Abstract
Background
Recent studies in mice have established that an endothelial cell protein, GPIHBP1, is essential for the lipolytic processing of triglyceride-rich lipoproteins.
Methods and Results
We report the discovery of a homozygous missense mutation in GPIHBP1 in a young boy with severe chylomicronemia. The mutation, p.C65Y, replaces a conserved cysteine in the GPIHBP1’s Ly6 domain with a tyrosine and is predicted to perturb protein structure by interfering with the formation of a disulfide bond. Studies with transfected CHO cells showed that GPIHBP1-C65Y reaches the cell surface but has lost the ability to bind LPL. When the GPIHBP1-C65Y homozygote was given an intravenous bolus of heparin, only trace amounts of LPL entered the plasma. We also observed very low levels of LPL in the postheparin plasma of a chylomicronemic subject who was homozygous for a different GPIHBP1 mutation (p.Q115P). When the GPIHBP1-Q115P homozygote was given a 6-h infusion of heparin, significant amounts of LPL appeared in the plasma, resulting in a fall in the plasma triglyceride levels from 1780 mg/dl to 120 mg/dl.
Conclusions
We identified a novel GPIHBP1 missense mutation (p.C65Y) associated with defective LPL binding in a young boy with severe chylomicronemia. We also show that homozygosity for the C65Y or Q115P mutations is associated with low levels of LPL in the postheparin plasma, demonstrating that GPIHBP1 is important for plasma triglyceride metabolism in humans.
Keywords: lipoprotein lipase, GPIHBP1, triglycerides
Introduction
Lipoprotein lipase (LPL) is essential for the lipolytic processing of triglyceride-rich lipoproteins. Partial deficiency of LPL leads to mild–moderate hypertriglyceridemia while a complete deficiency of LPL results in severe hypertriglyceridemia (chylomicronemia).1 LPL acts within capillaries, and for many years, LPL was thought to be bound to the surface of endothelial cells by interacting with heparan sulfate proteoglycans (HSPGs).2 Recently, GPIHBP1 (glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1) was identified as an endothelial cell player in lipolysis.3–5 Mice lacking GPIHBP1 (Gpihbp1−/−) display severe chylomicronemia, even on a low-fat diet.4 GPIHBP1 binds LPL and chylomicrons and is found on the surface of capillary endothelial cells in tissues where the lipolytic processing of lipoproteins occurs (e.g., adipose tissue, skeletal muscle and heart).4 Based on these findings, GPIHBP1 was proposed to be the “platform for lipolysis” along the luminal surface of capillaries.
During the past year, evidence for the involvement of GPIHBP1 in lipolysis has accumulated. Weinstein et al.6 showed that LPL release into the plasma after an injection of heparin is abnormal in Gpihbp1−/− mice. Following an intravenous injection of ~50 U of heparin into Gpihbp1−/− mice, the total amount of LPL entering the plasma was virtually normal, but the LPL entered the plasma with delayed kinetics. This observation led to speculation that the intravascular pool of LPL might be abnormally low in the setting of GPIHBP1 deficiency. Interestingly, the LPL that entered the plasma of Gpihbp1−/− mice after multiple intravenous injections of heparin was enzymatically active and quickly lowered plasma triglyceride levels. More recently, Sonnenburg et al.7 made two other important observations—that LPL is stabilized when it is bound to GPIHBP1 and that it is more resistant to inactivation by angiopoetin-like protein 4.
Beigneux, Franssen, and coworkers8 provided the first evidence that GPIHBP1 is important for the processing of triglyceride-rich lipoproteins in humans. They identified a homozygous missense mutation in GPIHBP1 (p.Q115P) in a young man with lifelong chylomicronemia. Using several different cell culture-based assays, they showed that this glutamine-to-proline substitution nearly abolished the ability of the GPIHBP1 to bind LPL.
In the current studies, we have continued to investigate the importance of GPIHBP1 for triglyceride metabolism in humans. We show that homozygosity for a GPIHBP1 missense mutation, p.C65Y, causes chylomicronemia. Like the Q115P mutation, the C65Y mutation abolishes the ability of GPIHBP1 to bind LPL. Additional studies revealed that the postheparin plasma LPL levels are abnormally low in humans with functionally defective GPIHBP1 proteins.
Methods
Subjects
A three-year-old boy living in the United Arab Emirates was diagnosed with chylomicronemia. The exons of LPL, APOA5, APOC2, LMF1, and GPIHBP1 were amplified and sequenced. Studies on the patient’s family were also performed. Also included in our studies was a young man with chylomicronemia who was homozygous for the Q115P mutation in GPIHBP1.8 Four healthy normolipidemic men and women were included as control subjects for the postheparin LPL study. Studies were approved by the Committees on Human Research at AMC, UCLA, and Tawam Hospital. All subjects gave informed consent.
Genomic DNA Analyses
Genomic DNA was prepared from blood leukocytes. The exons of GPIHBP1, LPL, APOA5, and APOC2 (along with ~50 bp of flanking introns) were enzymatically amplified as described.8 To facilitate DNA sequencing, an M13 tail was added to each primer (forward: 5′-GTTGTAAAACGACGGCCACT-3′; reverse: 5′-CACAGGAAACAGCTATGACC-3′). All amplicons were sequenced in both directions.
Biochemical Measurements
Blood was collected after a 12-h fast and placed on ice. Plasma was isolated by centrifugation and stored at −80° C. Total plasma cholesterol, triglycerides, high-density lipoprotein (HDL) cholesterol, and low-density lipoprotein (LDL) cholesterol levels were measured with commercial kits (Wako, Neuss, Germany and Randox, Crumlin, UK). Plasma apo-B, apo-CII, and apo-CIII levels were measured with commercial assays (Randox). Plasma apo-B48 levels were determined with an ELISA (Shibayagi, Ishihara, Japan). Size-fractionation of plasma lipoproteins was performed by fast protein liquid chromatography (FPLC); online triglyceride measurements were obtained with a commercial assay (Biomerieux, Dorval, Canada).8, 9 Prior to the FPLC fractionation, the most buoyant lipoproteins in the proband’s plasma were removed by centrifugation at 10,000 rpm for 10 min at 4°C (to prevent clogging of the column). Plasma LPL levels were measured after an intravenous injection of heparin (MW 6500 Da, Leo Pharma, Breda, the Netherlands; 50 IU/kg body weight). Blood was collected into heparin-lithium tubes at baseline and 1, 2, 3, 6, 9, 12, 15, and 18 minutes after heparin and put on ice. LPL and hepatic lipase (HL) activity levels were measured as previously described.10 HL activity was measured after inhibiting LPL activity with a monoclonal antibody against human LPL (5D2; a gift from Dr. John Brunzell, University of Washington, Seattle, USA) for 2 h at 4°C. 1 mU is equivalent to 1 nmol fatty acid released per minute. Plasma LPL and HL mass levels were measured with an ELISA (LPL: Daiichi, Tokyo, Japan; HL, as described by Bensadoun et al.11). Heparin infusion studies were performed as described earlier with a bolus injection of 2280 U heparin/m2 followed by a continuous infusion of heparin (1984 U/m2/h).12 After collecting blood samples, tetrahydrolipstatin was added to block lipolysis, and LPL mass and triglycerides were measured.
