68). These changes are likely to be downstream to other pathological pathways that are altered in Alzheimer's disease, although it should be noted that levels of C20 and C24-ceramide were significantly elevated long before symptoms or a cellular Alzheimer phenotype emerged (69).
CerS5
CerS5 (also known as LASS5 or TRH4 (10)) has been extensively studied, perhaps due to its robust activity and due to its ability to synthesize C16-ceramide, which is often considered to be an important pro-apoptotic ceramide. CerS5 was also the first mammalian CerS to be purified (35). The activity of purified CerS5 was totally dependent on exogenously-added phospholipids. However, no other subunits co-purified with CerS5, demonstrating that mammalian CerS, at least CerS5, do not require additional subunits for their activity, unlike yeast Lag1 which requires Lip1 (14,34,70). Interestingly, when human CerS5 was expressed in a yeast Δlag1Δlac1ΔLip1 mutant (71), it was able to synthesize C16-ceramides and the mutant survived in the absence of the C26-ceramides that are normally found in yeast. This may suggest that the function of Lip1 is somehow related to the synthesis of C26-ceramides by Lag1/Lac1.
Localization
CerS5 is the main CerS detected in lung epithelia. Knock-down studies in lung epithelium using CerS5 siRNA or using fumonisin B1 reduced total CerS activity by 45% or 78%, respectively (72), demonstrating that CerS5 indeed contributes significantly to ceramide synthesis in lung. In brain, CerS5 mRNA is detected in most cells within the gray and white matter (37).
Chemotherapeutic drugs
In a study examining the sensitivity of cells to chemotherapeutic drugs, siRNA to CerS1 prevented gemcitabine/doxorubicin-induced caspase-9 activation, whereas CerS4 or CerS5 siRNA did not have any effect (43). In another study, neither CerS4 nor CerS5 were translocated to the Golgi apparatus upon exposure to chemotherapeutic drugs, although CerS1 did translocate (42); moreover, cells transfected with CerS5 showed an increase in sensitivity to doxorubicin and vincristine, but not to cisplatin or carboplatin.
Apoptosis
De novo ceramide synthesis is an important trigger for Bax activation in hypoxia/reoxygenation. Upon knocking down acid-sphingomyelinase and CerS5 activities in NT-2 cells, Bax localization to mitochondria was attenuated (73). CerS5 expression was also elevated following hypoxia/reoxygenation, suggesting that CerS5 may be important in promoting ceramide up-regulation following hypoxia/reoxygenation, which results in apoptosis.
Regulation
A splice variant of CerS5 (with early termination of translation in exon 10) is expressed in lymphoma and other tumor cells and may be involved in tumor recognition by the immune system (74). Unlike the CerS2 splice variant, TMSG, which is truncated at its N-terminus is therefore unlikely to be active, the CerS5 splice variant results in loss of part of the C-terminus, which is probably not involved in catalytic activity but may rather regulate interaction with other proteins. Similarly, in response to p53 up-regulation, C16-ceramide levels were increased in leukemia and colon cancer cells, as were levels of CerS5 mRNA in the leukemia cells, but not in the colon cancer cells (75). Unfortunately, the promoter region of CerS5 has not been characterized, so the molecular details of how its expression (and the expression of its splice variants) is transcriptionally up-regulated is unknown. Nevertheless, CerS5 looks like a promising candidate as a major player in the regulation of cancer and of cell death pathways.
CerS6
CerS6 (also known as LASS6), which synthesizes C14- and C16-ceramides (36), shows high homology to CerS5; however, much less is known about CerS6 than CerS5. Some studies have suggested that CerS6 is involved in cancer etiology. For instance, a microarray study implied that CerS6 plays a role in cancer differentiation and early embryonic development (76), whereas another study demonstrated that CerS6 expression was regulated in an estrogen receptor dependent manner (77), and CerS6 mRNA levels were elevated in breast cancer tissue (55,56). Upon overexpression of CerS6, drug-induced CD95 activation and tumor cell killing were enhanced, whereas knockdown of CerS6 in colon adenocarcinoma cells suppressed CD95 activation (78). Knocking down CerS6 expression also suppressed CD95 activation in hepatoma, pancreatic, and ovarian cancer cells. Knocking-down CerS6 resulted in a specific decrease in intracellular C16-ceramide, and protected colon adenocarcinoma cells against TRAIL-mediated apoptosis and interfered with translocation of active caspase-3 into the nucleus, whereas increasing CerS6 expression sensitized the cells to TRAIL (79). Down-regulation of CerS6 in HNSCC resulted in the induction of ER stress, while overexpression of CerS6 increased HNSCC tumor development and growth (80). Treatment with an endocannabinoid analogue led to increased levels of C16- C18- C24- and C24:1-ceramides. There was also transcriptional induction of CerS6 and CerS3, effects which were attenuated using a brain cannabinoid receptor antagonist. Simultaneous knocking-down of CerS6 and CerS3 abrogated the endocannabinoid analogue-induced accumulation of C16 and C24 (81).
