Abstract
Nuclear receptor Rev-erbα (NR1D1), previously considered to be an orphan nuclear receptor, is a receptor for heme, which promotes transcriptional repression via recruitment of the NCoR-HDAC3 corepressor complex. Rev-erbα gene regulation is circadian, and Rev-erbα comprises a critical negative limb of the core circadian clock by directly repressing the expression of the positive clock component, Bmal1. Rev-erbα also regulates the metabolic gene pathway, thus serving as a heme sensor for coordination of circadian and metabolic pathways.
Introduction to Rev-erbα
Rev-erbα (NR1D1) was discovered in 1989, and as such was one of the first orphan nuclear receptors (NRs) to be described [Lazar et al., 1990b; Miyajima et al., 1989]. Rev-erbα is now recognized to be the first member of an interesting NR subfamily that includes the highly related Rev-erbβ [Bonnelye et al., 1994; Dumas et al., 1994; Retnakaran et al., 1994], as well as the ROR subfamily of orphan receptors (ROR) [Giguere et al., 1994]. A Rev-erbα homologue, called E75, is present in Drosophila [Segraves and Hogness, 1990]. The 3’-end of mouse Rev-erbα gene overlaps with that of mouse TRα2 gene [Lazar et al., 1990b; Miyajima et al., 1989]. It has been suggested that the presence of Rev-erbα transcripts may influence the alternative splicing of TRα by an antisense mechanism [Lazar et al., 1990a; Munroe and Lazar, 1991]. In mammals, Rev-erbα expression is highly regulated in both its tissue distribution and developmental profile. Indeed, knockout of Rev-erbα without perturbation of the TRα gene leads to a modest phenotype of reproductive abnormalities and delayed cerebellar development in mice [Chomez et al., 2000]. More importantly, Rev-erbα knockout mice show changes in their circadian rhythm of activity, which is characterized by a shorter period length and greater light-induced phase responsiveness than that of control mice [Preitner et al., 2002].
Rev-erbα is a potent repressor of gene transcription
Many NRs activate gene expression in the presence of a natural or pharmaceutical ligand, whose binding leads to a receptor conformation that allows tight interaction with transcriptional coactivators [McKenna and O'Malley, 2002]. In the absence of ligand, several NRs repress transcription, via recruitment of corepressors that specifically recognize the unliganded state [Hu and Lazar, 2000; Privalsky, 2004]. Binding of ligand thus induces a conformational switch from the repressed to an activated state [Glass and Rosenfeld, 2000]. The major ligand-dependent conformational change is a movement of a C-terminal helix, called H12, towards the hydrophobic core of the NR, making contact with and stabilizing the ligand binding [Wurtz et al., 1996]. H12 is required for coactivator binding [Halachmi et al., 1994], and forms part of the coactivator binding surface of the NR [Feng et al., 1998; Shiau et al., 1998]. The remainder of the coactivator binding surface overlaps with the region of the NR that is bound by corepressor [Hu and Lazar, 1999; Marimuthu et al., 2002; Nagy et al., 1999; Perissi et al., 1999; Xu et al., 2002]. Indeed, whereas deletion of H12 abolishes coactivator binding, it markedly increases corepressor binding to TR, RAR, and RXR [Schulman et al., 1996; Zhang et al., 1999]. This is particularly relevant to Rev-erbα, which is unique among NRs in that it completely lacks H12 and is a potent and constitutive transcriptional repressor [Harding and Lazar, 1995].
