Abstract
Identifying binding sites and target genes of transcription factors is a major biologic problem. The most commonly used current technique, chromatin immunoprecipitation (ChIP), is dependent on a high quality antibody for each protein of interest, which is not always available, and is also cumbersome, involving sequential cross-linking and reversal of cross-linking. We have developed a novel strategy to study protein DNA binding sites in vivo, which we term DamIP. By tethering a mutant form of E. coli DNA adenine methyltransferase to the target protein, the fusion protein introduces N-6-adenosine methylation to sequences proximal to the protein binding sites. DNA fragments with this modification, which is absent in eukaryotes, are detected using an antibody directed against methylated adenosine. For an initial test of the method we used human estrogen receptor α (hERα), one of the best studied transcription factors. We found that expression of Dam-hERα fusion proteins in MCF-7 cells introduces adenosine methylation near a series of known direct hERα binding sites. Specific methylation tags are also found at indirect hERα binding sites, including both primary binding sites for the ER interactors AP-1 and SP1, and promoters that are activated by upstream ER bound enhancers. DamIP provides a new tool for the study of DNA interacting protein function in vivo.
Introduction
Regulation of gene expression is a complex process controlled by a large number of different proteins that interact with DNA directly or indirectly. Study of how and where these proteins bind to the genome is essential for understanding this process. Historically, many methods have been developed to study DNA-protein interactions [Moss and Leblanc, 2009]. Chromatin immunoprecipitation (ChIP) is a widely utilized method that involves chemical cross-linking of DNA and protein followed by enriching the DNA-protein complexes with an antibody specifically directed against the protein of interest [Collas, 2009]. Combined with DNA microarray or massive parallel sequencing technologies, ChIP makes it possible to profile the occupancy of DNA interacting proteins at a genome wide scale [Ren et al., 2000; Robertson et al., 2007]. However, the ChIP assay is rather cumbersome and is limited by its dependence on a high quality antibody for each protein of interest.
Recently, van Steensel and colleagues developed a new strategy to study protein DNA interactions [Orian et al., 2009; van Steensel et al., 2001; van Steensel and Henikoff, 2000]. DNA adenine methyltransferase (Dam) from E. coli, which specifically methylates the adenine residue in a GATC recognition sequence, is attached to the protein of interest through a short linker. When expressed in cells, the fusion proteins bind to genomic DNA and introduce N-6-adenine methylation to nearby GATC tetramers. Locations of methylation can be identified with methylation sensitive restriction enzymes Dpn I and Dpn II. This method, named Dam IDentification (DamID), has been successfully applied to several eukaryotic model systems, e.g. budding yeast, plant, fruit fly, and cultured mammalian cells [Bianchi-Frias et al., 2004; Orian et al., 2003; Venkatasubrahmanyam et al., 2007; Weber et al., 2005; Zhang et al., 2007].
The high activity of E. coli Dam creates a signal to noise problem for the DamID approach. In addition, the tetrameric Dam recognition occurs on average once in every 256 nucleotides in the genome and may not be present near specific DNA binding sites of interest, which limits its resolution. DNA adenine methyltransferase has been extensively studied and its target sequence recognition is determined by several key amino acid residues in the catalytic pocket [Horton et al., 2006; Horton et al., 2005]. Previously described mutations of these residues decrease both the activity of the enzyme and the specificity for the GATC tetramer, thereby increasing the frequency of potential methylation sites and addressing both concerns. Here we describe a new method using such a mutant form of DNA adenine methyltransferase, combined with an antibody that specifically recognizes N-6-methylated DNA [Lopez et al., 2003]. The mutant Dam is linked to the protein of interest, and the fusion protein introduces N-6-adenosine methylation to sequences adjacent to specific DNA binding sites. Methylated DNA fragments are enriched by immunoprecipitation and detected by quantitative real time PCR (qPCR) or other methods. We have used hERα in an initial test of this method, and have found that Dam-hERα fusion protein can be used to specifically identify both direct and indirect hERα DNA binding sites.
Reagents and instruments
QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), Fugene HD (Roche, Indianapolis, IN), 17 β-estradiol, Fulvestrant, DNase-free RNase A (Sigma, St. Louis, MO), A/G plus agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA), proteinase K (Invitrogen, Carlsbad, CA), anti-N-6-methyladenosine antibody (Megabase Research, Lincoln, NE), QiaQuick PCR purification kit (Qiagen, Valencia, CA), Branson sonifier 250, (Branson Corporation, Danbury, CT), StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA).