GPIHBP1 Constructs and Cell Transfections
A human GPIHBP1 expression vector containing an internal S-protein tag, as well as mutant GPIHBP1 expression vectors with Q115P and G56R mutations have been described previously.4, 8, 13 In this study, we used the QuickChange kit (Stratagene, La Jolla, Ca, USA) to introduce the C65Y mutation into the human GPIHBP1 expression vector. Transient transfections of CHO-pgs-745 cells14 were performed by electroporation with the Nucleofector II apparatus (Lonza, Basel, Switzerland).
Western Blots
Proteins were size-fractionated on 4–12% Bis-Tris SDS-polyacrylamide gels and electrophoretically transferred to nitrocellulose membranes. The antibody dilutions for western blots were 1:1,000 for a goat polyclonal antibody against the S-protein tag (Abcam, Cambridge, MA, USA); 1:100 for a mouse monoclonal antibody against human GPIHBP1; 1:200 for a mouse monoclonal antibody against the V5 tag (Invitrogen, Carlsbad, CA, USA); 1:500 for a rabbit polyclonal antibody against β-actin (Abcam); 1:5000 for an IRdye800-conjugated donkey anti-goat IgG (Li-Cor; Lincoln, NE, USA); 1:2000 for an IRdye680-conjugated donkey anti-rabbit IgG (Li-Cor); and 1:500 for an IRdye800-conjugated donkey anti-mouse IgG (Li-Cor). Antibody binding was detected with an Odyssey infrared scanner (Li-Cor).
Assessing the Ability of GPIHBP1 to Reach the Cell Surface
To assess GPIHBP1 expression at the cell surface, we used three approaches. First, fluorescence microscopy was used to detect GPIHBP1 at the surface of CHO pgsA-745 cells that had been transfected with wild-type and mutant GPIHBP1 constructs. For these studies, CHO pgsA-745 cells were electroporated with 2.0 μg of plasmid DNA and plated on coverslips in 24-well plates. The cells were fixed in 3.0% paraformaldehyde, blocked with blocking buffer (PBS containing 1 mM MgCl2, 1 mM CaCl2, and 10% donkey serum), and incubated with a FITC-conjugated goat antiserum against the S-protein tag (Abcam; 1:400). In some experiments, the cells were permeabilized with 0.2% Triton X-100. After washing, cell nuclei were stained with DAPI. Images were obtained with an Axiovert 200M microscope equipped with a 40× objective, an AxioCam MRm, and an ApoTome, and were processed with the AxioVision 4.2 software (all from Zeiss).
Second, we assessed the release of GPIHBP1 from the surface of transfected cells after they had been incubated with a phosphatidylinositol-specific phospholipase C (PIPLC). CHO pgsA-745 cells (1 × 106 cells) were electroporated with 2.0 μg of plasmid DNA and seeded into duplicate wells of a 24-well plate. The next day, the cells were incubated with PIPLC (5 U/ml) in serum-free medium for 1 h at 37° C. At the end of the incubation, the medium was harvested and cell extracts were collected in RIPA buffer containing complete mini EDTA-free protease inhibitors (Roche).
Third, we used western blotting to assess the amount of GPIHBP1 on the surface of cells relative to the total amount of GPIHBP1 in cell extracts. CHO pgsA-745 cells (2 × 106 cells) were electroporated with 4.0 μg of plasmid DNA and then seeded into four wells of a 24-well plate. 24 h after the electroporation, the cells were incubated with a goat polyclonal antibody against the S-protein tag (1:400 for 2 h at 4° C) in ice-cold PBS containing 1 mM MgCl2 and 1 mM CaCl2 (PBS/Mg/Ca). At the end of the incubation, cells were washed six times in ice-cold PBS/Mg/Ca and cell extracts were prepared. To assess the amount of GPIHBP1 on the cell surface, western blots were performed on cell extracts with an IRdye680-conjugated donkey anti-goat IgG (Li-Cor, 1:800). To assess the total amount of GPIHBP1 in cells, we simultaneously performed a western blot with a mouse monoclonal antibody against GPIHBP1 (1:100).4 The binding of the mouse monoclonal antibody to GPIHBP1 was detected with an IRdye800-conjugated donkey anti-mouse IgG (Li-Cor, 1:2000). The intensity of each band was quantified with an Odyssey infrared scanner (Li-Cor). The ratio of GPIHBP1 on the cell surface (680-nm channel) compared to total GPIHBP1 (800-nm channel) was determined for each GPIHBP1 construct and expressed as a percentage of the ratio observed with wild-type GPIHBP1.
Assessing the Ability of GPIHBP1 to Bind LPL
To assess LPL binding to GPIHBP1, we incubated GPIHBP1-expressing cells with a V5-tagged human LPL. After washing the cells, we used western blots to assess the amount of LPL bound to cells. For these studies, 5 × 106 CHO pgsA-745 cells were electroporated with 5.0 μg of plasmid DNA and seeded into triplicate wells of a 24-well tissue culture plate. 24 h after the electroporation, cells were incubated for 2 h at 4° C with V5-tagged human LPL (400 μl/well). In some wells, heparin (final concentration, 500 U/ml) was included in the incubation medium. At the end of the incubation period, cells were washed six times in ice-cold PBS/Mg/Ca, and the cell extracts were collected in RIPA buffer containing complete mini EDTA-free protease inhibitors (Roche). The amounts of LPL and GPIHBP1 in cell extracts were assessed by western blotting.
Results
A Novel Missense Mutation Involving a Highly Conserved Cysteine in GPIHBP1
A three-year-old boy with chylomicronemia was examined. The patient was noted to have chylomicronemia at one year of age during a bout of pancreatitis. The fasting plasma triglyceride levels were 4005 mg/dl (45 mmol/l). On physical examination, the child was developing normally, but his body weight was less than the 10th percentile for his age. Fundoscopy revealed lipemia retinalis; there was no hepatosplenomegaly; and there were no eruptive xanthomas.