The role of the NCoR/HDAC3 complex in repression by Rev-erbα
Rev-erbα interacts strongly with Nuclear Receptor CoRepressor (NCoR) [Zamir et al., 1996], which it binds preferentially over Silencing Mediator for Retinoid and Thyroid Receptors (SMRT) when bound to target DNA [Hu et al., 2001; Zamir et al., 1997a]. Knockdown of NCoR attenuates repression by Rev-erbα [Ishizuka and Lazar, 2003]. The class I histone deacetylase (HDAC), HDAC3, interacts stably and quantitatively with endogenous NCoR [Guenther et al., 2000; Li et al., 2000; Zhang et al., 2002]. The catalytic activity of HDAC3 actually requires interaction with NCoR or SMRT [Guenther et al., 2001; Zhang et al., 2002], and the HDAC3 activity of cellular N-CoR complexes is lost after knockdown of HDAC3, suggesting that HDAC3 is the major HDAC associated with N-CoR [Ishizuka and Lazar, 2005]. HDAC3 is required for the repressive activity of Rev-erbα [Ishizuka and Lazar, 2003], and is recruited by Rev-erbα to endogenous target genes whose activity is de-repressed in the absence of HDAC3 [Yin and Lazar, 2005]. The role of other HDACs in Rev-erbα function is unclear at this time.
Determinants of genes that are targets for Rev-erbα repression
Like other NRs, the Rev-erbα DBD consists of two zinc-ordered modules creating a helix-turn-helix structure that recognizes the specific hexameric sequence "AGGTCA" [Umesono and Evans, 1989]. Rev-erbα binds DNA as a monomer, but only to extended half-sites in which an A/T-rich sequence is 5' to the AGGTCA [Harding and Lazar, 1995]. ROR, a constitutively active orphan NR, has a very similar DBD that binds DNA as a monomer to almost identical DNA sequences, called ROR elements (ROREs) [Giguere et al., 1995]. Structural analysis revealed that the C-terminal extension of the Rev-erbα DBD interacts with the A/T-rich 5' extension of the AGGTCA half-site that is required for high affinity binding [Zhao et al., 1998].
Many NRs bind to direct repeats of the "AGGTCA" site as a heterodimer with RXR [Mangelsdorf and Evans, 1995], and the spacing of the AGGTAC "half-sites" is a critical determinant of binding affinity and specificity [Umesono et al., 1991]. Unlike many other orphan NRs, Rev-erbα does not heterodimerize with RXR. Rev-erbα does bind specifically as a homodimer to tandem repeats spaced by two base pairs ("DR2"), but only when the 5' half-site of the DR2 is flanked by the A/T-rich sequence favored by the monomer; this is referred to as a Rev-DR2 [Harding and Lazar, 1995]. Unlike RXR heterodimers, the Rev-erbα homodimer is not stable in the absence of DNA, but does bind cooperatively to the DR2 [Harding and Lazar, 1995].
Rev-erbα is a potent repressor on the cooperatively bound Rev-DR2 [Harding and Lazar, 1995; Zamir et al., 1997b]. In addition, two Rev-erbα monomers bound independently to relatively widely spaced ROREs can also recruit NCoR to repress transcription [Harding and Lazar, 1995; Zamir et al., 1997b]. However, a single Rev-erbα molecule bound to a lone RORE cannot recruit NCoR [Zamir et al., 1997b]. This is because the stoichiometry of productive NCoR binding is two Rev-erbα molecules to one of NCoR [Zamir et al., 1997b], which is also true for other NRs such as TR [Cohen et al., 1998; Jeannin et al., 1998; Zamir et al., 1997b]. Although monomeric Rev-erbα cannot actively repress transcription, it can nevertheless function as a repressor by competing with constitutively active ROR for ROREs [Forman et al., 1994; Harding and Lazar, 1995; Retnakaran et al., 1994].