Concluding Comments
In this review we have attempted to summarize recent progress on the CerS. Although there have been significant advances in our knowledge of the CerS over the past 3 or 4 years, much information is still lacking. How are they regulated, both transcriptionally and also post-translationally? Although the genes are located on different chromosomes, are there any mechanisms for the coordinate regulation of their transcription; this appears likely since many studies show changes in ceramide levels that are synthesized by different CerS. Do the CerS proteins interact with each other, or with other proteins in the ER? Why is CerS1 apparently unique in its ability to be translocated to the Golgi apparatus prior to its degradation? Does CerS2 indeed act as a ‘house-keeping’ gene? Does CerS3 really play an important function in skin permeability? What is the relationship between CerS5 and 6, which have similar substrate specificities but appear to have quite different biological functions? These, and many other questions are likely to be the focus of intense studies in the years ahead, not only to researchers in the SL field, but also to those interested in topics as diverse as cancer biology and degenerative disease of the central nervous system.
Acknowledgments
Work in the Futerman laboratory on the CerS proteins is supported by the Israel Science Foundation (1404/07), the National Institutes of Health (GM076217), the Minerva Foundation and the U.S.-Israel Binational Science Foundation. ML is partially supported by the Center for Absorption in Science, The Israeli Ministry of Absorption, The State of Israel.
Abbreviations
- CerS
ceramide synthase
- ER
endoplasmic reticulum
- Hox
homeobox
- LASS
longevity assurance
- S1P
sphingosine 1-phosphate
- SL
sphingolipid
References
- 1.Pettus BJ, Chalfant CE, Hannun YA. Ceramide in apoptosis: an overview and current perspectives. Biochim Biophys Acta. 2002;1585:114–125. doi: 10.1016/s1388-1981(02)00331-1. [DOI] [PubMed] [Google Scholar]
- 2.Kolesnick R. The therapeutic potential of modulating the ceramide/sphingomyelin pathway. J Clin Invest. 2002;110:3–8. doi: 10.1172/JCI16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Futerman AH, Riezman H. The ins and outs of sphingolipid synthesis. Trends Cell Biol. 2005;15:312–318. doi: 10.1016/j.tcb.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 4.Hanada K. Discovery of the molecular machinery CERT for endoplasmic reticulum-to-Golgi trafficking of ceramide. Mol Cell Biochem. 2006;286:23–31. doi: 10.1007/s11010-005-9044-z. [DOI] [PubMed] [Google Scholar]
- 5.Dolgachev V, Farooqui MS, Kulaeva OI, Tainsky MA, Nagy B, Hanada K, Separovic D. De novo ceramide accumulation due to inhibition of its conversion to complex sphingolipids in apoptotic photosensitized cells. J Biol Chem. 2004;279:23238–23249. doi: 10.1074/jbc.M311974200. [DOI] [PubMed] [Google Scholar]
- 6.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 7.Pewzner-Jung Y, Ben-Dor S, Futerman AH. When do Lasses (longevity assurance genes) become CerS (ceramide synthases)?: Insights into the regulation of ceramide synthesis. J Biol Chem. 2006;281:25001–25005. doi: 10.1074/jbc.R600010200. [DOI] [PubMed] [Google Scholar]
- 8.Venkataraman K, Riebeling C, Bodennec J, Riezman H, Allegood JC, Sullards MC, Merrill AH, Jr, Futerman AH. Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a fumonisin B1-independent manner in mammalian cells. J Biol Chem. 2002;277:35642–35649. doi: 10.1074/jbc.M205211200. [DOI] [PubMed] [Google Scholar]
- 9.Guillas I, Jiang JC, Vionnet C, Roubaty C, Uldry D, Chuard R, Wang J, Jazwinski SM, Conzelmann A. Human homologues of LAG1 reconstitute Acyl-CoA-dependent ceramide synthesis in yeast. J Biol Chem. 2003;278:37083–37091. doi: 10.1074/jbc.M307554200. [DOI] [PubMed] [Google Scholar]