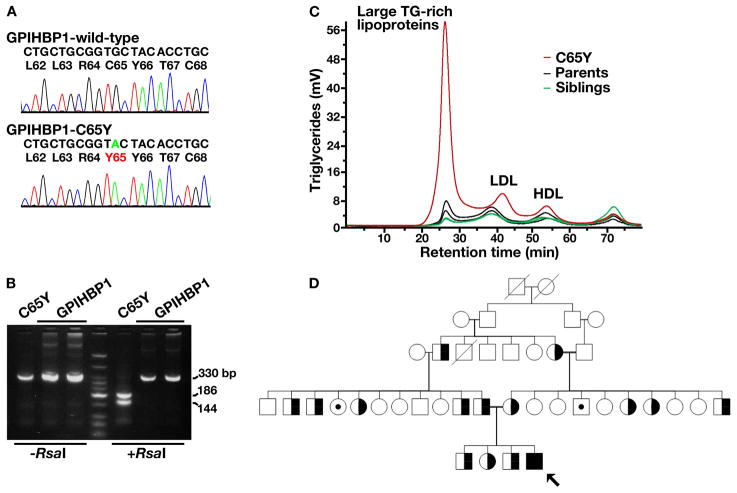
Sequence variations in LPL, APOC2, LMF1, and APOA5 were excluded by sequencing all of the exons of those genes. Sequencing of the coding regions of GPIHBP1 led to the identification of a homozygous G to A transversion in exon 3 (c.194G>A), a mutation that changed a conserved cysteine at residue 65 to a tyrosine (p.C65Y; Figure 1A). The presence of this mutation was confirmed by restriction endonuclease digestion of a DNA fragment amplified from the genomic DNA of the C65Y homozygote (Figure 1B).
Figure 1. Identification of a homozygous mutation in GPIHBP1 (p.C65Y) in a young boy with chylomicronemia.
A. DNA sequence of exon 3 of GPIHBP1 from a normolipidemic control subject (wild-type) and the p.C65Y homozygote. Nucleotide and amino acid sequences are shown above each chromatogram. The arrow indicates the nucleotide substitution (c.194G>A); this mutation creates an RsaI site.
B. RsaI digestion confirming the C65Y mutation. A 330-bp fragment of GPIHBP1 was amplified from genomic DNA with primers 5′-CATCTGAGCAGTGGGTGCTGG-3′ and 5′-AGGTGGCTCTGCAGGGCTC-3′. RsaI cleaves the amplified DNA fragment from the C65Y homozygote.
C. Distribution of triglycerides within the lipoproteins of the proband (C65Y), his parents, and his siblings. Plasma lipoproteins were size-fractionated by Fast Protein Liquid Chromatography with a Superose 6 HR column. This study showed that nearly all of the triglycerides in the proband’s plasma were in large lipoprotein particles.
D. Pedigree. The proband, indicated by the black arrow, is homozygous for the cysteine-to-tyrosine substitution at residue 65. Subjects with open symbols were not available for analysis. C65Y heterozygotes are denoted by half-filled symbols; symbols with small darkened circles indicate individuals without any GPIHBP1 mutation. Parental consanguinity is indicated by a double line. Crossed over means deceased.
The proband’s hyperlipidemia was partially responsive to diet, as is generally the case with patients with chylomicronemia.15 The plasma triglyceride level fell from ~4000 to 1575 mg/dl (17.7 mmol/l) when he was placed on a low-fat diet. As expected, fractionation of the plasma by FPLC revealed that the vast majority of the triglycerides were in large lipoproteins (Figure 1C). Plasma apo-B48 levels were elevated, as were plasma levels of apo-CII, apo-CIII, and total apo-B (Table 1).
Table 1.
Fasting plasma lipid and apolipoprotein levels
| Lipid and apolipoprotein levels | C65Y homozygote | Father | Mother | Siblings (n = 3) |
|---|---|---|---|---|
| TG, mg/dl | 1575 | 147 | 94 | 65 ± 14 |
| TC, mg/dl | 126 | 181 | 217 | 129 ± 12 |
| LDLc, mg/dl | 83 | 126 | 126 | 78 ± 6 |
| HDLc, mg/dl | 19.5 | 35.5 | 73 | 47.9 ± 6.7 |
| Apo-B, mg/dl | 63 | 102 | 93 | 54 ± 3 |
| Apo-B48, μg/ml | 125 | 2.1 | 2.6 | 5.2 ± 0.8 |
| Apo-CII, mg/dl | 25.5 | 5.3 | 2.8 | 1.7 ± 1.3 |
| Apo-CIII, mg/dl | 34.5 | 8.2 | 9.1 | 6.3 ± 1.1 |
Fasting plasma concentrations of triglycerides (TG), total cholesterol (TC), low-density cholesterol (LDLc), high-density lipoprotein cholesterol (HDLc), and apolipoproteins in the C65Y homozygote, his parents, and siblings. Data are presented as mean ± SD.
The proband is the youngest of four children; his three siblings were healthy and all were heterozygous for the C65Y mutation. The parents, both heterozygotes, were first cousins (Figure 1D). Heterozygosity for the C65Y mutation was not associated with abnormal plasma lipid or apolipoprotein levels (Table 1 and Figure 1C).