Modulation of the repressive activity of Rev-erbα
For other corepressor-binding NRs, repressive activity is regulated by ligand binding, which destabilizes the binding of the corepressor complex in addition to stabilizing NR-coactivator complexes. Remarkably, Rev-erbα binding to NCoR is actually stabilized by molecular heme, while depletion of intracellular heme abolishes the interaction between Rev-erbα and N-CoR protein. In 2007, two groups independently found that heme binds directly to Rev-erbα with a 1:1 stoichiometry, and that this binding is specific, saturable, reversible, and functional [Raghuram et al., 2007; Yin et al., 2007], thereby fitting the criteria for an NR ligand. Thus, Rev-erbα is no longer an “orphan” receptor. In addition to its endogenous ligand, a recent study indicates that, like other NRs, Rev-erbα can be targeted by other small molecules that have the potential to be used for therapeutic purposes [Meng et al., 2008]. It should be noted that the highly related Rev-erbβ can also bind heme, and the structure and function of heme-bound Rev-erbβ is sensitive to the presence of nitric oxide (NO) [Marvin et al., 2009; Reinking et al., 2005]. At this time, it is not clear if Rev-erbα transcriptional activity is regulated by NO [Raghuram et al., 2007; Yin et al., 2007]. In addition, it should be pointed out that the sensing of heme concentration by Rev-erbα contrasts with the stoichiometric role of heme in regulating the circadian neuronal PAS-domain protein 2 (NPAS2), whose heme-dependent binding of carbon monoxide results in an inhibition of DNA-binding activity [Dioum et al., 2002].
Regulation of Rev-erbα protein stability
The cellular levels of Rev-erbα are also major determinants of whether Rev-erbα target genes will be repressed in a given cell. We have recently identified a pathway whereby Rev-erbα protein levels are regulated by GSK3β-dependent phosphorylation, which prevents the rapid proteasomal degradation of Rev-erbα [Yin et al., 2006]. Phosphorylation stabilizes Rev-erbα protein until serum-induced phosphorylation of serine 9 (ser9-p) inhibits GSK3β enzymatic activity [Cohen and Frame, 2001; Frame and Cohen, 2001; Frame et al., 2001], leading to Rev-erbα ubiquitination and degradation via the 26S proteasome. Phosphorylation-directed proteolysis often involves a class of E3 ubiquitin ligases called SCF (Skp1-Cullin1-F box) complexes [Petroski and Deshaies, 2005]; however, the E3 ligase(s) responsible for Rev-erbα degradation is(are) not known at this time. Note that the GSK3β-dependent stabilization of Rev-erbα is opposite of this more usual scenario, whereby phosphorylated GSK3β substrates are specifically targeted to proteasomal degradation [Doble and Woodgett, 2003].
Role of Rev-erbα in circadian biology
Genetic and biochemical analysis revealed that 24 h circadian rhythms are present throughout the animal kingdom [Panda et al., 2002]. In mammals, circadian rhythm is a fundamental regulatory factor for many aspects of behavior and physiology, including sleep/wake cycles, blood pressure, body temperature and metabolism. Disruption in circadian rhythms leads to increased incidence of many diseases, such as cancer, metabolic disease, and mental illness [Gachon et al., 2004]. Cellular rhythms are generated and maintained through interconnected transcriptional feedback of clock genes. The cycle starts when two PAS-HLH proteins, BMAL1 and CLOCK, heterodimerize to activate a number of circadian genes including Per and Cry, which feedback and negatively regulate the activity of Bmal1/Clock [Takahashi et al., 2008].
Rev-erbα is also transcriptionally activated by BMAL1/CLOCK, initiating a second negative feedback loop that represses the transcription of the Bmal1 gene. This function of Rev-erbα is mediated by recruitment of the NCoR/HDAC3 complex to tandem Rev-erbα binding sites in the Bmal1 gene promoter [Yin and Lazar, 2005]. By repressing Bmal1 gene expression, Rev-erbα thereby represents an important link between the positive and negative loops of the circadian clock. Indeed, mice lacking Rev-erbα manifest a distinctive pattern of circadian rhythm [Preitner et al., 2002]. Under constant darkness, Rev-erbα null animals exhibit a significantly shorter circadian period length and an aberrant phase-shifting response to light stimuli. Although it has been thought that such circadian phenotype is due to loss of Rev-erbα-dependent Bmal1 gene regulation, the exact mechanism remains unclear. The GSK3β-mediated phosphorylation of Rev-erbα is also involved in initiation and synchronization of the cell autonomous circadian clock [Yin et al., 2006]. It is of interest that lithium, a widely used and effective treatment of bipolar disorder that also has effects on circadian rhythm, is a potent GSK3β inhibitor that induces Rev-erbα protein degradation and upregulation of Rev-erbα gene targets including Bmal1 [Pardee et al., 2009; Yin et al., 2006]. Indeed, inhibition of GSK3β by either small chemical inhibitor or siRNA knockdown consistently causes a strong short circadian period phenotype, a similar circadian behavior in Rev-erbα null mice [Hirota et al., 2008].
