Abstract
Peroxisome proliferator-activated receptor γ1 (PPARγ1) and liver X receptor α (LXRα) are nuclear receptors that play pivotal roles in macrophage cholesterol homeostasis and inflammation; key biological processes in atherogenesis. The activation of PPARγ1 and LXRα by natural or synthetic ligands results in the transactivation of ABCA1, ABCG1, and ApoE; integral players in cholesterol efflux and reverse cholesterol transport. In this review, we describe the structure, isoforms, expression pattern, and functional specificity of PPARs and LXRs. Control of PPARs and LXRs transcriptional activity by coactivators and corepressors is also highlighted. The specific roles that PPARγ1 and LXRα play in inducing macrophage cholesterol efflux mediators and antagonizing macrophage inflammatory responsiveness are summarized. Finally, this review focuses on the recently reported regulatory functions that adipocyte enhancer-binding protein 1 (AEBP1) exerts on PPARγ1 and LXRα transcriptional activity in the context of macrophage cholesterol homeostasis and inflammation.
Structure of PPARs and LXRs
As nuclear hormone receptors, peroxisome proliferator-activated receptors (PPARs) possess a canonical domain structure similar to that of other members of the nuclear hormone receptor superfamily. At the N-terminus, PPARs harbor a ligand-independent transactivation (AF-1) sub-domain within the A/B domain, followed by a DNA binding domain (DBD) containing two zinc finger motifs, ligand binding domain (LBD), and a ligand-dependent transactivation (AF-2) domain towards the C-terminus. DBD and LBD are the most conserved domains among different isoforms of PPARs. LBD serves complex functions since it does not only mediate ligand binding, but it also mediates interaction with RXR, as well as coactivators and corepressors, in a highly specific manner [Chen et al., 1996; Gearing et al., 1993].
Structurally, liver X receptors (LXRs) are similar to other members of the nuclear hormone superfamily. LXRs contain a poorly characterized N-terminus that has AF-1 domain, followed by a central DNA binding domain (DBD), and a relatively large C-terminus containing the ligand-binding domain (LBD) and AF-2 ligand-dependent domain [Chawla et al., 2001c]. DBD of LXRs contains two highly conserved zinc finger motifs, characteristic of other orphan nuclear receptors, which is required for physical contact between LXR-RXR heterodimers and LXR response elements (LXREs) in the promoters of target genes. The LBD of LXRs confer ligand specificity, heterodimerization with RXRs, as well as interactions with coactivators and corepressors [Renaud et al., 1995].
Isoforms, expression, and functional specificity of PPARs and LXRs
PPARα, PPARβ/δ, and PPARγ are three isoforms encoded by three different genes in eukaryotic cells, and these three isoforms constitute the PPAR subfamily of the orphan nuclear hormone receptor superfamily. PPARs are traditionally known as orphan nuclear receptors due to the initial lack of knowledge about their physiological ligands, which are now known to include a wide range of biomolecules. Whereas PPARα and PPARδ can be activated by a wide range of saturated and unsaturated fatty acids [Amri et al., 1995; Forman et al., 1997; Gottlicher et al., 1992; Kliewer et al., 1997; Yu et al., 1995], PPARγ prefers polyunsaturated fatty acids as ligands [Xu et al., 1999]. Fibrates, thiazolidinediones (TZDs) (e.g., rosiglitazone, pioglitazone, ciglitazone, and troglitazone), and α-substituted carboxylic acids (e.g., L-165041) are potent synthetic agonists for PPARα [Willson et al., 2000], PPARγ [Berger et al., 1996; Lehmann et al., 1995; Willson et al., 1996], and PPARβ/δ [Berger et al., 1999], respectively.
PPARs are ligand-activated transcription factors that regulate the expression of a wide range of genes whose products are critically involved in lipid metabolism. PPARs are thought to be ubiquitously expressed, with differential expression patterns among the three isoforms. PPARα, the first PPAR to be identified, is expressed in many tissues and cells including the liver, kidney, skeletal muscle, heart, brown adipose tissue, monocytes, endothelial cells, and vascular smooth muscle cells [Braissant et al., 1996; Issemann and Green, 1990]. PPARβ/δ is expressed in a wide range of tissues and cells, but its expression seems to be highest in the brain, skin, and adipose tissue [Braissant et al., 1996]. Interestingly, the PPARγ gene is transcribed into three different mRNA molecules: PPARγ1 and PPARγ2, which are transcribed from the same promoter by differential promoter usage and subsequent alternative mRNA splicing [Zhu et al., 1995], and PPARγ3, which is transcribed from an independent promoter [Fajas et al., 1998]. Yet, these three mRNA transcripts give rise to only two PPARγ proteins, PPARγ1 and PPARγ2, due to the fact that PPARγ3 mRNA is translated into a protein that is identical to PPARγ1 [Fajas et al., 1998]. PPARγ2 protein, whose expression is restricted to colon and adipose tissue [Fajas et al., 1997; Fajas et al., 1998; Tontonoz et al., 1994a], has 30 extra amino acid residues at its N-terminus compared to PPARγ1, which is ubiquitously expressed.
Upon ligand binding, PPARs become activated and they heterodimerize with RXR, which also has three isoforms designated RXRα, RXRβ, and RXRγ, all of which are activated by retinoic acid [Mangelsdorf et al., 1992]. PPAR-RXR obligate heterodimers bind to PPAR response elements (PPREs; direct repeats of AGGNCA separated by 1 or 2 nucleotides, DR1 and DR2 elements, respectively) within the promoter regions of their target genes, leading to gene transactivation. Such PPREs have been identified in the promoter region of several genes involved in lipid metabolism including aP2 [Tontonoz et al., 1994b], phosphenolpyruvate carboxykinase (PEPCK) [Tontonoz et al., 1995], lipoprotein lipase (LPL) [Schoonjans et al., 1996], CD36 [Sfeir et al., 1997], LXRα/β [Chawla et al., 2001b], and ApoE [Galetto et al., 2001].
LXRs are members of the orphan nuclear receptor superfamily that were first identified in the liver, hence their name [Apfel et al., 1994; Willy et al., 1995]. Two isoforms have already been characterized, namely LXRα and LXRβ, the latter being ubiquitously expressed [Song et al., 1994], while the expression of the former is more restricted in the kidney, spleen, adipose tissue, lung, intestine, skeletal muscle, and macrophages [Apfel et al., 1994; Peet et al., 1998a; Willy et al., 1995]. It is believed that intracellular cholesterol leads to the production of LXRs specific physiological ligands, oxysterols [Repa and Mangelsdorf, 2002]. 24(S),25-epoxycholesterol, 24(S)-hydroxycholesterol, and 22(R)-hydroxycholesterol are the most abundant and potent oxysterols capable of activating LXRs in the cell [Janowski et al., 1996]. Potent and specific pharmacological LXR agonists, such as T0901317 and GW3965, have been synthesized. Similar to PPARs, ligand-bound LXRs tend to form heterodimers with their obligate partner RXR, and activated LXR-RXR heterodimers are capable of binding to specific DNA binding sites known as LXREs, which consist of two direct repeats (AGGTCA) separated by four nucleotides (DR4 elements) [Willy et al., 1995]. LXREs have been identified in the promoter regions of several genes regulated by LXRs including ABCA1 [Costet et al., 2000; Schwartz et al., 2000], ABCG1 [Venkateswaran et al., 2000b], PPARγ [Seo et al., 2004], and ApoE [Laffitte et al., 2001].
Coactivation and corepression of PPARs and LXRs
As their names suggest, coactivators and corepressors are transcription modulators that allow transactivation and repression of target genes, respectively, by means of associating with transcription factors that regulate expression of such genes [Edwards et al., 2002; Rosen and Spiegelman, 2001]. Like other nuclear hormone receptors, PPARs are involved in protein-protein interactions with coactivators and corepressors, and such interactions are crucial for mediating physical association between PPAR-RXR heterodimers with chromatin and the basic transcription machinery [Rosen and Spiegelman, 2001]. PPAR coactivators include CBP/p300 [Debril et al., 2004; Flanagan et al., 2005; Ko et al., 2000; Lemon et al., 2001; Salma et al., 2004; Zhu et al., 1996], steroid receptor coactivator (SRC)-1 [Gelman et al., 1999; Kung et al., 2000; Lickert et al., 2004; Lim et al., 2004; Yao et al., 1998; Zhu et al., 1996], steroid receptor coactivator (SRC)-2 [Gelman et al., 1999; Lim et al., 2004], steroid receptor coactivator (SRC)-3 [Lim et al., 2004; Mizukami and Taniguchi, 1997], PPARγ coactivator (PGC)-1α [Li et al., 1997; Louet et al., 2006; Puigserver et al., 1998], PPARγ coactivator (PGC)-1β [Qi et al., 1999], PPAR binding protein (PBP or TRAP220) [Lim et al., 2004; Puigserver and Spiegelman, 2003; Surapureddi et al., 2002; Zhu et al., 1997], PPAR interacting protein (PRIP) [Goo et al., 2003; Lee et al., 1999], PRIC285 [Kim et al., 2003; Ko et al., 2000], PRIC320 [Lee et al., 2001], BAF60c [Debril et al., 2004], and FK614 [Fujimura et al., 2005]. Nuclear receptor corepressor (NCoR) [Guan et al., 2005; Horlein et al., 1995; Yu et al., 2005], silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) [Chen et al., 1996; Yu et al., 2005], small heterodimer partner (SHP) [Nishizawa et al., 2002; Shin and Osborne, 2008; Yamagata et al., 2007], receptor-interacting protein 140 (RIP140) [Debevec et al., 2007; Lim et al., 2004], and SIRT-1 [Picard et al., 2004] are among the well-characterized corepressors that interact with PPAR-RXR heterodimers, inhibiting transcriptional transactivation driven by active PPAR-RXR homodimers. DSS-AHC on X chromosome gene 1 (DAX-1), an atypical nuclear receptor, has been recently shown to function as a transcriptional corepressor of PPARγ by competing with the PPARγ coactivator (PGC)-1 leading to abrogated adipogenesis in 3T3-L1 cells [Kim et al., 2008].
Upon binding of PPAR-RXR heterodimers to PPREs of target genes, coactivators with histone acetylase activity bind to the ligand- and DNA-bound PPAR-RXR heterodimer. Such binding is thought to cause chromatin remodeling, giving access to other coactivators such as PBP, which connect the PPAR-RXR complex to the basic transcription machinery, leading to gene transactivation. In contrast, corepressors bind to ligand- and DNA-bound PPAR-RXR heterodimers and allow recruitment of histone deacetylases and/or conformational alterations that ultimately confer a condensed, inactive chromatin structure, leading to transcriptional repression [Chen and Li, 1998; Glass and Rosenfeld, 2000; Hu and Lazar, 2000; Rosenfeld and Glass, 2001].