GPIHBP1-C65Y Reaches the Cell Surface But Cannot Bind LPL
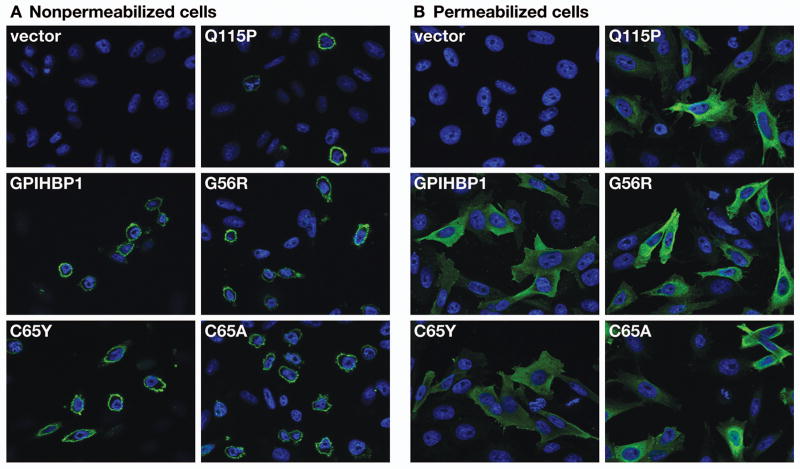
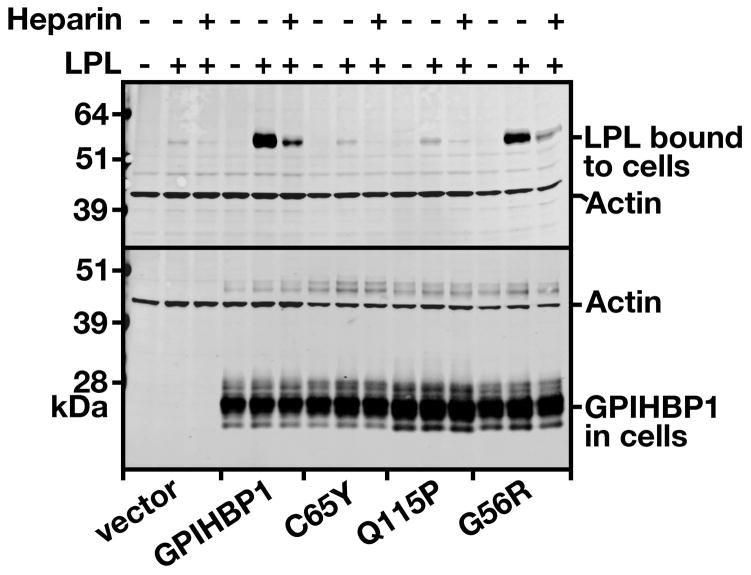
To determine if the C65Y mutation interferes with the ability of the molecule to reach the cell surface, we generated an expression vector for GPIHBP1-C65Y and electroporated it into CHO pgsA-745. In parallel, we performed an experiment with a GPIHBP1-C65A mutant.16 We used two previously characterized mutant GPIHBP1 proteins as controls, GPIHBP1-Q115P (which cannot bind LPL8) and GPIHBP1-G56R (which binds LPL normally13). By fluorescence microscopy, the amount of GPIHBP1-C65Y and GPIHBP1-C65A at the surface of nonpermeabilized cells appeared to be similar to that observed with cells expressing wild-type GPIHBP1 (Figure 2A). The amounts of GPIHBP1-Q115P and GPIHBP1-G56R proteins on the cell surface also appeared to be normal (Figure 2A). Fluorescence microscopy of permeabilized cells indicated that all of the cells expressed similar amounts of GPIHBP1 (Figure 2B).
Figure 2. Detection of the GPIHBP1-C65Y mutant at the cell surface.
CHO pgsA-745 cells were electroporated with an empty vector or an expression vector for wild-type human GPIHBP1, GPIHBP1-C65Y, GPIHBP1-C65A16, GPIHBP1-Q115P, or GPIHBP1-G56R. All constructs contained an amino-terminal S-protein tag. The expression of GPIHBP1 was determined in nonpermeabilized cells (A) and in permeabilized cells (B) by immunofluorescence microscopy with a FITC-conjugated goat antiserum against the S-protein tag. Cell nuclei were visualized with DAPI (blue).
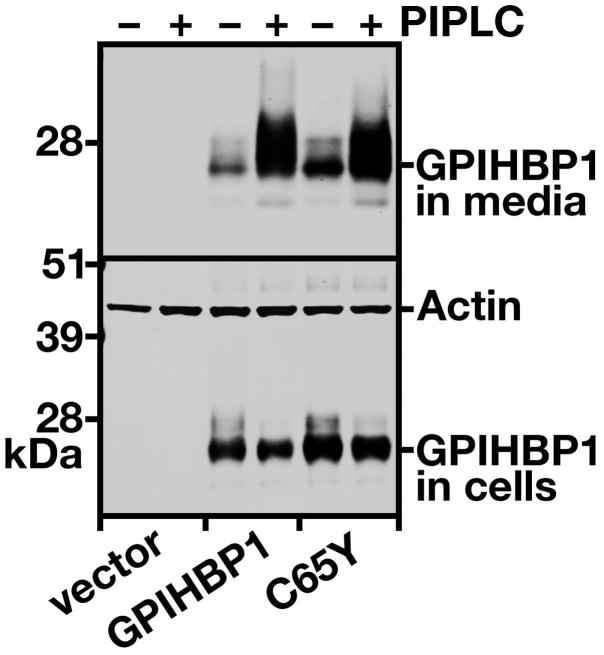
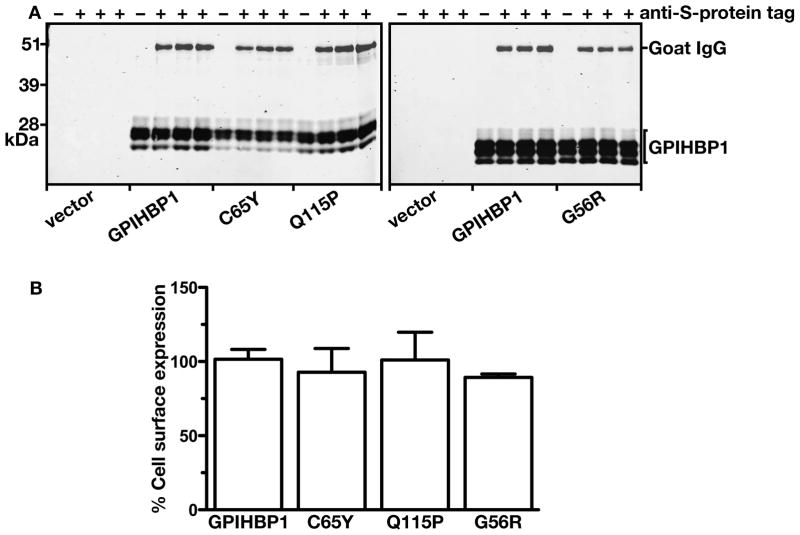
We also assessed the amount of GPIHBP1 at the cell surface by two other techniques. First, we examined the amount of GPIHBP1 that could be released into the medium after incubating live cells with PIPLC. PIPLC released similar amounts of wild-type GPIHBP1 and GPIHBP1-C65Y, as judged by western blotting (Figure 3), strongly suggesting that the C65Y mutation had little or no effect on the ability of the protein to reach the cell surface. Second, we tested the ability of an anti-S-protein tag antibody to bind to GPIHBP1 at the surface of live cells, and comparing that level of binding to the total amount of GPIHBP1 detectable in cells. Again, the amount of GPIHBP1-C65Y at the surface of cells was similar to the amount of GPIHBP1 at the surface of cells expressing wild-type GPIHBP1, GPIHBP1-Q115P, and GPIHBP1-G56R (Figure 4A). When the amount of GPIHBP1 at the cell surface was quantified and normalized to the total amount of GPIHBP1 in cells, we observed no differences among any of the different GPIHBP1 constructs (Figure 4B). Thus, the mutant GPIHBP1 proteins were no different than wild-type GPIHBP1 in terms of reaching the cell surface.