- 10.Riebeling C, Allegood JC, Wang E, Merrill AH, Jr, Futerman AH. Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J Biol Chem. 2003;278:43452–43459. doi: 10.1074/jbc.M307104200. [DOI] [PubMed] [Google Scholar]
- 11.D'Mello NP, Childress AM, Franklin DS, Kale SP, Pinswasdi C, Jazwinski SM. Cloning and characterization of LAG1, a longevity-assurance gene in yeast. J Biol Chem. 1994;269:15451–15459. [PubMed] [Google Scholar]
- 12.Jiang JC, Kirchman PA, Zagulski M, Hunt J, Jazwinski SM. Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 13.Barz WP, Walter P. Two endoplasmic reticulum (ER) membrane proteins that facilitate ER-to-Golgi transport of glycosylphosphatidylinositol-anchored proteins. Mol Biol Cell. 1999;10:1043–1059. doi: 10.1091/mbc.10.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guillas I, Kirchman PA, Chuard R, Pfefferli M, Jiang JC, Jazwinski SM, Conzelmann A. C26-CoA-dependent ceramide synthesis of Saccharomyces cerevisiae is operated by Lag1p and Lac1p. EMBO J. 2001;20:2655–2665. doi: 10.1093/emboj/20.11.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SJ. Expression of growth/differentiation factor 1 in the nervous system: conservation of a bicistronic structure. Proc Natl Acad Sci U S A. 1991;88:4250–4254. doi: 10.1073/pnas.88.10.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winter E, Ponting CP. TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem Sci. 2002;27:381–383. doi: 10.1016/s0968-0004(02)02154-0. [DOI] [PubMed] [Google Scholar]
- 17.Venkataraman K, Futerman AH. Do longevity assurance genes containing Hox domains regulate cell development via ceramide synthesis? FEBS Lett. 2002;528:3–4. doi: 10.1016/s0014-5793(02)03248-9. [DOI] [PubMed] [Google Scholar]
- 18.Spassieva S, Seo JG, Jiang JC, Bielawski J, Alvarez-Vasquez F, Jazwinski SM, Hannun YA, Obeid LM. Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J Biol Chem. 2006;281:33931–33938. doi: 10.1074/jbc.M608092200. [DOI] [PubMed] [Google Scholar]
- 19.Kageyama-Yahara N, Riezman H. Transmembrane topology of ceramide synthase in yeast. Biochem J. 2006;398:585–593. doi: 10.1042/BJ20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahiri S, Lee H, Mesicek J, Fuks Z, Haimovitz-Friedman A, Kolesnick RN, Futerman AH. Kinetic characterization of mammalian ceramide synthases: Determination of K(m) values towards sphinganine. FEBS letters. 2007;581:5289–5294. doi: 10.1016/j.febslet.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Lahiri S, Park H, Laviad EL, Lu X, Bittman R, Futerman AH. Ceramide Synthesis Is Modulated by the Sphingosine Analog FTY720 via a Mixture of Uncompetitive and Noncompetitive Inhibition in an Acyl-CoA Chain Length-de pend ent Manner. J Biol Chem. 2009;284:16090–16098. doi: 10.1074/jbc.M807438200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–271. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring WJ, Affolter M, Burglin T. Homeodomain proteins. Annu Rev Biochem. 1994;63:487–526. doi: 10.1146/annurev.bi.63.070194.002415. [DOI] [PubMed] [Google Scholar]
- 24.Mesika A, Ben-Dor S, Laviad EL, Futerman AH. A new functional motif in Hox domain-containing ceramide synthases: identification of a novel region flanking the Hox and TLC domains essential for activity. J Biol Chem. 2007;282:27366–27373. doi: 10.1074/jbc.M703487200. [DOI] [PubMed] [Google Scholar]
- 25.Mandon EC, Ehses I, Rother J, van Echten G, Sandhoff K. Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J Biol Chem. 1992;267:11144–11148. [PubMed] [Google Scholar]