Similar to PPARs, LXRs are involved in protein-protein interactions with coactivators and corepressors, which upon ligand binding to LXRs, take advantage of conformational changes that allow their recruitment [Edwards et al., 2002]. Coactivators and corepressors of LXRα lead to transcriptional activation and repression of LXR target genes by means of chromatin remodeling [Edwards et al., 2002]. Apparently, LXRs interact with coactivators (PGC-1, SRC-1, and CBP/p300) and corepressors (NCoR, SMRT, and SHP) that bind PPARs [Astapova et al., 2008; Brendel et al., 2002; Ghisletti et al., 2009; Hu et al., 2003; Phelan et al., 2008; Unno et al., 2005; Wagner et al., 2003]. Recently, RIP-140 has be demonstrated to be a vital coregulator for LXR activity, serving as a coactivator or a corepressor of LXR transcriptional activity in the liver depending on the target genes and metabolic processes [Herzog et al., 2007].
Some studies have also suggested that corepressors are constitutively bound to PPARs and LXRs, and upon ligand binding, conformational changes force simultaneous dissociation of corepressors and recruitment of coactivators [Albers et al., 2006; Edwards et al., 2002; Glass and Rosenfeld, 2000]. Interaction between nuclear receptors and their coactivators requires multiple LXXLL motifs located within NR boxes of coactivators [Heery et al., 1997; Le Douarin et al., 1996; McKenna and O'Malley, 2002; Torchia et al., 1997]. Slight differences within such NR boxes are critical determinants of nuclear hormone receptor (NHR)-coactivator specificity [Chen et al., 2000; Ding et al., 1998; Li et al., 2007; McInerney et al., 1998; Torchia et al., 1997]. Likewise, corepressors of NHRs contain small peptide motifs (CoRNR boxes) that mediate protein-protein interaction with NHRs, and subsequently transcriptional repression of target genes [Hu and Lazar, 1999]. Despite their remarkable sequence homology, CoRNR boxes within different corepressors have unique sequences, an important determinant in NHR-corepressor specificity [Cohen et al., 2001; Hodgson et al., 2008; Hu et al., 2001].
With regard to gene expression regulation by estrogen receptor (ER), differential recruitment of coactivators and corepressors has been proposed as an explanation for the target gene and cell type selectivity [Shang and Brown, 2002]. Likewise, mounting evidence suggest that the differential recruitment and interaction of coactivators and corepressors may be a crucial determinant in modulating the expression of target genes by PPARγ [Burgermeister et al., 2006; Cock et al., 2004; DiRenzo et al., 1997; Fujimura et al., 2005; Fujimura et al., 2006; Miller and Etgen, 2003; Oberfield et al., 1999; Rangwala and Lazar, 2002; Wigren et al., 2003; Zhang et al., 2007] and LXR [Albers et al., 2006; Jaye et al., 2005; Miao et al., 2004; Phelan et al., 2008; Quinet et al., 2004; Schmidt et al., 2006; Traves et al., 2007; Williams et al., 2003].
SUMOylation-mediated transrepression of PPARγ and LXRs target genes
SUMOylation is posttranslational modification process by which a small ubiquitin-like modifier (SUMO) (~20 kDa) is covalently conjugated to lysine residues on target proteins [Dohmen, 2004; Hay, 2005; Mabb and Miyamoto, 2007]. SUMOylation involves three enzymatic steps that proceed sequentially, ultimately leading to SUMO conjugation to the target protein by forming an isopeptide bond between SUMO and the ε-amino group of a lysine side chain [Liu and Shuai, 2008]. Modification of transcription factors by SUMOylation has been proposed as a mechanism to modulate the transactivation and/or transrepression potential of several transcription factors [Kotaja et al., 2002; Leuenberger et al., 2009; Ling et al., 2004; Nishida and Yasuda, 2002; Rytinki and Palvimo, 2009]. In 2004, and using different cell models, three research groups have independently reported that PPARγ2 is subject to SUMOylation via conjugation with SUMO-1 at K107 in the AF-1 domain, and that PPARγ2 SUMOylation significantly inhibits its transcriptional activity [Floyd and Stephens, 2004; Ohshima et al., 2004; Yamashita et al., 2004]. A year later, Glass and colleagues have proposed SUMOylation of PPARγ and LXRs as a molecular mechanism that underlies the corepressor-dependent transrepression of PPARγ and LXRs target genes in macrophages [Ghisletti et al., 2007; Pascual et al., 2005]. According to the proposed mechanism, ligand binding triggers SUMOylation of the LBD of PPARγ and LXRs, subsequently leading to PPARγ and LXRs recruitment to corepressor complexes (e.g., NCoR and HDAC3) on target genes. As a consequence, signal-dependent removal of corepressor complexes, which is mediated by ubiquitination and proteasome degradation, is interfered with in a way that prevents gene transactivation. Hence, the promoters of target genes remain occupied by the corepressor complexes, and the target genes are forced to settle in a repressed state.
In a study performed by Pascual and colleagues, it was demonstrated that PIAS1, a SUMO E3 ligase, is critical in PPARγ1-dependent transrepression of iNOS in macrophages [Pascual et al., 2005]. In that study, it was also shown that suppressed expression of Ubc9, the rate-limiting E2 ligase in the SUMOylation pathway, is associated with impaired PPARγ1-dependent transrepression of iNOS in macrophages [Pascual et al., 2005]. Interestingly, the potential of rosiglitazone to retain the repressor NCoR on the iNOS promoter in the presence of LPS is dependent on PIAS1 and Ubc9 [Pascual et al., 2005]. Using site-directed mutagenesis, Pascual and colleagues have also shown that K365, unlike K77, is the major SUMOylation site in PPARγ1 and that it is crucial for mediating transrepression of iNOS [Pascual et al., 2005]. These findings clearly indicate that PIAS1/Ubc9-mediated SUMOylation is an essential process involved in transrepression of PPARγ1 target genes in macrophages.
Intriguingly, this proposed mechanism of transrepression is not unique to PPARγ, since LXR transrepression of inflammatory target genes also utilizes a SUMOylation-dependent pathway [Ghisletti et al., 2007]. Yet, the key players involved in SUMOylation-dependent transrepression by PPARγ and LXRs are not identical. Specifically, while PPARγ is SUMOylated by SUMO-1, LXR is SUMOylated by SUMO-2 and SUMO-3 [Ghisletti et al., 2007]. Additionally, while PIAS1 serves as the main SUMO E3 ligase in SUMOylation-dependent transrepression by PPARγ, SUMOylation-dependent transrepression by LXR requires HDAC4 as the main SUMO E3 ligase [Ghisletti et al., 2007]. Like PPARγ, knockdown of Ubc9 leads to a significant impairment of LXR ligand-dependent transrepression of iNOS in LPS-treated macrophages due to impaired LXR recruitment to the iNOS promoter leading to retained NCoR binding [Ghisletti et al., 2007]. Site-directed mutagenesis experiments revealed that K328 and K434 in LXRα and K410 and K448 in LXRβ are the key SUMOylation target sites, and that lysine to arginine substitutions of these residues is accompanied by significant impairment of LXR ligand-dependent transrepression of iNOS in macrophages [Ghisletti et al., 2007]. Ghisletti and colleagues also concluded that the parallel SUMOylation-dependent transrepression pathways mediated by PPARγ and LXRs are themselves subject to regulation, and can be overridden by specific signals in a gene-specific manner. It is worth mentioning that interference with SUMOylation has no effect on the transactivation potential of PPARγ and LXRs towards their target genes in macrophages [Ghisletti et al., 2007; Pascual et al., 2005]. These findings provide a plausible explanation of the similar, but functionally distinctive potential of PPARγ and LXRs to regulate a specific set of target genes involved in key physiological processes such as inflammation and metabolic homeostasis. In a recent study, Jennewein and colleagues have demonstrated that SUMOylation of PPARγ in apoptotic cells prevents LPS-induced NCoR removal from κB binding sites within the promoters of pro-inflammatory genes, mediating transrepression of pro-inflammatory cytokines [Jennewein et al., 2008]. Taken together, there is mounting experimental evidence indicating that SUMOylation of PPARγ and LXRs on key lysine residues mediates transrepression of PPARγ and LXRs target genes in many cell types, and that this regulation can be differentially controlled to fine-tune the cellular outcomes in response to ligand binding.
Role of PPARγ1 and LXRα in macrophage cholesterol homeostasis
PPARγ1 and LXRα are known to be potent sterol and fatty acid sensors that play fundamental roles in lipid metabolism. PPARγ/LXRα signaling pathways are involved in various key biological processes that are implicated in many conditions such as obesity, diabetes mellitus, atherosclerosis, and inflammatory diseases [Cao et al., 2004; Walczak and Tontonoz, 2002]. Both PPARγ1 and LXRα are expressed abundantly in macrophages, especially in lipid-laden foam cells within atherosclerotic lesions [Ricote et al., 1998a; Tontonoz et al., 1998; Venkateswaran et al., 2000b].
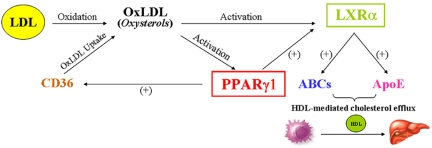
Upon uptake of oxLDL by macrophages, many intermediates such as oxidized fatty acids and oxysterols are formed, providing macrophages with PPARγ1 and LXRα natural ligands [Fu et al., 2001b; Janowski et al., 1999; Nagy et al., 1998]. Thus, PPARγ1 and LXRα become ligand-bound and heterodimerize with their obligate RXR molecules to become transcriptionally active. In fact, oxLDL does not only lead to PPARγ1 and LXRα activation, but it also leads to increased expression of these genes [Ricote et al., 1998a; Tontonoz et al., 1998]. In turn, PPARγ1 and LXRα signaling pathways are turned on, leading to the transactivation of a cascade of genes whose products are critically involved in cholesterol efflux in macrophages (Figure 1).
Figure 1. PPARγ1 and LXRα are key regulators of macrophage cholesterol homeostasis.
Upon uptake of oxLDL by macrophages, oxysterols are synthesized, which allows activation of PPARγ1 and LXRα. Once activated, PPARγ1 and LXRα not only induce the expression of each other, but they also induce the expression of many ABC transporters, as well as that of ApoE. ABCs and ApoE play integral roles in transferring excess cholesterol to its acceptor, HDL, and this initiates the process of reverse cholesterol transport (RCT). In RCT, excess peripheral cholesterol is scavenged by tissue macrophages, which process cholesterol and transport it to the liver via HDL for excretion.