Role of Rev-erbα in metabolic regulation
Rev-erbα is most highly expressed in metabolic tissues, including adipose tissue, liver and muscle [Burke et al., 1996; Lazar et al., 1989]. Little is known about the physiological functions of Rev-erbα in adipose tissue, although Rev-erbα mRNA is induced in two of the most well-studied models of adipogenesis, 3T3-L1 and 3T3-F442A cells [Chawla and Lazar, 1993]. Overexpression of Rev-erbα has been shown to enhance adipogenesis [Fontaine et al., 2003], and Rev-erbα is required for adipogenesis [Wang and Lazar, 2008]. The biological role of Rev-erbα in muscle is also not well understood, though it may be involved in muscle fiber type switching [Pircher et al., 2005].
More is known about the metabolic function of Rev-erbα in liver [Duez and Staels, 2009; Le Martelot et al., 2009]. Besides its function in regulating core clock genes, Rev-erbα also regulates time-specific expression of circadian output genes important for normal hepatic physiology. Rev-erbα has been shown to repress expression of apolipoprotein CIII (apoC-III) [Coste and Rodriguez, 2002; Raspe et al., 2002] and, consistent with this, mice lacking Rev-erbα have elevated apoC-III levels and a large increase in serum triglyceride and VLDL levels [Raspe et al., 2002]. Rev-erbα also represses the gluconeogenic gene, glucose 6-phosphatase, in hepatocytes [Yin et al., 2007]. Rev-erbα also regulates bile acid metabolism via both direct and indirect mechanisms [Duez et al., 2008; Le Martelot et al., 2009]. More recently, Rev-erbα has been demonstrated to regulate the expression of mir-122, a highly abundant liver-specific microRNA [Gatfield et al., 2009], further supporting a critical role for Rev-erbα in liver.
Rev-erbα also regulates the synthesis of its ligand, heme. The rate limiting enzyme in heme synthesis, ALAS1, has a circadian rhythm and is positively regulated by two circadian transcriptional regulators, the bHLH protein NPAS2 and the coactivator, PGC-1α [Handschin et al., 2005; Kaasik and Lee, 2004; Liu et al., 2007]. We recently showed that Rev-erbα directly represses PGC-1α, thus creating a feedback loop in which heme promotes Rev-erbα repression of PGC-1α, thereby reducing ALAS1 gene expression and heme biosynthesis. Conversely, low heme levels reduce Rev-erbα repression, enhancing PGC-1α stimulation of heme synthesis via transcriptional activation of the rate limiting enzyme, ALAS1 [Wu et al., 2009]. Rev-erbα thus serves as a sensor that functions to maintain intracellular heme levels within a limited range under normal physiological conditions.
Summary and conclusions
The biological significance of repression by NRs and their corepressors is increasingly apparent. Rev-erbα, which lacks H12 and thus is an obligate repressor, has been a superb model for understanding the mechanisms of repression. Moreover, the critical role of Rev-erbα in the core circadian clock provides a powerful and compelling validation of the biological importance of active gene repression and the corepressor complexes. Rev-erbα also regulates metabolic pathways, and is thus a molecular link between circadian rhythm and metabolic physiology. The function of Rev-erbα as a heme sensor serves to maintain heme homeostasis, while regulating metabolic and circadian processes that may be affected by ambient heme concentrations including the oxidative metabolism of the cell (Figure 1). Compounds that modulate Rev-erbα activity thus have the potential to contribute to or even control the crosstalk between circadian and metabolic processes, which is of great significance due to the marked rise in obesity, diabetes, and sleep disorders that plague advanced societies.