Methods
Plasmid construction
DNA adenine methyltransferase open reading frame was amplified from E. coli genomic DNA and inserted into a pCMX vector. To generate the lysine 9 to alanine mutant dam (DamK9A), AAG was changed to GCG with QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). A 3xFlag tag sequence was attached at the N-terminus or C-terminus as a linker to generate pCMX-N-DamK9A or pCMX-C-DamK9A vectors. A human estrogen receptor α ORF was cloned and inserted into either pCMX-N-DamK9A or pCMX-C-DamK9A vectors.
Cell culture, transfection and reporter assay
HeLa or MCF-7 cells were maintained in DMEM media supplemented with 10% fetal bovine serum and 1x nonessential amino acids (Invitrogen, Carlsbad, CA). HeLa cells were transfected with lipofectamine 2000 (Invitrogen, Carlsbad, CA) and MCF-7 cells were transfected with Fugene HD ((Roche, Indianapolis, IN) following manufacturers’ protocols.
DamIP
Twenty four hours after transfection, cells were treated with 100nM 17 β-estradiol for another 24 hours. Cells were then collected and washed twice with phosphate buffered saline (PBS). Cell pellets were resuspended with lysis buffer (150mM NaCl, 10mM Tris pH8, 25mM EDTA pH8, 0.5% SDS) and briefly sonicated to reduce viscosity. DNase-free RNase A (Sigma, St. Louis, MO) was added to final concentration of 10ug/mL and samples were incubated at 37°C for 30 minutes. Proteinase K was then added followed by overnight incubation at 50°C. Genomic DNA was extracted from the deproteinized lysate by phenol/chloroform extraction and ethanol precipitation. Purified DNA was resuspended in TE buffer and sonicated on ice until majority of the fragments were around 500 base pairs.
Five micrograms of sonicated DNA and 5pg of control plasmid DNA were mixed in TE buffer and heated for 10 minutes in a boiling water bath and quenched on ice for 5 minutes. The control DNA plasmid contains a sequence completely unrelated with mammalian genomes and is fully methylated by growth in the standard DH5α stain. The DNA solution was mixed with 0.11 volume of 10x DamIP buffer (100mM Na-Phosphate pH 7.0, 3M NaCl, 0.5% Triton X-100). Five micrograms of anti-N-6-methyladenosine antibody (Megabase Research, Lincoln, NE) was added to the DNA solution and rotated for one hour at room temperature. Buffer balanced A/G plus agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were added to each sample and rotated for another one hour at room temperature. Beads were washed with 1xDamIP buffer 5 times and treated with proteinase K overnight. Bound DNA was finally purified with QiaQuick PCR purification kit (Qiagen, Valencia, CA) and eluted in TE buffer. Eluted DNA was analyzed by real-time quantitative PCR. PCR primer sequences are available upon request. Normalized IP% is calculated as the ratio of precipitated DNA over input DNA and normalized with precipitation efficiency determined by the control DNA signal (assume fully methylated plasmid DNA is precipitated with efficiency of 1). All data are presented as mean of three independent experiments +/- SEM.
Results
Expression and activity of Dam fusion proteins
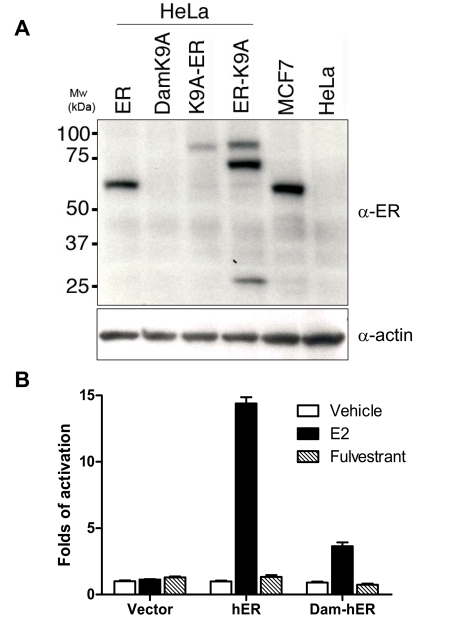
For proof of principle, we set out to test our new method with the most widely studied nuclear receptor, human estrogen receptor α (hERα). We fused an E. coli DNA adenosine methyltransferase K9A mutant to both the amino- and carboxyl-termini of hERα. The N-terminal fusion protein expresses as a single band in HeLa cells (Figure 1A). We further tested their transcriptional activities in transiently transfected HeLa cells. The N-terminal fusion protein, but not the C-terminal fusion protein, retains ligand responsiveness, although its activity is only approximately 20% of that of wild type hERα (Figure 1B and data not shown). We used the N-terminal fusion DamK9A-hERα in the following experiments.