ABCs are direct downstream targets of PPARγ1 and LXRα in macrophages
Members of the ATP-binding cassette (ABC) family of proteins are among the most extensively studied downstream targets of the PPARγ1-LXRα signaling pathway initiated by lipid loading in macrophages. ABCs are transmembrane proteins with two ATP-binding domains and 12 membrane-spanning domains, and they function as efficient cholesterol transporters by forming channel-like structures in the plasma membrane [Decottignies and Goffeau, 1997]. ABCs are involved in active transport of cholesterol from inside the cell onto HDL particles. In macrophages, ABCA1 and ABCG1 play major roles in HDL-mediated and ApoAI-mediated cholesterol efflux [Brooks-Wilson et al., 1999; Lawn et al., 1999]. Lipid loading of macrophages increases the expression of ABCA1 [Langmann et al., 1999] and ABCG1 [Klucken et al., 2000; Venkateswaran et al., 2000b]. The importance of ABCA1 function as a cholesterol efflux mediator is illustrated by Tangier disease, a genetic disorder characterized by extremely low plasma HDL levels and remarkable accumulation of cholesterol in macrophages localized in the tonsils, spleen, liver, and intestine [Serfaty-Lacrosniere et al., 1994]. Numerous studies have demonstrated that familial Tangier disease mainly results from mutations in the ABCA1 gene [Altilia et al., 2003; Bodzioch et al., 1999; Brooks-Wilson et al., 1999; Guan et al., 2004; Hooper et al., 2009; Maekawa et al., 2009; Rust et al., 1998; Singaraja et al., 2006]. Moreover, macrophages isolated from Tangier patients express significantly lower levels of ABCG1 [Lorkowski et al., 2001]. Interestingly, pharmacological activation of PPARγ1 and/or LXRα in macrophages cannot overcome ABCA1 deficiency, which completely abolishes cholesterol efflux [Chinetti et al., 2001]. Experimental evidence exists indicating that reduced levels of ABCG1 correlate with inhibited cholesterol efflux in macrophages [Mauldin et al., 2008; Wang et al., 2004]. Clearly, ABCs are involved in the first step of reverse cholesterol transport (RCT), and they are also involved in the control of total plasma HDL levels, major events associated with atherogenesis. Interestingly, about 40% of Tangier patients develop symptoms of atherosclerosis at one point in their life [Serfaty-Lacrosniere et al., 1994].
Studies have shown that PPARγ1 and LXRα are involved in a positive, reciprocal relationship, in which the activation of one of them leads to the upregulation of the other [Chawla et al., 2001b; Seo et al., 2004]. However, experimental evidence suggests that ABCA1 and ABCG1 are not direct targets of PPARγ1 in macrophages, and that PPARγ1 activation leads to induction of ABCA1 and ABCG1 levels via LXRα upregulation [Chawla et al., 2001b]. Actually, Chawla and colleagues have shown that LXRα activation results in marked induction of ABCA1 and ABCG1 in PPARγ1-deficient macrophages, indicating that PPARγ1 is dispensable for LXRα-mediated upregulation of ABC proteins in macrophages [Chawla et al., 2001b]. In concert, PPARγ1 deficiency had no effect on the ability of 22(R)-hydroxycholesterol, an LXR agonist, to induce ABCA1 and ABCG1 expression in macrophages [Akiyama et al., 2002]. In addition, LXRα-deficient macrophages display diminished cholesterol efflux due to lack of upregulation of ABCA1 [Repa et al., 2000b], and ABCG1 [Laffitte et al., 2001], confirming that PPARγ1 is insufficient in upregulating ABCA1 and ABCG1 in the absence of LXRα. However, studies using PPARγ1 conditional knockout mice revealed that PPARγ1-deficient macrophages have reduced LXRα, ABCA1, and ABCG1 levels, and thus, diminished cholesterol efflux, and they develop into foam cells [Akiyama et al., 2002; Chawla et al., 2001b]. These studies suggest a complex, regulatory loop implicating PPARγ1 and LXRα as key, upstream players in a signaling pathway that culminates in upregulation of ABC proteins and cholesterol clearance from macrophages.
It is important to note that both forms of LXR, LXRα and LXRβ, have been shown to play comparable roles in macrophages with regard to ABCA1 induction and cholesterol efflux [Costet et al., 2000; Joseph et al., 2004; Mak et al., 2002; Peet et al., 1998b; Repa et al., 2000a; Schwartz et al., 2000; Venkateswaran et al., 2000a; Walczak et al., 2004]. Consistently, in vivo studies have shown that LXRα and LXRβ exert comparable atheroprotective effects, in which both forms must be deleted to induce atherogenesis in mice [Schuster et al., 2002]. Along the same line, another study has demonstrated that ablation of both LXRα and LXRβ is essential to promote atherosclerosis in ApoE-/- and LDLR-/- mice [Tangirala et al., 2002]. Although a few studies have recently suggested partial differential effects of LXRα and LXRβ with regard to regulation of macrophage cholesterol efflux regulators [Lund et al., 2006], non-redundant roles of these two forms of LXR are still unclear. The development and use of LXRα-selective and LXRβ-selective agonists [Lund et al., 2006; Miao et al., 2004; Quinet et al., 2006; Szewczyk et al., 2006] is ongoing in an attempt to provide a lucid answer to the question whether LXRα and LXRβ play redundant roles in macrophage cholesterol homeostasis and atherogenesis.
ApoE is a direct downstream target of PPARγ1 and LXRα
An extensive body of literature demonstrates that ApoE, which is expressed abundantly in macrophages, is a prominent player in macrophage cholesterol efflux and foam cell formation [Basu et al., 1983; Basu et al., 1982; Dory, 1989; Lin et al., 1999; Mazzone and Reardon, 1994; Zhang et al., 1996b]. ApoE-deficient macrophages display severely diminished ability to efflux cholesterol and other lipids to HDL particles or lipid-free apolipoproteins [Langer et al., 2000; Mazzone, 1996; Van Eck et al., 2000], indicative of a key role of ApoE in RCT. Noteworthy, the PPARγ1-LXRα-ABC signaling pathway described above also leads to upregulation of ApoE in macrophages, as suggested by many independent studies. It was shown that basal expression of ApoE is attenuated in PPARγ1-deficient macrophages, indicating that PPARγ1 positively regulates ApoE expression [Akiyama et al., 2002]. In addition, treatment of THP-1 macrophages with ciglitazone, a potent PPARγ agonist, results in a significant increase in ApoE levels [Galetto et al., 2001], indicating that PPARγ1 activity positively correlates with ApoE expression in macrophages. However, treatment of PPARγ1-deficient macrophages with 22(R)-hydroxycholesterol, a natural LXRα agonist, induces ApoE expression, and thus, bypasses PPARγ1 dependency [Akiyama et al., 2002]. Another line of evidence indicates that 22(R)-hydroxycholesterol and synthetic LXR agonist T0901317 upregulate ApoE expression in wildtype, but not LXRα-deficient, macrophages, confirming that ApoE upregulation in macrophages is LXRα-dependent, consistent with the fact that the ApoE promoter contains consensus LXRE [Laffitte et al., 2001]. Despite the presence of PPRE within the promoter region of ApoE [Galetto et al., 2001], solid evidence indicating a direct PPARγ1-mediated upregulatory effect on ApoE expression is lacking. Collectively, these studies suggest that activation of PPARγ1 leads to induction of ApoE expression via LXRα upregulation, and that ApoE is a major component of the PPARγ1-LXRα signaling paradigm. Noteworthy, defects in any component of the PPARγ1-LXRα signaling pathway in macrophages render cholesterol efflux defective, and thus, result in the transformation of macrophages into lipid-laden foam cells.
AEBP1 impedes macrophage cholesterol homeostasis by suppressing PPARγ1 and LXRα
AEBP1 gene is located on chromosome 7 and 11 in the human and mouse genomes, respectively. Through alternative splicing, AEBP1 gene is transcribed to yield two related, but distinct, transcripts. One transcript is translated into an 82-kDa protein, AEBP1, while the other transcript is translated into a 175-kDa protein, ACLP (aortic carboxypeptidase-like protein), which has an additional 380-amino acid coding sequence at its N-terminus [Ro et al., 2001]. ACLP was initially identified in human aortic smooth muscle cells, and its protein expression can only be detected in the aorta [Layne et al., 1998] and adipose tissue [Layne et al., 2001]. Cell fractionation and immunofluorescent staining experiments revealed that ACLP is excluded from the nucleus and localized in the perinuclear space, indicative of its entry into the secretory pathway [Layne et al., 2001]. Unlike ACLP, which is targeted to the extracellular matrix (ECM) [Layne et al., 2001], due to the presence a lysine- and proline-rich 11-amino acid repeating motif and a signal sequence [Layne et al., 1998], AEBP1 is an intracellular protein that exists in the cytoplasm and the nucleus [Majdalawieh et al., 2007; Park et al., 1999]. AEBP1 protein is composed of three main domains: a discoidin-like domain (DLD) at its N-terminus, a central carboxypeptidase (CP) domain, and a structurally uncharacterized C-terminal DNA-binding domain [He et al., 1995]. The C-terminal domain of AEBP1 is divided into three distinct subdomains: a lysine- and arginine-rich basic region, a serine-, threonine-, and proline-rich region, and a glutamate-rich acidic region towards the end. Unlike ACLP, AEBP1 is ubiquitously expressed in many tissues and cells, and its expression seems to be highest in white and brown adipose tissues, liver, lung, spleen, brain, and macrophages [Majdalawieh and Ro, 2009; Majdalawieh et al., 2006; Majdalawieh et al., 2007; Ro et al., 2001].
According to the model proposed by Chawla and colleagues, uptake of oxLDL leads to PPARγ1 and LXRα activation in macrophages [Chawla et al., 2001b]. LXRα directly upregulates ABCA1, ABCG1, and ApoE expression, promoting cholesterol clearance from macrophages. Studies from our laboratory have demonstrated that AEBP1 modulates macrophage cholesterol homeostasis by its ability to downregulate PPARγ1 and LXRα expression and transcriptional activity [Majdalawieh and Ro, 2009; Majdalawieh et al., 2006]. Both endogenous and exogenous overexpression of AEBP1 have been accompanied by significant reduction in PPARγ1 and LXRα levels [Majdalawieh and Ro, 2009; Majdalawieh et al., 2006]. Indeed, mutagenesis analysis revealed that PPARγ1 and LXRα repression by AEBP1 is DNA-binding-dependent, in which the C-terminus of AEBP1 is crucial for such AEBP1 suppressive effects [Majdalawieh et al., 2006]. Consistent with its ability to repress PPARγ1 and LXRα, AEBP1 overexpression and ablation lead to decreased and increased levels of ABCA1, ABCG1, and ApoE in macrophages, respectively [Majdalawieh and Ro, 2009; Majdalawieh et al., 2006]. As expected, inhibited expression of ABCA1, ABCG1, and ApoE via PPARγ1, and LXRα transcriptional repression by AEBP1, results in inefficient cholesterol efflux from macrophages [Majdalawieh et al., 2006]. Indeed, macrophages that overexpress AEBP1 (AEBP1TG macrophages) accumulate considerable amounts of lipids in their cytoplasmic compartments compared to their control counterparts (AEBP1NT macrophages) [Majdalawieh et al., 2006], indicating that sustained lipid accumulation is a direct indication of disrupted cholesterol efflux in macrophages that express significantly decreased levels of ABCA1, ABCG1, and ApoE due to AEBP1 overexpression. These findings strengthen the model proposing that activation of PPARγ1 and LXRα is essential for upregulated surface expression of ABC transporters and ApoE, as well as successive removal of accumulated lipids in macrophages [Chawla et al., 2001b; Laffitte et al., 2001]. Thereby, negative regulation of ApoE by AEBP1 is consistent with AEBP1-mediated transcriptional repression of PPARγ1 and LXRα in macrophages.