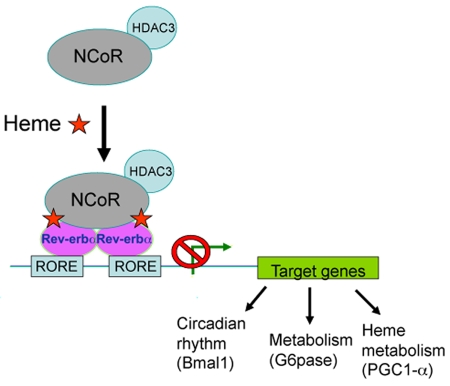
Figure 1. Rev-erbα coordinates circadian rhythm and metabolic pathways in a heme-dependent mode.
The binding elements of Rev-erbα, called ROREs, are present in core clock genes and also in important metabolic genes. Heme, a physiological ligand of Rev-erbα, promotes recruitment of the NCoR-HDAC3 corepressor complex to Rev-erbα homodimers bound to target genes and enhances Rev-erbα-mediated repression of those target genes. Heme binding to Rev-erbα induces the feedback inhibition of its own biosynthesis.
Acknowledgments
This work was supported by NIH grants DK45586 and DK062434 (NURSA) to M.A.L. and DK077449 to L.Y.
References
- Bonnelye E., Vanacker J. M., Desbiens X., Begue A., Stehelin D., Laudet V. Rev-erb β, a new member of the nuclear receptor superfamily, is expressed in the nervous system during chicken development. Cell Growth Differ. 1994;5:1357–65. [PubMed] [Google Scholar]
- Burke L., Downes M., Carozzi A., Giguere V., Muscat G. E. Transcriptional repression by the orphan steroid receptor RVR/Rev-erb β is dependent on the signature motif and helix 5 in the E region: functional evidence for a biological role of RVR in myogenesis. Nucleic Acids Res. 1996;24:3481–9. doi: 10.1093/nar/24.18.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A., Lazar M. A. Induction of Rev-ErbA α, an orphan receptor encoded on the opposite strand of the α-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268:16265–9. [PubMed] [Google Scholar]
- Chomez P., Neveu I., Mansen A., Kiesler E., Larsson L., Vennstrom B., Arenas E. Increased cell death and delayed development in the cerebellum of mice lacking the rev-erbA(α) orphan receptor. Development. 2000;127:1489–98. doi: 10.1242/dev.127.7.1489. [DOI] [PubMed] [Google Scholar]
- Cohen P., Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–76. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- Cohen R. N., Wondisford F. E., Hollenberg A. N. Two separate NCoR (nuclear receptor corepressor) interaction domains mediate corepressor action on thyroid hormone response elements. Mol Endocrinol. 1998;12:1567–81. doi: 10.1210/mend.12.10.0188. [DOI] [PubMed] [Google Scholar]
- Coste H., Rodriguez J. C. Orphan nuclear hormone receptor Rev-erbalpha regulates the human apolipoprotein CIII promoter. J Biol Chem. 2002;277:27120–9. doi: 10.1074/jbc.M203421200. [DOI] [PubMed] [Google Scholar]
- Dioum E. M., Rutter J., Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A., McKnight S. L. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–7. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- Doble B. W., Woodgett J. R. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–86. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duez H., van der Veen J. N., Duhem C., Pourcet B., Touvier T., Fontaine C., Derudas B., Bauge E., Havinga R., Bloks V. W., Wolters H., van der Sluijs F. H., Vennstrom B., Kuipers F., Staels B. Regulation of bile acid synthesis by the nuclear receptor Rev-erbalpha. Gastroenterology. 2008;135:689–98. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- Duez H., Staels B. Rev-erb-α: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107:1972–80. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas B., Harding H. P., Choi H. S., Lehmann K. A., Chung M., Lazar M. A., Moore D. D. A new orphan member of the nuclear hormone receptor superfamily closely related to Rev-Erb. Mol Endocrinol. 1994;8:996–1005. doi: 10.1210/mend.8.8.7997240. [DOI] [PubMed] [Google Scholar]