Figure 1. Expression and transcriptional activities of DamK9A-hERα fusion protein.
(A). HeLa cells were transfected with various plasmids as indicated and cell lysate was analyzed by Western blot. MCF-7 and untransfected HeLa cell lysate were loaded as positive and negative control, respectively. The same blot was detected with anti-β-actin antibody as loading control. Molecular weights (Mw) are indicated as kilodaltons (kDa). (B). Fusion protein transcriptional activities in HeLa cells. Cells were co-transfected with various plasmids and luciferase reporter vectors. Luciferase activities were measured 24 hours after ligand treatment. Data are presented as mean +/- SEM.
DamIP in MCF-7 cells
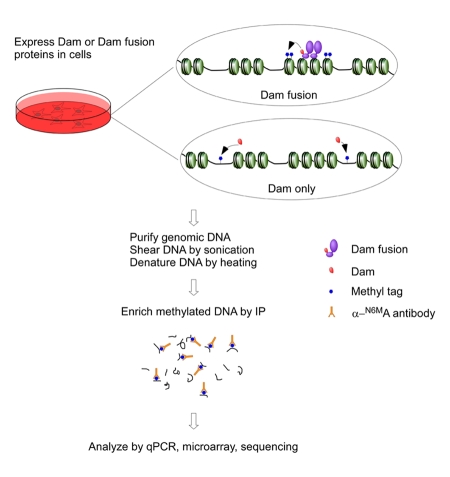
Upon activation by agonist ligands, ERα regulates the expression of most target genes by direct binding to specific estrogen response elements (EREs). As described in the Methods section and outlined in Figure 2, we transiently transfected MCF-7 cells with DamK9A or DamK9A-hER and performed DamIP. Genomic DNA was harvested, sonicated and denatured before immunoprecipitation using an anti-N-6-methyladenosine antibody (α-N6MA). Enriched DNA fragments were analyzed by real-time quantitative PCR. We checked several regions containing known ERα binding sites and several regions without ERα binding sites [Carroll et al., 2006; Labhart et al., 2005]. As shown in Figure 3A, DamK9A-hERα introduces much more methylation into fragments that contain known ERα binding sites than Dam protein alone. In the regions without known ERα binding sites, methylation introduced by DamK9A-hERα is comparable with Dam protein alone (Figure 3B). DamK9A-hERα is clearly able to specifically introduce N-6-adenosine methylation into the genome around known ERα binding sites.
Figure 2. Illustration of the procedure of DamIP.
DamK9A or DamK9A fusion proteins are expressed in cells. Genomic DNA is purified, sonicated and denatured before mixed with anti-N-6-methyladenine antibodies. Methylated DNA recognized by the antibody is enriched and analyzed by various methods, e.g. qPCR, microarray and next-generation sequencing. Details are described in the Methods section.
Figure 3. RT-qPCR analysis of DamIP.
Primers were designed to amplify genomic regions with or without known ERE. DNA enriched with DamIP from MCF-7 cells transfected with DamK9A-hER (Dam-ER) or Dam alone was analyzed by qPCR. Data are presented as mean +/- SEM.
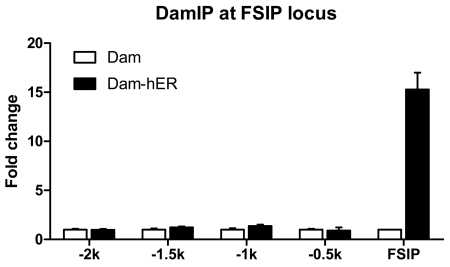
DamID uses wild type Dam protein, which methylates the tetrameric sequence GATC that occurs on average once in the genome in every 256 bp. Thus, the resolution of DamID may be limited by the absence of potential methylation target sequences in proximity to the binding sites. In addition, the high activity of the native enzyme introduces methyl tags relatively far from the binding sites. The DamK9A mutant, which loses the recognition of the first G and methylates the target sequence “ATC” [Horton et al., 2006] should have four fold more potential methylation target sequences and decreased activity, both potentially increasing resolution. We tested methylation around the well characterized FSIP ERE [Carroll et al., 2006; Labhart et al., 2005]. Remarkably, the robust DamK9A-hERα FSIP signal was not detected in fragments more than 500bp away from the known ERα binding region. We conclude that DamIP has good resolution, comparable to that of the conventional ChIP assay (Figure 4).
