Abstract
The peroxisome proliferator-activated receptor α (PPARα, or NR1C1) is a nuclear hormone receptor activated by a structurally diverse array of synthetic chemicals known as peroxisome proliferators. Endogenous activation of PPARα in liver has also been observed in certain gene knockout mouse models of lipid metabolism, implying the existence of enzymes that either generate (synthesize) or degrade endogenous PPARα agonists. For example, substrates involved in fatty acid oxidation can function as PPARα ligands. PPARα serves as a xenobiotic and lipid sensor to regulate energy combustion, hepatic steatosis, lipoprotein synthesis, inflammation and liver cancer. Mainly, PPARα modulates the activities of all three fatty acid oxidation systems, namely mitochondrial and peroxisomal β-oxidation and microsomal ω-oxidation, and thus plays a key role in energy expenditure. Sustained activation of PPARα by either exogenous or endogenous agonists leads to the development of hepatocellular carcinoma resulting from sustained oxidative and possibly endoplasmic reticulum stress and liver cell proliferation. PPARα requires transcription coactivator PPAR-binding protein (PBP)/mediator subunit 1(MED1) for its transcriptional activity.
Introduction
Peroxisome proliferator-activated receptor (PPAR) α (NR1C1), β/δ (NR1C2), and γ (NR1C3), are ligand-activated transcription factors that are members of the nuclear-hormone receptor (NR) superfamily [Bookout et al., 2006; Issemann and Green, 1990; Michalik et al., 2006]. The foundation for the discovery and designation of the PPAR subfamily of nuclear receptors in the 1990s [Dreyer et al., 1992; Issemann and Green, 1990], is the cumulative work over the preceding 25 years with peroxisome proliferators (PPs), a group of structurally diverse chemicals that lower serum lipids, and induce massive proliferation of peroxisomes in liver cells, with associated coordinated transcriptional activation of peroxisomal fatty acid β-oxidation system genes [Reddy et al., 1980; Reddy and Krishnakantha, 1975; Reddy et al., 1976; Reddy et al., 1986b]. A receptor for these chemicals was originally postulated in the mid 1980s [Lalwani et al., 1987; Lalwani et al., 1983; Rao and Reddy, 1987; Reddy and Lalwani, 1983] to account for the cell-specific highly reproducible pleiotropic responses [Reddy and Chu, 1996]. Chronic exposure to PPs results in the development of liver cancer in rats and mice, although these agents are nonmutagenic [Rao and Reddy, 1987; Reddy et al., 1980; Reddy et al., 1976]. It was therefore envisaged that a receptor-mediated mechanism is responsible for both the immediate and long-term effects of PPs, including the development of liver cancer [Rao and Reddy, 1987; Reddy and Chu, 1996]. A search for a receptor for these ligands was underway [Lalwani et al., 1987], and this culminated in the cloning of a receptor activated by PPs by Issemann and Green [Issemann and Green, 1990]. This receptor was appropriately designated PP-Activated Receptor or PPAR [Issemann and Green, 1990]. Subsequent identification of two other members of this family, namely PPARβ/δ and PPARγ, resulted in designating the original receptor activated by PPs as PPARα [Dreyer et al., 1992]. Gene knockout studies in mice have unequivocally established that PPARα is indeed responsible for the PP-induced array of pleiotropic responses including liver cancer in rodents [Lee et al., 1995].
All three members of the PPAR subfamily function as sensors for fatty acids and fatty acid derivatives, and thus control important metabolic pathways involved in lipid and energy metabolism [Chawla et al., 2001; Krey et al., 1997]. In the liver, PPARα is activated by synthetic PPs and by both saturated and polyunsaturated fatty acids and their derivatives, and regulates fatty acid oxidation systems to increase energy combustion. PPARα also plays a role in lipoprotein synthesis, inflammatory responses and the development of cancer in the rodent liver [Lee et al., 1995; Mei et al., 2009; Mukherjee et al., 2008]. PPARβ/δ also enhances fatty acid catabolism and energy uncoupling in skeletal muscle and adipose tissue as well as recently shown in the liver where it may participate in the hepatic response to starvation [Sanderson et al., 2009; Wang et al., 2003]. Additionally, PPARβ/δ may play a protective role in the liver by downregulating inflammatory signals in circumstances of liver damage [Nagasawa et al., 2006; Shan et al., 2008a; Shan et al., 2008b]. Together, PPARα and PPARβ/δ are active participants in energy burning, whereas PPARγ, which has two isoforms, PPARγ1 and PPARγ2, is critical in regulating adipocyte differentiation and energy storage by adipocytes [Kliewer et al., 2001; Tontonoz et al., 1994; Zhu et al., 1995]. PPARγ can also induce lipid accumulation in other cell types, such as hepatocytes, when overexpressed [Yu et al., 2003]. In essence, PPARγ regulates anabolic lipid metabolism, while PPARα and PPARβ/δ function as catabolic regulators of energy [Chawla et al., 1994; Tontonoz et al., 1994; Wang et al., 2003].
The PPARα gene and protein structure
The human PPARα (NR1C1) gene, which spans ~93.2 kb, is located on chromosome 22 at position 22q12-q13.1 and it encodes a protein of 468 amino acids [Sher et al., 1993]. The mouse PPARα gene, located on chromosome 15E2, also encodes a protein of 468 amino acids [Issemann and Green, 1990]. As in the mouse, the human PPARα gene encoded mRNA is derived from 8 exons with a 5' untranslated region encoded by exons 1, 2, and part of exon 3 [Desvergne and Wahli, 1999; Sher et al., 1993]. The remainder of exon 3 and exons 4-8 contribute to the coding region of PPARα [Desvergne and Wahli, 1999; Sher et al., 1993]. The last 232 bp of exon 8 contribute to the 3'-untranslated region [Vohl et al., 2000].
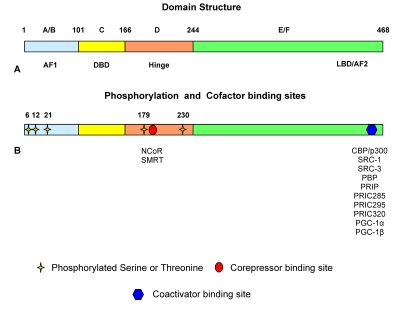
PPARα shares common structural characteristics with the other members of the NR family [Desvergne and Wahli, 1999]. It displays five distinguishable domains designated the A/B, C, D, E and F domains, respectively (Figure 1). The PPARα N-terminal A/B domain contains an activation function-1 (AF-1) region. This region has a low level of basal transcriptional activity and functions independent of ligand-binding [Desvergne and Wahli, 1999]. The specificity of gene transcription is granted by the isoform-specific sequence of the A/B domain of the receptor [Hummasti and Tontonoz, 2006]. Chimeric proteins generated by fusion with the A/B domains of other receptor proteins attenuate the specificity of target gene activation [Hummasti and Tontonoz, 2006]. The A/B domain is followed by a DNA-binding domain (C) encompassing amino acids 101-166 in the human that contains two very highly conserved zinc finger motifs and architectural elements capable of sequence-specific binding to DNA [Desvergne and Wahli, 1999].
Figure 1. Schematic view of the PPARα structure.
Domain structure of the PPARα protein (A) with the ligand-independent activation function 1 (AF1) domain or A/B domain shown in blue, the DNA-binding domain (DBD) or C domain shown in yellow, the hinge or D domain shown in orange, and the ligand-binding domain (LBD) or E domain together with the activation function 2 (AF2) or F domain shown in green. (B) Schematic representation of the PPARα protein with phosphorylation sites labeled with yellow stars, the corepressor site labeled with a red circle, and the coactivator binding site shown with a blue hexagon.
Next to the DNA-binding domain is a flexible hinge domain (D), which connects the DNA-binding domain and the ligand-binding domain (E) [Desvergne and Wahli, 1999]. This hinge region binds corepressor proteins, with the characteristic LXXXIXXXL repressor motif, to the receptor in its quiescent, unliganded state [Dowell et al., 1999]. The corepressor-binding site partly overlaps with the coactivator-binding site. The ligand binding domain in the human PPARα protein extends from amino acids 280-468 and contains an activation function-2 (AF-2) region composed of two α-helices flanking one four-stranded β-sheet [Desvergne and Wahli, 1999]. The AF-2 domain is repressed until ligand-binding occurs. Following ligand-binding, the AF-2 domain undergoes a conformational shift, which allows the formation of hydrogen bonds between Tyr-314 and Tyr-464, as well as the formation of a charge clamp between Glu-462 and Lys-292 [Xu et al., 2001]. This conformational change in the protein allows interaction of the receptor with the LXXLL (L, leucine; X, any amino acid) motifs located in coactivator proteins [Xu et al., 2001]. PPARα, like other nuclear receptors, will undergo conformational adjustment upon binding to a ligand to achieve the coregulator exchanges and activation of the target genes [Xu et al., 2001].
The PPAR ligand binding cavity, with a total volume of 1300 to 1400 A°, is larger than that found in other NRs [Desvergne and Wahli, 1999; Xu et al., 2001]. It is defined by 34 amino acid residues and 80% of these residues are highly conserved across the three PPAR subtypes [Dreyer et al., 1992]. PPARα and PPARγ ligand binding cavities are similar in size, but the PPARβ/δ pocket is significantly narrower in the region adjacent to the AF-2 helix and does not readily accommodate bulky polar heads [Cronet et al., 2001]. Similarity between PPARα and PPARγ ligand binding pockets may account for some degree of flexibility in ligand-receptor interaction between these two isoforms and their ability to bind a wide range of synthetic and natural lipophilic compounds [Xu et al., 2001]. Comparison of the crystal structures of the ligand binding domains showed that a single amino acid, tyrosine 314 in PPARα and histidine 323 in PPARγ, accounts for the subtype selectivity for classes of ligands [Xu et al., 2001]. The PPARα pocket is more lipophilic than that of PPARγ and this may account for the greater affinity of PPARα to bind the more lipophilic-saturated fatty acids [Xu et al., 2001].
Expression of PPARα
PPARα belongs to the type II nuclear receptor family and like the other two PPAR isotypes, is localized in the nucleus. This differs from type I nuclear receptors, which are cytosolic and translocate to the nucleus upon ligand-binding. High levels of PPARα expression are found in tissues with active fatty acid catabolism, such as liver, heart, kidney, brown adipose tissue, muscle, small intestine and the large intestine [Bookout et al., 2006; Braissant et al., 1996; Issemann and Green, 1990]. Similar tissue expression profiles of PPARα have been found in rodents and human [Bookout et al., 2006]. PPARα exerts a dominant role in fatty acid catabolism and ketone body synthesis in the liver [Kersten et al., 1999]. PPARβ/δ also participates in energy catabolism, but its effects are more prominent in muscle and heart, though it also regulates some genes in the liver [Sanderson et al., 2009]. In contrast, PPARγ is expressed abundantly in white adipose tissue, where it is involved in lipid storage and in brown adipose tissue, where it participates in energy dissipation [Tontonoz et al., 1994].
While PPARα regulates many target genes, the expression of the PPARα gene itself is under the control of other transcription factors [Pineda Torra et al., 2002]. In rodents, PPARα is regulated by various physiological conditions such as stress, hormones (including growth hormone), glucocorticoids, insulin, and leptin [Inoue et al., 2008; Lee et al., 2002]. PPARα expression is also reported to be related to aging [Poynter and Daynes, 1998], and is induced during brown adipocyte differentiation [Kelly et al., 1998]. Expression levels of PPARα in mouse liver are increased during starvation and this leads to enhanced expression of PPARα target genes in liver to catabolize influxed fatty acids [Kersten et al., 1999]. Human PPARα is also regulated at the transcriptional level by nuclear receptors, such as hepatocyte nuclear factor 4 (HNF4), and chicken ovalbumin upstream promoter-transcription factor II (COUP-TFII) [Pineda Torra et al., 2002]. HNF4 positively regulates PPARα expression via a direct repeat 1 (DR1) element, which is composed of the consensus sequence AGG(A/T)CA separated with a single nucleotide spacing between two repeats [Pineda Torra et al., 2002]. On the other hand, the DR1 element in the human PPARα promoter is antagonized by the orphan receptor COUP-TFII [Pineda Torra et al., 2002]. Furthermore, PPARα appears to modulate its own expression [Corton et al., 2000]. In human liver, PPARα mRNA levels may vary significantly among individuals and are considered lower than the levels noted in rodent livers, but this may in part be related to variations in liver sample procurement for the frozen archival liver bank [Palmer et al., 1998; Walgren et al., 2000]. PPARα transcript levels are also induced during macrophage differentiation by high glucose levels [Rigamonti et al., 2008]. PPARα is also regulated by the ubiquitin proteasomal degradation system [Genini et al., 2008].
Posttranslational modifications
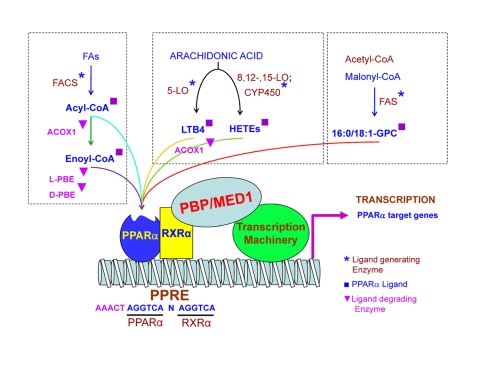
A truncated PPARα isoform resulting from a posttranslational exon skipping mechanism has been detected in human, swine, and jerboas [El Kebbaj et al., 2009; Gervois et al., 1999]. This truncated form harbors dominant negative transcriptional repressive activity on PPARα. In addition, PPARα activity is regulated by posttranslational modifications such as phosphorylation (Figure 1). PPARα was first shown to be phosphorylated in primary rat adipocytes in culture [Shalev et al., 1996]. Insulin treatment increases PPARα phosphorylation and nearly doubles its transcriptional activity in CV-1 and HepG2 cells in culture [Shalev et al., 1996]. This insulin-mediated phosphorylation of PPARα involves amino acids Ser12 and Ser21 [Juge-Aubry et al., 1999]. Stress stimuli exert an increase in PPARα phosphorylation in rat neonatal cardiac myocytes by the p38 mitogen-activated protein kinase (MAPK) pathway [Barger et al., 2001]. p38 MAPK phosphorylates Ser 6, 12 and 21 residues located within the N-terminal A/B domain of the protein and this phosphorylation significantly enhanced ligand-dependent transactivation of PPARα [Barger et al., 2001]. PPARα is also reported to be phosphorylated by protein kinase C (PKC). Inhibition of PKC activity impairs ligand-activated PPARα transactivation activity, but enhances PPARα transrepression activity. Purified PPARα protein is phosphorylated in vitro by recombinant PKCα and -βII. These data demonstrate that the PKC signaling pathway may act as a molecular switch for the transactivation and transrepression properties of PPARα, which involve phosphorylation of Ser179 and Ser230 [Blanquart et al., 2004]. This PKC phosphorylation of PPARα is believed to play an important role in statin-mediated acute anti-inflammatory effects, though the importance of phosphorylation in PPARα signaling has never been clearly demonstrated in vivo [Paumelle et al., 2006].
PPARα ligands
PPARα ligands fall into two general categories: synthetic xenobiotics (exogenous) and biological molecules (endogenous). Synthetic ligands, referred to as PPs, include hypolipidemic drugs such as clofibrate, fenofibrate, gemfibrozil, bezafibrate, ciprofibrate, nafenopin, methyl clofenapate, tibric acid, and Wy-14,643 (pirinixic acid) used in the treatment of dyslipidemias, and industrial phthalate-monoester plasticizers, such as di-(2-ethylhexyl)-phthalate (DEHP), and di-(2-ethylhexyl) adipate (DEHA) used in the manufacture of polyvinyl chloride plastics [Lalwani et al., 1983; Maloney and Waxman, 1999]. Certain herbicides, pesticides, industrial solvents, food flavoring agents, and leukotriene D4 receptor antagonists also function as PPARα ligands [Maloney and Waxman, 1999]. These structurally-diverse chemicals induce highly reproducible and similar pleiotropic responses in the rat and mouse liver. Proliferation of peroxisomes in liver parenchymal cells and profound transcriptional activation of fatty acid oxidation system genes are the hallmarks of PP-induced pleiotropic responses in the rat and mouse liver.
Endogenous or naturally-occurring biological molecules that serve as PPARα ligands include fatty acids and fatty acid derivatives (Figure 2) [Devchand et al., 1996; Hostetler et al., 2005]. Endogenous PPARα activators, in most part, have been identified using in vitro approaches [Forman et al., 1997; Gottlicher et al., 1992; Hostetler et al., 2005; Krey et al., 1997]. Both dietary saturated and unsaturated fatty acids function as direct ligands for PPARα [Hostetler et al., 2005]. Alternately, the ultimate ligands for this receptor may be generated during fatty acid catabolism and new fatty acid synthesis in vivo [Chakravarthy et al., 2009] (Figure 2). Several enzymes such as the 8-, 12-, 15- and 5-lipoxygeneases, the cyclooxygenases and cytochrome P450s utilize fatty acids as substrates to produce putative PPARα ligands [Brash, 1999; Crisafulli and Cuzzocrea, 2009; Ng et al., 2007]. Although many synthetic ligands activate PPARα efficiently, it appears that PPARα has evolved primarily for lipid sensing to regulate energy combustion [Reddy, 2004; Reddy and Rao, 2006].