PPARγ1 induction of lipid uptake via CD36 and lipid efflux via LXRα-ABCs raises the question of whether the net effect of PPARγ1 activation would be to promote or impede foam cell formation. Although PPARγ1 induces CD36 upregulation, forcing macrophages to uptake and accumulate lipids, it concurrently induces expression of LXRα, ABCs, ApoE, and lipoprotein lipase (LPL), crucial factors favoring macrophage cholesterol efflux [Akiyama et al., 2002]. Meaningfully, a bone marrow transplantation experiment revealed that the PPARγ1-LXRα-ABC efflux pathway dominates in vivo [Chawla et al., 2001b]. Consistently, AEBP1’s regulatory function in macrophages supports a protective role of PPARγ1 against foam cell formation since PPARγ1 downregulation in AEBP1TG macrophages is accompanied by decreased levels of not only LXRα, ABCA1, ABCG1, and ApoE, but also CD36.
Many studies have shown that lipopolysaccharide (LPS) treatment in macrophages is accompanied by significant reduction in PPARγ1 levels [Miksa et al., 2007; Welch et al., 2003; Zhou et al., 2008]. Consistently, LPS was shown to induce foam cell formation due to disrupted cholesterol clearance from macrophages [Funk et al., 1993; Kalayoglu and Byrne, 1998a; Kalayoglu and Byrne, 1998b; Oiknine and Aviram, 1992]. Recently, AEBP1 was shown to be critical in mediating LPS-suppressive effects on PPARγ1 and LXRα expression in macrophages [Majdalawieh and Ro, 2009]. In agreement, AEBP1 ablation is accompanied by attenuated LPS-mediated suppression of PPARγ1 and LXRα expression in AEBP1-deficient macrophages [Majdalawieh and Ro, 2009]. This regulatory role of AEBP1 seems to be physiologically significant given that induction of foam cell formation by LPS is dependent on AEBP1, in which AEBP1-deficient macrophages are rendered protective against LPS-induced foam cell formation [Majdalawieh and Ro, 2009]. Interestingly, LPS treatment was shown to induce AEBP1 expression in macrophages [Majdalawieh and Ro, 2009], which may serve as a mechanism that explains the regulatory role that AEBP1 plays in mediating LPS-suppressive effects on PPARγ1 and LXRα in macrophages.
Anti-inflammatory roles of PPARγ1 and LXRα
Aside from its imperative role in controlling macrophage cholesterol homeostasis, PPARγ1 possesses potent anti-inflammatory functions in macrophages [Lee and Evans, 2002; Rizzo and Fiorucci, 2006; Zelcer and Tontonoz, 2006]. A variety of PPARγ synthetic agonists have been used to demonstrate that PPARγ1 exerts anti-inflammatory effects in macrophages due to their ability to suppress the expression of a wide range of pro-inflammatory genes. Treatment of murine peritoneal macrophages with 15-deoxy-Δ12,14-prostaglandin or BRL 49653, specific PPARγ agonists, results in marked reduction in IFNγ-induced and PMA-induced expression of iNOS and MMP-9 (also known as gelatinase B), respectively [Ricote et al., 1998b]. Moreover, PMA-induced expression of IL-1β, IL-6, and TNFα is significantly reduced in primary human monocytes treated with two PPARγ agonists, 15-deoxy-Δ12,14-prostaglandin and troglitazone [Jiang et al., 1998]. 15-deoxy-Δ12,14-prostaglandin and troglitazone also lead to inhibited TNFα promoter-driven expression in the human monocyte/macrophage cell line U937 [Jiang et al., 1998]. Numerous studies have shown that activation of PPARγ1 in macrophages also leads to blocked expression of IL-12 [Alleva et al., 2002; Chung et al., 2000], iNOS [Bernardo et al., 2000; Fahmi et al., 2001; Petrova et al., 1999] and COX-2 [Tsubouchi et al., 2001]. PPARγ1 activation has also been shown to suppress TGFβ expression [Fu et al., 2001a; Guo et al., 2004; Lee et al., 2005; Lee et al., 2006; Maeda et al., 2005; Zhao et al., 2006]. However, the effort of these studies to present PPARγ1 as a potent anti-inflammatory mediator is hampered by experiments performed in PPARγ1-deficient macrophages. Chawla and colleagues have demonstrated that LPS treatment leads to equivalent induction of IL-6 and TNFα in wildtype and PPARγ1-deficient macrophages [Chawla et al., 2001a]. Most importantly, PPARγ1 expression in macrophages or lack of it had no effect on the ability of 15-deoxy-Δ12,14-prostaglandin to inhibit LPS-induced expression of IL-6 and TNFα [Chawla et al., 2001a]. Alternatively, Hinz and colleagues demonstrated that 15-deoxy-Δ12,14-prostaglandin and ciglitazone resulted in reduced LPS-induced expression of IL-6, TNFα, and COX-2 in human monocytes in presence of the PPARγ antagonist, bisphenol A diglycidyl ether (BADGE) [Hinz et al., 2003]. Other studies have also demonstrated that PPARγ1-independent anti-inflammatory effects exerted by PPARγ agonists in vitro and in vivo [Brunmair et al., 2001; Chawla et al., 2001a; Lennon et al., 2002; Niino et al., 2001; Reilly et al., 2000]. These findings raised questions about the exact role of PPARγ1 in macrophage inflammatory responsiveness. Later studies have shown that PPARγ agonists exert anti-inflammatory functions in macrophages via PPARγ1-dependent and PPARγ1-independent mechanisms depending on the concentration of agonists and the nature of the inflammatory signal. When PPARγ agonists are used at low receptor-specific concentrations, their anti-inflammatory function seems to be dependent on PPARγ1 expression [Welch et al., 2003]. At high agonist concentrations, however, PPARγ1-independent mechanism(s), possibly involving PPARβ/δ, manifests anti-inflammatory effects of such PPARγ agonists in macrophages [Welch et al., 2003].
A strong line of evidence indicates the existence of a vital crosstalk between macrophage cholesterol homeostasis and inflammation based on recent studies demonstrating a direct role of LXRα in mediating anti-inflammatory effects in macrophages. Using murine peritoneal macrophages, the LXR agonists T0901317 and GW3965 have been shown to be very effective in inhibiting LPS-induced expression of a wide range of pro-inflammatory mediators including IL-1β, IL-6, iNOS, COX-2, MCP-1, MCP-3, MIP-1β, IP-10, and MMP-9 [Joseph et al., 2003]. Although lack of expression of either LXRα or LXRβ in macrophages did not interfere with LXR agonist-mediated inhibition of LPS-induced expression of these pro-inflammatory mediators, macrophages that lack both isoforms (i.e., LXRα-/-LXRβ-/- macrophages) were unresponsive to the anti-inflammatory effects exerted by LXR agonists [Joseph et al., 2003]. This data indicate that the loss of one LXR isoform can be compensated by the other isoform, and that LXR agonists exert their anti-inflammatory effects in macrophages in an LXR-dependent mechanism. Interestingly, in vivo experiments revealed that LXR-mediated anti-inflammatory functions are not restricted to macrophages, since LPS-induced hepatic expression of IL-6, TNFα, and iNOS, as well as LPS-induced aortic expression of IL-6, iNOS, and MMP-9 is significantly elevated in LXRα-/-LXRβ-/- mice compared to wildtype mice [Joseph et al., 2003].
Consistently, LPS-, IL-1β-, and TNFα-induced expression of MMP-9 is markedly inhibited by T0901317 and GW3965 in murine peritoneal macrophages [Castrillo et al., 2003]. Furthermore, T0901317 and GW3965 treatment results in reduced LPS-induced expression levels of tissue factor (TF) and osteopontin (OPN), pro-inflammatory, pro-atherogenic mediators, in murine peritoneal macrophages and RAW 264.7 macrophages, respectively [Ogawa et al., 2005; Terasaka et al., 2005]. In vivo, LXR agonists are also capable of reducing LPS-induced TF expression levels in the aorta, kidney, and lung [Terasaka et al., 2005]. Activation of LXR has also been shown to reduce inflammation in the aortae of atherosclerotic mice [Joseph et al., 2002] and in a mouse model of contact dermatitis [Fowler et al., 2003]. Recently, the LXR agonists have been shown to suppress lung inflammatory responses via inhibiting the expression of inflammatory genes in alveolar macrophages [Birrell et al., 2007; Smoak et al., 2008].
NF-κB role in PPARγ1- and LXRα-mediated anti-inflammation
Despite the strong evidence behind the anti-inflammatory effects of selective PPARγ and LXR agonists, the exact mechanism underlying such effects is poorly understood. Strong evidence suggests that activation of PPARγ1 and/or LXRα interferes with NF-κB, STAT, and AP-1 activity in macrophages [Bonfield et al., 2008; Castrillo et al., 2003; Chang et al., 2007; Chinetti et al., 1998; Joseph et al., 2003; Park et al., 2009; Ricote et al., 1998b; Straus et al., 2000; Welch et al., 2003; Zhou and Waxman, 1999]. Mechanisms leading to inhibited NF-κB activity by PPARγ agonists in macrophages include inhibited IKK activity, and thus, decreased IκBα phosphorylation [Straus et al., 2000; Wang et al., 2007; Zingarelli et al., 2003], covalent modifications of NF-κB subunits leading to abrogated NF-κB-DNA interaction [Chung et al., 2000], and induced NF-κB nuclear export [Kelly et al., 2004]. Indeed, it was shown that PPARγ1 is capable of interfering with NF-κB activity by interacting with NF-κB subunits, rendering NF-κB transcriptionally inactive [Chung et al., 2000]. Furthermore, PPARγ agonists may be capable of modulating NF-κB activity by means of alkylation of cysteine residues located in the DNA-binding domains of NF-κB subunits (C62 in p50 and C38 in p65) [Straus et al., 2000]. It was also suggested that PPARγ agonists lead to enhanced MAPK activation, and subsequently more phosphorylation of PPARγ1, which is more efficient in interacting with NF-κB when phosphorylated [Chen et al., 2003]. However, other studies have demonstrated that PPARγ agonists impede NF-κB signaling via PPARγ1-independent mechanisms. In a study performed in RAW 264.7 macrophages, which lack endogenous PPARγ1, 15-deoxy-Δ12,14-prostaglandin treatment resulted in significant inhibition of NF-κB activity [Straus et al., 2000], suggesting that PPARγ1 is not required for PPARγ agonist-mediated anti-inflammatory effects in macrophages. Hence, it is not surprising that the importance of PPARγ1 involvement in NF-κB inhibition by PPARγ agonists is very controversial. Interestingly, many studies have also shown that activation of IKK/NF-κB pathway results in abrogated PPARγ1 transcriptional activity [Gao et al., 2006; Ruan et al., 2003; Suzawa et al., 2003; Torti et al., 1989; Zhang et al., 1996a].