Figure 4. DamIP has great resolution.
A series of primers were designed to amplify the regions upstream of the FSIP locus that contains known ERE. These targeted regions are about 500bp from each other and designated as -2k, -1.5k, -1k, -0.5k, respectively. The specific modification introduced by DamK9A-hER is only observed at the FSIP locus.
DamIP detects indirect ER regulation
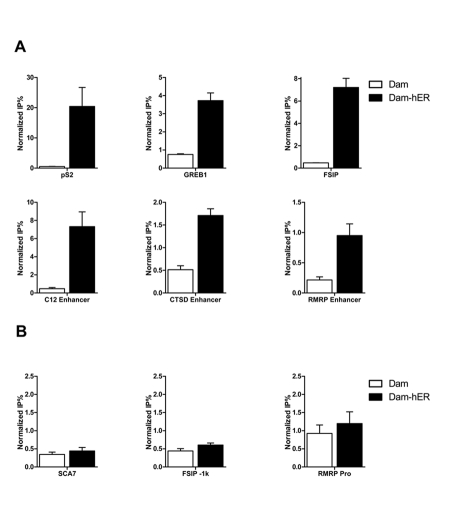
In addition to direct binding to DNA, ERα can indirectly regulate transcription by interacting with transcriptional factors on their sites, including Sp1 and AP-1 (jun/fos) [Nilsson et al., 2001]. We investigated two known sites of indirect ERα regulation. As shown in Figure 5C, DamK9A-hERα but not Dam alone specifically introduced methylation in these regions.
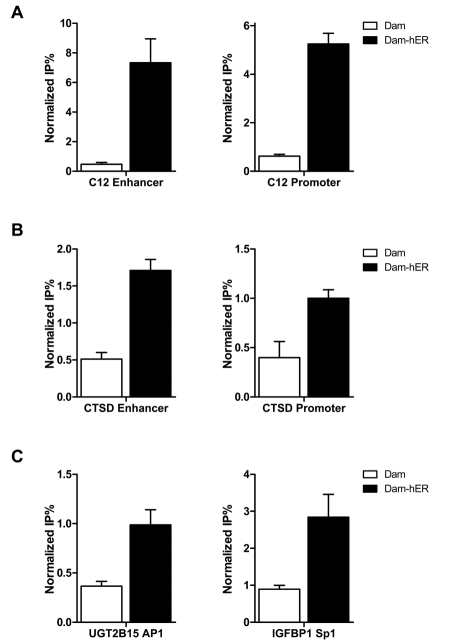
Figure 5. DamIP identifies indirect regulation by ER.
ER can regulate gene expression through long range chromosome looping (A, B) or binding with other transcriptional factors (C). Carbonic anhydrase XII (C12) and cathepsin D (CTSD) enhancers contain functional EREs but not promoter regions, however, DamIP indicates ER communicates with the ERE-less promoter regions during transcription. ER regulation through interaction with AP1 (C, left panel) or Sp1 (C, right panel) can also be detected by DamIP.
Some ERα binding sites are located several thousand base pairs upstream of the transcription starting sites. Regulation of gene expression through these distal enhancers may require communication between the enhancers and proximal promoters via long range chromosome looping [Barnett et al., 2008; Pan et al., 2008]. Carbonic anhydrase XII (CA12) gene encodes a zinc metalloenzyme, which is overexpressed in certain cancers, including breast cancer. ERα strongly regulates the expression of C12 through a distal ERα binding site located about 6kb upstream of the transcription start site. Communication between the distal enhancer and proximal promoter after ER activation is not apparent in standard ChIP, but can by detected by the chromatin conformation capture assay [Barnett et al., 2008]. We analyzed the distal enhancer and proximal promoter regions of CA12 gene with DamIP. As shown in Figure 5A, DamK9A-hERα specifically introduced methylation at the ERE containing distal enhancer region, but Dam protein alone did not. For the proximal promoter region, which does not contain a known ERE, DamK9A-hERα also introduced increased methylation compared with Dam protein alone, indicating indirect interaction/communication of ERα with this region.