- 26.Hirschberg K, Rodger J, Futerman AH. The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem J. 1993;290(Pt 3):751–757. doi: 10.1042/bj2900751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH, Jr, Futerman AH. Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem. 2008;283:5677–5684. doi: 10.1074/jbc.M707386200. [DOI] [PubMed] [Google Scholar]
- 28.Shimeno H, Soeda S, Sakamoto M, Kouchi T, Kowakame T, Kihara T. Partial purification and characterization of sphingosine N-acyltransferase (ceramide synthase) from bovine liver mitochondrion-rich fraction. Lipids. 1998;33:601–605. doi: 10.1007/s11745-998-0246-2. [DOI] [PubMed] [Google Scholar]
- 29.Siskind LJ. Mitochondrial ceramide and the induction of apoptosis. J Bioenerg Biomembr. 2005;37:143–153. doi: 10.1007/s10863-005-6567-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Novgorodov SA, Chudakova D, Zhu H, Bielawska A, Bielawski J, Obeid LM, Kindy MS, Gudz TI. JNK3 signaling pathway activates ceramide synthase leading to mitochondrial dysfunction. J Biol Chem. 2007;282:25940–25949. doi: 10.1074/jbc.M701812200. [DOI] [PubMed] [Google Scholar]
- 31.Becker KP, Kitatani K, Idkowiak-Baldys J, Bielawski J, Hannun YA. Selective inhibition of juxtanuclear translocation of protein kinase C betaII by a negative feedback mechanism involving ceramide formed from the salvage pathway. J Biol Chem. 2005;280:2606–2612. doi: 10.1074/jbc.M409066200. [DOI] [PubMed] [Google Scholar]
- 32.Kitatani K, Idkowiak-Baldys J, Bielawski J, Taha TA, Jenkins RW, Senkal CE, Ogretmen B, Obeid LM, Hannun YA. Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J Biol Chem. 2006;281:36793–36802. doi: 10.1074/jbc.M608137200. [DOI] [PubMed] [Google Scholar]
- 33.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. Proc Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vallee B, Riezman H. Lip1p: a novel subunit of acyl-CoA ceramide synthase. EMBO J. 2005;24:730–741. doi: 10.1038/sj.emboj.7600562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lahiri S, Futerman AH. LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J Biol Chem. 2005;280:33735–33738. doi: 10.1074/jbc.M506485200. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani Y, Kihara A, Igarashi Y. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem J. 2006;398:531–538. doi: 10.1042/BJ20060379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker I, Wang-Eckhardt L, Yaghootfam A, Gieselmann V, Eckhardt M. Differential expression of (dihydro)ceramide synthases in mouse brain: oligodendrocyte-specific expression of CerS2/Lass2. Histochem Cell Biol. 2008;129:233–241. doi: 10.1007/s00418-007-0344-0. [DOI] [PubMed] [Google Scholar]
- 38.Schulz A, Mousallem T, Venkataramani M, Persaud-Sawin DA, Zucker A, Luberto C, Bielawska A, Bielawski J, Holthuis JC, Jazwinski SM, Kozhaya L, Dbaibo GS, Boustany RM. The CLN9 protein, a regulator of dihydroceramide synthase. J Biol Chem. 2006;281:2784–2794. doi: 10.1074/jbc.M509483200. [DOI] [PubMed] [Google Scholar]
- 39.Koybasi S, Senkal CE, Sundararaj K, Spassieva S, Bielawski J, Osta W, Day TA, Jiang JC, Jazwinski SM, Hannun YA, Obeid LM, Ogretmen B. Defects in cell growth regulation by C18:0-ceramide and longevity assurance gene 1 in human head and neck squamous cell carcinomas. J Biol Chem. 2004;279:44311–44319. doi: 10.1074/jbc.M406920200. [DOI] [PubMed] [Google Scholar]
- 40.Saddoughi SA, Song P, Ogretmen B. Roles of bioactive sphingolipids in cancer biology and therapeutics. Subcell Biochem. 2008;49:413–440. doi: 10.1007/978-1-4020-8831-5_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, Bielawski J, Day TA, Gillespie MB, Sinha D, Norris JS, Hannun YA, Ogretmen B. Clinical relevance of ceramide metabolism in the pathogenesis of human head and neck squamous cell carcinoma (HNSCC): attenuation of C(18)-ceramide in HNSCC tumors correlates with lymphovascular invasion and nodal metastasis. Cancer Lett. 2007;256:101–111. doi: 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Min J, Mesika A, Sivaguru M, Van Veldhoven PP, Alexander H, Futerman AH, Alexander S. (Dihydro)ceramide synthase 1 regulated sensitivity to cisplatin is associated with the activation of p38 mitogen-activated protein kinase and is abrogated by sphingosine kinase 1. Mol Cancer Res. 2007;5:801–812. doi: 10.1158/1541-7786.MCR-07-0100. [DOI] [PubMed] [Google Scholar]