Figure 3. Release of the GPIHBP1-C65Y mutant from the surface of cells with a phosphatidylinositol-specific phospholipase C (PIPLC).
CHO pgsA-745 cells were electroporated with empty vector, an expression vector for wild-type human GPIHBP1, or an expression vector for GPIHBP1-C65Y. 24 h later, the cells were incubated with PIPLC (5 U/ml) in serum-free medium for 1 h at 37° C. The amounts of GPIHBP1 in the cell culture media and the cell lysates were assessed by western blotting with a goat antibody against the S-protein tag. Actin was used as a loading control.
Figure 4. Assessing the amounts of GPIHBP1 at the surface of cells relative to the total amounts of GPIHBP1 in the cell with a goat antibody against the S-protein tag and a mouse monoclonal antibody against human GPIHBP1.
(A) Western blot analysis of GPIHBP1 at the cell surface. CHO pgsA-745 cells were electroporated with an empty vector or an expression vector encoding wild-type human GPIHBP1, GPIHBP1-C65Y, GPIHBP1-Q115P, or GPIHBP1-G56R. All constructs contained an amino-terminal S-protein tag. The next day, the cells were incubated for 2 h at 4° C with a goat antiserum against the S-protein tag. After washing the cells six times in ice-cold PBS, cell extracts were prepared for western blotting with a IR680-conjugated donkey antibody against goat IgG and a mouse monoclonal antibody against human GPIHBP1. The mouse monoclonal antibody was detected with an IR800-conjugated donkey antibody against mouse IgG.
(B) Quantification of the western blot. The signal corresponding to the goat anti-S-protein tag IgG was normalized to the signal for the monoclonal antibody against human GPIHBP1 and expressed relative to the ratio for wild-type GPIHBP1 (set at 100%).
We next assessed the ability of wild-type and mutant GPIHBP1 proteins to bind LPL. Human LPL bound avidly to cells expressing wild type human GPIHBP1, but there was little if any LPL binding to cells expressing GPIHBP1-C65Y. Consistent with earlier studies,8, 13 GPIHBP1-G56R bound LPL avidly but GPIHBP1-Q115P did not (Figure 5).
Figure 5. Assessing the binding of human V5-tagged LPL to GPIHBP1-C65Y.
CHO pgsA-745 cells were electroporated with empty vector, an expression vector for wild-type human GPIHBP1, GPIHBP1-C65Y, GPIHBP1-Q115P, or GPIHBP1-G56R. 24 h later, the cells were incubated for 2 h at 4° C with human V5-tagged LPL with or without heparin (500 U/ml). At the end of the incubation, the cells were washed six times with ice-cold PBS, and cell extracts were prepared for western blotting. The amount of LPL bound to the cells was determined with a mouse monoclonal against the V5 tag; the levels of GPIHBP1 expression were assessed with a goat antibody against the S-protein tag. Actin was used as a loading control.
Release of LPL by Heparin in Humans with GPIHBP1 Defects
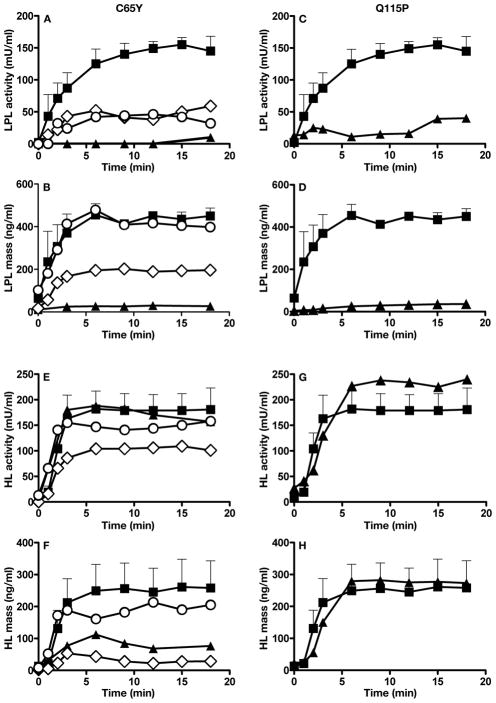
When Gpihbp1−/− mice are given an intravenous bolus of heparin, LPL is released into the plasma, but with delayed kinetics.6 The plasma LPL levels in Gpihbp1−/− mice are low during the first few minutes after a heparin bolus, but after 15 minutes, the LPL levels are similar to those of wild-type mice. Whether the pattern of LPL release after heparin is perturbed in humans with GPIHBP1 mutations is unknown. To address this issue, we assessed the release of LPL into the plasma after an intravenous bolus of heparin in the GPIHBP1-C65Y homozygote (Figure 6A–B) and the GPIHBP1-Q115P homozygote (Figure 6C–D). Following heparin, LPL activity and mass levels were very low in each of multiple plasma samples obtained over an 18-min time span (Figure 6A–D). The parents of the GPIHBP1-C65Y homozygote, both heterozygous carriers, had higher postheparin LPL mass and activity levels, as did unrelated control subjects (Figure 6A–B). Postheparin HL mass and activity were similar in the GPIHBP1-C65Y homozygote (Figure 6E–F), his heterozygous parents (Figure 6E–F), and the GPIHBP1-Q115P homozygote (Figure 6G–H).
Figure 6. The appearance of LPL and HL in the plasma after heparin.
A–B. LPL in the plasma after an intravenous bolus of heparin in the subject homozygous for a C65Y mutation in GPIHBP1, his heterozygous parents, and four normolipidemic controls. LPL activity (A) and mass (B) in postheparin plasma were measured during an 18-min sampling period after an intravenous injection of heparin (50 U/kg body weight). Proband (▲); father (○) mother (◇); and controls (■). For the controls, data are expressed as mean ± SEM.
C–D. LPL in the plasma after an intravenous bolus of heparin in the subject homozygous for a Q115P mutation in GPIHBP1 and normolipidemic controls. LPL activity (C) and mass (D) in postheparin plasma was measured during an 18-min sampling period after an intravenous injection of heparin (50 U/kg body weight). Proband (▲); and controls (■). For the controls (n = 4), data are expressed as mean ± SEM.