Figure 2. Biological ligands of PPARα.
Diagram illustrating different known biological ligands of PPARα. Metabolic sources for ligands are grouped in the boxes together with ligand generating enzymes marked with an asterisk, ligands marked with a square and ligand degrading enzymes marked with a triangle. The PPARα and RXRα heterodimer are shown bound to a PPRE sequence in the promoter of a target gene with associated coactivator proteins forming a complex with the cellular transcription machinery.
Evidence suggests that PPARα senses certain endogenous lipid metabolic intermediates as ligands and participates in their metabolism (degradation) by inducing downstream lipid metabolism-related genes [Fan et al., 1998]. The first in vivo evidence came from studies with fatty acyl-CoA oxidase 1 (ACOX1) gene disrupted mice, in that mice deficient in this first and rate-limiting enzyme of the fatty acid β-oxidation system reveal profound activation of PPARα in liver [Fan et al., 1996; Fan et al., 1998; Yeldandi et al., 2000]. Animals deficient for the ACOX1 have high levels of the very long chain fatty acids and since very-long chain fatty acyl-CoAs are incapable of entering the fatty acid oxidation pathway due to ACOX1 deficiency, these unmetabolized substrates act as ligands to hyperactivate PPARα [Fan et al., 1998; Yeldandi et al., 2000]. Recent studies with other gene knockout mouse models suggest that enzymes such as ACOX1, enoyl-CoA hydratase/L-3-hydroxyacyl-CoA dehydrogenase (L-PBE/MFP), D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase (D-PBE/MFP2) and sterol carrier protein x (SCPx) are necessary for the degradative metabolism of endogenously-generated PPARα ligands [Ellinghaus et al., 1999; Fan et al., 1998; Jia et al., 2003; Martens et al., 2008; Qi et al., 1999a; Seedorf et al., 1998], while other enzymes such as fatty-acid synthase (FAS), fatty acyl-CoA synthetase (FACS) and certain lipoxygenases (LO), are required for PPARα ligand generation [Chakravarthy et al., 2009; Martin et al., 2000; Yu and Reddy, 2007]. It is implied that substrates of ACOX1, such as very long-chain fatty acyl-CoAs, when left unmetabolized in the absence of this enzyme, function as potent endogenous ligands of PPARα [Fan et al., 1998; Yeldandi et al., 2000]. It is known that long-chain acyl-CoA esters can act as regulatory molecules in vivo [Faergeman and Knudsen, 1997; Fan et al., 1996; Fan et al., 1998; Hostetler et al., 2005]. PPARα exhibits high affinity for the CoA thioesters of the common (C20-C24) very long chain fatty acids and these CoA thioesters of very-long chain and branched-chain fatty acids are more potent PPARα ligands than the free fatty acids [Hostetler et al., 2005]. Further evidence that fatty acids are not the direct in vivo ligands for PPARα comes from mice with disrupted fatty acyl-CoA synthetase gene [Li et al., 2009]. Fatty acyl-CoA synthetases activate long-chain fatty acids to acyl-CoAs, thereby providing substrates for downstream fatty acid β-oxidation pathways [Li et al., 2009]. Mice with a disrupted fatty acyl-CoA synthetase 1 gene [Li et al., 2009], or deficient for the adrenoleukodystrophy protein gene (X-ALD) [Heinzer et al., 2003; Reddy and Hashimoto, 2001], fail to convert fatty acids to acyl-CoAs to serve as substrates for fatty acid β-oxidation vis à vis ligands for PPARα [Heinzer et al., 2003; Li et al., 2009]. If long-chain fatty acids function as PPARα ligands in vivo, one would have expected increased PPARα target gene activation in X-ALD and fatty acyl-CoA synthetase null mice, but that does not appear to be the case in these null livers [Heinzer et al., 2003; Li et al., 2009]. Failure to induce peroxisome proliferation in liver by dietary lipid overload also indicates that fatty acids per se are not effective PPARα ligands [Fan et al., 1998].
Experiments with liver-specific inactivation of fatty-acid synthase (FAS) suggest that products of FAS serve as endogenous activators of PPARα in adult liver [Chakravarthy et al., 2007]. FAS-deficient mice reveal a phenotype resembling PPARα deficiency [Chakravarthy et al., 2007]. Recently, it has been demonstrated that FAS generates 1-palmityl-2-oleolyl-sn-glycerol-3-phosphocholine (16:0/18:1-GPC), which appears to be a physiologically relevant PPARα ligand [Chakravarthy et al., 2009].
Leukotriene B4 (LTB4) is another well known putative PPARα ligand, which connects the receptor to inflammation and immune responses. 19- and 20-hydroxyeicosatetraenoic acid (19- and 20-HETE) are the major products of cytochrome P450 catalyzed conversion of arachidonic acid and play roles in the cardiovascular system, kidney function, cellular proliferation and inflammation. HETEs bind to and activate PPARα in HepG2 cells transfected with human PPARα, but do not show any transactivation activity when the cells are transfected with the unrelated nuclear receptor TRβ [Ng et al., 2007]. Oleylethanolamide (OEA), a naturally-occurring lipid has been identified as a high affinity PPARα ligand regulating PPARα activity and lipid metabolism. Finding a way of regulating PPARα activity through modulating endogenous ligand, and therefore achieving control of energy metabolism and body weight, may have the potential to generate new therapies.
A high degree of interest presently exists in discovering new partial agonists of PPARα and of other PPAR isoforms that are selective for beneficial effects of PPAR activation and avoid problems with dose-related toxicity and other negative side effects of PPAR activation [Chang et al., 2007]. Direct glucose binding with the receptor has also been shown to occur and may play an important regulatory role in PPARα activity, particularly in the setting of diabetes [Hostetler et al., 2008].
PPARα agonists, including fibrates such as fenofibrate, gemfibrozil and less commonly clofibrate, have been in use for several decades as therapeutic agents to decrease plasma levels of triglycerides and VLDL by enhancing lipid uptake and catabolism and to increase plasma levels of HDL-C [Bays and Stein, 2003; Lefebvre et al., 2006; Marx et al., 2004]. The fibrate class of PPARα ligands increase the production in the liver of the apolipoproteins A-I and A-II, which are major components of HDL-C [Bays and Stein, 2003]. The search for agents that display potent and selective binding to the PPARα isotype that support robust recruitment of transcription coactivators is expected to result in the identification of new classes of ligands [Kane et al., 2009]. These novel ligands, including LY518674, a novel PPARα ligand recently reported by Nissen et al., have the traditional benefits of reducing serum triglycerides and increasing serum HDL-C, but may confer additional benefits such as reduction of serum LDL-C, especially when paired with other treatments. Novel therapeutics such as this may hold the key to better treatments for lipid disorders such as atherogenic dyslipidemia, which lead to other metabolic and cardiac problems [Nissen et al., 2007]. Other current therapeutic benefits of potential PPARα agonists are under investigation for use in the prevention of atherosclerosis, cardiomyopathies, ischemia-reperfusion injuries, prevention of angiogenesis, anti-proliferation of tumorigenic cells and others [Bulhak et al., 2009; Crisafulli and Cuzzocrea, 2009; Gizard et al., 2008; Massip-Salcedo et al., 2008; Panigrahy et al., 2008; Patel et al., 2009; Ramanan et al., 2008; Staels et al., 2008; Wang et al., 2009].
Other PPARα ligands with environmental impact, in particular phthalate ester plasticizers widely used in the manufacture of polyvinyl chloride plastics, and industrial solvents such as trichloroethylene have been in use for over 50 years. These are known contaminants of ground water and estuaries, but the impact on human health is unclear. More recently, certain herbicides have been shown to function as PPARα ligands, implying that these agents also constitute a major category of environmental contaminants that can activate this receptor, and thus raising issues about long-term or lifelong exposure.
PPARα heterodimerization with RXR, binding to DNA elements and PPARα-regulated genes
PPARα, like many non-steroid members of the NR family, functions as an obligate heterodimer with another nuclear receptor, retinoid X receptor (RXR; NR2B) [Kliewer et al., 1992]. The PPARα/RXR heterodimers can form independent of the PPARα ligand and these unliganded heterodimers recruit corepressor protein complex and inhibit target gene transcription [DiRenzo et al., 1997]. Upon ligand binding, the corepressor complex is released from the PPARα/RXR heterodimer, and the coactivator complexes will be recruited to the promoter region of target genes to initiate transcription [Dowell et al., 1999]. The PPARα/RXR heterodimers bind to a specific DNA sequence element called a peroxisome proliferator response element (PPRE), which was initially characterized using the PPAR responsive promoter of the rat Acox1 gene [Chandra et al., 2008; Kliewer et al., 1992; Osumi et al., 1991; Tugwood et al., 1992; van der Meer et al., 2010]. This PPRE element is located in the promoter region of target genes [Tugwood et al., 1992; van der Meer et al., 2010; Zhang et al., 1992]. The PPRE is of the direct repeat 1 (DR1) category, which is composed of two copies of the consensus hexamer sequence AGG(A/T)CA separated by a single base pair (Figure 2). Neither PPARα homodimers nor monomers bind to the PPRE. PPARα target genes usually contain one or more PPRE in the promoter and preferentially bind PPARα/RXR heterodimers [Chu et al., 1995; Tugwood et al., 1992]. Occasionally, PPRE may be in the proximal transcribed regions of certain target genes [Di-Poi et al., 2002]. The PPRE element contains an additional AACT motif positioned 5’ to the DR-1 [Juge-Aubry et al., 1997]. This upstream extended hexamer of the DR1 interacts with PPARs, whereas the downstream hexamer is known to interact with RXR of the PPAR/RXR heterodimer [Desvergne and Wahli, 1999; Juge-Aubry et al., 1999]. Of the three PPARs, PPARγ binds more strongly than do PPARα and PPARβ/δ, implying that conservation of the 5’-flanking extension is more important for the binding of PPARα and PPARβ/δ [Juge-Aubry et al., 1997]. The nature of this upstream extension may confer isotype specificity [IJpenberg et al., 1997].
The presence of PPRE upstream of the fatty acyl-CoA oxidase 1 (ACOX1) gene [Tugwood et al., 1992], and peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase (L-PBE) gene [Zhang et al., 1992] were the first described. Interestingly, these were the first two genes identified to be transcriptionally activated by PPs [Reddy et al., 1986a; Reddy and Hashimoto, 2001; Tugwood et al., 1992; Zhang et al., 1992]. Over the years, many other genes that possess PPREs have been characterized as PPARα target genes (Supplementary File 1 and Supplementary File 2). cDNA microarray profiling and comparative proteomic data have revealed several known PPARα target genes and some new genes that are upregulated in liver by the PPARα ligand, Wy-14,643 [Cherkaoui-Malki et al., 2001; Chu et al., 2004; Kane et al., 2009; Nielsen et al., 2006]. These fall into several functional categories including lipid metabolism, xenobiotic metabolism, inflammation and others.
RXR, the heterodimerization partner of PPARs, is also a partner of many other NRs [Bugge et al., 1992; Juge-Aubry et al., 1995; Mangelsdorf et al., 1995]. This promiscuous centrality of RXR in NR functioning, raises the possibility of competition between NRs for this common binding partner [Juge-Aubry et al., 1995]. Reciprocal negative interaction between the PPARα and thyroid hormone receptor signaling pathways is indirect, in that it is mostly due to sequestration of RXR, the common heterodimeric partner [Chu et al., 1995; Juge-Aubry et al., 1995]. A similar competition can also occur among PPAR isotypes with RXR in different cell types, but the in vivo effects of this competition and limiting amounts of RXR protein remain unclear [Jow and Mukherjee, 1995].
PPARα knockout phenotype
PPARα knockout mice were generated by the Gonzalez group [Lee et al., 1995]. These mice have demonstrated unequivocally that PPARα is indeed the bona fide receptor responsible for mediating the pleiotropic responses induced by synthetic peroxisome proliferators [Lee et al., 1995]. In this knockout mouse model, the ligand binding domain was disrupted by homologous recombination. Mice homozygous for the mutation did not express PPARα protein and yet are viable and fertile, with no detectable gross phenotypic defects [Lee et al., 1995]. PPARα null mice display normal basal levels of the inducible peroxisomal β-oxidation enzymes in liver, but generally lower levels of mitochondrial fatty acid metabolizing enzymes [Aoyama et al., 1998]. PPARα knockout mice do not reveal PP-induced pleiotropic responses when challenged with the classical peroxisome proliferators, clofibrate and Wy-14,643 [Lee et al., 1995]. Subsequently, it was shown that PPARα knockout mice fail to develop liver tumors when chronically exposed to these PPARα ligands, indicating that PP-induced hepatocarcinogenesis is receptor mediated, as previously proposed [Gonzalez and Shah, 2008; Hays et al., 2005; Peters et al., 1997; Rao and Reddy, 1987]. In essence, these observations clearly established that PPARα is the central player in mediating the pleiotropic responses of peroxisome proliferators [Lee et al., 1995]. Numerous studies with PPARα null mice during the past 15 years have demonstrated the critical roles played by this receptor in energy metabolism, hepatic steatosis, inflammation, cardiac pathophysiology, cell cycle alterations and hepatocarcinogenesis [Lefebvre et al., 2006].
PPARα repressors and coactivators
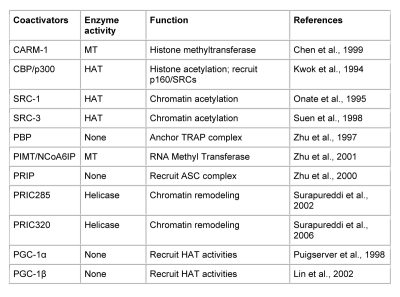
Transcriptional activation of genes is a complex process, which involves the participation of many transcription coregulators [Glass and Rosenfeld, 2000]. PPARs, like other nuclear receptors, interact with coactivators such as SRC-1 (steroid receptor coactivator-1) [Onate et al., 1995; Zhu et al., 1996], or corepressors such as N-CoR (nuclear corepressor) [Chen and Evans, 1995; Dowell et al., 1999], and SMRT (silencing mediator for retinoid and thyroid hormone receptors) [Chen and Evans, 1995; Dowell et al., 1999; Hu and Lazar, 1999]. PPARα-interacting coactivators and corepressors augment or repress, respectively, the PPARα transactivation activity [Feige et al., 2006; Yu and Reddy, 2007; Zoete et al., 2007]. Since the cloning of SRC-1 [Onate et al., 1995] 15 years ago, over 300 coactivators/coregulators have been identified, with new members still being added to this expanding spectrum [Lonard and O'Malley, 2006]. PPARα is known to interact with some of these coregulators (Table 1). These include CBP/p300, three members of the SRC/p160 family, PBP/MED1 (PPAR-binding protein/TRAP220/DRIP205/mediator subunit 1), PRIP/NCoA6 (ASC2/RAP250/TRBP/NRC) [Zhu et al., 2000a], PRIC285, PRIC295, PRIC320, PGC-1α, and PGC-1β, as well as coactivator-associated proteins PIMT (NCoA6IP) and CARM-1 (Table 1). The PPARα-interacting coregulator (PRIC) complex isolated from rat liver nuclear extracts reveals the presence of many of these coregulators, presumably forming one mega complex [Surapureddi et al., 2002]. This diversity raises several issues about the evolutionary importance of the versatility and complexity of coregulatory molecules, their relative abundance in various cell types, and their affinity for a given nuclear receptor in orchestrating transcription in gene-, cell-, and developmental stage-specific transcription.
Table 1. Some known PPARα coactivator proteins.
Coactivator proteins known to interact with activated PPARα, with their enzymatic activities and functions. Acronyms used: (MT) Methyltransferase, (HAT) Histone acetyltransferase. References identifying each protein as a coregulator of PPARα are provided in the reference section.
In the absence of a specific ligand, PPARα interacts with corepressors NCoR and SMRT, but the importance of this interaction for PPARα action is not well understood [Feige et al., 2006]. Homozygous deletion of NCoR or SMRT in mice is embryonic lethal, indicating that they cannot fully compensate for each other during development [Ghisletti et al., 2009; Jepsen et al., 2008; Jepsen et al., 2000]. Furthermore, another corepressor, the Receptor Interacting Protein 140 (RIP140), which can interact with PPARα, is known to repress the activity of NRs by competing with coactivators and by recruiting downstream effectors such as histone deacetylases (HDACs) [White et al., 2004]. Interestingly, the phenotype of RIP140 knockout mice suggests a role for this corepressor in PPARα signaling, as these mice exhibit resistance to high-fat diet-induced obesity resulting from upregulation of genes involved in energy dissipation [Leonardsson et al., 2004].