The pro-inflammatory role of AEBP1 is independent of its ability to suppress PPARγ1 and LXRα in macrophages
The ability of AEBP1 to induce macrophage inflammatory responsiveness leading to enhanced expression of pro-inflammatory mediators (e.g., IL-6, TNFα, MCP-1, and iNOS) [Majdalawieh et al., 2006] has been attributed to its potential to promote NF-κB activity [Majdalawieh et al., 2007]. Since interference with NF-κB activity has been proposed as a mechanism underlying the anti-inflammatory effects of PPARγ1 and LXRα [Castrillo et al., 2003; Chang et al., 2007; Chinetti et al., 1998; Joseph et al., 2003; Park et al., 2009; Ricote et al., 1998b; Straus et al., 2000; Welch et al., 2003; Zhou and Waxman, 1999], it is plausible for one to speculate that AEBP1-mediated suppression of PPARγ1 and LXRα may be a direct cause of ABEP1’s ability to upregulate NF-κB activity in macrophages. However, this possibility is ruled out in light of mutagenesis analyses showing that the C-terminus mutant form of AEBP1, which is incapable of suppressing PPARγ1 and LXRα [Majdalawieh et al., 2006], is as effective as full-length AEBP1 in promoting NF-κB activity [Majdalawieh et al., 2007]. In agreement, the N-terminus mutant form of AEBP1 is capable of suppressing PPARγ1 and LXRα [Majdalawieh et al., 2006], yet it has no upregulatory effect on NF-κB activity [Majdalawieh et al., 2007]. Collectively, AEBP1’s ability to promote macrophage inflammatory responsiveness via NF-κB upregulation is not attributed to AEBP1-mediated suppression of PPARγ1 and LXRα in macrophages.
Concluding remarks
Experimental evidence suggests that AEBP1 manifests itself as a critical regulator of macrophage cholesterol homeostasis and macrophage inflammatory responsiveness, and thus, as a potent pro-atherogenic factor. The pro-atherogenic properties exhibited by AEBP1 seem to be a byproduct of a vital interplay of its ability to antagonize PPARγ1 and LXRα cholesterol efflux functions in macrophages and its ability to promote macrophage inflammatory responsiveness via upregulated NF-κB transcriptional activity due to AEBP1-mediated attenuation of IκBα inhibitory function. Bone marrow transplantation experiments using ApoE-/- mice and analysis of atherosclerotic lesion formation in AEBP1-/-/LDLR-/- hybrid mice are currently underway to elucidate the pro-atherogenic potential of AEBP1 in vivo. Interestingly, AEBP1 expression was previously shown to be negligible in monocytes and that its expression is significantly upregulated during monocyte differentiation into macrophages [Majdalawieh and Ro, 2009]. Interestingly, monocytes differentiation is also accompanied by upregulation of PPARγ1 and LXRα expression [Kohro et al., 2000; Langmann et al., 2005; Marx et al., 1998; Quinet et al., 2004; von Knethen et al., 2007; Whitney et al., 2001; Zhu et al., 1998]. Given that AEBP1 level is hardly detectable in monocytes and considering the parallel upregulation of AEBP1, as well as PPARγ1 and LXRα expression, during monocyte differentiation, we believe that AEBP1 plays vital regulatory roles in macrophages, but not in monocytes. AEBP1 is proposed to serve as an indispensable regulator in the PPARγ1-LXRα signaling pathway that is critically involved in cholesterol clearance from macrophages; the initial step in reverse cholesterol transport. Under normal physiological conditions, AEBP1 fine-tunes the expression and transcriptional activity of PPARγ1 and LXRα in macrophages so that cholesterol homeostasis is maintained. Similarly, and given the vital roles of PPARγ1 and LXRs in macrophage inflammation, AEBP1 may have evolved to serve as a check point for regulating the expression of various pro-inflammatory and anti-inflammatory mediators in macrophages. Noteworthy, cholesterol and oxysterol accumulation in macrophages has been shown to trigger apoptosis of lipid-laden macrophages [Colles et al., 1996; Fazio et al., 2001; Lordan et al., 2009; Panini and Sinensky, 2001; Tabas, 2002]. It is intriguing to propose that foam cell formation may be induced via upregulating AEBP1 expression, and thus impeding cholesterol efflux through PPARγ1 and LXRα downregulation, in cases where macrophage apoptosis is physiologically desired.
Recently, AEBP1 expression was shown to be induced by LPS, and that LPS-induced downregulation of PPARγ1 and LXRα, leading to foam cell formation, is largely mediated by AEBP1 [Majdalawieh and Ro, 2009]. This suggests that Gram positive bacteria-induced atherosclerosis may be mediated by LPS ability to manipulate AEBP1 expression in macrophages, leading to attenuated PPARγ1-LXRα signaling and subsequently foam cell formation and atherosclerosis. The regulatory role that AEBP1 plays in controlling macrophage cholesterol homeostasis and metabolism is consistent with the documented effects of AEBP1 in adiposity and energy metabolism [Ro et al., 2007; Zhang et al., 2005].
Because the entire AEBP1 amino acid sequence is encoded in ACLP, both proteins seem to play similar roles in key biological processes, in which both proteins promote proliferation of preadipocytes and inhibit their differentiation into mature, fat-filled adipocytes [Gagnon et al., 2002; He et al., 1995]. Nonetheless, AEBP1 and ACLP play very distinctive and unique roles in other biological processes [Layne et al., 1998; Layne et al., 2001; Majdalawieh and Ro, 2009; Majdalawieh et al., 2006; Majdalawieh et al., 2007; Ro et al., 2007; Schissel et al., 2009; Zhang et al., 2005]. Notably, none of the regulatory roles that AEBP1 plays in macrophages can be exercised by ACLP since it is an extracellular matrix-associated secretory protein that is excluded from the nuclear and cytoplasmic compartments of mouse aortic smooth muscle cells [Layne et al., 2001] and it is not expressed in primary macrophages or J774 macrophage cell line [Majdalawieh et al., 2007]. Thus, although ACLP shares the exact C-terminal DNA-binding domain of AEBP1, it is not possible for ACLP to function as a transcriptional repressor due to its absence in the nucleus, and hence, it is inconceivable that ACLP can repress the expression of PPARγ1 and LXRα in macrophages. Likewise, since ACLP is not a cytosolic protein, it is inconceivable that ACLP can have any effect on IκBα function or NF-κB activity, despite the fact that both AEBP1 and ACLP share DLD; the domain that mediates AEBP1-IκBα interaction in macrophages [Majdalawieh et al., 2007]. Moreover, it is important to emphasize that the repressed expression of PPARγ1 and LXRα, the disrupted cholesterol efflux, and the enhanced inflammatory responsiveness associated with AEBP1TG macrophages [Majdalawieh et al., 2006] can only be attributed to AEBP1, not ACLP, since AEBP1 targeted overexpression is driven by a transgene carrying AEBP1 cDNA [Zhang et al., 2005]. Thus, despite the fact that they are transcribed from the same gene and share identical structural domains, AEBP1 and ACLP display differential expression pattern, cellular localization, and physiological function.
Clearly, further in vitro and in vivo studies are needed to shed more light on the exact molecular and physiological mechanisms by which AEBP1 manifests its regulatory effects on pivotal factors involved in macrophage biology and metabolism. Finally, we anticipate that AEBP1 may be considered as a potential molecular target to manipulate macrophage behavior and develop therapeutic strategies for the prevention or treatment of metabolic and inflammatory conditions.