Another known ER target gene cathepsin D (CTSD) has multiple EREs in the distal enhancer region, and conventional ChIP detects ERα recruitment after ligand treatment at the enhancer regions but not the promoter [Carroll et al., 2006]. DamIP shows ERα interaction at both regions (Figure 5B), indicating that ER may regulate cathepsin D expression through a similar chromatin looping mechanism as CA12. This result is consistent with the finding that the distal ERE enhancer regulates CTSD expression through long range chromosomal looping mechanism [Bretschneider et al., 2008].
Discussion
Here we describe a novel method to identify DNA binding elements in vivo. Our method uses a mutant form of E. coli Dam joined to the protein of interest, which can introduce N-6-adenosine methylation into genomic DNA around sites bound by the fusion protein. This modification is not present in mammalian DNA, and the specifically tagged DNA can be enriched with antibody and analyzed by various assays. The effectiveness of this technology is clearly validated by the results described here, which suggest that it is very sensitive. Eleven out of 12 known ER binding sites that we tested showed significant enrichment compared with control. We are in the process of using massive parallel sequencing to analyze DamIP samples, which will provide a genome-wide comparison of DamIP with conventional ChIP.
The introduction of the Dam mutation was important for two reasons. The first is that wild type Dam is a very active enzyme and its expression in mammalian cells can rapidly result in methylation of the whole genome. The second is that the GATC specificity of the wild type enzyme limits the potential resolution of the technique. We screened several Dam mutants with decreased activity and specificity in DamIP and found that the DamK9A mutant showed the best signal to noise ratio. This mutant decreases specificity for the initial G [Horton et al., 2006], increasing the frequency of potential sites by fourfold.
There are several technical issues that confronted the development of the DamIP technology, and others that must be addressed in its potential application to other proteins of interest. During the preparation of plasmids for transient transfection, we noticed that even those propagated in Dam/Dcm negative strains are methylated. This is likely due to the leaky expression of Dam proteins driven by the mammalian promoter in the plasmid. Fragments of these methylated plasmids are recognized and precipitated by the α-N6MA antibody. These fragments did not interfere with qPCR analysis in our study, but may cause potential background problems in certain analysis methods, e.g. microarray. This potential problem could be easily circumvented using a virus-based expression system or generating stable cell lines. However, methylated plasmids can also be useful in DamIP. We prepared a fully methylated plasmid containing a completely unrelated sequence from D. discoideum. A constant amount of this control plasmid equivalent to that of a single copy gene was added to each sample prior to immunoprecipitation. Analyzing the amount of the control plasmid in the final enriched DNA provides an internal control to monitor the precipitation efficiency of individual samples.
During the development and optimization of DamIP, we tested a variety of different salt/detergent concentrations and antibody binding beads to identify the best immunoprecipitation condition. With all of these efforts, we still observed some background binding with unmethylated DNA. However, this background was largely eliminated by generating single strand DNA conjugated agarose that was pre-incubated with the anti-N-6-methyladenosine antibody.
As shown in Figure 1, the fusion protein DamK9A-hERα showed reduced transcriptional activities compared with the wild type hERα. This was partially due to the reduced expression of the fusion protein, but more detailed studies indicated decreased activity of DamK9A-hERα and other fusion proteins tested. The basis for this decreased activity is not clear, but could reflect either simple steric effects or more specific impact of the Dam protein itself. As with other fusion approaches, it is essential to determine the activity of the fusion proteins and may be necessary to employ a range of amino- and carboxyl-terminal fusion strategies.
DamIP relies on an antibody against N-6-methyladenine. We initially obtained an antibody from Megabase Research, which worked very well for the studies described here. Unfortunately, this company no longer exists and the antibody is no longer available. However, several groups have generated similar antibodies and another company (SYSY, Goettingen, Germany) also provides α-N6MA antibody that has good affinity against N6MA and works well in DamIP. We are now generating our own antibody according to published protocols [Erlanger and Beiser, 1964; Lopez et al., 2003] and the crude sera also works.