E–F. HL in the plasma after an intravenous bolus of heparin in the subject homozygous for a C65Y mutation in GPIHBP1, his heterozygous parents, and four normolipidemic controls. HL activity (E) and mass (F) in postheparin plasma were measured during an 18-min sampling period after an intravenous injection of heparin (50 U/kg body weight). Proband (▲); father (○); mother (◇); and controls (■). For the controls, data are expressed as mean ± SEM.
G–H. HL in the plasma after an intravenous bolus of heparin in the subject homozygous for a Q115P mutation in GPIHBP1, and normolipidemic controls. HL activity (G) and mass (H) in postheparin plasma was measured during an 18-min sampling period after an intravenous injection of heparin (50 U/kg body weight). Proband (▲); and controls (■). For the (n = 4), data are expressed as mean ± SEM.
In Gpihbp1−/− mice, the release of LPL into the plasma with heparin injections lowered plasma triglyceride levels.6 The same was the case in the human Q115P homozygote. During a 6-h heparin infusion, plasma triglyceride concentrations gradually fell from 1780 to 534 mg/dl (20 to 6 mmol/l) (Figure 7A). A heparin infusion in normal subjects lowered triglyceride levels acutely but then the triglyceride levels stabilized (Figure 7B). In normal subjects, LPL levels peaked very quickly following the bolus of heparin and remained high during the continuous heparin infusion (Figure 7C). In the Q115P homozygote, there was no sudden increase in LPL levels with the bolus of heparin, but LPL gradually appeared in the plasma during the heparin infusion.
Figure 7. LPL mass and triglycerides in the plasma during a continuous infusion of heparin in the Q115P homozygote and controls.
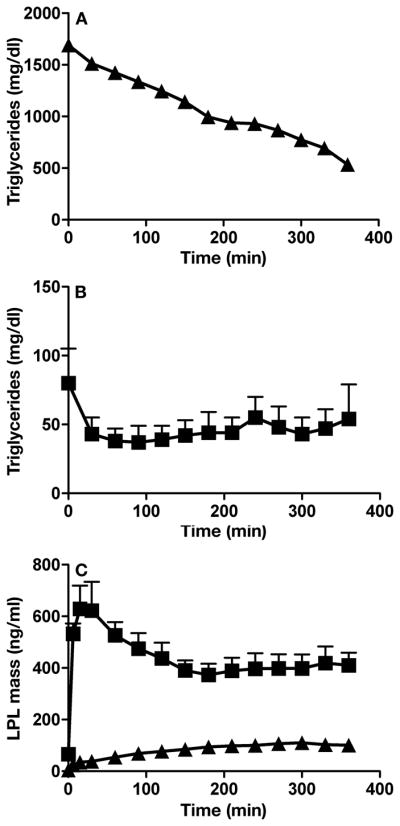
We analyzed LPL mass and triglyceride levels in multiple samples taken every 30 min. (A) Plasma triglycerides in the Q115P homozygote. (B) Plasma triglycerides in normolipidemic control subjects. For the experiments shown in Panels A and B, an inhibitor of lipolysis, tetrahydrolipstatin, was added to the blood after they were collected. Thus, the fall in plasma triglycerides observed in Panel A could not be ascribed to continuing lipolysis occurring in the tube ex vivo. (C) Plasma LPL mass measurements in the normolipidemic controls and the Q115 P homozygote. Proband (▲); and controls (■). For the controls (n = 7), data are expressed as mean ± SEM.
Discussion
We identified a novel homozygous missense mutation in GPIHBP1, C65Y, in a young boy with chylomicronemia. Heterozygous carriers of the mutation were normolipidemic. A series of immunocytochemical, biochemical, and western blot studies revealed that the cysteine-to-tyrosine substitution had little if any effect on the ability of GPIHBP1 to reach the cell surface. However, the mutation abolished the ability of the protein to bind LPL. Cells expressing wild-type GPIHBP1 or GPIHBP1-G56R bound LPL avidly, while cells expressing GPIHBP1-C65Y did not. The C65Y mutation represents the second example of a GPIHBP1 mutation that causes defective LPL binding. Beigneux, Franssen, and coworkers8 previously showed that a young man with severe lifelong chylomicronemia was homozygous for another GPIHBP1 missense mutation (Q115P). Like GPIHBP1-C65Y, GPIHBP1-Q115P reached the cell surface but was unable to bind LPL. While preparing a revised version of this manuscript, Dr. Gunilla Olivecrona and coworkers described a family in which three siblings had severe chylomicronemia; all three siblings had two mutant GPIHBP1 alleles.17 One of the mutant alleles contained a C65S mutation, while the other had a C68G mutation. These mutations abolished LPL binding but had little or no effect on GPIHBP1 trafficking to the plasma membrane. In light of these new findings, there are now four different missense mutations in GPIHBP1’s Ly6 domain that have been associated with defective LPL binding. These findings strongly support the notion that the Ly6 domain is functionally important for LPL binding.
One of the important findings in the current paper is that the heterozygotes in the kindred had normal lipid levels. That finding strongly suggests that half-normal amounts of a functional GPIHBP1 are sufficient for normal lipolytic processing of triglyceride-rich lipoproteins.
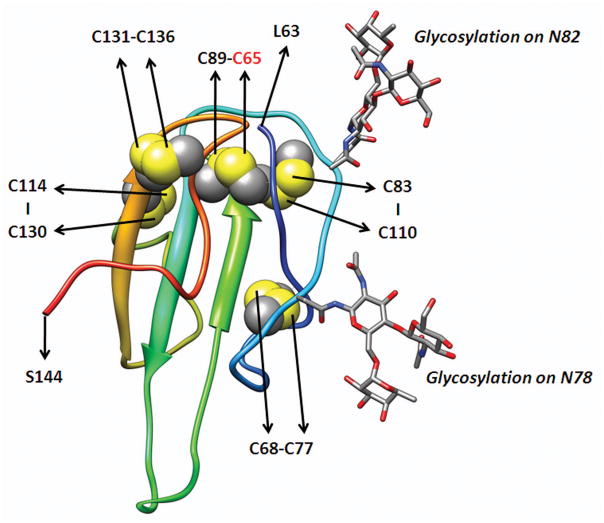
GPIHBP1 is a member of a family of proteins containing a cysteine-rich Ly6 domain. The Ly6 domain contains either 8 or 10 cysteines arranged in a characteristic spacing pattern.18, 19 The structures of several Ly6 proteins, for example CD59 and UPAR (urokinase-type plasminogen activator receptor), have been solved by NMR and X-ray crystallographic studies.20, 21 These structures have demonstrated that each of the cysteines of the Ly6 domain is disulfide-bonded. In UPAR and CD59, the first cysteine of the Ly6 domain forms a disulfide bond with the fifth cysteine, the second with the third, the fourth with the sixth, the seventh with the eight, and the ninth with the tenth. The five disulfide bonds of the Ly6 domain create a three-fingered structural motif.22 At the current time, the structure of GPIHBP1 is not known, but we have generated a likely model of GPIHBP1’s Ly6 domain based on the known structure of CD59 (Figure 8).