- Feng W., Ribeiro R. C., Wagner R. L., Nguyen H., Apriletti J. W., Fletterick R. J., Baxter J. D., Kushner P. J., West B. L. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–9. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- Fontaine C., Dubois G., Duguay Y., Helledie T., Vu-Dac N., Gervois P., Soncin F., Mandrup S., Fruchart J. C., Fruchart-Najib J., Staels B. The orphan nuclear receptor Rev-Erbalpha is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARgamma-induced adipocyte differentiation. J Biol Chem. 2003;278:37672–80. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Chen J., Blumberg B., Kliewer S. A., Henshaw R., Ong E. S., Evans R. M. Cross-talk among ROR α 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8:1253–61. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- Frame S., Cohen P., Biondi R. M. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell. 2001b;7:1321–7. doi: 10.1016/s1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- Frame S., Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001a;359:1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F., Nagoshi E., Brown S. A., Ripperger J., Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–12. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gatfield D., Le Martelot G., Vejnar C. E., Gerlach D., Schaad O., Fleury-Olela F., Ruskeepaa A. L., Oresic M., Esau C. C., Zdobnov E. M., Schibler U. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 2009;23:1313–26. doi: 10.1101/gad.1781009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V., McBroom L. D., Flock G. Determinants of target gene specificity for ROR α 1: monomeric DNA binding by an orphan nuclear receptor. Mol Cell Biol. 1995;15:2517–26. doi: 10.1128/mcb.15.5.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V., Tini M., Flock G., Ong E., Evans R. M., Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR α, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–53. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Guenther M. G., Lane W. S., Fischle W., Verdin E., Lazar M. A., Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–57. [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G., Barak O., Lazar M. A. The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol Cell Biol. 2001;21:6091–101. doi: 10.1128/MCB.21.18.6091-6101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halachmi S., Marden E., Martin G., MacKay H., Abbondanza C., Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–8. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- Handschin C., Lin J., Rhee J., Peyer A. K., Chin S., Wu P. H., Meyer U. A., Spiegelman B. M. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell. 2005;122:505–15. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Lazar M. A. The monomer-binding orphan receptor Rev-Erb represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15:4791–802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Lewis W. G., Liu A. C., Lee J. W., Schultz P. G., Kay S. A. A chemical biology approach reveals period shortening of the mammalian circadian clock by specific inhibition of GSK-3beta. Proc Natl Acad Sci U S A. 2008;105:20746–51. doi: 10.1073/pnas.0811410106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Li Y., Lazar M. A. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol Cell Biol. 2001;21:1747–58. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Ishizuka T., Lazar M. A. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol Cell Biol. 2003;23:5122–31. doi: 10.1128/MCB.23.15.5122-5131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T., Lazar M. A. The nuclear receptor corepressor deacetylase activating domain is essential for repression by thyroid hormone receptor. Mol Endocrinol. 2005;19:1443–51. doi: 10.1210/me.2005-0009. [DOI] [PubMed] [Google Scholar]
- Jeannin E., Robyr D., Desvergne B. Transcriptional regulatory patterns of the myelin basic protein and malic enzyme genes by the thyroid hormone receptors alpha1 and beta1. J Biol Chem. 1998;273:24239–48. doi: 10.1074/jbc.273.37.24239. [DOI] [PubMed] [Google Scholar]
- Kaasik K., Lee C. C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–71. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Darling D. S., Chin W. W. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA α transcriptional unit. Mol Cell Biol. 1989;9:1128–36. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar M. A., Hodin R. A., Cardona G., Chin W. W. Gene expression from the c-erbA α/Rev-ErbA α genomic locus. Potential regulation of alternative splicing by opposite strand transcription. J Biol Chem. 1990b;265:12859–63. [PubMed] [Google Scholar]