Compared with conventional ChIP assay, our method has two major advantages: 1) no need for protein-specific antibodies; 2) no need for formaldehyde crosslinking. Conventional ChIP requires a high quality antibody against the protein of interest to allow satisfactory enrichment over the background, and such antibodies are not always available. In contrast, our method only needs the antibody against N-6-methyladenine. A potentially tricky and cumbersome step of the conventional ChIP assay is the crosslinking of protein-DNA complexes with formaldehyde. In addition to the toxicity of formaldehyde, crosslinking with formaldehyde is difficult under certain conditions, especially when the ChIP assay is applied to whole tissue or organs. In general, DamIP shows comparable or better enrichment compared with conventional ChIP. When using lentiviral transfection to express Dam-ER proteins in MCF-7 cells, we observed more than 100 fold enrichment at the FSIP ERE by DamIP compared with about 17 fold by conventional ChIP assay [Labhart et al., 2005].
In DamIP, the methyl tags are covalently attached to the genomic DNA. We assume that this covalent modification is essentially permanent in eukaryotic cells, although there is no mechanism to replicate it and it is possible that it could be recognized as DNA damage and repaired. However, the modification is not affected by harsh treatments to remove proteins prior to DamIP, and most of the procedures can be conveniently performed at room temperature. This property of DamIP makes it particularly suitable for in vivo identification of DNA binding sites.
A limitation of DamIP relative to ChIP is the reliance on expression of the exogenous fusion protein rather than the endogenous protein. In this, DamIP resembles other techniques that rely on fusion proteins, for example the BirA fusions that rely on the high affinity avidin-biotin interaction rather than antibodies to purify crosslinked, specifically bound DNA fragments [Kim et al., 2009].
An important qualitative difference between DamIP and ChIP is that conventional ChIP gives a snap-shot of DNA-protein binding profile at the time of crosslinking, while DamIP provides an cumulative profile of DNA-protein interaction over time. Thus, DamIP cannot be used for time course studies such as those used to follow events after estradiol administration [Metivier et al., 2003]. In fact, we observed a low level of specific methylation at ERα binding sites in MCF7 cells even in the absence of estradiol or in the presence of antagonist. We believe this is the result of cumulative methylation when ERα briefly interacts with its target in the presence of antagonist or in the absence of ligands, which is hard to detect with conventional ChIP assay.
However, DamIP could be used to identify an increasing range of primary and potential secondary targets over time after ligand addition. For example, recruitment of ERα at the proximal CTSD promoter is usually hard to detect by conventional ChIP probably because the recruitment is transient and time-dependent [Bretschneider et al., 2008; Carroll et al., 2006; Shang et al., 2000]. However, DamIP can readily detect this interaction (Figure 5B), potentially as the result of cumulative ERα recruitment profile over the period of ligand treatment. Particularly when combined with tetracycline [Corbel and Rossi, 2002] or ecdysone [No et al., 1996] inducible systems, DamIP will provide another option to study DNA-protein interactions in vivo. With the development and implementation of next generation massive parallel sequencing technology and bioinformatics, DamIP will provide an excellent tool to study and identify binding sites or target genes of transcriptional factors.
Acknowledgments
This work was supported by the NURSA grant U19 DK62434.
Abbreviations
- CA12
carbonic anhydrase XII
- ChIP
chromatin immunoprecipitation
- CTSD
cathepsin D
- Dam
DNA adenine methyltransferase
- DamID
Dam identification
- DamIP
Dam immunoprecipitation
- ERE
estrogen response element
- ERα
estrogen receptor α
- FSIP
fibrous sheath interacting protein1
- IGFBP1
insulin-like growth factor binding protein 1
- RMRP
RNA component of mitochondrial RNA processing endoribonuclease
- SEM
standard error of mean
- UGT2B15
UDP glycosyltransferase 2B15
References
- Barnett D. H., Sheng S., Charn T. H., Waheed A., Sly W. S., Lin C. Y., Liu E. T., Katzenellenbogen B. S. Estrogen receptor regulation of carbonic anhydrase XII through a distal enhancer in breast cancer. Cancer Res. 2008;68:3505–15. doi: 10.1158/0008-5472.CAN-07-6151. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D., Orian A., Delrow J. J., Vazquez J., Rosales-Nieves A. E., Parkhurst S. M. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider N., Sara Kangaspeska, Seifert M., Reid G., Gannon F., Denger S. E2-mediated cathepsin D (CTSD) activation involves looping of distal enhancer elements. Mol Oncol. 2008;2:182–90. doi: 10.1016/j.molonc.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Collas P. The State-of-the-Art of Chromatin Immunoprecipitation. Methods Mol Biol. 2009;567:1–25. doi: 10.1007/978-1-60327-414-2_1. [DOI] [PubMed] [Google Scholar]