Figure 8. Model for the Ly6 domain of human GPIHBP1 based on the known structure of CD59.
The amino acid sequence of GPIHBP1 was aligned to NCBI conserved domain cd000117 with the Conserved Domain Database Search Service, v2.17 (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).23 Using the NMR structure of CD59 (PDB code 1CDR) as a template, 20 models of GPIHBP1 structure were built with the loop model procedure of MODELER 9v6.24, 25 The model with the best DOPE (Discrete Optimized Protein Energy) score was selected. Human GPIHBP1 has two putative N-linked glycosylation sites (Asn-78, Asn-82). The locations of the 10 cysteines and the predicted disulfide bonds (C65–C89, C68–C77, C83–C110, C114–C130, and C131–C136) are shown.
Precisely how the C65Y substitution alters the structure of GPIHBP1 is not known. However, this mutation eliminates the first cysteine of the Ly6 domain and therefore would interfere with the disulfide bond between the first and fifth cysteine (C89) of the Ly6 domain. We suspect that the elimination of the C65–C89 disulfide bond disrupts the three-fingered structure of GPIHBP1’s Ly6 domain.
The fact that the elimination of a cysteine in GPIHBP1’s Ly6 domain would have important consequences for protein function is intriguing but not completely unexpected. Eliminating specific cysteines in the Ly6 domain of CD59 altered the ability of that protein to inhibit complement activation.26 Also, amino acid substitutions involving two different cysteines in the Ly6 domain of another Ly6 protein, SLURP1, leads to a severe skin disease in humans (Mal de Meleda).27 Finally, the observation that a C65Y mutation interferes with protein function and actually lead to chylomicronemia is perfectly consistent with extremely recent site-directed mutagenesis studies indicating that cysteine-to-alanine substitutions in GPIHBP1 interfere with the ability of the protein to bind LPL.16
A novel finding of this study is that the release of LPL into the plasma after an injection of heparin is abnormal in humans with GPIHBP1 mutations. When the C65Y and Q115P homozygotes were given a bolus of heparin, LPL mass and activity levels in the plasma remained very low over the entire 18 min of observation. It is intriguing that the LPL levels were lower in the two heterozygous carriers than in unrelated control subjects. However, the significance of this finding is uncertain, given that their lipid levels were entirely normal.
This observation of very low postheparin LPL levels in the two human homozygotes in this study was different from results in Gpihbp1−/− mice. Weinstein et al.6 showed that, following a bolus of heparin, LPL levels were abnormally low for several minutes; however, by the 15-min time point, the plasma LPL levels in Gpihbp1−/− were similar to those in control mice. In the human homozygotes, LPL levels were low initially and remained low. What accounts for the differences between mice and humans? We do not know the answer to this question, but one potential explanation relates to differences in the doses of heparin that were used. Most of the studies in mice involved a bolus injection of ~2000 IU/kg, whereas the humans were given 50 IU/kg. Had the human subjects been given a much higher dose of heparin, it is conceivable that we would have observed higher levels of LPL in their plasma. However, safety concerns related to the anticoagulant properties of heparin make it impossible to explore this possibility.
The origin of the LPL that is released into the plasma by heparin is not fully understood. Weinstein et al.8 speculated that there are two pools of heparin-releasable LPL. One pool is located in the lumen of capillaries, bound to GPIHBP1, and another pool is located in the subendothelial spaces, perhaps bound to heparan-sulfate proteoglycans.28 They suggested that the delayed release of LPL into the plasma of the Gpihbp1−/− mice was due to the absence of the luminal pool of LPL. In Gpihbp1−/− mice, the tissue stores of LPL were entirely normal. In wild-type mice, the tissue stores of LPL fell very little after an intravenous bolus of heparin, suggesting that most of the heparin-releasable LPL may be located in the subendothelial compartment. Whether GPIHBP1 deficiency in humans affects tissue LPL stores is unknown and needs to be investigated in the future.
In conclusion, we identified a homozygous missense mutation in GPIHBP1 in a patient with severe chylomicronemia. Cell culture studies revealed that the mutant protein reached the cell surface but could not bind LPL. An important and novel finding of the current studies is that GPIHBP1 defects in humans are associated with very low levels of LPL in postheparin plasma. Together with the Q115P studies reported earlier8 and the very recent mutagenesis studies on GPIHBP116, our current studies provide strong evidence that GPIHBP1 is important for the lipolytic processing of triglyceride-rich lipoproteins in humans.
Acknowledgments
We thank the patients, their family, and the controls for participating in the study; J. H. Levels for help with the FPLC experiments; and U. Beisiegel (Hamburg, Germany) for sending us the patient’s genomic DNA.
Funding Sources
This work was supported by a grant from the Dutch Heart Foundation 2008B070 (RF) and HL094732-01 (APB); HL090553 and HL087228 (SGY).