The binding of ligand to a nuclear receptor influences first, the release of corepressors and beginning recruitment of coactivator complexes, such as members of the SRC-1/p160 family, which exhibit histone acetyltransferase activity required to facilitate chromatin remodeling. Subsequent docking of other coregulators, either singly, or as preassembled multisubunit protein complexes, including mediator complex (MED), facilitates interaction of ligand-activated receptor with RNA polymerase and the general basal transcription machinery to enhance the transcription of a specific set of genes [Dotson et al., 2000; Glass and Rosenfeld, 2000; Shang et al., 2000]. As indicated above, coactivators contain an LXXLL motif that forms two turns of α-helix and binds to a hydrophobic cleft on the surface of the nuclear receptor [Voegel et al., 1998]. Data on the identification and characterization of coactivators have been derived mostly from in vitro observations, but there is a paucity of information about the in vivo cell- and gene-specific functional roles of individual coactivators [Yu and Reddy, 2007].
Coactivator gene-knockout mice: PPARα signaling requires PBP/MED1
Gene knockout mouse models have established that some of the coactivators are essential for embryonic growth and survival, while others are not critical for mouse embryogenesis [Reddy et al., 2006; Yu and Reddy, 2007]. For example, disruption of the PBP/MED1 gene results in embryonic lethality in the mouse around gestational day 11.5 [Crawford et al., 2002; Ito et al., 2000; Landles et al., 2003; Zhu et al., 2000b]. Likewise, germ-line deletion of the PRIP/ASC2/NRC/RAP250/NCoA6 gene [Antonson et al., 2003; Qi et al., 2004], and the PRIP-interacting protein PIMT/TGS1 (NCoA6IP) gene in the mouse also result in embryonic lethality [Jia et al., 2009b; Monecke et al., 2009]. In contrast, null mutation of coactivators SRC-1 [Qi et al., 1999b], SRC-2 [Chopra et al., 2008], and SRC-3, PGC-1, PRIC285 [Viswakarma et al., 2009], and CARM1 result in a viable phenotype with or without overt functional changes.
Studies on the role of germ-line deletion of coactivators SRC-1, SRC-2, and SRC-3 in the mouse revealed that none of these are essential for PPARα ligand-induced pleiotropic responses in liver ([Qi et al., 1999b] and Jia, Y., Xu, J., and Reddy, J.K., unpublished observations). These p160 family members are among the coactivators considered important for nuclear receptor function, but the in vivo results indicate their redundancy at least for PPARα function in liver [Qi et al., 1999b]. It is likely that other coactivators with histone acetyltransferase activity may be adequate to facilitate chromatin remodeling in the absence of SRC-1 and other members of the p160 family [Qi et al., 1999b]. Further studies are needed to ascertain the effects of loss of two or more members of this p160 family in PPARα function. Combined loss of p/CIP/SRC-3 and SRC-1 function has been shown to lead to a specific block in brown fat development, in part due to a failure of induction of PPARγ target genes [Wang et al., 2006]. Loss of both p/CIP/SRC-3 and SRC-1 also leads to increased food intake on both regular chow and a high-fat diet, indicating excess energy burning [Wang et al., 2006]. Whether loss of these two coactivators results in strongly increased energy expenditure by regulating PPARα- [Reddy and Hashimoto, 2001; Reddy and Rao, 2006] and PPARβ/δ-dependent transcriptional activities [Wang et al., 2003] remains to be investigated.
To study the cell- and gene-specific functions of coactivator genes such as PBP/MED1 and PRIP/NCoA6, which when germ-line disrupted, lead to embryonic lethality in the mouse, it becomes necessary to generate conditional null mice using the Cre-loxP strategy [Jia et al., 2005; Jia et al., 2004; Jia et al., 2009a; Matsumoto et al., 2007; Sarkar et al., 2007]. Conditional deletion of PBP/MED1 gene in liver results in the abrogation of PPARα ligand-induced pleiotropic effects, indicating that PBP/MED1 is essential for PPARα signaling [Jia et al., 2005; Matsumoto et al., 2007]. When fed a diet containing the PPARα ligands, Wy-14,643 or ciprofibrate, PBP/MED1 liver null mice revealed no peroxisome proliferation in PBP/MED1-deficient hepatocytes [Jia et al., 2004]. The presence of an occasional PBP/MED1-positive hepatocyte in these livers that escaped Cre-excision provided a characteristic visual contrast, in that such cells displayed the expected response to PPARα ligands, whereas the response of PBP/MED1-deficient hepatocytes was similar to that observed in cells that lacked PPARα [Jia et al., 2004]. PPARα ligand-induced liver cell proliferation was abolished in PBP/MED1 null hepatocytes and in these livers, residual PBP/MED1-expressing cells displayed a proliferative advantage [Jia et al., 2004; Matsumoto et al., 2007]. Furthermore, PBP/MED1 is essential for partial hepatectomy-related liver regeneration and PPARα ligand Wy-14,643-induced receptor-mediated hepatocarcinogenesis [Matsumoto et al., 2007]. PBP/MED1-deficient hepatocytes also fail to give rise to liver tumors following the administration of the genotoxic carcinogen diethylnitrosamine [Matsumoto et al., 2010].
Deletion of the PBP/MED1 gene in liver also abolished the responses induced by constitutive androstane receptor (CAR) activators, phenobarbital or 1, 4-bis[2-(3,5-dichloropyridyloxy)]benzene (TCPOBOP) and of acetaminophen-induced hepatotoxicity [Jia et al., 2005; Jia et al., 2009a]. In contrast, liver conditional knockout of the PRIP/NCoA6 gene had no effect on PPARα and CAR signaling in liver [Sarkar et al., 2007]. Mice with floxed PBP/MED1 [Jia et al., 2005; Jia et al., 2009a; Matsumoto et al., 2007], PRIP/NCoA6 [Antonson et al., 2003; Qi et al., 2004], PIMT/TGS1/NCoA6IP [Jia et al., 2009b], PRIC285 [Viswakarma et al., 2009]; and PRIC320 (Viswakarma, N. and Reddy, J.K., unpublished observation) alleles should serve as valuable tools for conditional gene disruption in cells of interest to further evaluate the functional roles of PPARα.
PPARα functions
PPARα and energy burning
Obesity, resulting from increased consumption of energy-rich foods high in saturated fats and sugars, and reduced physical activity, has reached epidemic proportions globally, with more than 1 billion adults overweight [WHO, 2006]. When consumption of energy far exceeds the combustion of calories, there is increased deposition of long-chain fatty acids, principally in the form of triglycerides in adipose and other tissues leading to obesity [Evans et al., 2004]. All three members of the PPAR subfamily of nuclear receptors have been shown to participate in energy metabolism in that PPARα and PPARβ/δ function mostly as catabolic regulators of energy expenditure, while PPARγ regulates anabolic metabolism in that it plays a role in energy storage [Chawla et al., 2001; Chawla et al., 1994; Cook et al., 2000; Evans et al., 2004; Krey et al., 1997; Tontonoz et al., 1994].
The liver is a central player in the whole body energy homeostasis by its ability to orchestrate fatty acid and glucose metabolism. Hepatic lipid metabolism principally involves three aspects: lipogenesis, oxidation of fatty acids and secretion of lipids. Lipogenesis consists of de novo fatty acid synthesis and subsequent conversion of these fatty acids into triglycerides. In the liver, lipogenesis is regulated by transcription factors, sterol regulatory element-binding protein (SREBP-1c), carbohydrate response-element binding protein (ChREBP) and PPARγ [Browning and Horton, 2004; Horton et al., 2002; Wang et al., 2003; Yu et al., 2003]. SREBP-1c regulates the expression of constellations of glycolytic and lipogenic genes, including stearoyl CoA desaturase (Scd-1) and fatty acid synthase (Fas) [Hebbachi et al., 2008; Horton et al., 2002; Reddy and Rao, 2006]. Emerging evidence suggests that PPARα also influences lipogenesis by increasing the transcription of Scd-1 and other lipogenic genes by regulating the primary transcription factors SREBP-1c and liver X receptor α (LXRα) [Hebbachi et al., 2008]. It is suggested that PPARα participates in the generation of an endogenous LXRα ligand, since a synthetic, non-steroidal ligand of LXRα has been found effective in the induction of lipogenic genes in mice deficient in PPARα [Hebbachi et al., 2008]. However, the role of PPARα in lipogenesis, albeit modest, is intriguing, as it appears paradoxical to the well-known role of this transcription factor in the regulation of fatty acid oxidation [Reddy and Hashimoto, 2001]. The involvement of PPARα in lipogenesis may suggest a fail-safe compensatory mechanism for the removal of important fatty acids, for example, during starvation [Hebbachi et al., 2008].
Liver plays an important role in fatty acid oxidation and this catabolic energy burning is regulated by PPARα [Reddy and Hashimoto, 2001; Reddy and Rao, 2006]. Oxidation of fatty acids occurs in three subcellular organelles, with the bulk of β-oxidation confined to mitochondria and peroxisomes, while CYP4A-catalyzed ω-oxidation takes place in the endoplasmic reticulum [Rao and Reddy, 2004; Reddy and Hashimoto, 2001]. Some of the key enzymes involved in these three fatty acid oxidation systems possess PPRE elements and are regulated by PPARα, though PPARβ/δ has also been shown to regulate some of these genes [Desvergne and Wahli, 1999; Reddy and Chu, 1996; Reddy and Hashimoto, 2001; Sanderson et al., 2009; van der Meer et al., 2010; Varanasi et al., 1996]. Fatty acids are converted into acyl-CoAs prior to oxidation.
Mitochondrial β-oxidation is primarily involved in the oxidation of the major portion of the short-(<C8), medium-(C8-C12) and long-(C12-C20) chain fatty acids, and in the process, constitutes the primary source of energy derived from fatty acids [Reddy and Hashimoto, 2001]. The PPARα-regulated enzymes, long chain acyl-CoA synthetases and carnitine palmitoyl transferase-1, are essential, respectively, for generating fatty acyl-CoA, and facilitating the entry of fatty acyl carnitine into mitochondria [Coleman et al., 2002]. The first step in mitochondrial β-oxidation is comprised of α-β-dehydrogenation of the fatty acyl-CoA ester by a family of four chain length specific straight chain acyl-CoA dehydrogenases [Hashimoto, 1999; Reddy and Hashimoto, 2001]. These include very-long chain, medium chain and short chain enzymes. A heterotrimeric protein, exhibiting 2-enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase, and 3-keto acyl-CoA thiolase activities, participates in the second, third and fourth steps in the mitochondrial β-oxidation pathway [Hashimoto, 1999; Reddy and Rao, 2006]. Mitochondrial β-oxidation results in the progressive shortening of fatty acids into acetyl-CoA subunits. These either condense into ketone bodies to serve as oxidizable energy substrates for extrahepatic tissues, especially during starvation, or enter into the tricarboxylic acid cycle for further oxidation to water and carbon dioxide [Reddy and Hashimoto, 2001; Reddy and Rao, 2006].
Peroxisomal β-oxidation is streamlined toward the metabolism, almost exclusively, of very long straight-chain fatty acids (>C20), 2-methyl branched fatty acids (e.g., pristanic acid that is generated by α-oxidation of the 3-methyl-branched fatty acid phytanic acid), prostanoids, dicarboxylic acids, and the C27 bile acid intermediates di- and trihydroxycoprostanoic acids [Ferdinandusse et al., 2009; Jia et al., 2003]. Substrates for peroxisomal β-oxidation are less abundant and relatively more toxic, and they are not processed by the mitochondrial β-oxidation system [Reddy and Rao, 2006]. However, peroxisomal β-oxidation is required to shorten the chain length of very long chain fatty acids and chain-shortened acyl-CoAs are shunted to mitochondria for the completion of β-oxidation [Reddy and Hashimoto, 2001]. The coordinate inducibility of peroxisomal β-oxidation system enzymes by peroxisome proliferators in now well established [Reddy and Hashimoto, 2001; Reddy et al., 1986b] and this induction is regulated by PPARα [Issemann and Green, 1990; Kliewer et al., 2001; Lee et al., 1995].
Peroxisomal β-oxidation consists of four metabolic steps and each metabolic conversion can be performed by at least two different enzymes [Hashimoto, 1999; Reddy and Hashimoto, 2001]. Some of these enzymes are inducible by peroxisome proliferators, while others seem refractive to induction by these PPARα ligands [Reddy and Hashimoto, 2001]. Straight-chain fatty acyl-CoAs serve as substrates for the classical inducible pathway, whereas the second non-inducible β-oxidation enzyme system acts on 2-methyl-branched fatty acyl-CoAs [Hashimoto, 1999]. In the inducible PPARα-regulated peroxisomal β-oxidation pathway, fatty acyl-CoA oxidase 1 (ACOX1) is responsible for the initial oxidation of very-long-chain fatty acyl-CoAs to their corresponding trans-2-enoyl-CoAs. On the other hand, the non-inducible fatty acyl-CoA oxidase 2 (ACOX2) serves as the first step in the metabolism of branched-chain fatty acyl-CoAs. The second and third reactions in the inducible β-oxidation system, hydration and dehydrogenation of enoyl-CoA esters to 3-ketoacyl-CoA are catalyzed by a single bifunctional enzyme enoyl-CoA hydratase/L-3-hydroxyacyl-CoA dehydrogenase (L-PBE/MFP1). Ketoacyl-CoAs generated by L-PBE/MFP1 are then converted by 3-ketoacyl-CoA thiolase (PTL) to acetyl-CoA and to an acyl-CoA that is two carbon atoms shorter than the original molecule. The shortened acyl-CoA reenters the β-oxidation cycle, and this process repeats for about 5 cycles, resulting in the removal of ten carbon atoms. The appropriately chain-shortened acyl-CoAs are then transported to mitochondria for the completion of β-oxidation [Hashimoto, 1999]. All three genes of this classical inducible peroxisomal β-oxidation system (ACOX1, L-PBE/MFP1, and PTL) are tightly regulated by PPARα. Of interest is that L-PBE/MFP1 protein is induced abundantly in the livers of rats and mice treated with peroxisome proliferators and accounts in part for the increase in peroxisome population [Jia et al., 2003; Reddy, 2004]. Although, L-PBE/MFP1 is highly inducible by both synthetic and natural PPARα ligands in liver, the functions carried out by this enzyme can be also performed highly effectively by the D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase (D-bi/multifunctional enzyme 2; D-PBE/MFP2) [Hashimoto, 1999; Jia et al., 2003]. 3-ketoacyl-CoAs generated by D-PBE/MFP2 of the non-inducible pathway are cleaved by the third enzyme of this system, sterol carrier protein X, which possesses thiolase activity [Hashimoto, 1999].
In addition to mitochondrial and peroxisomal β-oxidative processing, fatty acids are also oxidized by microsomal ω-oxidation carried out by CYP4A enzymes that are regulated by PPARα [Reddy and Hashimoto, 2001]. The first step of ω-hydroxylation of saturated and unsaturated fatty acids occurs in the endoplasmic reticulum and the resulting ω-hydroxy fatty acid is then dehydrogenated to yield highly toxic dicarboxylic acids in the cytosol. Dicarboxylic acids are converted to dicarboxylyl-CoAs and enter peroxisomes for further metabolism by the classical inducible β-oxidative enzymes [Ferdinandusse et al., 2004; Reddy and Rao, 2006]. Dicarboxylic acids are unique in that they serve as substrates for the peroxisomal β-oxidation system and also as ligands for PPARα [Hashimoto, 1999]. It would appear that dicarboxylic fatty acids streamline their own metabolism by inducing PPARα activation and thus regulating all three fatty acid oxidation enzyme pathways [Reddy and Hashimoto, 2001].
In summary, the PPARα regulates all three fatty acid oxidation systems. Gene knockout mouse models have provided evidence to indicate that substrates for peroxisomal β-oxidation function as PPARα ligands to enhance energy burning. In this regard, hyperactivation of PPARα by pharmacological intervention might prove to be useful as an adjuvant to exercise in combating obesity in individuals who are refractory to modulating energy consumption. PPARα ligands do not appear to cause weight loss independent of PPARα activation.