Abbreviations
- ABC
ATP-binding cassette
- ACLP
aortic carboxypeptidase-like protein
- AEBP1
adipocyte enhancer-binding protein-1
- AF
activation function
- aP2
gene encoding adipocyte lipid-binding protein (ALBP)
- Apo-AI
apolipoprotein AI
- ApoE
apolipoprotein E
- c/EBP
CCAAT-enhancer binding protein
- CHO
Chinese hamster ovary
- CoRNR
corepressor for nuclear receptor
- COX-2
cyclooxygenase 2
- DAX-1
DSS-AHC on X chromosome gene 1
- DBD
DNA binding domain
- DR
direct repeat
- ER
estrogen receptor
- HDAC
histone deacetylase
- HDL
high density lipoprotein
- IL
interleukin
- IFNγ
interferon γ
- IκB
inhibitor of NF-κB
- IKK
IκB kinase
- iNOS
inducible nitric oxide synthase
- IP-10
IFN-inducible protein 10
- LBD
ligand binding domain
- LDLR
low density lipoprotein receptor
- LPL
lipoprotein lipase
- LPS
lipopolysaccharide
- LXRα
liver X receptor α
- LXRE
LXR response element
- MAPK
mitogen-activated protein kinase
- MCP-1
monocyte chemoattractant protein-1
- MIP-1β
macrophage inhibitory protein β
- MMP
matrix metalloproteinase
- NcoR
nuclear receptor corepressor
- NF-κB
nuclear factor κ B
- NHR
nuclear hormone receptor
- NT
non-transgenic
- OPN
osteopontin
- OxLDL
oxidized low density lipoprotein
- PEPCK
phosphenolpyruvate carboxykinase
- PGC
PPARγ coactivator
- PIAS1
protein inhibitor of activated STAT1
- PMA
phorbol-12-myristate-13-acetate
- PPARγ
peroxisome proliferator-activated receptor γ
- PPRE
PPAR response element
- RCT
reverse cholesterol transport
- RIP140
receptor-interacting protein 140
- RXR
retinoid X receptor
- SHP
small heterodimer partner
- SIRT-1
sirtuin
- SMRT
silencing mediator of retinoic acid and thyroid hormone receptor
- SRC
steroid receptor coactivator
- SUMO
small ubiquitin-like modifier
- TF
tissue factor
- TG
transgenic
- TGFβ
tumor growth factor β
- TNFα
tumor necrosis factor α
- Ubc9
ubiquitin-conjugating enzyme 9
References
- Akiyama T. E., Sakai S., Lambert G., Nicol C. J., Matsusue K., Pimprale S., Lee Y. H., Ricote M., Glass C. K., Brewer H. B., Jr., Gonzalez F. J. Conditional disruption of the peroxisome proliferator-activated receptor γ gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol Cell Biol. 2002;22:2607–19. doi: 10.1128/MCB.22.8.2607-2619.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers M., Blume B., Schlueter T., Wright M. B., Kober I., Kremoser C., Deuschle U., Koegl M. A novel principle for partial agonism of liver X receptor ligands. Competitive recruitment of activators and repressors. J Biol Chem. 2006;281:4920–30. doi: 10.1074/jbc.M510101200. [DOI] [PubMed] [Google Scholar]
- Alleva D. G., Johnson E. B., Lio F. M., Boehme S. A., Conlon P. J., Crowe P. D. Regulation of murine macrophage proinflammatory and anti-inflammatory cytokines by ligands for peroxisome proliferator-activated receptor-γ: counter-regulatory activity by IFN-γ. J Leukoc Biol. 2002;71:677–85. [PubMed] [Google Scholar]
- Altilia S., Pisciotta L., Garuti R., Tarugi P., Cantafora A., Calabresi L., Tagliabue J., Maccari S., Bernini F., Zanotti I., Vergani C., Bertolini S., Calandra S. Abnormal splicing of ABCA1 pre-mRNA in Tangier disease due to a IVS2 +5G>C mutation in ABCA1 gene. J Lipid Res. 2003;44:254–64. doi: 10.1194/jlr.M200248-JLR200. [DOI] [PubMed] [Google Scholar]
- Amri E. Z., Bonino F., Ailhaud G., Abumrad N. A., Grimaldi P. A. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J Biol Chem. 1995;270:2367–71. doi: 10.1074/jbc.270.5.2367. [DOI] [PubMed] [Google Scholar]
- Apfel R., Benbrook D., Lernhardt E., Ortiz M. A., Salbert G., Pfahl M. A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor subfamily. Mol Cell Biol. 1994;14:7025–35. doi: 10.1128/mcb.14.10.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astapova I., Lee L. J., Morales C., Tauber S., Bilban M., Hollenberg A. N. The nuclear corepressor, NCoR, regulates thyroid hormone action in vivo. Proc Natl Acad Sci U S A. 2008;105:19544–9. doi: 10.1073/pnas.0804604105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S. K., Ho Y. K., Brown M. S., Bilheimer D. W., Anderson R. G., Goldstein J. L. Biochemical and genetic studies of the apoprotein E secreted by mouse macrophages and human monocytes. J Biol Chem. 1982;257:9788–95. [PubMed] [Google Scholar]
- Basu S. K., Goldstein J. L., Brown M. S. Independent pathways for secretion of cholesterol and apolipoprotein E by macrophages. Science. 1983;219:871–3. doi: 10.1126/science.6823554. [DOI] [PubMed] [Google Scholar]
- Berger J., Leibowitz M. D., Doebber T. W., Elbrecht A., Zhang B., Zhou G., Biswas C., Cullinan C. A., Hayes N. S., Li Y., Tanen M., Ventre J., Wu M. S., Berger G. D., Mosley R., Marquis R., Santini C., Sahoo S. P., Tolman R. L., Smith R. G., Moller D. E. Novel peroxisome proliferator-activated receptor (PPAR) γ and PPARdelta ligands produce distinct biological effects. J Biol Chem. 1999;274:6718–25. doi: 10.1074/jbc.274.10.6718. [DOI] [PubMed] [Google Scholar]
- Berger J., Bailey P., Biswas C., Cullinan C. A., Doebber T. W., Hayes N. S., Saperstein R., Smith R. G., Leibowitz M. D. Thiazolidinediones produce a conformational change in peroxisomal proliferator-activated receptor-γ: binding and activation correlate with antidiabetic actions in db/db mice. Endocrinology. 1996;137:4189–95. doi: 10.1210/endo.137.10.8828476. [DOI] [PubMed] [Google Scholar]
- Bernardo A., Levi G., Minghetti L. Role of the peroxisome proliferator-activated receptor-γ (PPAR-γ) and its natural ligand 15-deoxy-Delta12, 14-prostaglandin J2 in the regulation of microglial functions. Eur J Neurosci. 2000;12:2215–23. doi: 10.1046/j.1460-9568.2000.00110.x. [DOI] [PubMed] [Google Scholar]
- Birrell M. A., Catley M. C., Hardaker E., Wong S., Willson T. M., McCluskie K., Leonard T., Farrow S. N., Collins J. L., Haj-Yahia S., Belvisi M. G. Novel role for the liver X nuclear receptor in the suppression of lung inflammatory responses. J Biol Chem. 2007;282:31882–90. doi: 10.1074/jbc.M703278200. [DOI] [PubMed] [Google Scholar]
- Bodzioch M., Orso E., Klucken J., Langmann T., Bottcher A., Diederich W., Drobnik W., Barlage S., Buchler C., Porsch-Ozcurumez M., Kaminski W. E., Hahmann H. W., Oette K., Rothe G., Aslanidis C., Lackner K. J., Schmitz G. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat Genet. 1999;22:347–51. doi: 10.1038/11914. [DOI] [PubMed] [Google Scholar]
- Bonfield T. L., Thomassen M. J., Farver C. F., Abraham S., Koloze M. T., Zhang X., Mosser D. M., Culver D. A. Peroxisome proliferator-activated receptor-γ regulates the expression of alveolar macrophage macrophage colony-stimulating factor. J Immunol. 2008;181:235–42. doi: 10.4049/jimmunol.181.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-α, -β, and -γ in the adult rat. Endocrinology. 1996;137:354–66. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- Brendel C., Schoonjans K., Botrugno O. A., Treuter E., Auwerx J. The small heterodimer partner interacts with the liver X receptor α and represses its transcriptional activity. Mol Endocrinol. 2002;16:2065–76. doi: 10.1210/me.2001-0194. [DOI] [PubMed] [Google Scholar]
- Brooks-Wilson A., Marcil M., Clee S. M., Zhang L. H., Roomp K., van Dam M., Yu L., Brewer C., Collins J. A., Molhuizen H. O., Loubser O., Ouelette B. F., Fichter K., Ashbourne-Excoffon K. J., Sensen C. W., Scherer S., Mott S., Denis M., Martindale D., Frohlich J., Morgan K., Koop B., Pimstone S., Kastelein J. J., Genest J., Jr., Hayden M. R. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat Genet. 1999;22:336–45. doi: 10.1038/11905. [DOI] [PubMed] [Google Scholar]
- Brunmair B., Gras F., Neschen S., Roden M., Wagner L., Waldhausl W., Furnsinn C. Direct thiazolidinedione action on isolated rat skeletal muscle fuel handling is independent of peroxisome proliferator-activated receptor-γ-mediated changes in gene expression. Diabetes. 2001;50:2309–15. doi: 10.2337/diabetes.50.10.2309. [DOI] [PubMed] [Google Scholar]
- Burgermeister E., Schnoebelen A., Flament A., Benz J., Stihle M., Gsell B., Rufer A., Ruf A., Kuhn B., Marki H. P., Mizrahi J., Sebokova E., Niesor E., Meyer M. A novel partial agonist of peroxisome proliferator-activated receptor-γ (PPARgamma) recruits PPARgamma-coactivator-1alpha, prevents triglyceride accumulation, and potentiates insulin signaling in vitro. Mol Endocrinol. 2006;20:809–30. doi: 10.1210/me.2005-0171. [DOI] [PubMed] [Google Scholar]
- Cao G., Liang Y., Jiang X. C., Eacho P. I. Liver X receptors as potential therapeutic targets for multiple diseases. Drug News Perspect. 2004;17:35–41. doi: 10.1358/dnp.2004.17.1.829024. [DOI] [PubMed] [Google Scholar]
- Castrillo A., Joseph S. B., Marathe C., Mangelsdorf D. J., Tontonoz P. Liver X receptor-dependent repression of matrix metalloproteinase-9 expression in macrophages. J Biol Chem. 2003;278:10443–9. doi: 10.1074/jbc.M213071200. [DOI] [PubMed] [Google Scholar]
- Chang L., Zhang Z., Li W., Dai J., Guan Y., Wang X. Liver-X-receptor activator prevents homocysteine-induced production of IgG antibodies from murine B lymphocytes via the ROS-NF-kappaB pathway. Biochem Biophys Res Commun. 2007;357:772–8. doi: 10.1016/j.bbrc.2007.04.016. [DOI] [PubMed] [Google Scholar]
- Chawla A., Boisvert W. A., Lee C. H., Laffitte B. A., Barak Y., Joseph S. B., Liao D., Nagy L., Edwards P. A., Curtiss L. K., Evans R. M., Tontonoz P. A PPAR γ-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol Cell. 2001b;7:161–71. doi: 10.1016/s1097-2765(01)00164-2. [DOI] [PubMed] [Google Scholar]
- Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001a;294:1866–70. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Chawla A., Barak Y., Nagy L., Liao D., Tontonoz P., Evans R. M. PPAR-γ dependent and independent effects on macrophage-gene expression in lipid metabolism and inflammation. Nat Med. 2001c;7:48–52. doi: 10.1038/83336. [DOI] [PubMed] [Google Scholar]
- Chen S., Johnson B. A., Li Y., Aster S., McKeever B., Mosley R., Moller D. E., Zhou G. Both coactivator LXXLL motif-dependent and -independent interactions are required for peroxisome proliferator-activated receptor γ (PPARgamma) function. J Biol Chem. 2000;275:3733–6. doi: 10.1074/jbc.275.6.3733. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Li H. Coactivation and corepression in transcriptional regulation by steroid/nuclear hormone receptors. Crit Rev Eukaryot Gene Expr. 1998;8:169–90. doi: 10.1615/critreveukargeneexpr.v8.i2.40. [DOI] [PubMed] [Google Scholar]
- Chen F., Wang M., O'Connor J. P., He M., Tripathi T., Harrison L. E. Phosphorylation of PPARgamma via active ERK1/2 leads to its physical association with p65 and inhibition of NF-kappabeta. J Cell Biochem. 2003;90:732–44. doi: 10.1002/jcb.10668. [DOI] [PubMed] [Google Scholar]