Footnotes
Disclosures
None
References
- 1.Wang H, Eckel RH. Lipoprotein Lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 2.Williams KJ. Molecular processes that handle - and mishandle - dietary lipids. J Clin Invest. 2008;118:3247–3259. doi: 10.1172/JCI35206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young SG, Davies BS, Fong LG, Gin P, Weinstein MM, Bensadoun A, Beigneux AP. GPIHBP1: an endothelial cell molecule important for the lipolytic processing of chylomicrons. Curr Opin Lipidol. 2007;18:389–396. doi: 10.1097/MOL.0b013e3281527914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beigneux AP, Davies BS, Gin P, Weinstein MM, Farber E, Qiao X, Peale F, Bunting S, Walzem RL, Wong JS, Blaner WS, Ding ZM, Melford K, Wongsiriroj N, Shu X, De Sauvage FJ, Ryan RO, Fong LG, Bensadoun A, Young SG. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beigneux AP, Davies BS, Bensadoun A, Fong LG, Young SG. GPIHBP1, a GPI-anchored protein required for the lipolytic processing of triglyceride-rich lipoproteins. J Lipid Res. 2009;50 (Suppl):S57–S62. doi: 10.1194/jlr.R800030-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstein MM, Yin L, Beigneux AP, Davies BS, Gin P, Estrada K, Melford K, Bishop JR, Esko JD, Dallinga-Thie GM, Fong LG, Bensadoun A, Young SG. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J Biol Chem. 2008;283:34511–34518. doi: 10.1074/jbc.M806067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sonnenburg WK, Yu D, Lee EC, Xiong W, Gololobov G, Key B, Gay J, Wilganowski N, Hu Y, Zhao S, Schneider M, Ding ZM, Zambrowicz BP, Landes G, Powell DR, Desai U. Glycosylphosphatidylinositol-anchored HDL-binding protein stabilizes lipoprotein lipase and prevents its inhibition by angiopoietin-like 3 and angiopoietin-like 4. J Lipid Res. 2009;50:2421–2429. doi: 10.1194/jlr.M900145-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beigneux AP, Franssen R, Bensadoun A, Gin P, Melford K, Peter J, Walzem RL, Weinstein MM, Davies BS, Kuivenhoven JA, Kastelein JJ, Fong LG, Dallinga-Thie GM, Young SG. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol. 2009;29:956–962. doi: 10.1161/ATVBAHA.109.186577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levels JH, Marquart JA, Abraham PR, Van den Ende AE, Molhuizen HO, van Deventer SJ, Meijers JC. Lipopolysaccharide is transferred from high-density to low-density lipoproteins by lipopolysaccharide-binding protein and phospholipid transfer protein. Infect Immun. 2005;73:2321–2326. doi: 10.1128/IAI.73.4.2321-2326.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastelein JJP, Jukema JW, Zwinderman AH, Clee S, Van Boven AJ, Jansen H, Rabelink TJ, Peters RJG, Lie KI, Liu G, Bruschke AVG, Hayden MR, REGRESS SG. Lipoprotein lipase activity is associated with severity of angina pectoris. Circulation. 2000;102:1629–1633. doi: 10.1161/01.cir.102.14.1629. [DOI] [PubMed] [Google Scholar]
- 11.Bensadoun A. Sandwich immunoassay for measurement of human hepatic lipase. Methods Enzymol. 1996;263:333–338. doi: 10.1016/s0076-6879(96)63025-0. [DOI] [PubMed] [Google Scholar]
- 12.Brunzell JD, Porte D, Jr, Bierman EL. Abnormal lipoprotein-lipase-mediated plasma triglyceride removal in untreated diabetes mellitus associated with hypertriglyceridemia. Metabolism. 1979;28:901–907. doi: 10.1016/0026-0495(79)90089-1. [DOI] [PubMed] [Google Scholar]
- 13.Gin P, Beigneux AP, Davies B, Young MF, Ryan RO, Bensadoun A, Fong LG, Young SG. Normal binding of lipoprotein lipase, chylomicrons, and apo-AV to GPIHBP1 containing a G56R amino acid substitution. Biochim Biophys Acta. 2007;1771:1464–1468. doi: 10.1016/j.bbalip.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esko JD, Stewart TE, Taylor WH. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc Natl Acad Sci U S A. 1985;82:3197–3201. doi: 10.1073/pnas.82.10.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brunzell JD. Clinical practice. Hypertriglyceridemia. N Engl J Med. 2007;357:1009–1017. doi: 10.1056/NEJMcp070061. [DOI] [PubMed] [Google Scholar]
- 16.Beigneux AP, Gin P, Davies BS, Weinstein MM, Bensadoun A, Fong LG, Young SG. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J Biol Chem. 2009;284:30240–30247. doi: 10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olivecrona G, Ehrenborg E, Semb H, Makoveichuk E, Lindberg A, Hayden MR, Gin P, Davies BSJ, Weinstein MM, Fong LG, Beigneux AP, Young SG, Olivecrona T, Hernell O. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J Lipid Res. 2009 doi: 10.1194/jlr.M002717. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fry BG, Wuster W, Kini RM, Brusic V, Khan A, Venkataraman D, Rooney AP. Molecular evolution and phylogeny of elapid snake venom three-finger toxins. J Mol Evol. 2003;57:110–129. doi: 10.1007/s00239-003-2461-2. [DOI] [PubMed] [Google Scholar]
- 19.Mallya M, Campbell RD, Aguado B. Characterization of the five novel Ly-6 superfamily members encoded in the MHC, and detection of cells expressing their potential ligands. Protein Sci. 2006;15:2244–2256. doi: 10.1110/ps.062242606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Fedarovich A, Tomlinson S, Davies C. Crystal structure of CD59: implications for molecular recognition of the complement proteins C8 and C9 in the membrane-attack complex. Acta Cryst. 2007;D63:714–721. doi: 10.1107/S0907444907015557. [DOI] [PubMed] [Google Scholar]
- 21.Kjaergaard M, Hansen LV, Jacobsen B, Gardsvoll H, Ploug M. Structure and ligand interactions of the urokinase receptor (uPAR) Front Biosci. 2008;13:5441–5461. doi: 10.2741/3092. [DOI] [PubMed] [Google Scholar]
- 22.Galat A, Gross G, Drevet P, Sato A, Menez A. Conserved structural determinants in three-fingered protein domains. FEBS J. 2008;275:3207–3225. doi: 10.1111/j.1742-4658.2008.06473.x. [DOI] [PubMed] [Google Scholar]
- 23.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 25.Fletcher CM, Harrison RA, Lachmann PJ, Neuhaus D. Structure of a soluble, glycosylated form of the human complement regulatory protein CD59. Structure. 1994;2:185–199. doi: 10.1016/s0969-2126(00)00020-4. [DOI] [PubMed] [Google Scholar]
- 26.Petranka J, Zhao J, Norris J, Tweedy NB, Ware RE, Sims PJ, Rosse WF. Structure-function relationships of the complement regulatory protein, CD59. Blood Cells Mol Dis. 1996;22:281–296. doi: 10.1006/bcmd.1996.0111. [DOI] [PubMed] [Google Scholar]
- 27.Charfeddine C, Mokni M, Ben MR, Elkares R, Bouchlaka C, Boubaker S, Ghedamsi S, Baccouche D, Ben OA, Dellagi K, Abdelhak S. A novel missense mutation in the gene encoding SLURP-1 in patients with Mal de Meleda from northern Tunisia. Br J Dermatol. 2003;149:1108–1115. doi: 10.1111/j.1365-2133.2003.05606.x. [DOI] [PubMed] [Google Scholar]
- 28.Williams KJ. Some things just have to be done in vivo: GPIHBP1, caloric delivery, and the generation of remnant lipoproteins. Arterioscler Thromb Vasc Biol. 2009;29:792–795. doi: 10.1161/ATVBAHA.109.187823. [DOI] [PubMed] [Google Scholar]