PPARα and hepatic steatosis
Fatty liver disease is the most common liver disease, as it encompasses a morphological spectrum of hepatic steatosis and steatohepatitis that progresses to cirrhosis and hepatocellular carcinoma [Reddy, 2001; Reddy and Rao, 2006; Zafrani, 2004]. Fatty liver disease occurs worldwide in those with excess alcohol consumption (alcoholic fatty liver disease), and those who are obese with or without added insulin resistance (non-alcoholic fatty liver disease) [Crabb et al., 2004; Evans et al., 2004; Hamaguchi et al., 2005; Reddy and Rao, 2006; Sozio and Crabb, 2008]. Several metabolic and genetic diseases that influence fatty acid metabolism also develop fatty liver disease [Browning and Horton, 2004; Reddy and Rao, 2006]. Fatty liver disease is the culmination of increased energy uptake, increased hepatic lipogenesis, decreased energy combustion and decreased hepatic secretion of liver triglycerides. PPARα gene knockout mice have provided valuable clues regarding the role of this transcription factor in energy metabolism by liver.
By virtue of its unique ability to orchestrate fatty acid oxidation, PPARα appears to play a significant role in the pathogenesis of hepatic steatosis. First, as indicated above, PPARα influences the expression of hepatic lipogenic genes by regulating the primary transcription factors SREBP-1c and liver X receptor α (LXRα) [Browning and Horton, 2004; Hebbachi et al., 2008]. Second, in conditions for increased demand for fatty acid oxidation, such as starvation, PPARα is essential for the upregulation of some of the enzymes necessary for this process, though PPARβ/δ has recently been shown to be important in regulating some of these genes independently of PPARα [Sanderson et al., 2009]. Under fasted conditions, PPARα senses the lipid influx into the liver and upregulates all three fatty acid oxidation systems to burn the energy and minimize hepatic steatosis [Hashimoto et al., 2000; Kersten et al., 1999; Leone et al., 1999]. PPARα null mice are unable to efficiently induce the expression of genes involved in the oxidization of fatty acids released from adipose tissue. Accordingly, PPARα-deficient mice exhibit hepatic steatosis, hyperlipidemia and hypothermia [Hashimoto et al., 2000; Kersten et al., 1999; Leone et al., 1999]. PPARα null mice also develop severe steatohepatitis compared to wild-type mice when maintained on a diet deficient in methionine and choline [Ip et al., 2003]. Furthermore, upon treatment with PPARα ligands there is an attenuation of methionine-choline deficient diet-induced steatohepatitis [Rao and Reddy, 2004].
PPARα null mice fed ethanol develop marked hepatomegaly, steatohepatitis, and liver cell death and proliferation, implying a role for decreased fatty acid oxidation in mice deficient in this transcription factor [Nakajima et al., 2004]. Furthermore, ethanol is known to inhibit fatty acid oxidation and this is attributed to ethanol inhibition of PPARα transcription [Sozio and Crabb, 2008]. Hypo-activity of PPARα might play a role in the severity of alcohol liver disease in the human. On the other hand, it has been shown that PPARα activation is essential for hepatitis C virus (HCV) core protein-induced hepatic steatosis and hepatocellular carcinoma in mice [Tanaka et al., 2008].
PPARα and hypolipidemic effects
PPARα ligands reduce VLDL production and enhance the catabolism of TG-rich particles, which indirectly decreases small dense LDL (sdLDL) particles, enhancing the formation of HDL particles and hepatic elimination of excess cholesterol [Lefebvre et al., 2006]. PPARα upregulates lipoprotein lipase transcription in liver and muscle and this leads to increased TG hydrolysis.
The activity of ApoC-III, which is an inhibitor of both LPL activity and remnant clearance, is lowered by PPARα agonists [Peters et al., 1997]. PPARα agonists induce ABCA1 and SR-BI expression in macrophages. HDL apolipoprotein genes apoA-I and apoA-II are activated by PPARα by direct transcriptional control [Desvergne and Wahli, 1999; Lefebvre et al., 2006; Peters et al., 1997]. Thus, PPARα activation by fibrates and other compounds, elicits a global normolipidemic response, by reducing TG-rich particle production, increasing their lipolysis, and promoting HDL metabolism [Lefebvre et al., 2006].
PPARα and inflammation
Evidence indicates that PPARα plays a beneficial role in reducing inflammation. Activation of this receptor appears to influence both acute and chronic inflammatory disorders involving neutrophils and macrophages. Leukotriene B4 (LTB4), a powerful chemotactic inflammatory eicosanoid, is an endogenous PPARα ligand (Figure 2). Like other PPARα ligands, it induces transcription of genes of the β- and ω-oxidation pathways that neutralize and degrade LTB4 itself to regulate the inflammatory response [Devchand et al., 1996; Ford-Hutchinson, 1990]. Absence of PPARα prolongs the LTB4-induced inflammatory response [Devchand et al., 1996]. Agents containing LTB4 or its precursor arachidonic acid, when applied to the ears of PPARα knockout and wild-type mice, showed that the inflammatory response was significantly prolonged in PPARα null mice, compared to the wild-type controls [Devchand et al., 1996]. These data established that PPARα regulates the duration of the inflammatory response, possibly through limiting cytokine expression and by inducing genes that metabolize LTB4 [Devchand et al., 1996]. Based on these observations, it is logical to surmise that PPARα ligands exert potential anti-inflammatory effects in modulating various inflammatory processes such as atherogenesis and hepatitis [Devchand et al., 1996; Ricote and Glass, 2007; Staels et al., 2008; Zandbergen and Plutzky, 2007].
Macrophages are mediators of inflammation in the vasculature and participate in the development of atherosclerotic plaques [Schwartz et al., 1985]. In the pathogenesis of atherosclerosis, PPARα activation could result initially in reduced leukocyte adhesion to activated endothelial cells of the arterial lumen, and subsequently in inhibiting the formation of macrophage foam cells by regulating expression of genes involved in reverse cholesterol transport and reactive oxygen species output [Chinetti et al., 2001; Zandbergen and Plutzky, 2007]. Accordingly, activation of PPARα may be beneficial in curtailing the inflammatory response and, in particular, the formation and progression of atherosclerotic plaques by minimizing lipoprotein oxidative modifications.
PPARα ligands significantly reduce the levels of pro-inflammatory cytokines such as interleukin-1 (IL-1), tumor necrosis factor-α (TNF-α), cyclooxygenase-2 (COX-2), and inducible nitric oxide synthase (iNOS) by inhibiting the translocation of the p65 subunit of nuclear factor κ-B (NF-κB) and decreasing phosphorylation of the c-jun subunit of AP-1 [Ramanan et al., 2008]. Loss of PPARα-mediated gene transcription in PPARα null macrophages resulted in enhanced MAPK phosphorylation, leading to increased NF-κB [Crisafulli and Cuzzocrea, 2009]. Several other studies have also demonstrated a similar, important role for PPARα in modulating inflammation in vascular endothelial cells, cartilage and bone tissue, kidney, adipose tissue, and glial cells in the central nervous system (CNS) [Kim et al., 2009; Kono et al., 2009; Murakami et al., 2007; Poleni et al., 2007; Still et al., 2008]. These studies may point toward potential new roles for PPARα and PPARα target genes as therapeutic targets in disorders involving inflammation such as atherosclerosis, joint disease, and autoimmune disorders such as multiple sclerosis [Arzuaga et al., 2007; Dushkin et al., 2007].
PPARα and liver cancer
The development of HCC in mice fed a diet containing nafenopin, a potent peroxisome proliferator, was first reported in 1976 [Reddy et al., 1976]. Subsequently, several hypolipidemic compounds and plasticizers such as DEHP and DEHA have also been shown to induce liver tumors in rats and mice [Kluwe et al., 1983; Rao and Reddy, 1987; Reddy et al., 1980]. Rodents exposed to peroxisome proliferators develop hepatomegaly, largely due to the early hyperplastic and hypertrophic growth of hepatocytes [Reddy et al., 1980; Reddy and Krishnakantha, 1975; Reddy et al., 1979]. With chronic exposure, hepatic adenomas and hepatocellular carcinomas develop in these rodents [Rao and Reddy, 1996; Reddy et al., 1980; Reddy et al., 1976]. The latency period and the incidence of liver tumors appear to correlate well with the effectiveness of a given compound to induce peroxisome proliferation-associated pleiotropic responses [Rao and Reddy, 1996]. Phenotypic features of peroxisome proliferator-induced liver tumors differ from those induced by classic genotoxic hepatocarcinogens [Rao and Reddy, 1996]. Since peroxisome proliferators are neither DNA damaging nor mutagenic, it was proposed that these compounds constitute a novel class of nongenotoxic hepatocarcinogens and this concept laid the foundation for the receptor-mediated hepatocarcinogenesis [Rao and Reddy, 1987; Reddy et al., 1980; Reddy and Lalwani, 1983; Yeldandi et al., 2000].
Peroxisome proliferators do not directly cause genetic damage. It became increasingly evident that hepatocarcinogenicity is due to metabolic alterations resulting from sustained receptor activation that contribute to oxidative stress induced DNA damage in liver [Peters et al., 2005; Rao and Reddy, 1987; Reddy and Lalwani, 1983; Rusyn et al., 2004; Yeldandi et al., 2000]. In normal liver, hydrogen peroxide is produced as a byproduct of many oxidative reactions [Yeldandi et al., 2000]. Of these, the catabolism of very long chain fatty acids, resulting from massive induction of the peroxisomal fatty acid oxidation system by peroxisome proliferators, has received considerable attention in peroxisome proliferator-induced liver carcinogenesis [Nemali et al., 1988; Reddy et al., 1982]. Peroxisomal catalase degrades hydrogen peroxide in normal liver, but in livers with peroxisome proliferation catalase expression increases ~2- to 5-fold in contrast to disproportionately large increases in hydrogen peroxide producing acyl-CoA oxidase and microsomal ω-oxidation enzymes [Chu et al., 2004; Nemali et al., 1988; Yeldandi et al., 2000]. This imbalance between the expression of enzymes capable of producing- and degrading- hydrogen peroxide and other reactive oxygen species in hepatocytes contribute to oxidative stress, lipid peroxidation and oxidative DNA damage [Rao and Reddy, 1987]. These changes influence lipofuscin accumulation, a product of the oxidation of unsaturated fatty acids, as well as hepatocellular proliferation [Peters et al., 2005; Rao and Reddy, 1996; Reddy et al., 1982]. DNA damage through oxidative stress and hepatocellular proliferation, together, are considered as possible mechanisms responsible for the development of hepatocellular carcinomas in rodents chronically exposed to peroxisome proliferators [Peters et al., 2005; Rao and Reddy, 1987; Reddy, 2004; Reddy et al., 1980]. Consistent with this concept is a concordant marked induction in the liver of genes specific for the long-patch base excision DNA repair following exposure to PPARα ligand, Wy-14,643 [Rusyn et al., 2004]. Long-patch base excision DNA repair is a predominant pathway that removes oxidized DNA lesions [Rusyn et al., 2004]. It is evident that DNA-damaging oxidants are generated by enzymes induced after activation of PPARα, such as those involved in fatty acid oxidation [Rusyn et al., 2004]. Based on these observations, it is concluded that chronic exposure to synthetic peroxisome proliferators results in sustained activation of PPARα and transcriptional activation of PPARα responsive genes that affect intermediary metabolism in liver [Reddy and Chu, 1996; Reddy and Hashimoto, 2001]. These metabolic changes, along with the anti-apoptotic effects of PPARα activation, contribute to oxidative DNA damage and increased hepatocellular proliferation leading to liver cancer development [Chen et al., 2009; Peters et al., 2005; Reddy and Chu, 1996].
Levels of regulation of PPARα transcriptional activity
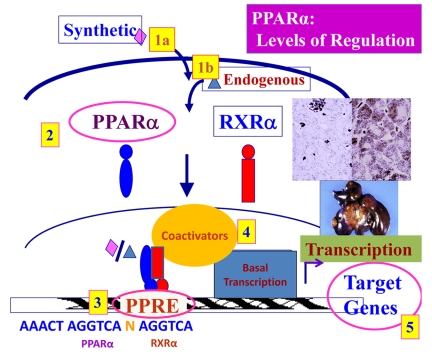
Although it has been well-demonstrated that peroxisome proliferation and liver cancer develop in rodent models, it is not clear that chronic administration of PPARα ligands leads to tumorigenesis in humans. However, many factors may contribute to this variability seen in species- and cell-specific responses to PPARα ligands. The levels of regulation of PPARα-induced responses (Figure 3) include: the potency of specific exogenous and endogenous PPARα ligands to activate PPARα [Fan et al., 1998; Reddy et al., 1976], differential expression of the PPARα and its heterodimerizing partner RXR [Palmer et al., 1998], species differences in the promoter regions of PPARα target genes [Reddy and Chu, 1996], differential expression of certain coactivator proteins necessary for PPARα-mediated transactivation and differing expression levels and activity of PPARα target genes [Jia et al., 2004; Yu and Reddy, 2007].
Figure 3. Levels of regulation of PPARα-mediated transcription.
Diagram illustrating the different factors which regulate the ability of PPARα to activate or repress transcription of target genes. Five major levels of regulation are: (1) the ligand, (2) the receptor expression, (3) the promoter, (4) the coactivator proteins and (5) the expression of target genes.
PPARα ligands show an intrinsic difference in their capability to induce maximal PPARα activation and peroxisome proliferation. Male F344 rats require feeding of DEHP at a 2% (w/w) dietary level for 30 days in order to achieve a significant degree of hepatomegaly and peroxisome proliferation, while rats fed for the same period on a diet containing only 0.01% ciprofibrate show a significant increase in liver to body weight ratio [Reddy et al., 1986a]. Ciprofibrate is more effective at inducing the classical peroxisome proliferative response when compared to phthalate ester plasticizers such as DEHP. Furthermore, it should be noted that hepatic peroxisome proliferation is inducible in the livers of several species including cats, chickens, pigeons and the nonhuman primates such as rhesus and cynomologus monkeys by ciprofibrate at dose levels that exceed therapeutic levels [Lalwani et al., 1985; Reddy et al., 1984]. It would appear that induction of peroxisome proliferation, in part, can be dose-dependent, but not a species-specific phenomenon.
Tissue levels of expression of the PPARα receptor may help explain the differences seen in response to treatment with PPARα agonists. In the mouse, liver, kidney and heart use fatty acid β-oxidation as a source of fuel. In these tissues, the basal levels of mitochondrial β-oxidation enzymes are similar. However, liver shows a much higher degree of induction of PPARα-regulated fatty acid oxidation enzymes than kidney or heart [Cook et al., 2000]. PPARα levels appear to be lower in human livers, as compared to rodent livers, and have been proposed to account for reduced response of human liver to peroxisome proliferation [Palmer et al., 1998]. Since these estimates were from post mortem livers, data on freshly obtained liver biopsy samples will be required for reliable comparison [Palmer et al., 1998; Reddy, 2004]. Adenoviral overexpression of human PPARα in PPARα-null mouse liver restored peroxisome proliferator-induced pleiotropic responses in liver, including peroxisome proliferation, induction of PPARα target genes and liver cell proliferation [Yu et al., 2001].
The PPARα transactivational response is also regulated by the nature of the PPRE sequence present in the promoter region of responsive genes. PPARα may regulate gene activity in conjunction with other transcription factors such as CCAAT/enhancer-binding protein (C/EBP), sterol regulatory element binding protein 1 (SREBP1), TATA-binding protein (TBP) and members of the signal transducer and regulator of transcription (STAT) family [van der Meer et al., 2010]. The presence or absence of motifs which bind these other transcription factors in the promoter regions of PPARα target genes had a significant influence on the ability of PPARα to transactivate or transrepress expression of those genes. Species differences in the promoter regions of PPARα target genes may account for some of the differences observed in response to treatment with PPARα ligands.
As previously discussed, coactivator proteins are required for effective transactivation of target genes by PPARα. The protein, PBP/MED1, when disrupted in mouse liver, results in the abrogation of peroxisome proliferation and other pleiotropic effects of treatment with PPARα ligands [Jia et al., 2004; Matsumoto et al., 2007]. The degree of expression of different coactivator proteins such as PBP/MED1, and the particular milieu of coactivators expressed in a particular tissue, organism, or species may therefore also contribute to the variable species response to treatment with PPARα ligands.
Recently, it has been shown that activation of PPARα represses the expression of the let-7C miRNA, which in turn releases the repression of c-myc expression [Shah et al., 2007]. Interestingly, mice expressing the human PPARα protein do not show this repression of the let-7C miRNA [Yang et al., 2008]. Accordingly, these mice also do not exhibit hepatocellular proliferation or development of tumors [Gonzalez and Shah, 2008; Morimura et al., 2006; Shah et al., 2007]. This induced expression of c-myc protein is proposed to be the mechanism contributing to PPARα ligand-induced hepatocellular proliferation [Shah et al., 2007]. Increased cell proliferative stimulus mediated by PPARα ligands in an intense oxidative stress inducing environment in livers with peroxisome proliferation, could act in concert and contribute to the development of liver tumors. Nonetheless, the level of human PPARα expression may be an issue for consideration in humanized PPARα mice, since adenoviral overexpression of human PPARα in PPARα-null mice showed robust liver cell proliferation in response to a synthetic PPARα ligand [Yu et al., 2001]. Accordingly, sustained activation of PPARα by synthetic ligands is hepatocarcinogenic, as observed in rodent models [Reddy and Chu, 1996].