- 43.Senkal CE, Ponnusamy S, Rossi MJ, Bialewski J, Sinha D, Jiang JC, Jazwinski SM, Hannun YA, Ogretmen B. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol Cancer Ther. 2007;6:712–722. doi: 10.1158/1535-7163.MCT-06-0558. [DOI] [PubMed] [Google Scholar]
- 44.Baran Y, Salas A, Senkal CE, Gunduz U, Bielawski J, Obeid LM, Ogretmen B. Alterations of ceramide/sphingosine 1-phosphate rheostat involved in the regulation of resistance to imatinib-induced apoptosis in K562 human chronic myeloid leukemia cells. J Biol Chem. 2007;282:10922–10934. doi: 10.1074/jbc.M610157200. [DOI] [PubMed] [Google Scholar]
- 45.Sridevi P, Alexander H, Laviad EL, Pewzner-Jung Y, Hannink M, Futerman AH, Alexander S. Ceramide synthase 1 is regulated by proteasomal mediated turnover. Biochim Biophys Acta. 2009;1793:1218–1227. doi: 10.1016/j.bbamcr.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sridevi P, Alexander H, Laviad EL, Min J, Mesika A, Hannink M, Futerman AH, Alexender S. Stress-induced ER to Golgi translocation of ceramide synthase 1 is dependent on proteasomal processing. Exp Cell Res. 2009 doi: 10.1016/j.yexcr.2009.1009.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai XF, Tao Z, Yan ZQ, Yang SL, Gong Y. Molecular cloning, characterisation and tissue-specific expression of human LAG3, a member of the novel Lag1 protein family. DNA Seq. 2003;14:79–86. doi: 10.1080/1042517021000041831. [DOI] [PubMed] [Google Scholar]
- 48.He XH, Tan WX, Wang JR, Hu J, Zhu HX, Wan DF, Gu JR. Chromosome localization and genomic organization of five novel human genes related to cell growth control. Yi Chuan. 2002;24:111–116. [PubMed] [Google Scholar]
- 49.Pan H, Qin WX, Huo KK, Wan DF, Yu Y, Xu ZG, Hu QD, Gu KT, Zhou XM, Jiang HQ, Zhang PP, Huang Y, Li YY, Gu JR. Cloning, mapping, and characterization of a human homologue of the yeast longevity assurance gene LAG1. Genomics. 2001;77:58–64. doi: 10.1006/geno.2001.6614. [DOI] [PubMed] [Google Scholar]
- 50.Farkash-Amar S, Lipson D, Polten A, Goren A, Helmstetter C, Yakhini Z, Simon I. Global organization of replication time zones of the mouse genome. Genome Res. 2008;18:1562–1570. doi: 10.1101/gr.079566.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma C, Ning J, You J, Liu L, Wang J, Cui X, Wu B, Zheng J. Primary functional identification of gene TMSG-1. Sci China C Life Sci. 2003;46:641–650. doi: 10.1360/02yc0159. [DOI] [PubMed] [Google Scholar]
- 52.Yu Y, Lu H, Pan H, Ma JH, Ding ZJ, Li YY. Expression of LASS2 controlled by LAG1 or ADH1 promoters cannot functionally complement Lag1p. Microbiol Res. 2006;161:203–211. doi: 10.1016/j.micres.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Mandala SM, Thornton R, Tu Z, Kurtz MB, Nickels J, Broach J, Menzeleev R, Spiegel S. Sphingoid base 1-phosphate phosphatase: a key regulator of sphingolipid metabolism and stress response. Proc Natl Acad Sci U S A. 1998;95:150–155. doi: 10.1073/pnas.95.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spassieva SD, Mullen TD, Townsend DM, Obeid LM. isruption of ceramide synthesis by CerS2 down-regulation leads to autophagy and the unfolded protein response. Biochem J. 2009 doi: 10.1042/BJ20090699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schiffmann S, Sandner J, Birod K, Wobst I, Angioni C, Ruckhaberle E, Kaufmann M, Ackermann H, Lotsch J, Schmidt H, Geisslinger G, Grosch S. Ceramide synthases and ceramide levels are increased in breast cancer tissue. Carcinogenesis. 2009;30:745–752. doi: 10.1093/carcin/bgp061. [DOI] [PubMed] [Google Scholar]