- Corbel S. Y., Rossi F. M. Latest developments and in vivo use of the Tet system: ex vivo and in vivo delivery of tetracycline-regulated genes. Curr Opin Biotechnol. 2002;13:448–52. doi: 10.1016/s0958-1669(02)00361-0. [DOI] [PubMed] [Google Scholar]
- Erlanger B. F., Beiser S. M. Antibodies Specific for Ribonucleosides and Ribonucleotides and Their Reaction with DNA. Proc Natl Acad Sci U S A. 1964;52:68–74. doi: 10.1073/pnas.52.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. R., Liebert K., Bekes M., Jeltsch A., Cheng X. Structure and substrate recognition of the Escherichia coli DNA adenine methyltransferase. J Mol Biol. 2006;358:559–70. doi: 10.1016/j.jmb.2006.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton J. R., Liebert K., Hattman S., Jeltsch A., Cheng X. Transition from nonspecific to specific DNA interactions along the substrate-recognition pathway of dam methyltransferase. Cell. 2005;121:349–61. doi: 10.1016/j.cell.2005.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Cantor A. B., Orkin S. H., Wang J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat Protoc. 2009;4:506–17. doi: 10.1038/nprot.2009.23. [DOI] [PubMed] [Google Scholar]
- Labhart P., Karmakar S., Salicru E. M., Egan B. S., Alexiadis V., O'Malley B. W., Smith C. L. Identification of target genes in breast cancer cells directly regulated by the SRC-3/AIB1 coactivator. Proc Natl Acad Sci U S A. 2005;102:1339–44. doi: 10.1073/pnas.0409578102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez O. J., Quintanar A., Padhye N. V., Nelson M. Genotyping of DNA using sequence-specific methyltransferases followed by immunochemical detection. J Immunoassay Immunochem. 2003;24:11–28. doi: 10.1081/IAS-120018466. [DOI] [PubMed] [Google Scholar]
- Metivier R., Penot G., Hubner M. R., Reid G., Brand H., Kos M., Gannon F. Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Moss T., Leblanc B., editors. Methods in Molecular Biology. New York: Humana Press; 2009. DNA-Protein Interactions: Principles and Protocols . [Google Scholar]
- Nilsson S., Makela S., Treuter E., Tujague M., Thomsen J., Andersson G., Enmark E., Pettersson K., Warner M., Gustafsson J. A. Mechanisms of estrogen action. Physiol Rev. 2001;81:1535–65. doi: 10.1152/physrev.2001.81.4.1535. [DOI] [PubMed] [Google Scholar]
- No D., Yao T. P., Evans R. M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 1996;93:3346–51. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A., Abed M., Kenyagin-Karsenti D., Boico O. DamID: A Methylation-Based Chromatin Profiling Approach. Methods Mol Biol. 2009;567:155–69. doi: 10.1007/978-1-60327-414-2_11. [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H. J., Li L., Sawado T., Williams E., Loo L. W., Cowley S. M., Yost C., Pierce S., Edgar B. A., Parkhurst S. M., Eisenman R. N. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 2003;17:1101–14. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y. F., Wansa K. D., Liu M. H., Zhao B., Hong S. Z., Tan P. Y., Lim K. S., Bourque G., Liu E. T., Cheung E. Regulation of estrogen receptor-mediated long range transcription via evolutionarily conserved distal response elements. J Biol Chem. 2008;283:32977–88. doi: 10.1074/jbc.M802024200. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Volkert T. L., Wilson C. J., Bell S. P., Young R. A. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu X., DiRenzo J., Lazar M. A., Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow J., Henikoff S. Chromatin profiling using targeted DNA adenine methyltransferase. Nat Genet. 2001;27:304–8. doi: 10.1038/85871. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–8. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Venkatasubrahmanyam S., Hwang W. W., Meneghini M. D., Tong A. H., Madhani H. D. Genome-wide, as opposed to local, antisilencing is mediated redundantly by the euchromatic factors Set1 and H2A.Z. Proc Natl Acad Sci U S A. 2007;104:16609–14. doi: 10.1073/pnas.0700914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M., Davies J. J., Wittig D., Oakeley E. J., Haase M., Lam W. L., Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Zhang X., Germann S., Blus B. J., Khorasanizadeh S., Gaudin V., Jacobsen S. E. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–71. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]