Additionally, PPARα activity is influenced by the ability of target genes, such as ACOX1 to efficiently metabolize and thus remove endogenous ligands in hepatocytes. In ACOX1 null mouse livers, PPARα activity is dramatically increased as a result of the accumulation of unmetabolized, endogenous ligands [Cook et al., 2000; Fan et al., 1998; Yeldandi et al., 2000]. As a result, species differences in the expression of PPARα target genes may also contribute to species-specific responses to treatment with PPARα ligands. ACOX1 null mouse livers display the hallmarks of PPARα activation such as hepatocellular regeneration accompanied by profound generalized spontaneous peroxisome proliferation, and increased mRNA levels of genes that are regulated by PPARα, which ultimately lead to liver tumor development [Fan et al., 1998]. Although ACOX1, a major hydrogen peroxide generating enzyme in peroxisomes, is absent in these livers, there appear to be other sources that may contribute to oxidative stress in liver such as cytochrome P450 family proteins induced by activated PPARα [Fan et al., 1998; Reddy and Hashimoto, 2001; Yeldandi et al., 2000]. These CYP4A proteins typically catalyze the addition of oxygen to a variety of substrate molecules and may produce reactive oxygen as a by-product [Reddy, 2004; Reddy and Hashimoto, 2001; Yeldandi et al., 2000]. Thus, it would be essential to consider that ACOX1 is not the only source of oxidative stress in PPARα-activated livers. Expression of long-patch base excision DNA repair genes was found to be upregulated significantly in livers of naïve ACOX1-null mice and was increased similarly to the effect of WY-14,643 [Rusyn et al., 2004]. These observations strongly support that sustained activation of PPARα in liver, either by synthetic or natural ligands, leads to endoplasmic reticulum and oxidative stress. Liver tumors that develop in ACOX1 null livers exhibit similar cDNA profiles as tumors resulting from long-term treatment with PPARα ligands, but not similar to tumors resulting from treatment with diethylnitrosamine, a genotoxic mutagen [Fan et al., 1998; Meyer et al., 2003]. These observations further support the novel nature of PPARα ligand-induced hepatocarcinogenesis.
PPARα polymorphism
Genetic variation and polymorphism of PPARα in the human have been under intensive study [Carlberg and Dunlop, 2006]. To date, at least 14 polymorphisms of PPARα have been reported (Figure 1) [Naito et al., 2006] and include P22R, D140Y [Hara et al., 2001], R127Q (rs1800204), R131Q [Sapone et al., 2000], L162V (rs1800206), R178G [Nielsen et al., 2003], V227A (rs1800234), A268V (rs1042311), D304N (rs1800242), G395A (rs2229245), D409T (rs1800243), Q413L (rs9615759), D140N and G395E [Naito et al., 2006]. Some of these variations were reportedly associated with human lipid metabolic changes. For example, PPARα-V227A is a major polymorphism in the Japanese population [Naito et al., 2006]. Its activity may be greater than that of wild-type PPARα, but is decreased by increased alcohol consumption, suggesting that alcoholic liver injury may affect the activity of this receptor [Naito et al., 2006]. Among the 4,248 subjects from 3 ethnic groups (2,899 Chinese, 761 Malay and 588 Asian Indians) genotyped for polymorphisms at the PPARA locus, allele frequencies for the V227A mutation were 0.04 in Chinese, 0.006 in Malays and 0.003 in Asian Indians [Chan et al., 2006]. This polymorphism is associated with lower serum concentrations of total cholesterol and triglycerides in Chinese women. The V227A polymorphism is believed to modulate the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentration. These observations suggest that genetic variation of PPARα may determine the response to changes in dietary PUFA intake [Chan et al., 2006]. Furthermore, L162V polymorphism in the PPARα gene, first identified in the Caucasian population [Sapone et al., 2000; Vohl et al., 2000], was associated with increased levels of serum apolipoprotein B and LDL cholesterol. The L162V polymorphism in PPARα may determine the response to PPARα ligands [Bosse et al., 2002]. The PPARα-L162V polymorphism alone or in interaction with dietary fat intake is associated with components of the metabolic syndrome and may contribute to variability in plasma lipoprotein and lipid response after modification of the dietary polyunsaturated to saturated fatty acids ratio [Paradis et al., 2005; Robitaille et al., 2004; Sapone et al., 2000; Vohl et al., 2000].
Conclusions
Studies with a structurally diverse group of chemicals designated as peroxisome proliferators laid the foundation for the discovery of a subfamily of nuclear receptors named peroxisome proliferator-activated receptor (PPAR) with three members: PPARα, PPARβ/δ and PPARγ. Of these, PPARα plays a critical role in fatty acid oxidation and is thus responsible for energy expenditure. Sustained activation of PPAR leads to the development of liver tumors in rats and mice, whereas PPARα deficiency leads to impaired response to fatty acid influx into liver and contributes to hepatic steatosis. Studies with gene knockout mouse models have yielded information on putative endogenous PPARα ligands that are either generated or degraded by certain enzymes during intermediary metabolism. Several nuclear receptor coactivators have been noted to interact with PPARα, but studies to-date indicate that the coactivator PBP/MED1 is essential for PPAR target gene activation, liver cell proliferation and liver tumorigenesis. As for future challenges, it remains to be determined the extent and nature of other endogenous PPAR agonists and other coactivators that influence PPARα function. Such information may provide clues for therapeutic intervention to enhance energy utilization, minimize the adverse effects of both alcoholic and nonalcoholic fatty liver disease and modulate liver cell proliferation.
Abbreviations
- ACOX1
peroxisomal fatty acyl-CoA oxidase 1
- CBP
cAMP response element-binding protein-binding protein
- CYP
cytochrome P450
- D-PBE
D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase (D-bifunctional enzyme)
- DRIP
vitamin D receptor-interacting protein(s)
- FACS
fatty acyl-CoA synthetase
- FAS
fatty acid synthase
- L-PBE
enoyl-CoA hydratase/L-3-hydroxyacyl-CoA dehydrogenase (L-bifunctional enzyme)
- MED1
mediator subunit 1
- PBP
PPAR-binding protein
- PIMT
PRIP-interacting protein with methyltransferase activity
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response element
- PRIC
PPARα-interacting coregulator complex
- PRIP
PPAR-interacting protein
- PTL
peroxisomal thiolase
- RXR
retinoid X receptor
- SRC-1
steroid receptor coactivator-1
- TRAP
thyroid hormone receptor-associated protein
Supplementary Material
References
- Antonson P., Schuster G. U., Wang L., Rozell B., Holter E., Flodby P., Treuter E., Holmgren L., Gustafsson J. A. Inactivation of the nuclear receptor coactivator RAP250 in mice results in placental vascular dysfunction. Mol Cell Biol. 2003;23:1260–8. doi: 10.1128/MCB.23.4.1260-1268.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARalpha) J Biol Chem. 1998;273:5678–84. doi: 10.1074/jbc.273.10.5678. [DOI] [PubMed] [Google Scholar]
- Arzuaga X., Reiterer G., Majkova Z., Kilgore M. W., Toborek M., Hennig B. PPARalpha ligands reduce PCB-induced endothelial activation: possible interactions in inflammation and atherosclerosis. Cardiovasc Toxicol. 2007;7:264–72. doi: 10.1007/s12012-007-9005-8. [DOI] [PubMed] [Google Scholar]
- Barger P. M., Browning A. C., Garner A. N., Kelly D. P. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor α: a potential role in the cardiac metabolic stress response. J Biol Chem. 2001;276:44495–501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- Bays H., Stein E. A. Pharmacotherapy for dyslipidaemia--current therapies and future agents. Expert Opin Pharmacother. 2003;4:1901–38. doi: 10.1517/14656566.4.11.1901. [DOI] [PubMed] [Google Scholar]
- Blanquart C., Mansouri R., Paumelle R., Fruchart J. C., Staels B., Glineur C. The protein kinase C signaling pathway regulates a molecular switch between transactivation and transrepression activity of the peroxisome proliferator-activated receptor α. Mol Endocrinol. 2004;18:1906–18. doi: 10.1210/me.2003-0327. [DOI] [PubMed] [Google Scholar]
- Bookout A. L., Jeong Y., Downes M., Yu R. T., Evans R. M., Mangelsdorf D. J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–99. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse Y., Pascot A., Dumont M., Brochu M., Prud'homme D., Bergeron J., Despres J. P., Vohl M. C. Influences of the PPAR α-L162V polymorphism on plasma HDL(2)-cholesterol response of abdominally obese men treated with gemfibrozil. Genet Med. 2002;4:311–5. doi: 10.1097/00125817-200207000-00010. [DOI] [PubMed] [Google Scholar]
- Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137:354–66. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- Brash A. R. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem. 1999;274:23679–82. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- Browning J. D., Horton J. D. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–52. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugge T. H., Pohl J., Lonnoy O., Stunnenberg H. G. RXR α, a promiscuous partner of retinoic acid and thyroid hormone receptors. Embo J. 1992;11:1409–18. doi: 10.1002/j.1460-2075.1992.tb05186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulhak A. A., Jung C., Ostenson C. G., Lundberg J. O., Sjoquist P. O., Pernow J. PPAR-α activation protects the type 2 diabetic myocardium against ischemia-reperfusion injury: involvement of the PI3-Kinase/Akt and NO pathway. Am J Physiol Heart Circ Physiol. 2009;296:H719–27. doi: 10.1152/ajpheart.00394.2008. [DOI] [PubMed] [Google Scholar]
- Carlberg C., Dunlop T. W. An integrated biological approach to nuclear receptor signaling in physiological control and disease. Crit Rev Eukaryot Gene Expr. 2006;16:1–22. doi: 10.1615/critreveukargeneexpr.v16.i1.10. [DOI] [PubMed] [Google Scholar]
- Chakravarthy M. V., Zhu Y., Lopez M., Yin L., Wozniak D. F., Coleman T., Hu Z., Wolfgang M., Vidal-Puig A., Lane M. D., Semenkovich C. F. Brain fatty acid synthase activates PPARalpha to maintain energy homeostasis. J Clin Invest. 2007;117:2539–52. doi: 10.1172/JCI31183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy M. V., Lodhi I. J., Yin L., Malapaka R. R., Xu H. E., Turk J., Semenkovich C. F. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–88. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T. P., Rastinejad F. Structure of the intact PPAR-γ-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–6. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Jaber L. A., Berlie H. D., O'Connell M. B. Evolution of peroxisome proliferator-activated receptor agonists. Ann Pharmacother. 2007;41:973–83. doi: 10.1345/aph.1K013. [DOI] [PubMed] [Google Scholar]
- Chan E., Tan C. S., Deurenberg-Yap M., Chia K. S., Chew S. K., Tai E. S. The V227A polymorphism at the PPARA locus is associated with serum lipid concentrations and modulates the association between dietary polyunsaturated fatty acid intake and serum high density lipoprotein concentrations in Chinese women. Atherosclerosis. 2006;187:309–15. doi: 10.1016/j.atherosclerosis.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–70. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chawla A., Schwarz E. J., Dimaculangan D. D., Lazar M. A. Peroxisome proliferator-activated receptor (PPAR) γ: adipose-predominant expression and induction early in adipocyte differentiation. Endocrinology. 1994;135:798–800. doi: 10.1210/endo.135.2.8033830. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Evans R. M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–7. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Chen H. H., Sue Y. M., Chen C. H., Hsu Y. H., Hou C. C., Cheng C. Y., Lin S. L., Tsai W. L., Chen T. W., Chen T. H. Peroxisome proliferator-activated receptor α plays a crucial role in L-carnitine anti-apoptosis effect in renal tubular cells. Nephrol Dial Transplant. 2009;24:3042–9. doi: 10.1093/ndt/gfp258. [DOI] [PubMed] [Google Scholar]
- Cherkaoui-Malki M., Meyer K., Cao W. Q., Latruffe N., Yeldandi A. V., Rao M. S., Bradfield C. A., Reddy J. K. Identification of novel peroxisome proliferator-activated receptor α (PPARalpha) target genes in mouse liver using cDNA microarray analysis. Gene Expr. 2001;9:291–304. doi: 10.3727/000000001783992533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., Brewer H. B., Fruchart J. C., Clavey V., Staels B. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med. 2001;7:53–8. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- Chopra A. R., Louet J. F., Saha P., An J., Demayo F., Xu J., York B., Karpen S., Finegold M., Moore D., Chan L., Newgard C. B., O'Malley B. W. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke's disease. Science. 2008;322:1395–9. doi: 10.1126/science.1164847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu R., Lin Y., Rao M. S., Reddy J. K. Cooperative formation of higher order peroxisome proliferator-activated receptor and retinoid X receptor complexes on the peroxisome proliferator responsive element of the rat hydratase-dehydrogenase gene. J Biol Chem. 1995;270:29636–9. doi: 10.1074/jbc.270.50.29636. [DOI] [PubMed] [Google Scholar]
- Chu R., Lim H., Brumfield L., Liu H., Herring C., Ulintz P., Reddy J. K., Davison M. Protein profiling of mouse livers with peroxisome proliferator-activated receptor α activation. Mol Cell Biol. 2004;24:6288–97. doi: 10.1128/MCB.24.14.6288-6297.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R. A., Lewin T. M., Van Horn C. G., Gonzalez-Baro M. R. Do long-chain acyl-CoA synthetases regulate fatty acid entry into synthetic versus degradative pathways? J Nutr. 2002;132:2123–6. doi: 10.1093/jn/132.8.2123. [DOI] [PubMed] [Google Scholar]
- Cook W. S., Yeldandi A. V., Rao M. S., Hashimoto T., Reddy J. K. Less extrahepatic induction of fatty acid β-oxidation enzymes by PPAR α. Biochem Biophys Res Commun. 2000;278:250–7. doi: 10.1006/bbrc.2000.3739. [DOI] [PubMed] [Google Scholar]
- Corton J. C., Anderson S. P., Stauber A. Central role of peroxisome proliferator-activated receptors in the actions of peroxisome proliferators. Annu Rev Pharmacol Toxicol. 2000;40:491–518. doi: 10.1146/annurev.pharmtox.40.1.491. [DOI] [PubMed] [Google Scholar]
- Crabb D. W., Galli A., Fischer M., You M. Molecular mechanisms of alcoholic fatty liver: role of peroxisome proliferator-activated receptor α. Alcohol. 2004;34:35–8. doi: 10.1016/j.alcohol.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Crawford S. E., Qi C., Misra P., Stellmach V., Rao M. S., Engel J. D., Zhu Y., Reddy J. K. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem. 2002;277:3585–92. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- Crisafulli C., Cuzzocrea S. The role of endogenous and exogenous ligands for the peroxisome proliferator-activated receptor α (PPAR-α) in the regulation of inflammation in macrophages. Shock. 2009;32:62–73. doi: 10.1097/shk.0b013e31818bbad6. [DOI] [PubMed] [Google Scholar]
- Cronet P., Petersen J. F., Folmer R., Blomberg N., Sjoblom K., Karlsson U., Lindstedt E. L., Bamberg K. Structure of the PPARalpha and -γ ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9:699–706. doi: 10.1016/s0969-2126(01)00634-7. [DOI] [PubMed] [Google Scholar]
- Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–88. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- Devchand P. R., Keller H., Peters J. M., Vazquez M., Gonzalez F. J., Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- Di-Poi N., Tan N. S., Michalik L., Wahli W., Desvergne B. Antiapoptotic role of PPARbeta in keratinocytes via transcriptional control of the Akt1 signaling pathway. Mol Cell. 2002;10:721–33. doi: 10.1016/s1097-2765(02)00646-9. [DOI] [PubMed] [Google Scholar]
