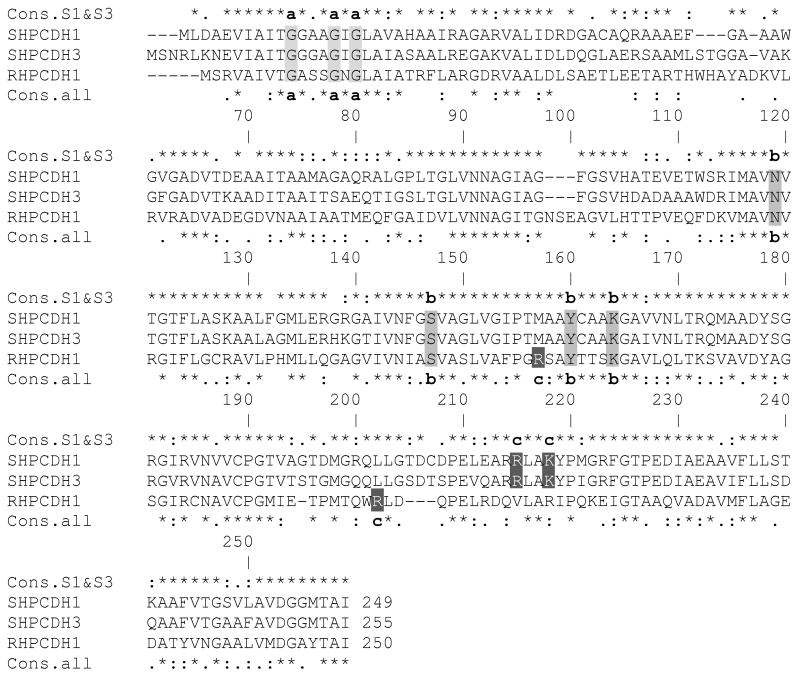
Figure 2.
Multiple-sequence alignment of S-HPCDH1, S-HPCDH3 and R-HPCDH1 enzymes from X. autotrophicus Py2. Abbreviations: Cons.S1&3, consensus amino acid alignment for S-HPCDH1 and S-HPCDH3; Cons.all, consensus of S-HPCDH1, S-HPCDH3 and R-HPCDH1 Letter designations: a, Classic GXXXGXG glycine-rich NAD+ binding motif. b, Catalytic tetrad residues of Asn, Ser, Tyr and Lys. c, Positively charged residues that have been shown (R-HPCDH1) or are proposed (S-HPCDH) to interact with the sulfonate group of CoM in the substrate. The alignment was generated using MULTALIN with default parameters, while the consensus was derived using ClustalW2. The following symbols mean that the residues are: (*) identical; (:) conserved; (.) semi-conserved.