- Chen J. D., Umesono K., Evans R. M. SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc Natl Acad Sci U S A. 1996;93:7567–71. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinetti G., Griglio S., Antonucci M., Torra I. P., Delerive P., Majd Z., Fruchart J. C., Chapman J., Najib J., Staels B. Activation of proliferator-activated receptors α and γ induces apoptosis of human monocyte-derived macrophages. J Biol Chem. 1998;273:25573–80. doi: 10.1074/jbc.273.40.25573. [DOI] [PubMed] [Google Scholar]
- Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., Brewer H. B., Fruchart J. C., Clavey V., Staels B. PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med. 2001;7:53–8. doi: 10.1038/83348. [DOI] [PubMed] [Google Scholar]
- Chung S. W., Kang B. Y., Kim S. H., Pak Y. K., Cho D., Trinchieri G., Kim T. S. Oxidized low density lipoprotein inhibits interleukin-12 production in lipopolysaccharide-activated mouse macrophages via direct interactions between peroxisome proliferator-activated receptor-γ and nuclear factor-κ B. J Biol Chem. 2000;275:32681–7. doi: 10.1074/jbc.M002577200. [DOI] [PubMed] [Google Scholar]
- Cock T. A., Houten S. M., Auwerx J. Peroxisome proliferator-activated receptor-γ: too much of a good thing causes harm. EMBO Rep. 2004;5:142–7. doi: 10.1038/sj.embor.7400082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R. N., Brzostek S., Kim B., Chorev M., Wondisford F. E., Hollenberg A. N. The specificity of interactions between nuclear hormone receptors and corepressors is mediated by distinct amino acid sequences within the interacting domains. Mol Endocrinol. 2001;15:1049–61. doi: 10.1210/mend.15.7.0669. [DOI] [PubMed] [Google Scholar]
- Colles S. M., Irwin K. C., Chisolm G. M. Roles of multiple oxidized LDL lipids in cellular injury: dominance of 7 β-hydroperoxycholesterol. J Lipid Res. 1996;37:2018–28. [PubMed] [Google Scholar]
- Costet P., Luo Y., Wang N., Tall A. R. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J Biol Chem. 2000;275:28240–5. doi: 10.1074/jbc.M003337200. [DOI] [PubMed] [Google Scholar]
- Debevec D., Christian M., Morganstein D., Seth A., Herzog B., Parker M., White R. Receptor interacting protein 140 regulates expression of uncoupling protein 1 in adipocytes through specific peroxisome proliferator activated receptor isoforms and estrogen-related receptor α. Mol Endocrinol. 2007;21:1581–92. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debril M. B., Gelman L., Fayard E., Annicotte J. S., Rocchi S., Auwerx J. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–86. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- Decottignies A., Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–45. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- Ding X. F., Anderson C. M., Ma H., Hong H., Uht R. M., Kushner P. J., Stallcup M. R. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12:302–13. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- DiRenzo J., Soderstrom M., Kurokawa R., Ogliastro M. H., Ricote M., Ingrey S., Horlein A., Rosenfeld M. G., Glass C. K. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Mol Cell Biol. 1997;17:2166–76. doi: 10.1128/mcb.17.4.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohmen R. J. SUMO protein modification. Biochim Biophys Acta. 2004;1695:113–31. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- Dory L. Synthesis and secretion of apoE in thioglycolate-elicited mouse peritoneal macrophages: effect of cholesterol efflux. J Lipid Res. 1989;30:809–16. [PubMed] [Google Scholar]
- Edwards P. A., Kast H. R., Anisfeld A. M. BAREing it all: the adoption of LXR and FXR and their roles in lipid homeostasis. J Lipid Res. 2002;43:2–12. [PubMed] [Google Scholar]
- Fahmi H., Di Battista J. A., Pelletier J. P., Mineau F., Ranger P., Martel-Pelletier J. Peroxisome proliferator--activated receptor γ activators inhibit interleukin-1beta-induced nitric oxide and matrix metalloproteinase 13 production in human chondrocytes. Arthritis Rheum. 2001;44:595–607. doi: 10.1002/1529-0131(200103)44:3<595::AID-ANR108>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Fajas L., Fruchart J. C., Auwerx J. PPARgamma3 mRNA: a distinct PPARgamma mRNA subtype transcribed from an independent promoter. FEBS Lett. 1998;438:55–60. doi: 10.1016/s0014-5793(98)01273-3. [DOI] [PubMed] [Google Scholar]
- Fajas L., Auboeuf D., Raspe E., Schoonjans K., Lefebvre A. M., Saladin R., Najib J., Laville M., Fruchart J. C., Deeb S., Vidal-Puig A., Flier J., Briggs M. R., Staels B., Vidal H., Auwerx J. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–89. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- Fazio S., Major A. S., Swift L. L., Gleaves L. A., Accad M., Linton M. F., Farese R. V., Jr. Increased atherosclerosis in LDL receptor-null mice lacking ACAT1 in macrophages. J Clin Invest. 2001;107:163–71. doi: 10.1172/JCI10310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J. F., Mi L. Z., Chruszcz M., Cymborowski M., Clines K. L., Kim Y., Minor W., Rastinejad F., Khorasanizadeh S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–5. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- Floyd Z. E., Stephens J. M. Control of peroxisome proliferator-activated receptor gamma2 stability and activity by SUMOylation. Obes Res. 2004;12:921–8. doi: 10.1038/oby.2004.112. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Chen J., Evans R. M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci U S A. 1997;94:4312–7. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A. J., Sheu M. Y., Schmuth M., Kao J., Fluhr J. W., Rhein L., Collins J. L., Willson T. M., Mangelsdorf D. J., Elias P. M., Feingold K. R. Liver X receptor activators display anti-inflammatory activity in irritant and allergic contact dermatitis models: liver-X-receptor-specific inhibition of inflammation and primary cytokine production. J Invest Dermatol. 2003;120:246–55. doi: 10.1046/j.1523-1747.2003.12033.x. [DOI] [PubMed] [Google Scholar]
- Fu X., Menke J. G., Chen Y., Zhou G., MacNaul K. L., Wright S. D., Sparrow C. P., Lund E. G. 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J Biol Chem. 2001a;276:38378–87. doi: 10.1074/jbc.M105805200. [DOI] [PubMed] [Google Scholar]
- Fujimura T., Sakuma H., Konishi S., Oe T., Hosogai N., Kimura C., Aramori I., Mutoh S. FK614, a novel peroxisome proliferator-activated receptor γ modulator, induces differential transactivation through a unique ligand-specific interaction with transcriptional coactivators. J Pharmacol Sci. 2005;99:342–52. doi: 10.1254/jphs.fp0050578. [DOI] [PubMed] [Google Scholar]
- Fujimura T., Sakuma H., Ohkubo-Suzuki A., Aramori I., Mutoh S. Unique properties of coactivator recruitment caused by differential binding of FK614, an anti-diabetic agent, to peroxisome proliferator-activated receptor γ. Biol Pharm Bull. 2006;29:423–9. doi: 10.1248/bpb.29.423. [DOI] [PubMed] [Google Scholar]
- Funk J. L., Feingold K. R., Moser A. H., Grunfeld C. Lipopolysaccharide stimulation of RAW 264.7 macrophages induces lipid accumulation and foam cell formation. Atherosclerosis. 1993;98:67–82. doi: 10.1016/0021-9150(93)90224-i. [DOI] [PubMed] [Google Scholar]
- Fu M., Zhang J., Zhu X., Myles D. E., Willson T. M., Liu X., Chen Y. E. Peroxisome proliferator-activated receptor γ inhibits transforming growth factor β-induced connective tissue growth factor expression in human aortic smooth muscle cells by interfering with Smad3. J Biol Chem. 2001b;276:45888–94. doi: 10.1074/jbc.M105490200. [DOI] [PubMed] [Google Scholar]
- Gagnon A., Abaiian K. J., Crapper T., Layne M. D., Sorisky A. Down-regulation of aortic carboxypeptidase-like protein during the early phase of 3T3-L1 adipogenesis. Endocrinology. 2002;143:2478–85. doi: 10.1210/endo.143.7.8875. [DOI] [PubMed] [Google Scholar]
- Galetto R., Albajar M., Polanco J. I., Zakin M. M., Rodriguez-Rey J. C. Identification of a peroxisome-proliferator-activated-receptor response element in the apolipoprotein E gene control region. Biochem J. 2001;357:521–7. doi: 10.1042/0264-6021:3570521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., He Q., Peng B., Chiao P. J., Ye J. Regulation of nuclear translocation of HDAC3 by IkappaBalpha is required for tumor necrosis factor inhibition of peroxisome proliferator-activated receptor γ function. J Biol Chem. 2006;281:4540–7. doi: 10.1074/jbc.M507784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gearing K. L., Gottlicher M., Teboul M., Widmark E., Gustafsson J. A. Interaction of the peroxisome-proliferator-activated receptor and retinoid X receptor. Proc Natl Acad Sci U S A. 1993;90:1440–4. doi: 10.1073/pnas.90.4.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelman L., Zhou G., Fajas L., Raspe E., Fruchart J. C., Auwerx J. p300 interacts with the N- and C-terminal part of PPARgamma2 in a ligand-independent and -dependent manner, respectively. J Biol Chem. 1999;274:7681–8. doi: 10.1074/jbc.274.12.7681. [DOI] [PubMed] [Google Scholar]
- Ghisletti S., Huang W., Jepsen K., Benner C., Hardiman G., Rosenfeld M. G., Glass C. K. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–93. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S., Huang W., Ogawa S., Pascual G., Lin M. E., Willson T. M., Rosenfeld M. G., Glass C. K. Parallel SUMOylation-dependent pathways mediate gene- and signal-specific transrepression by LXRs and PPARgamma. Mol Cell. 2007;25:57–70. doi: 10.1016/j.molcel.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Goo Y. H., Sohn Y. C., Kim D. H., Kim S. W., Kang M. J., Jung D. J., Kwak E., Barlev N. A., Berger S. L., Chow V. T., Roeder R. G., Azorsa D. O., Meltzer P. S., Suh P. G., Song E. J., Lee K. J., Lee Y. C., Lee J. W. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–9. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlicher M., Widmark E., Li Q., Gustafsson J. A. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc Natl Acad Sci U S A. 1992;89:4653–7. doi: 10.1073/pnas.89.10.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J. Z., Tamasawa N., Brunham L. R., Matsui J., Murakami H., Suda T., Ochiai S., Tsutsui M., Kudou K., Satoh K., Hayden M. R. A case of Tangier disease with a novel mutation in the C-terminal region of ATP-binding cassette transporter A1. Am J Med Genet A. 2004;130A:398–401. doi: 10.1002/ajmg.a.30284. [DOI] [PubMed] [Google Scholar]
- Guan H. P., Ishizuka T., Chui P. C., Lehrke M., Lazar M. A. Corepressors selectively control the transcriptional activity of PPARgamma in adipocytes. Genes Dev. 2005;19:453–61. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Koya D., Isono M., Sugimoto T., Kashiwagi A., Haneda M. Peroxisome proliferator-activated receptor-γ ligands inhibit TGF-β 1-induced fibronectin expression in glomerular mesangial cells. Diabetes. 2004;53:200–8. doi: 10.2337/diabetes.53.1.200. [DOI] [PubMed] [Google Scholar]