- Lazar M. A., Jones K. E., Chin W. W. Isolation of a cDNA encoding human Rev-ErbA α: transcription from the noncoding DNA strand of a thyroid hormone receptor gene results in a related protein that does not bind thyroid hormone. DNA Cell Biol. 1990a;9:77–83. doi: 10.1089/dna.1990.9.77. [DOI] [PubMed] [Google Scholar]
- Le Martelot G., Claudel T., Gatfield D., Schaad O., Kornmann B., Sasso G. L., Moschetta A., Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J., Wang J., Nawaz Z., Liu J. M., Qin J., Wong J. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li S., Liu T., Borjigin J., Lin J. D. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–81. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Evans R. M. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–50. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Marimuthu A., Feng W., Tagami T., Nguyen H., Jameson J. L., Fletterick R. J., Baxter J. D., West B. L. TR surfaces and conformations required to bind nuclear receptor corepressor. Mol Endocrinol. 2002;16:271–86. doi: 10.1210/mend.16.2.0777. [DOI] [PubMed] [Google Scholar]
- Marvin K. A., Reinking J. L., Lee A. J., Pardee K., Krause H. M., Burstyn J. N. Nuclear receptors homo sapiens Rev-erbbeta and Drosophila melanogaster E75 are thiolate-ligated heme proteins which undergo redox-mediated ligand switching and bind CO and NO. Biochemistry. 2009;48:7056–71. doi: 10.1021/bi900697c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–5. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- Meng Q. J., McMaster A., Beesley S., Lu W. Q., Gibbs J., Parks D., Collins J., Farrow S., Donn R., Ray D., Loudon A. Ligand modulation of REV-ERBalpha function resets the peripheral circadian clock in a phasic manner. J Cell Sci. 2008;121:3629–35. doi: 10.1242/jcs.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima N., Horiuchi R., Shibuya Y., Fukushige S., Matsubara K., Toyoshima K., Yamamoto T. Two erbA homologs encoding proteins with different T3 binding capacities are transcribed from opposite DNA strands of the same genetic locus. Cell. 1989;57:31–9. doi: 10.1016/0092-8674(89)90169-4. [DOI] [PubMed] [Google Scholar]
- Munroe S. H., Lazar M. A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991;266:22083–6. [PubMed] [Google Scholar]
- Nagy L., Kao H. Y., Love J. D., Li C., Banayo E., Gooch J. T., Krishna V., Chatterjee K., Evans R. M., Schwabe J. W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–16. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S., Hogenesch J. B., Kay S. A. Circadian rhythms from flies to human. Nature. 2002;417:329–35. doi: 10.1038/417329a. [DOI] [PubMed] [Google Scholar]
- Pardee K. I., Xu X., Reinking J., Schuetz A., Dong A., Liu S., Zhang R., Tiefenbach J., Lajoie G., Plotnikov A. N., Botchkarev A., Krause H. M., Edwards A. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBbeta. PLoS Biol. 2009;7:e43. doi: 10.1371/journal.pbio.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Staszewski L. M., McInerney E. M., Kurokawa R., Krones A., Rose D. W., Lambert M. H., Milburn M. V., Glass C. K., Rosenfeld M. G. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski M. D., Deshaies R. J. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- Pircher P., Chomez P., Yu F., Vennstrom B., Larsson L. Aberrant expression of myosin isoforms in skeletal muscles from mice lacking the rev-erbAalpha orphan receptor gene. Am J Physiol Regul Integr Comp Physiol. 2005;288:R482–90. doi: 10.1152/ajpregu.00690.2003. [DOI] [PubMed] [Google Scholar]
- Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Privalsky M. L. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annu Rev Physiol. 2004;66:315–60. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Raghuram S., Stayrook K. R., Huang P., Rogers P. M., Nosie A. K., McClure D. B., Burris L. L., Khorasanizadeh S., Burris T. P., Rastinejad F. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–13. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspe E., Duez H., Mansen A., Fontaine C., Fievet C., Fruchart J. C., Vennstrom B., Staels B. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43:2172–9. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- Reinking J., Lam M. M., Pardee K., Sampson H. M., Liu S., Yang P., Williams S., White W., Lajoie G., Edwards A., Krause H. M. The Drosophila nuclear receptor e75 contains heme and is gas responsive. Cell. 2005;122:195–207. doi: 10.1016/j.cell.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Retnakaran R., Flock G., Giguere V. Identification of RVR, a novel orphan nuclear receptor that acts as a negative transcriptional regulator. Mol Endocrinol. 1994;8:1234–44. doi: 10.1210/mend.8.9.7838156. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Juguilon H., Evans R. M. Activation and repression by nuclear hormone receptors: hormone modulates an equilibrium between active and repressive states. Mol Cell Biol. 1996;16:3807–13. doi: 10.1128/mcb.16.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segraves W. A., Hogness D. S. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–19. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–37. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. The genetics of mammalian circadian order and disorder: implications for physiology and disease. Nat Rev Genet. 2008;9:764–75. doi: 10.1038/nrg2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K., Evans R. M. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–46. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami K. K., Thompson C. C., Evans R. M. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–66. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Lazar M. A. Bifunctional role of Rev-erbalpha in adipocyte differentiation. Mol Cell Biol. 2008;28:2213–20. doi: 10.1128/MCB.01608-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N., Yin L., Hanniman E. A., Joshi S., Lazar M. A. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbalpha. Genes Dev. 2009;23:2201–9. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtz J. M., Bourguet W., Renaud J. P., Vivat V., Chambon P., Moras D., Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- Xu H. E., Stanley T. B., Montana V. G., Lambert M. H., Shearer B. G., Cobb J. E., McKee D. D., Galardi C. M., Plunket K. D., Nolte R. T., Parks D. J., Moore J. T., Kliewer S. A., Willson T. M., Stimmel J. B. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–7. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- Yin L., Wang J., Klein P. S., Lazar M. A. Nuclear receptor Rev-erbalpha is a critical lithium-sensitive component of the circadian clock. Science. 2006;311:1002–5. doi: 10.1126/science.1121613. [DOI] [PubMed] [Google Scholar]
- Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., Waitt G. M., Parks D. J., Pearce K. H., Wisely G. B., Lazar M. A. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–9. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Yin L., Lazar M. A. The orphan nuclear receptor Rev-erbalpha recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–9. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- Zamir I., Harding H. P., Atkins G. B., Horlein A., Glass C. K., Rosenfeld M. G., Lazar M. A. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–65. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I., Dawson J., Lavinsky R. M., Glass C. K., Rosenfeld M. G., Lazar M. A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci U S A. 1997a;94:14400–5. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir I., Zhang J., Lazar M. A. Stoichiometric and steric principles governing repression by nuclear hormone receptors. Genes Dev. 1997b;11:835–46. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- Zhang J., Hu X., Lazar M. A. A novel role for helix 12 of retinoid X receptor in regulating repression. Mol Cell Biol. 1999;19:6448–57. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kalkum M., Chait B. T., Roeder R. G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol Cell. 2002;9:611–23. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Khorasanizadeh S., Miyoshi Y., Lazar M. A., Rastinejad F. Structural elements of an orphan nuclear receptor-DNA complex. Mol Cell. 1998;1:849–61. doi: 10.1016/s1097-2765(00)80084-2. [DOI] [PubMed] [Google Scholar]