- DiRenzo J., Soderstrom M., Kurokawa R., Ogliastro M. H., Ricote M., Ingrey S., Horlein A., Rosenfeld M. G., Glass C. K. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Mol Cell Biol. 1997;17:2166–76. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotson M. R., Yuan C. X., Roeder R. G., Myers L. C., Gustafsson C. M., Jiang Y. W., Li Y., Kornberg R. D., Asturias F. J. Structural organization of yeast and mammalian mediator complexes. Proc Natl Acad Sci U S A. 2000;97:14307–10. doi: 10.1073/pnas.260489497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell P., Ishmael J. E., Avram D., Peterson V. J., Nevrivy D. J., Leid M. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor α interacting protein. J Biol Chem. 1999;274:15901–7. doi: 10.1074/jbc.274.22.15901. [DOI] [PubMed] [Google Scholar]
- Dreyer C., Krey G., Keller H., Givel F., Helftenbein G., Wahli W. Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell. 1992;68:879–87. doi: 10.1016/0092-8674(92)90031-7. [DOI] [PubMed] [Google Scholar]
- Dushkin M. I., Khoshchenko O. M., Posokhova E. N., Schvarts YSh. Agonists of PPAR-α, PPAR-γ, and RXR inhibit the formation of foam cells from macrophages in mice with inflammation. Bull Exp Biol Med. 2007;144:713–6. doi: 10.1007/s10517-007-0413-3. [DOI] [PubMed] [Google Scholar]
- El Kebbaj Z., Andreoletti P., Mountassif D., Kabine M., Schohn H., Dauca M., Latruffe N., El Kebbaj M. S., Cherkaoui-Malki M. Differential regulation of peroxisome proliferator-activated receptor (PPAR)-alpha1 and truncated PPARalpha2 as an adaptive response to fasting in the control of hepatic peroxisomal fatty acid β-oxidation in the hibernating mammal. Endocrinology. 2009;150:1192–201. doi: 10.1210/en.2008-1394. [DOI] [PubMed] [Google Scholar]
- Ellinghaus P., Wolfrum C., Assmann G., Spener F., Seedorf U. Phytanic acid activates the peroxisome proliferator-activated receptor α (PPARalpha) in sterol carrier protein 2-/ sterol carrier protein x-deficient mice. J Biol Chem. 1999;274:2766–72. doi: 10.1074/jbc.274.5.2766. [DOI] [PubMed] [Google Scholar]
- Evans R. M., Barish G. D., Wang Y. X. PPARs and the complex journey to obesity. Nat Med. 2004;10:355–61. doi: 10.1038/nm1025. [DOI] [PubMed] [Google Scholar]
- Faergeman N. J., Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323 ( Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C. Y., Pan J., Chu R., Lee D., Kluckman K. D., Usuda N., Singh I., Yeldandi A. V., Rao M. S., Maeda N., Reddy J. K. Hepatocellular and hepatic peroxisomal alterations in mice with a disrupted peroxisomal fatty acyl-coenzyme A oxidase gene. J Biol Chem. 1996;271:24698–710. doi: 10.1074/jbc.271.40.24698. [DOI] [PubMed] [Google Scholar]
- Fan C. Y., Pan J., Usuda N., Yeldandi A. V., Rao M. S., Reddy J. K. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor α natural ligand metabolism. J Biol Chem. 1998;273:15639–45. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- Feige J. N., Gelman L., Michalik L., Desvergne B., Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog Lipid Res. 2006;45:120–59. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Ferdinandusse S., Denis S., Faust P. L., Wanders R. J. Bile acids: the role of peroxisomes. J Lipid Res. 2009;50:2139–47. doi: 10.1194/jlr.R900009-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S., Denis S., Van Roermund C. W., Wanders R. J., Dacremont G. Identification of the peroxisomal β-oxidation enzymes involved in the degradation of long-chain dicarboxylic acids. J Lipid Res. 2004;45:1104–11. doi: 10.1194/jlr.M300512-JLR200. [DOI] [PubMed] [Google Scholar]
- Ford-Hutchinson A. W. Leukotriene B4 in inflammation. Crit Rev Immunol. 1990;10:1–12. [PubMed] [Google Scholar]
- Forman B. M., Chen J., Evans R. M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci U S A. 1997;94:4312–7. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genini D., Carbone G. M., Catapano C. V. Multiple Interactions between Peroxisome Proliferators-Activated Receptors and the Ubiquitin-Proteasome System and Implications for Cancer Pathogenesis. PPAR Res. 2008;2008:195065. doi: 10.1155/2008/195065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervois P., Torra I. P., Chinetti G., Grotzinger T., Dubois G., Fruchart J. C., Fruchart-Najib J., Leitersdorf E., Staels B. A truncated human peroxisome proliferator-activated receptor α splice variant with dominant negative activity. Mol Endocrinol. 1999;13:1535–49. doi: 10.1210/mend.13.9.0341. [DOI] [PubMed] [Google Scholar]
- Ghisletti S., Huang W., Jepsen K., Benner C., Hardiman G., Rosenfeld M. G., Glass C. K. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–93. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizard F., Nomiyama T., Zhao Y., Findeisen H. M., Heywood E. B., Jones K. L., Staels B., Bruemmer D. The PPARalpha/p16INK4a pathway inhibits vascular smooth muscle cell proliferation by repressing cell cycle-dependent telomerase activation. Circ Res. 2008;103:1155–63. doi: 10.1161/CIRCRESAHA.108.186205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Gonzalez F. J., Shah Y. M. PPARalpha: mechanism of species differences and hepatocarcinogenesis of peroxisome proliferators. Toxicology. 2008;246:2–8. doi: 10.1016/j.tox.2007.09.030. [DOI] [PubMed] [Google Scholar]
- Gottlicher M., Widmark E., Li Q., Gustafsson J. A. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1992;89:4653–7. doi: 10.1073/pnas.89.10.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaguchi M., Kojima T., Takeda N., Nakagawa T., Taniguchi H., Fujii K., Omatsu T., Nakajima T., Sarui H., Shimazaki M., Kato T., Okuda J., Ida K. The metabolic syndrome as a predictor of nonalcoholic fatty liver disease. Ann Intern Med. 2005;143:722–8. doi: 10.7326/0003-4819-143-10-200511150-00009. [DOI] [PubMed] [Google Scholar]
- Hara M., Wang X., Paz V. P., Iwasaki N., Honda M., Iwamoto Y., Bell G. I. Identification of three missense mutations in the peroxisome proliferator-activated receptor α gene in Japanese subjects with maturity-onset diabetes of the young. J Hum Genet. 2001;46:285–8. doi: 10.1007/s100380170080. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Cook W. S., Qi C., Yeldandi A. V., Reddy J. K., Rao M. S. Defect in peroxisome proliferator-activated receptor α-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J Biol Chem. 2000;275:28918–28. doi: 10.1074/jbc.M910350199. [DOI] [PubMed] [Google Scholar]
- Hashimoto T. Peroxisomal β-oxidation enzymes. Neurochem Res. 1999;24:551–63. doi: 10.1023/a:1022540030918. [DOI] [PubMed] [Google Scholar]
- Hays T., Rusyn I., Burns A. M., Kennett M. J., Ward J. M., Gonzalez F. J., Peters J. M. Role of peroxisome proliferator-activated receptor-α (PPARalpha) in bezafibrate-induced hepatocarcinogenesis and cholestasis. Carcinogenesis. 2005;26:219–27. doi: 10.1093/carcin/bgh285. [DOI] [PubMed] [Google Scholar]
- Hebbachi A. M., Knight B. L., Wiggins D., Patel D. D., Gibbons G. F. Peroxisome proliferator-activated receptor α deficiency abolishes the response of lipogenic gene expression to re-feeding: restoration of the normal response by activation of liver X receptor α. J Biol Chem. 2008;283:4866–76. doi: 10.1074/jbc.M709471200. [DOI] [PubMed] [Google Scholar]
- Heinzer A. K., Watkins P. A., Lu J. F., Kemp S., Moser A. B., Li Y. Y., Mihalik S., Powers J. M., Smith K. D. A very long-chain acyl-CoA synthetase-deficient mouse and its relevance to X-linked adrenoleukodystrophy. Hum Mol Genet. 2003;12:1145–54. doi: 10.1093/hmg/ddg126. [DOI] [PubMed] [Google Scholar]
- Horton J. D., Goldstein J. L., Brown M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetler H. A., Huang H., Kier A. B., Schroeder F. Glucose directly links to lipid metabolism through high affinity interaction with peroxisome proliferator-activated receptor α. J Biol Chem. 2008;283:2246–54. doi: 10.1074/jbc.M705138200. [DOI] [PubMed] [Google Scholar]
- Hostetler H. A., Petrescu A. D., Kier A. B., Schroeder F. Peroxisome proliferator-activated receptor α interacts with high affinity and is conformationally responsive to endogenous ligands. J Biol Chem. 2005;280:18667–82. doi: 10.1074/jbc.M412062200. [DOI] [PubMed] [Google Scholar]
- Hummasti S., Tontonoz P. The peroxisome proliferator-activated receptor N-terminal domain controls isotype-selective gene expression and adipogenesis. Mol Endocrinol. 2006;20:1261–75. doi: 10.1210/me.2006-0025. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- IJpenberg A., Jeannin E., Wahli W., Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J Biol Chem. 1997;272:20108–17. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- Inoue J., Satoh S., Kita M., Nakahara M., Hachimura S., Miyata M., Nishimaki-Mogami T., Sato R. PPARalpha gene expression is up-regulated by LXR and PXR activators in the small intestine. Biochem Biophys Res Commun. 2008;371:675–8. doi: 10.1016/j.bbrc.2008.04.100. [DOI] [PubMed] [Google Scholar]
- Ip E., Farrell G. C., Robertson G., Hall P., Kirsch R., Leclercq I. Central role of PPARalpha-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology. 2003;38:123–32. doi: 10.1053/jhep.2003.50307. [DOI] [PubMed] [Google Scholar]
- Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–50. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Ito M., Yuan C. X., Okano H. J., Darnell R. B., Roeder R. G. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell. 2000;5:683–93. doi: 10.1016/s1097-2765(00)80247-6. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Hermanson O., Onami T. M., Gleiberman A. S., Lunyak V., McEvilly R. J., Kurokawa R., Kumar V., Liu F., Seto E., Hedrick S. M., Mandel G., Glass C. K., Rose D. W., Rosenfeld M. G. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–63. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- Jepsen K., Gleiberman A. S., Shi C., Simon D. I., Rosenfeld M. G. Cooperative regulation in development by SMRT and FOXP1. Genes Dev. 2008;22:740–5. doi: 10.1101/gad.1637108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Viswakarma N., Fu T., Yu S., Rao M. S., Borensztajn J., Reddy J. K. Conditional ablation of mediator subunit MED1 (MED1/PPARBP) gene in mouse liver attenuates glucocorticoid receptor agonist dexamethasone-induced hepatic steatosis. Gene Expr. 2009b;14:291–306. doi: 10.3727/105221609788681213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Viswakarma N., Matsumoto K., Pyper S., Rao M. S., Reddy J. K. Early Embryonic Lethality of Mice with Disrupted Transcription Cofactor PIMT/NCoA6IP Gene. FASEB J. 2009a;23:739. [Google Scholar]
- Jia Y., Qi C., Zhang Z., Hashimoto T., Rao M. S., Huyghe S., Suzuki Y., Van Veldhoven P. P., Baes M., Reddy J. K. Overexpression of peroxisome proliferator-activated receptor-α (PPARalpha)-regulated genes in liver in the absence of peroxisome proliferation in mice deficient in both L- and D-forms of enoyl-CoA hydratase/dehydrogenase enzymes of peroxisomal β-oxidation system. J Biol Chem. 2003;278:47232–9. doi: 10.1074/jbc.M306363200. [DOI] [PubMed] [Google Scholar]
- Jia Y., Qi C., Kashireddi P., Surapureddi S., Zhu Y. J., Rao M. S., Le Roith D., Chambon P., Gonzalez F. J., Reddy J. K. Transcription coactivator PBP, the peroxisome proliferator-activated receptor (PPAR)-binding protein, is required for PPARalpha-regulated gene expression in liver. J Biol Chem. 2004;279:24427–34. doi: 10.1074/jbc.M402391200. [DOI] [PubMed] [Google Scholar]
- Jia Y., Guo G. L., Surapureddi S., Sarkar J., Qi C., Guo D., Xia J., Kashireddi P., Yu S., Cho Y. W., Rao M. S., Kemper B., Ge K., Gonzalez F. J., Reddy J. K. Transcription coactivator peroxisome proliferator-activated receptor-binding protein/mediator 1 deficiency abrogates acetaminophen hepatotoxicity. Proc Natl Acad Sci U S A. 2005;102:12531–6. doi: 10.1073/pnas.0506000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jow L., Mukherjee R. The human peroxisome proliferator-activated receptor (PPAR) subtype NUC1 represses the activation of hPPAR α and thyroid hormone receptors. J Biol Chem. 1995;270:3836–40. doi: 10.1074/jbc.270.8.3836. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry C., Pernin A., Favez T., Burger A. G., Wahli W., Meier C. A., Desvergne B. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5'-flanking region. J Biol Chem. 1997;272:25252–9. doi: 10.1074/jbc.272.40.25252. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry C. E., Gorla-Bajszczak A., Pernin A., Lemberger T., Wahli W., Burger A. G., Meier C. A. Peroxisome proliferator-activated receptor mediates cross-talk with thyroid hormone receptor by competition for retinoid X receptor. Possible role of a leucine zipper-like heptad repeat. J Biol Chem. 1995;270:18117–22. doi: 10.1074/jbc.270.30.18117. [DOI] [PubMed] [Google Scholar]
- Juge-Aubry C. E., Hammar E., Siegrist-Kaiser C., Pernin A., Takeshita A., Chin W. W., Burger A. G., Meier C. A. Regulation of the transcriptional activity of the peroxisome proliferator-activated receptor α by phosphorylation of a ligand-independent trans-activating domain. J Biol Chem. 1999;274:10505–10. doi: 10.1074/jbc.274.15.10505. [DOI] [PubMed] [Google Scholar]
- Kane C. D., Stevens K. A., Fischer J. E., Haghpassand M., Royer L. J., Aldinger C., Landschulz K. T., Zagouras P., Bagley S. W., Hada W., Dullea R., Hayward C. M., Francone O. L. Molecular characterization of novel and selective peroxisome proliferator-activated receptor α agonists with robust hypolipidemic activity in vivo. Mol Pharmacol. 2009;75:296–306. doi: 10.1124/mol.108.051656. [DOI] [PubMed] [Google Scholar]
- Kelly L. J., Vicario P. P., Thompson G. M., Candelore M. R., Doebber T. W., Ventre J., Wu M. S., Meurer R., Forrest M. J., Conner M. W., Cascieri M. A., Moller D. E. Peroxisome proliferator-activated receptors γ and α mediate in vivo regulation of uncoupling protein (UCP-1, UCP-2, UCP-3) gene expression. Endocrinology. 1998;139:4920–7. doi: 10.1210/endo.139.12.6384. [DOI] [PubMed] [Google Scholar]
- Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–98. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. H., Won Y. S., Kim D. Y., Kim B., Kim E. Y., Yoon M., Oh G. T. Signal crosstalk between estrogen and peroxisome proliferator-activated receptor α on adiposity. BMB Rep. 2009;42:91–5. doi: 10.5483/bmbrep.2009.42.2.091. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A., Umesono K., Noonan D. J., Heyman R. A., Evans R. M. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–4. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer S. A., Xu H. E., Lambert M. H., Willson T. M. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog Horm Res. 2001;56:239–63. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- Kluwe W. M., Haseman J. K., Huff J. E. The carcinogenicity of di(2-ethylhexyl) phthalate (DEHP) in perspective. J Toxicol Environ Health. 1983;12:159–69. doi: 10.1080/15287398309530414. [DOI] [PubMed] [Google Scholar]
- Kono K., Kamijo Y., Hora K., Takahashi K., Higuchi M., Kiyosawa K., Shigematsu H., Gonzalez F. J., Aoyama T. PPAR{α} attenuates the proinflammatory response in activated mesangial cells. Am J Physiol Renal Physiol. 2009;296:F328–36. doi: 10.1152/ajprenal.00484.2007. [DOI] [PubMed] [Google Scholar]
- Krey G., Braissant O., L'Horset F., Kalkhoven E., Perroud M., Parker M. G., Wahli W. Fatty acids, eicosanoids, and hypolipidemic agents identified as ligands of peroxisome proliferator-activated receptors by coactivator-dependent receptor ligand assay. Mol Endocrinol. 1997;11:779–91. doi: 10.1210/mend.11.6.0007. [DOI] [PubMed] [Google Scholar]
- Lalwani N. D., Fahl W. E., Reddy J. K. Detection of a nafenopin-binding protein in rat liver cytosol associated with the induction of peroxisome proliferation by hypolipidemic compounds. Biochem Biophys Res Commun. 1983;116:388–93. doi: 10.1016/0006-291x(83)90534-x. [DOI] [PubMed] [Google Scholar]
- Lalwani N. D., Reddy M. K., Ghosh S., Barnard S. D., Molello J. A., Reddy J. K. Induction of fatty acid β-oxidation and peroxisome proliferation in the liver of rhesus monkeys by DL-040, a new hypolipidemic agent. Biochem Pharmacol. 1985;34:3473–82. doi: 10.1016/0006-2952(85)90720-8. [DOI] [PubMed] [Google Scholar]