- 56.Erez-Roman R, Pienik R, Futerman AH. Increased ceramide synthase 2 and 6 mRNA levels in breast cancer tissues and correlation with sphingosine kinase expression. Biochem Biophys Res Commun. 2009 doi: 10.1016/j.bbrc.2009.1011.1035. [DOI] [PubMed] [Google Scholar]
- 57.Bonzon-Kulichenko E, Schwudke D, Gallardo N, Molto E, Fernandez-Agullo T, Shevchenko A, Andres A. Central leptin regulates total ceramide content and sterol regulatory element binding protein-1C proteolytic maturation in rat white adipose tissue. Endocrinology. 2009;150:169–178. doi: 10.1210/en.2008-0505. [DOI] [PubMed] [Google Scholar]
- 58.Wang X, Carre W, Saxton AM, Cogburn LA. Manipulation of thyroid status and/or GH injection alters hepatic gene expression in the juvenile chicken. Cytogenet Genome Res. 2007;117:174–188. doi: 10.1159/000103178. [DOI] [PubMed] [Google Scholar]
- 59.Harvey S. Thyroid hormones inhibit growth hormone secretion in domestic fowl (Gallus domesticus) J Endocrinol. 1983;96:329–334. doi: 10.1677/joe.0.0960329. [DOI] [PubMed] [Google Scholar]
- 60.Pewzner-Jung Y, Park H, Laviad EL, Silva LC, Lahiri S, Stiban J, Erez-Roman R, Brugger B, Sachsenheimer T, Wieland FT, Prieto M, Merrill AH, Futerman AH. A critical role for ceramide synthase 2 in liver homeostasis. I. Alterations in lipid metabolic pathways. J Biol Chem. 2010 doi: 10.1074/jbc.M1109.077594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pewzner-Jung Y, Brenner O, Braunn S, Laviad EL, Ben -Dor S, Feldmesser E, Horn-Saban S, Amann-Zalcenstein D, Raanan C, Berkutzki T, Erez-Roman R, Ben-David O, Levy M, Holzman D, Park H, Nyska A, Merrill AH, Futerman AH. A critical role for ceramide synthase 2 in liver homeostasis. II. Insights into molecular changes leading to hepatopathy. J Biol Chem. 2010 doi: 10.1074/jbc.M1109.077610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, Gieselmann V, Sandhoff K, Willecke K. Adult Ceramide Synthase 2 (Cers2) deficient mice exhibit myelin sheath defects, cerebellar degeneration and hepatocarcinomas. J Biol Chem. 2009;284:33549–33560. doi: 10.1074/jbc.M109.031971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rabionet M, van der Spoel AC, Chuang CC, von Tumpling-Radosta B, Litjens M, Bouwmeester D, Hellbusch CC, Korner C, Wiegandt H, Gorgas K, Platt FM, Grone HJ, Sandhoff R. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J Biol Chem. 2008;283:13357–13369. doi: 10.1074/jbc.M800870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mizutani Y, Kihara A, Chiba H, Tojo H, Igarashi Y. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J Lipid Res. 2008;49:2356–2364. doi: 10.1194/jlr.M800158-JLR200. [DOI] [PubMed] [Google Scholar]
- 65.Han X, Cheng H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J Lipid Res. 2005;46:163–175. doi: 10.1194/jlr.D400022-JLR200. [DOI] [PubMed] [Google Scholar]
- 66.Ponec M, Weerheim A, Kempenaar J, Mulder A, Gooris GS, Bouwstra J, Mommaas AM. The formation of competent barrier lipids in reconstructed human epidermis requires the presence of vitamin C. J Invest Dermatol. 1997;109:348–355. doi: 10.1111/1523-1747.ep12336024. [DOI] [PubMed] [Google Scholar]
- 67.Coderch L, Lopez O, de la Maza A, Parra JL. Ceramides and skin function. Am J Clin Dermatol. 2003;4:107–129. doi: 10.2165/00128071-200304020-00004. [DOI] [PubMed] [Google Scholar]
- 68.Wang G, Silva J, Dasgupta S, Bieberich E. Long-chain ceramide is elevated in presenilin 1 (PS1M146V) mouse brain and induces apoptosis in PS1 astrocytes. Glia. 2008;56:449–456. doi: 10.1002/glia.20626. [DOI] [PubMed] [Google Scholar]