- Lalwani N. D., Alvares K., Reddy M. K., Reddy M. N., Parikh I., Reddy J. K. Peroxisome proliferator-binding protein: identification and partial characterization of nafenopin-, clofibric acid-, and ciprofibrate-binding proteins from rat liver. Proc Natl Acad Sci U S A. 1987;84:5242–6. doi: 10.1073/pnas.84.15.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landles C., Chalk S., Steel J. H., Rosewell I., Spencer-Dene B., Lalani el N., Parker M. G. The thyroid hormone receptor-associated protein TRAP220 is required at distinct embryonic stages in placental, cardiac, and hepatic development. Mol Endocrinol. 2003;17:2418–35. doi: 10.1210/me.2003-0097. [DOI] [PubMed] [Google Scholar]
- Lee Y., Yu X., Gonzales F., Mangelsdorf D. J., Wang M. Y., Richardson C., Witters L. A., Unger R. H. PPAR α is necessary for the lipopenic action of hyperleptinemia on white adipose and liver tissue. Proc Natl Acad Sci U S A. 2002;99:11848–53. doi: 10.1073/pnas.182420899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. S., Pineau T., Drago J., Lee E. J., Owens J. W., Kroetz D. L., Fernandez-Salguero P. M., Westphal H., Gonzalez F. J. Targeted disruption of the α isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol. 1995;15:3012–22. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre P., Chinetti G., Fruchart J. C., Staels B. Sorting out the roles of PPAR α in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–80. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardsson G., Steel J. H., Christian M., Pocock V., Milligan S., Bell J., So P. W., Medina-Gomez G., Vidal-Puig A., White R., Parker M. G. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A. 2004;101:8437–42. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone T. C., Weinheimer C. J., Kelly D. P. A critical role for the peroxisome proliferator-activated receptor α (PPARalpha) in the cellular fasting response: the PPARalpha-null mouse as a model of fatty acid oxidation disorders. Proc Natl Acad Sci U S A. 1999;96:7473–8. doi: 10.1073/pnas.96.13.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. O., Ellis J. M., Paich H. A., Wang S., Gong N., Altshuller G., Thresher R. J., Koves T. R., Watkins S. M., Muoio D. M., Cline G. W., Shulman G. I., Coleman R. A. Liver-specific loss of long chain acyl-CoA synthetase-1 decreases triacylglycerol synthesis and β-oxidation and alters phospholipid fatty acid composition. J Biol Chem. 2009;284:27816–26. doi: 10.1074/jbc.M109.022467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonard D. M., O'Malley B. W. The expanding cosmos of nuclear receptor coactivators. Cell. 2006;125:411–4. doi: 10.1016/j.cell.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Maloney E. K., Waxman D. J. trans-Activation of PPARalpha and PPARgamma by structurally diverse environmental chemicals. Toxicol Appl Pharmacol. 1999;161:209–18. doi: 10.1006/taap.1999.8809. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens K., Ver Loren van Themaat E., van Batenburg M. F., Heinaniemi M., Huyghe S., Van Hummelen P., Carlberg C., Van Veldhoven P. P., Van Kampen A., Baes M. Coordinate induction of PPAR α and SREBP2 in multifunctional protein 2 deficient mice. Biochim Biophys Acta. 2008;1781:694–702. doi: 10.1016/j.bbalip.2008.07.010. [DOI] [PubMed] [Google Scholar]
- Martin G., Poirier H., Hennuyer N., Crombie D., Fruchart J. C., Heyman R. A., Besnard P., Auwerx J. Induction of the fatty acid transport protein 1 and acyl-CoA synthase genes by dimer-selective rexinoids suggests that the peroxisome proliferator-activated receptor-retinoid X receptor heterodimer is their molecular target. J Biol Chem. 2000;275:12612–8. doi: 10.1074/jbc.275.17.12612. [DOI] [PubMed] [Google Scholar]
- Marx N., Duez H., Fruchart J. C., Staels B. Peroxisome proliferator-activated receptors and atherogenesis: regulators of gene expression in vascular cells. Circ Res. 2004;94:1168–78. doi: 10.1161/01.RES.0000127122.22685.0A. [DOI] [PubMed] [Google Scholar]
- Massip-Salcedo M., Zaouali M. A., Padrissa-Altes S., Casillas-Ramirez A., Rodes J., Rosello-Catafau J., Peralta C. Activation of peroxisome proliferator-activated receptor-α inhibits the injurious effects of adiponectin in rat steatotic liver undergoing ischemia-reperfusion. Hepatology. 2008;47:461–72. doi: 10.1002/hep.21935. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Yu S., Jia Y., Ahmed M. R., Viswakarma N., Sarkar J., Kashireddy P. V., Rao M. S., Karpus W., Gonzalez F. J., Reddy J. K. Critical role for transcription coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein/TRAP220 in liver regeneration and PPARalpha ligand-induced liver tumor development. J Biol Chem. 2007;282:17053–60. doi: 10.1074/jbc.M701956200. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Huang J., Viswakarma N., Bai L., Jia Y., Zhu Y. T., Yang G., Borensztajn J., Rao M. S., Zhu Y. J., Reddy J. K. Transcription coactivator PBP/MED1-deficient hepatocytes are not susceptible to diethylnitrosamine-induced hepatocarcinogenesis in the mouse. Carcinogenesis. 2010;31:318–25. doi: 10.1093/carcin/bgp306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei C. L., He P., Cheng B., Liu W., Wang Y. F., Wan J. J. Chlamydia pneumoniae induces macrophage-derived foam cell formation via PPAR α and PPAR γ-dependent pathways. Cell Biol Int. 2009;33:301–8. doi: 10.1016/j.cellbi.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Meyer K., Lee J. S., Dyck P. A., Cao W. Q., Rao M. S., Thorgeirsson S. S., Reddy J. K. Molecular profiling of hepatocellular carcinomas developing spontaneously in acyl-CoA oxidase deficient mice: comparison with liver tumors induced in wild-type mice by a peroxisome proliferator and a genotoxic carcinogen. Carcinogenesis. 2003;24:975–84. doi: 10.1093/carcin/bgg040. [DOI] [PubMed] [Google Scholar]
- Michalik L., Auwerx J., Berger J. P., Chatterjee V. K., Glass C. K., Gonzalez F. J., Grimaldi P. A., Kadowaki T., Lazar M. A., O'Rahilly S., Palmer C. N., Plutzky J., Reddy J. K., Spiegelman B. M., Staels B., Wahli W. International Union of Pharmacology. LXI. Peroxisome proliferator-activated receptors. Pharmacol Rev. 2006;58:726–41. doi: 10.1124/pr.58.4.5. [DOI] [PubMed] [Google Scholar]
- Monecke T., Dickmanns A., Ficner R. Structural basis for m7G-cap hypermethylation of small nuclear, small nucleolar and telomerase RNA by the dimethyltransferase TGS1. Nucleic Acids Res. 2009;37:3865–77. doi: 10.1093/nar/gkp249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura K., Cheung C., Ward J. M., Reddy J. K., Gonzalez F. J. Differential susceptibility of mice humanized for peroxisome proliferator-activated receptor α to Wy-14,643-induced liver tumorigenesis. Carcinogenesis. 2006;27:1074–80. doi: 10.1093/carcin/bgi329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee R., Locke K. T., Miao B., Meyers D., Monshizadegan H., Zhang R., Search D., Grimm D., Flynn M., O'Malley K. M., Zhang L., Li J., Shi Y., Kennedy L. J., Blanar M., Cheng P. T., Tino J., Srivastava R. A. Novel peroxisome proliferator-activated receptor α agonists lower low-density lipoprotein and triglycerides, raise high-density lipoprotein, and synergistically increase cholesterol excretion with a liver X receptor agonist. J Pharmacol Exp Ther. 2008;327:716–26. doi: 10.1124/jpet.108.143271. [DOI] [PubMed] [Google Scholar]
- Murakami K., Bujo H., Unoki H., Saito Y. Effect of PPARalpha activation of macrophages on the secretion of inflammatory cytokines in cultured adipocytes. Eur J Pharmacol. 2007;561:206–13. doi: 10.1016/j.ejphar.2006.12.037. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Inada Y., Nakano S., Tamura T., Takahashi T., Maruyama K., Yamazaki Y., Kuroda J., Shibata N. Effects of bezafibrate, PPAR pan-agonist, and GW501516, PPARdelta agonist, on development of steatohepatitis in mice fed a methionine- and choline-deficient diet. Eur J Pharmacol. 2006;536:182–91. doi: 10.1016/j.ejphar.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Naito H., Yamanoshita O., Kamijima M., Katoh T., Matsunaga T., Lee C. H., Kim H., Aoyama T., Gonzalez F. J., Nakajima T. Association of V227A PPARalpha polymorphism with altered serum biochemistry and alcohol drinking in Japanese men. Pharmacogenet Genomics. 2006;16:569–77. doi: 10.1097/01.fpc.0000220565.90466.79. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Kamijo Y., Tanaka N., Sugiyama E., Tanaka E., Kiyosawa K., Fukushima Y., Peters J. M., Gonzalez F. J., Aoyama T. Peroxisome proliferator-activated receptor α protects against alcohol-induced liver damage. Hepatology. 2004;40:972–80. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- Nemali M. R., Usuda N., Reddy M. K., Oyasu K., Hashimoto T., Osumi T., Rao M. S., Reddy J. K. Comparison of constitutive and inducible levels of expression of peroxisomal β-oxidation and catalase genes in liver and extrahepatic tissues of rat. Cancer Res. 1988;48:5316–24. [PubMed] [Google Scholar]
- Ng V. Y., Huang Y., Reddy L. M., Falck J. R., Lin E. T., Kroetz D. L. Cytochrome P450 eicosanoids are activators of peroxisome proliferator-activated receptor α. Drug Metab Dispos. 2007;35:1126–34. doi: 10.1124/dmd.106.013839. [DOI] [PubMed] [Google Scholar]
- Nielsen E. M., Hansen L., Echwald S. M., Drivsholm T., Borch-Johnsen K., Ekstrom C. T., Hansen T., Pedersen O. Evidence for an association between the Leu162Val polymorphism of the PPARalpha gene and decreased fasting serum triglyceride levels in glucose tolerant subjects. Pharmacogenetics. 2003;13:417–23. doi: 10.1097/01.fpc.0000054105.48725.5c. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Grontved L., Stunnenberg H. G., Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Mol Cell Biol. 2006;26:5698–714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen S. E., Nicholls S. J., Wolski K., Howey D. C., McErlean E., Wang M. D., Gomez E. V., Russo J. M. Effects of a potent and selective PPAR-α agonist in patients with atherogenic dyslipidemia or hypercholesterolemia: two randomized controlled trials. Jama. 2007;297:1362–73. doi: 10.1001/jama.297.12.1362. [DOI] [PubMed] [Google Scholar]
- Onate S. A., Tsai S. Y., Tsai M. J., O'Malley B. W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–7. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- Osumi T., Wen J. K., Hashimoto T. Two cis-acting regulatory sequences in the peroxisome proliferator-responsive enhancer region of rat acyl-CoA oxidase gene. Biochem Biophys Res Commun. 1991;175:866–71. doi: 10.1016/0006-291x(91)91645-s. [DOI] [PubMed] [Google Scholar]
- Palmer C. N., Hsu M. H., Griffin K. J., Raucy J. L., Johnson E. F. Peroxisome proliferator activated receptor-α expression in human liver. Mol Pharmacol. 1998;53:14–22. [PubMed] [Google Scholar]
- Panigrahy D., Kaipainen A., Huang S., Butterfield C. E., Barnes C. M., Fannon M., Laforme A. M., Chaponis D. M., Folkman J., Kieran M. W. PPARalpha agonist fenofibrate suppresses tumor growth through direct and indirect angiogenesis inhibition. Proc Natl Acad Sci U S A. 2008;105:985–90. doi: 10.1073/pnas.0711281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis A. M., Fontaine-Bisson B., Bosse Y., Robitaille J., Lemieux S., Jacques H., Lamarche B., Tchernof A., Couture P., Vohl M. C. The peroxisome proliferator-activated receptor α Leu162Val polymorphism influences the metabolic response to a dietary intervention altering fatty acid proportions in healthy men. Am J Clin Nutr. 2005;81:523–30. doi: 10.1093/ajcn.81.2.523. [DOI] [PubMed] [Google Scholar]
- Patel N. S., di Paola R., Mazzon E., Britti D., Thiemermann C., Cuzzocrea S. Peroxisome proliferator-activated receptor-α contributes to the resolution of inflammation after renal ischemia/reperfusion injury. J Pharmacol Exp Ther. 2009;328:635–43. doi: 10.1124/jpet.108.146191. [DOI] [PubMed] [Google Scholar]
- Paumelle R., Blanquart C., Briand O., Barbier O., Duhem C., Woerly G., Percevault F., Fruchart J. C., Dombrowicz D., Glineur C., Staels B. Acute antiinflammatory properties of statins involve peroxisome proliferator-activated receptor-α via inhibition of the protein kinase C signaling pathway. Circ Res. 2006;98:361–9. doi: 10.1161/01.RES.0000202706.70992.95. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Cheung C., Gonzalez F. J. Peroxisome proliferator-activated receptor-α and liver cancer: where do we stand? J Mol Med. 2005;83:774–85. doi: 10.1007/s00109-005-0678-9. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Cattley R. C., Gonzalez F. J. Role of PPAR α in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis. 1997;18:2029–33. doi: 10.1093/carcin/18.11.2029. [DOI] [PubMed] [Google Scholar]
- Pineda Torra I., Jamshidi Y., Flavell D. M., Fruchart J. C., Staels B. Characterization of the human PPARalpha promoter: identification of a functional nuclear receptor response element. Mol Endocrinol. 2002;16:1013–28. doi: 10.1210/mend.16.5.0833. [DOI] [PubMed] [Google Scholar]
- Poleni P. E., Bianchi A., Etienne S., Koufany M., Sebillaud S., Netter P., Terlain B., Jouzeau J. Y. Agonists of peroxisome proliferators-activated receptors (PPAR) α, β/δ or γ reduce transforming growth factor (TGF)-β-induced proteoglycans' production in chondrocytes. Osteoarthritis Cartilage. 2007;15:493–505. doi: 10.1016/j.joca.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Poynter M. E., Daynes R. A. Peroxisome proliferator-activated receptor α activation modulates cellular redox status, represses nuclear factor-kappaB signaling, and reduces inflammatory cytokine production in aging. J Biol Chem. 1998;273:32833–41. doi: 10.1074/jbc.273.49.32833. [DOI] [PubMed] [Google Scholar]
- Qi C., Zhu Y., Pan J., Usuda N., Maeda N., Yeldandi A. V., Rao M. S., Hashimoto T., Reddy J. K. Absence of spontaneous peroxisome proliferation in enoyl-CoA Hydratase/L-3-hydroxyacyl-CoA dehydrogenase-deficient mouse liver. Further support for the role of fatty acyl CoA oxidase in PPARalpha ligand metabolism. J Biol Chem. 1999a;274:15775–80. doi: 10.1074/jbc.274.22.15775. [DOI] [PubMed] [Google Scholar]
- Qi C., Zhu Y., Pan J., Yeldandi A. V., Rao M. S., Maeda N., Subbarao V., Pulikuri S., Hashimoto T., Reddy J. K. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor α-regulated gene expression. Proc Natl Acad Sci U S A. 1999b;96:1585–90. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi C., Kashireddy P., Zhu Y. T., Rao S. M., Zhu Y. J. Null mutation of peroxisome proliferator-activated receptor-interacting protein in mammary glands causes defective mammopoiesis. J Biol Chem. 2004;279:33696–701. doi: 10.1074/jbc.M401266200. [DOI] [PubMed] [Google Scholar]
- Ramanan S., Kooshki M., Zhao W., Hsu F. C., Robbins M. E. PPARalpha ligands inhibit radiation-induced microglial inflammatory responses by negatively regulating NF-kappaB and AP-1 pathways. Free Radic Biol Med. 2008;45:1695–704. doi: 10.1016/j.freeradbiomed.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M. S., Reddy J. K. Hepatocarcinogenesis of peroxisome proliferators. Ann N Y Acad Sci. 1996;804:573–87. doi: 10.1111/j.1749-6632.1996.tb18646.x. [DOI] [PubMed] [Google Scholar]
- Rao M. S., Reddy J. K. Peroxisome proliferation and hepatocarcinogenesis. Carcinogenesis. 1987;8:631–6. doi: 10.1093/carcin/8.5.631. [DOI] [PubMed] [Google Scholar]
- Rao M. S., Reddy J. K. PPARalpha in the pathogenesis of fatty liver disease. Hepatology. 2004;40:783–6. doi: 10.1002/hep.20453. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Lalwani N. D. Carcinogenesis by hepatic peroxisome proliferators: evaluation of the risk of hypolipidemic drugs and industrial plasticizers to humans. Crit Rev Toxicol. 1983;12:1–58. doi: 10.3109/10408448309029317. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Reddy M. K., Usman M. I., Lalwani N. D., Rao M. S. Comparison of hepatic peroxisome proliferative effect and its implication for hepatocarcinogenicity of phthalate esters, di(2-ethylhexyl) phthalate, and di(2-ethylhexyl) adipate with a hypolipidemic drug. Environ Health Perspect. 1986a;65:317–27. doi: 10.1289/ehp.8665317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy J. K., Lalwani N. D., Reddy M. K., Qureshi S. A. Excessive accumulation of autofluorescent lipofuscin in the liver during hepatocarcinogenesis by methyl clofenapate and other hypolipidemic peroxisome proliferators. Cancer Res. 1982;42:259–66. [PubMed] [Google Scholar]