- 69.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc Natl Acad Sci U S A. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schorling S, Vallee B, Barz WP, Riezman H, Oesterhelt D. Lag1p and Lac1p are essential for the Acyl-CoA-dependent ceramide synthase reaction in Saccharomyces cerevisae. Mol Biol Cell. 2001;12:3417–3427. doi: 10.1091/mbc.12.11.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cerantola V, Vionnet C, Aebischer OF, Jenny T, Knudsen J, Conzelmann A. Yeast sphingolipids do not need to contain very long chain fatty acids. Biochem J. 2007;401:205–216. doi: 10.1042/BJ20061128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xu Z, Zhou J, McCoy DM, Mallampalli RK. LASS5 is the predominant ceramide synthase isoform involved in de novo sphingolipid synthesis in lung epithelia. J Lipid Res. 2005;46:1229–1238. doi: 10.1194/jlr.M500001-JLR200. [DOI] [PubMed] [Google Scholar]
- 73.Jin J, Hou Q, Mullen TD, Zeidan YH, Bielawski J, Kraveka JM, Bielawska A, Obeid LM, Hannun YA, Hsu YT. Ceramide generated by sphingomyelin hydrolysis and the salvage pathway is involved in hypoxia/reoxygenation-induced Bax redistribution to mitochondria in NT-2 cells. J Biol Chem. 2008;283:26509–26517. doi: 10.1074/jbc.M801597200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Hall T, Wolpert EZ, van Veelen P, Laban S, van der Veer M, Roseboom M, Bres S, Grufman P, de Ru A, Meiring H, de Jong A, Franken K, Teixeira A, Valentijn R, Drijfhout JW, Koning F, Camps M, Ossendorp F, Karre K, Ljunggren HG, Melief CJ, Offringa R. Selective cytotoxic T-lymphocyte targeting of tumor immune escape variants. Nat Med. 2006;12:417–424. doi: 10.1038/nm1381. [DOI] [PubMed] [Google Scholar]
- 75.Panjarian S, Kozhaya L, Arayssi S, Yehia M, Bielawski J, Bielawska A, Usta J, Hannun YA, Obeid LM, Dbaibo GS. De novo N-palmitoylsphingosine synthesis is the major biochemical mechanism of ceramide accumulation following p53 up-regulation. Prostaglandins Other Lipid Mediat. 2008;86:41–48. doi: 10.1016/j.prostaglandins.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 76.Weinmann A, Galle PR, Teufel A. LASS6, an additional member of the longevity assurance gene family. Int J Mol Med. 2005;16:905–910. [PubMed] [Google Scholar]
- 77.Ruckhaberle E, Rody A, Engels K, Gaetje R, von Minckwitz G, Schiffmann S, Grosch S, Geisslinger G, Holtrich U, Karn T, Kaufmann M. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res Treat. 2008;112:41–52. doi: 10.1007/s10549-007-9836-9. [DOI] [PubMed] [Google Scholar]
- 78.Walker T, Mitchell C, Park MA, Yacoub A, Graf M, Rahmani M, Houghton PJ, Voelkel-Johnson C, Grant S, Dent P. Sorafenib and vorinostat kill colon cancer cells by CD95-dependent and -independent mechanisms. Mol Pharmacol. 2009;76:342–355. doi: 10.1124/mol.109.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.White-Gilbertson S, Mullen T, Senkal C, Lu P, Ogretmen B, Obeid L, Voelkel-Johnson C. Ceramide synthase 6 modulates TRAIL sensitivity and nuclear translocation of active caspase-3 in colon cancer cells. Oncogene. 2009;28:1132–1141. doi: 10.1038/onc.2008.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. Antiapoptotic roles of ceramide-synthase-6-generated C16-ceramide via selective regulation of the ATF6/CHOP arm of ER-stress-response pathways. FASEB J. 2009 doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gustafsson K, Sander B, Bielawski J, Hannun YA, Flygare J. Potentiation of cannabinoid-induced cytotoxicity in mantle cell lymphoma through modulation of ceramide metabolism. Mol Cancer Res. 2009;7:1086–1098. doi: 10.1158/1541-7786.MCR-08-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]