- Reddy J. K., Krishnakantha T. P. Hepatic peroxisome proliferation: induction by two novel compounds structurally unrelated to clofibrate. Science. 1975;190:787–9. doi: 10.1126/science.1198095. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Rao S., Moody D. E. Hepatocellular carcinomas in acatalasemic mice treated with nafenopin, a hypolipidemic peroxisome proliferator. Cancer Res. 1976;36:1211–7. [PubMed] [Google Scholar]
- Reddy J. K., Azarnoff D. L., Hignite C. E. Hypolipidaemic hepatic peroxisome proliferators form a novel class of chemical carcinogens. Nature. 1980;283:397–8. doi: 10.1038/283397a0. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Lalwani N. D., Qureshi S. A., Reddy M. K., Moehle C. M. Induction of hepatic peroxisome proliferation in nonrodent species, including primates. Am J Pathol. 1984;114:171–83. [PMC free article] [PubMed] [Google Scholar]
- Reddy J. K., Rao M. S. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006a;290:G852–8. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Rao M. S., Azarnoff D. L., Sell S. Mitogenic and carcinogenic effects of a hypolipidemic peroxisome proliferator, [4-chloro-6-(2,3-xylidino)-2-pyrimidinylthio]acetic acid (Wy-14, 643), in rat and mouse liver. Cancer Res. 1979;39:152–61. [PubMed] [Google Scholar]
- Reddy J. K. Nonalcoholic steatosis and steatohepatitis. III. Peroxisomal β-oxidation, PPAR α, and steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2001b;281:G1333–9. doi: 10.1152/ajpgi.2001.281.6.G1333. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Guo D., Jia Y., Yu S., Rao M.S. Nuclear receptor transcriptional coactivators in development and metabolism. Advances in Developmental Biology. 2006b;16:389–420. [Google Scholar]
- Reddy J. K., Hashimoto T. Peroxisomal β-oxidation and peroxisome proliferator-activated receptor α: an adaptive metabolic system. Annu Rev Nutr. 2001a;21:193–230. doi: 10.1146/annurev.nutr.21.1.193. [DOI] [PubMed] [Google Scholar]
- Reddy J. K., Chu R. Peroxisome proliferator-induced pleiotropic responses: pursuit of a phenomenon. Ann N Y Acad Sci. 1996;804:176–201. doi: 10.1111/j.1749-6632.1996.tb18616.x. [DOI] [PubMed] [Google Scholar]
- Reddy J. K. Peroxisome proliferators and peroxisome proliferator-activated receptor α: biotic and xenobiotic sensing. Am J Pathol. 2004;164:2305–21. doi: 10.1016/s0002-9440(10)63787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy J. K., Goel S. K., Nemali M. R., Carrino J. J., Laffler T. G., Reddy M. K., Sperbeck S. J., Osumi T., Hashimoto T., Lalwani N. D. Transcription regulation of peroxisomal fatty acyl-CoA oxidase and enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase in rat liver by peroxisome proliferators. Proc Natl Acad Sci U S A. 1986b;83:1747–51. doi: 10.1073/pnas.83.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M., Glass C. K. PPARs and molecular mechanisms of transrepression. Biochim Biophys Acta. 2007;1771:926–35. doi: 10.1016/j.bbalip.2007.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti E., Chinetti-Gbaguidi G., Staels B. Regulation of macrophage functions by PPAR-α, PPAR-γ, and LXRs in mice and men. Arterioscler Thromb Vasc Biol. 2008;28:1050–9. doi: 10.1161/ATVBAHA.107.158998. [DOI] [PubMed] [Google Scholar]
- Robitaille J., Brouillette C., Houde A., Lemieux S., Perusse L., Tchernof A., Gaudet D., Vohl M. C. Association between the PPARalpha-L162V polymorphism and components of the metabolic syndrome. J Hum Genet. 2004;49:482–9. doi: 10.1007/s10038-004-0177-9. [DOI] [PubMed] [Google Scholar]
- Rusyn I., Asakura S., Pachkowski B., Bradford B. U., Denissenko M. F., Peters J. M., Holland S. M., Reddy J. K., Cunningham M. L., Swenberg J. A. Expression of base excision DNA repair genes is a sensitive biomarker for in vivo detection of chemical-induced chronic oxidative stress: identification of the molecular source of radicals responsible for DNA damage by peroxisome proliferators. Cancer Res. 2004;64:1050–7. doi: 10.1158/0008-5472.can-03-3027. [DOI] [PubMed] [Google Scholar]
- Sanderson L. M., Degenhardt T., Koppen A., Kalkhoven E., Desvergne B., Muller M., Kersten S. Peroxisome proliferator-activated receptor β/δ (PPARbeta/δ) but not PPARalpha serves as a plasma free fatty acid sensor in liver. Mol Cell Biol. 2009;29:6257–67. doi: 10.1128/MCB.00370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapone A., Peters J. M., Sakai S., Tomita S., Papiha S. S., Dai R., Friedman F. K., Gonzalez F. J. The human peroxisome proliferator-activated receptor α gene: identification and functional characterization of two natural allelic variants. Pharmacogenetics. 2000;10:321–33. doi: 10.1097/00008571-200006000-00006. [DOI] [PubMed] [Google Scholar]
- Sarkar J., Qi C., Guo D., Ahmed M. R., Jia Y., Usuda N., Viswakarma N., Rao M. S., Reddy J. K. Transcription coactivator PRIP, the peroxisome proliferator-activated receptor (PPAR)-interacting protein, is redundant for the function of nuclear receptors PParalpha and CAR, the constitutive androstane receptor, in mouse liver. Gene Expr. 2007;13:255–69. doi: 10.3727/000000006780666948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C. J., Valente A. J., Sprague E. A., Kelley J. L., Suenram C. A., Rozek M. M. Atherosclerosis as an inflammatory process. The roles of the monocyte-macrophage. Ann N Y Acad Sci. 1985;454:115–20. doi: 10.1111/j.1749-6632.1985.tb11849.x. [DOI] [PubMed] [Google Scholar]
- Seedorf U., Raabe M., Ellinghaus P., Kannenberg F., Fobker M., Engel T., Denis S., Wouters F., Wirtz K. W., Wanders R. J., Maeda N., Assmann G. Defective peroxisomal catabolism of branched fatty acyl coenzyme A in mice lacking the sterol carrier protein-2/sterol carrier protein-x gene function. Genes Dev. 1998;12:1189–201. doi: 10.1101/gad.12.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah Y. M., Morimura K., Yang Q., Tanabe T., Takagi M., Gonzalez F. J. Peroxisome proliferator-activated receptor α regulates a microRNA-mediated signaling cascade responsible for hepatocellular proliferation. Mol Cell Biol. 2007;27:4238–47. doi: 10.1128/MCB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalev A., Siegrist-Kaiser C. A., Yen P. M., Wahli W., Burger A. G., Chin W. W., Meier C. A. The peroxisome proliferator-activated receptor α is a phosphoprotein: regulation by insulin. Endocrinology. 1996;137:4499–502. doi: 10.1210/endo.137.10.8828512. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shan W., Palkar P. S., Murray I. A., McDevitt E. I., Kennett M. J., Kang B. H., Isom H. C., Perdew G. H., Gonzalez F. J., Peters J. M. Ligand activation of peroxisome proliferator-activated receptor β/δ (PPARbeta/δ) attenuates carbon tetrachloride hepatotoxicity by downregulating proinflammatory gene expression. Toxicol Sci. 2008b;105:418–28. doi: 10.1093/toxsci/kfn142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W., Nicol C. J., Ito S., Bility M. T., Kennett M. J., Ward J. M., Gonzalez F. J., Peters J. M. Peroxisome proliferator-activated receptor-β/δ protects against chemically induced liver toxicity in mice. Hepatology. 2008a;47:225–35. doi: 10.1002/hep.21925. [DOI] [PubMed] [Google Scholar]
- Sher T., Yi H. F., McBride O. W., Gonzalez F. J. cDNA cloning, chromosomal mapping, and functional characterization of the human peroxisome proliferator activated receptor. Biochemistry. 1993;32:5598–604. doi: 10.1021/bi00072a015. [DOI] [PubMed] [Google Scholar]
- Sozio M., Crabb D. W. Alcohol and lipid metabolism. Am J Physiol Endocrinol Metab. 2008;295:E10–6. doi: 10.1152/ajpendo.00011.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B., Maes M., Zambon A. Fibrates and future PPARalpha agonists in the treatment of cardiovascular disease. Nat Clin Pract Cardiovasc Med. 2008;5:542–53. doi: 10.1038/ncpcardio1278. [DOI] [PubMed] [Google Scholar]
- Still K., Grabowski P., Mackie I., Perry M., Bishop N. The peroxisome proliferator activator receptor α/δ agonists linoleic acid and bezafibrate upregulate osteoblast differentiation and induce periosteal bone formation in vivo. Calcif Tissue Int. 2008;83:285–92. doi: 10.1007/s00223-008-9175-9. [DOI] [PubMed] [Google Scholar]
- Surapureddi S., Yu S., Bu H., Hashimoto T., Yeldandi A. V., Kashireddy P., Cherkaoui-Malki M., Qi C., Zhu Y. J., Rao M. S., Reddy J. K. Identification of a transcriptionally active peroxisome proliferator-activated receptor α -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc Natl Acad Sci U S A. 2002;99:11836–41. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N., Moriya K., Kiyosawa K., Koike K., Gonzalez F. J., Aoyama T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J Clin Invest. 2008;118:683–94. doi: 10.1172/JCI33594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B. M. Stimulation of adipogenesis in fibroblasts by PPAR γ 2, a lipid-activated transcription factor. Cell. 1994;79:1147–56. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Tugwood J. D., Issemann I., Anderson R. G., Bundell K. R., McPheat W. L., Green S. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5' flanking sequence of the rat acyl CoA oxidase gene. Embo J. 1992;11:433–9. doi: 10.1002/j.1460-2075.1992.tb05072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meer D. L., Degenhardt T., Vaisanen S., de Groot P. J., Heinaniemi M., de Vries S. C., Muller M., Carlberg C., Kersten S. Profiling of promoter occupancy by PPAR{α} in human hepatoma cells via ChIP-chip analysis. Nucleic Acids Res. 2010:(in press) . doi: 10.1093/nar/gkq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanasi U., Chu R., Huang Q., Castellon R., Yeldandi A. V., Reddy J. K. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J Biol Chem. 1996;271:2147–55. doi: 10.1074/jbc.271.4.2147. [DOI] [PubMed] [Google Scholar]
- Viswakarma N., Matsumoto K., Jia Y., Rao M. S., Reddy J. K. Mice lacking transcription cofactor PRIC285 reveal attenuation of liver regeneration but are viable and develop normally. FASEB J. 2009;23:117. [Google Scholar]
- Voegel J. J., Heine M. J., Tini M., Vivat V., Chambon P., Gronemeyer H. The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. Embo J. 1998;17:507–19. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohl M. C., Lepage P., Gaudet D., Brewer C. G., Betard C., Perron P., Houde G., Cellier C., Faith J. M., Despres J. P., Morgan K., Hudson T. J. Molecular scanning of the human PPARa gene: association of the L162v mutation with hyperapobetalipoproteinemia. J Lipid Res. 2000;41:945–52. [PubMed] [Google Scholar]
- Walgren J. E., Kurtz D. T., McMillan J. M. Expression of PPAR(α) in human hepatocytes and activation by trichloroacetate and dichloroacetate. Res Commun Mol Pathol Pharmacol. 2000;108:116–32. [PubMed] [Google Scholar]
- Wang Z., Qi C., Krones A., Woodring P., Zhu X., Reddy J. K., Evans R. M., Rosenfeld M. G., Hunter T. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 2006;3:111–22. doi: 10.1016/j.cmet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Wang Y., Feng W., Xue W., Tan Y., Hein D. W., Li X. K., Cai L. Inactivation of GSK-3beta by metallothionein prevents diabetes-related changes in cardiac energy metabolism, inflammation, nitrosative damage, and remodeling. Diabetes. 2009;58:1391–402. doi: 10.2337/db08-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. X., Lee C. H., Tiep S., Yu R. T., Ham J., Kang H., Evans R. M. Peroxisome-proliferator-activated receptor δ activates fat metabolism to prevent obesity. Cell. 2003;113:159–70. doi: 10.1016/s0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- White J. H., Fernandes I., Mader S., Yang X. J. Corepressor recruitment by agonist-bound nuclear receptors. Vitam Horm. 2004;68:123–43. doi: 10.1016/S0083-6729(04)68004-6. [DOI] [PubMed] [Google Scholar]
- World Health Organization; 2006. Obesity and Overweight http://www.who.int/mediacentre/factsheets/fs311/en/index.html. [Google Scholar]
- Xu H. E., Lambert M. H., Montana V. G., Plunket K. D., Moore L. B., Collins J. L., Oplinger J. A., Kliewer S. A., Gampe R. T., Jr., McKee D. D., Moore J. T., Willson T. M. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001;98:13919–24. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Nagano T., Shah Y., Cheung C., Ito S., Gonzalez F. J. The PPAR α-humanized mouse: a model to investigate species differences in liver toxicity mediated by PPAR α. Toxicol Sci. 2008;101:132–9. doi: 10.1093/toxsci/kfm206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeldandi A. V., Rao M. S., Reddy J. K. Hydrogen peroxide generation in peroxisome proliferator-induced oncogenesis. Mutat Res. 2000;448:159–77. doi: 10.1016/s0027-5107(99)00234-1. [DOI] [PubMed] [Google Scholar]
- Yu S., Matsusue K., Kashireddy P., Cao W. Q., Yeldandi V., Yeldandi A. V., Rao M. S., Gonzalez F. J., Reddy J. K. Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor gamma1 (PPARgamma1) overexpression. J Biol Chem. 2003;278:498–505. doi: 10.1074/jbc.M210062200. [DOI] [PubMed] [Google Scholar]
- Yu S., Cao W. Q., Kashireddy P., Meyer K., Jia Y., Hughes D. E., Tan Y., Feng J., Yeldandi A. V., Rao M. S., Costa R. H., Gonzalez F. J., Reddy J. K. Human peroxisome proliferator-activated receptor α (PPARalpha) supports the induction of peroxisome proliferation in PPARalpha-deficient mouse liver. J Biol Chem. 2001;276:42485–91. doi: 10.1074/jbc.M106480200. [DOI] [PubMed] [Google Scholar]
- Yu S., Reddy J. K. Transcription coactivators for peroxisome proliferator-activated receptors. Biochim Biophys Acta. 2007;1771:936–51. doi: 10.1016/j.bbalip.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Zafrani E. S. Non-alcoholic fatty liver disease: an emerging pathological spectrum. Virchows Arch. 2004;444:3–12. doi: 10.1007/s00428-003-0943-7. [DOI] [PubMed] [Google Scholar]
- Zandbergen F., Plutzky J. PPARalpha in atherosclerosis and inflammation. Biochim Biophys Acta. 2007;1771:972–82. doi: 10.1016/j.bbalip.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Marcus S. L., Sajjadi F. G., Alvares K., Reddy J. K., Subramani S., Rachubinski R. A., Capone J. P. Identification of a peroxisome proliferator-responsive element upstream of the gene encoding rat peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase. Proc Natl Acad Sci U S A. 1992;89:7541–5. doi: 10.1073/pnas.89.16.7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Calandra C., Rao M. S., Reddy J. K. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor γ. Gene Expr. 1996;6:185–95. [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Jia Y., Nye J. S., Rao M. S., Reddy J. K. Deletion of PBP/PPARBP, the gene for nuclear receptor coactivator peroxisome proliferator-activated receptor-binding protein, results in embryonic lethality. J Biol Chem. 2000b;275:14779–82. doi: 10.1074/jbc.C000121200. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Kan L., Qi C., Kanwar Y. S., Yeldandi A. V., Rao M. S., Reddy J. K. Isolation and characterization of peroxisome proliferator-activated receptor (PPAR) interacting protein (PRIP) as a coactivator for PPAR. J Biol Chem. 2000a;275:13510–6. doi: 10.1074/jbc.275.18.13510. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Korenberg J. R., Chen X. N., Noya D., Rao M. S., Reddy J. K. Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPAR γ) gene: alternative promoter use and different splicing yield two mPPAR γ isoforms. Proc Natl Acad Sci U S A. 1995;92:7921–5. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoete V., Grosdidier A., Michielin O. Peroxisome proliferator-activated receptor structures: ligand specificity, molecular switch and interactions with regulators. Biochim Biophys Acta. 2007;1771:915–25. doi: 10.1016/j.bbalip.2007.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.