- Hay R. T. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- He G. P., Muise A., Li A. W., Ro H. S. A eukaryotic transcriptional repressor with carboxypeptidase activity. Nature. 1995;378:92–6. doi: 10.1038/378092a0. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Herzog B., Hallberg M., Seth A., Woods A., White R., Parker M. G. The nuclear receptor cofactor, receptor-interacting protein 140, is required for the regulation of hepatic lipid and glucose metabolism by liver X receptor. Mol Endocrinol. 2007;21:2687–97. doi: 10.1210/me.2007-0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B., Brune K., Pahl A. 15-Deoxy-δ(12,14)-prostaglandin J2 inhibits the expression of proinflammatory genes in human blood monocytes via a PPAR-γ-independent mechanism. Biochem Biophys Res Commun. 2003;302:415–20. doi: 10.1016/s0006-291x(03)00195-5. [DOI] [PubMed] [Google Scholar]
- Hodgson M. C., Shen H. C., Hollenberg A. N., Balk S. P. Structural basis for nuclear receptor corepressor recruitment by antagonist-liganded androgen receptor. Mol Cancer Ther. 2008;7:3187–94. doi: 10.1158/1535-7163.MCT-08-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper A. J., Robertson K., Ng L., Kattampallil J. S., Latchem D., Willsher P. C., Thom J., Baker R. I., Burnett J. R. A novel ABCA1 nonsense mutation, R1270X, in Tangier disease associated with an unrecognised bleeding tendency. Clin Chim Acta. 2009;409:136–9. doi: 10.1016/j.cca.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Horlein A. J., Naar A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C. K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Hu X., Li Y., Lazar M. A. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol Cell Biol. 2001;21:1747–58. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Li S., Wu J., Xia C., Lala D. S. Liver X receptors interact with corepressors to regulate gene expression. Mol Endocrinol. 2003;17:1019–26. doi: 10.1210/me.2002-0399. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–6. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Issemann I., Green S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature. 1990;347:645–50. doi: 10.1038/347645a0. [DOI] [PubMed] [Google Scholar]
- Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. An oxysterol signalling pathway mediated by the nuclear receptor LXR α. Nature. 1996;383:728–31. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- Janowski B. A., Grogan M. J., Jones S. A., Wisely G. B., Kliewer S. A., Corey E. J., Mangelsdorf D. J. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci U S A. 1999;96:266–71. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaye M. C., Krawiec J. A., Campobasso N., Smallwood A., Qiu C., Lu Q., Kerrigan J. J., De Los Frailes Alvaro M., Laffitte B., Liu W. S., Marino J. P., Jr., Meyer C. R., Nichols J. A., Parks D. J., Perez P., Sarov-Blat L., Seepersaud S. D., Steplewski K. M., Thompson S. K., Wang P., Watson M. A., Webb C. L., Haigh D., Caravella J. A., Macphee C. H., Willson T. M., Collins J. L. Discovery of substituted maleimides as liver X receptor agonists and determination of a ligand-bound crystal structure. J Med Chem. 2005;48:5419–22. doi: 10.1021/jm050532w. [DOI] [PubMed] [Google Scholar]
- Jennewein C., Kuhn A. M., Schmidt M. V., Meilladec-Jullig V., von Knethen A., Gonzalez F. J., Brune B. Sumoylation of peroxisome proliferator-activated receptor γ by apoptotic cells prevents lipopolysaccharide-induced NCoR removal from kappaB binding sites mediating transrepression of proinflammatory cytokines. J Immunol. 2008;181:5646–52. doi: 10.4049/jimmunol.181.8.5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C., Ting A. T., Seed B. PPAR-γ agonists inhibit production of monocyte inflammatory cytokines. Nature. 1998;391:82–6. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- Joseph S. B., Bradley M. N., Castrillo A., Bruhn K. W., Mak P. A., Pei L., Hogenesch J., O'Connell R M., Cheng G., Saez E., Miller J. F., Tontonoz P. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Joseph S. B., Castrillo A., Laffitte B. A., Mangelsdorf D. J., Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–9. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Joseph S. B., McKilligin E., Pei L., Watson M. A., Collins A. R., Laffitte B. A., Chen M., Noh G., Goodman J., Hagger G. N., Tran J., Tippin T. K., Wang X., Lusis A. J., Hsueh W. A., Law R. E., Collins J. L., Willson T. M., Tontonoz P. Synthetic LXR ligand inhibits the development of atherosclerosis in mice. Proc Natl Acad Sci U S A. 2002;99:7604–9. doi: 10.1073/pnas.112059299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalayoglu M. V., Byrne G. I. A Chlamydia pneumoniae component that induces macrophage foam cell formation is chlamydial lipopolysaccharide. Infect Immun. 1998b;66:5067–72. doi: 10.1128/iai.66.11.5067-5072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalayoglu M. V., Byrne G. I. Induction of macrophage foam cell formation by Chlamydia pneumoniae. J Infect Dis. 1998a;177:725–9. doi: 10.1086/514241. [DOI] [PubMed] [Google Scholar]
- Kelly D., Campbell J. I., King T. P., Grant G., Jansson E. A., Coutts A. G., Pettersson S., Conway S. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-γ and RelA. Nat Immunol. 2004;5:104–12. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- Kim S. W., Park K., Kwak E., Choi E., Lee S., Ham J., Kang H., Kim J. M., Hwang S. Y., Kong Y. Y., Lee K., Lee J. W. Activating signal cointegrator 2 required for liver lipid metabolism mediated by liver X receptors in mice. Mol Cell Biol. 2003;23:3583–92. doi: 10.1128/MCB.23.10.3583-3592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G. S., Lee G. Y., Nedumaran B., Park Y. Y., Kim K. T., Park S. C., Lee Y. C., Kim J. B., Choi H. S. The orphan nuclear receptor DAX-1 acts as a novel transcriptional corepressor of PPARgamma. Biochem Biophys Res Commun. 2008;370:264–8. doi: 10.1016/j.bbrc.2008.03.098. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A., Sundseth S. S., Jones S. A., Brown P. J., Wisely G. B., Koble C. S., Devchand P., Wahli W., Willson T. M., Lenhard J. M., Lehmann J. M. Fatty acids and eicosanoids regulate gene expression through direct interactions with peroxisome proliferator-activated receptors α and γ. Proc Natl Acad Sci U S A. 1997;94:4318–23. doi: 10.1073/pnas.94.9.4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucken J., Buchler C., Orso E., Kaminski W. E., Porsch-Ozcurumez M., Liebisch G., Kapinsky M., Diederich W., Drobnik W., Dean M., Allikmets R., Schmitz G. ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci U S A. 2000;97:817–22. doi: 10.1073/pnas.97.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohro T., Nakajima T., Wada Y., Sugiyama A., Ishii M., Tsutsumi S., Aburatani H., Imoto I., Inazawa J., Hamakubo T., Kodama T., Emi M. Genomic structure and mapping of human orphan receptor LXR α: upregulation of LXRa mRNA during monocyte to macrophage differentiation. J Atheroscler Thromb. 2000;7:145–51. doi: 10.5551/jat1994.7.145. [DOI] [PubMed] [Google Scholar]
- Kotaja N., Karvonen U., Janne O. A., Palvimo J. J. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol Cell Biol. 2002;22:5222–34. doi: 10.1128/MCB.22.14.5222-5234.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko L., Cardona G. R., Chin W. W. Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc Natl Acad Sci U S A. 2000;97:6212–7. doi: 10.1073/pnas.97.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung A. L., Rebel V. I., Bronson R. T., Ch'ng L. E., Sieff C. A., Livingston D. M., Yao T. P. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–7. [PMC free article] [PubMed] [Google Scholar]
- Laffitte B. A., Repa J. J., Joseph S. B., Wilpitz D. C., Kast H. R., Mangelsdorf D. J., Tontonoz P. LXRs control lipid-inducible expression of the apolipoprotein E gene in macrophages and adipocytes. Proc Natl Acad Sci U S A. 2001;98:507–12. doi: 10.1073/pnas.021488798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer C., Huang Y., Cullen P., Wiesenhutter B., Mahley R. W., Assmann G., von Eckardstein A. Endogenous apolipoprotein E modulates cholesterol efflux and cholesteryl ester hydrolysis mediated by high-density lipoprotein-3 and lipid-free apolipoproteins in mouse peritoneal macrophages. J Mol Med. 2000;78:217–27. doi: 10.1007/s001090000096. [DOI] [PubMed] [Google Scholar]
- Langmann T., Liebisch G., Moehle C., Schifferer R., Dayoub R., Heiduczek S., Grandl M., Dada A., Schmitz G. Gene expression profiling identifies retinoids as potent inducers of macrophage lipid efflux. Biochim Biophys Acta. 2005;1740:155–61. doi: 10.1016/j.bbadis.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Langmann T., Klucken J., Reil M., Liebisch G., Luciani M. F., Chimini G., Kaminski W. E., Schmitz G. Molecular cloning of the human ATP-binding cassette transporter 1 (hABC1): evidence for sterol-dependent regulation in macrophages. Biochem Biophys Res Commun. 1999;257:29–33. doi: 10.1006/bbrc.1999.0406. [DOI] [PubMed] [Google Scholar]
- Lawn R. M., Wade D. P., Garvin M. R., Wang X., Schwartz K., Porter J. G., Seilhamer J. J., Vaughan A. M., Oram J. F. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J Clin Invest. 1999;104:R25–31. doi: 10.1172/JCI8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layne M. D., Endege W. O., Jain M. K., Yet S. F., Hsieh C. M., Chin M. T., Perrella M. A., Blanar M. A., Haber E., Lee M. E. Aortic carboxypeptidase-like protein, a novel protein with discoidin and carboxypeptidase-like domains, is up-regulated during vascular smooth muscle cell differentiation. J Biol Chem. 1998;273:15654–60. doi: 10.1074/jbc.273.25.15654. [DOI] [PubMed] [Google Scholar]
- Layne M. D., Yet S. F., Maemura K., Hsieh C. M., Bernfield M., Perrella M. A., Lee M. E. Impaired abdominal wall development and deficient wound healing in mice lacking aortic carboxypeptidase-like protein. Mol Cell Biol. 2001;21:5256–61. doi: 10.1128/MCB.21.15.5256-5261.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B., Nielsen A. L., Garnier J. M., Ichinose H., Jeanmougin F., Losson R., Chambon P. A possible involvement of TIF1 α and TIF1 β in the epigenetic control of transcription by nuclear receptors. Embo J. 1996;15:6701–15. [PMC free article] [PubMed] [Google Scholar]
- Lee S. K., Anzick S. L., Choi J. E., Bubendorf L., Guan X. Y., Jung Y. K., Kallioniemi O. P., Kononen J., Trent J. M., Azorsa D., Jhun B. H., Cheong J. H., Lee Y. C., Meltzer P. S., Lee J. W. A nuclear factor, ASC-2, as a cancer-amplified transcriptional coactivator essential for ligand-dependent transactivation by nuclear receptors in vivo. J Biol Chem. 1999;274:34283–93. doi: 10.1074/jbc.274.48.34283. [DOI] [PubMed] [Google Scholar]
- Lee S. J., Yang E. K., Kim S. G. Peroxisome proliferator-activated receptor-γ and retinoic acid X receptor α represses the TGFbeta1 gene via PTEN-mediated p70 ribosomal S6 kinase-1 inhibition: role for Zf9 dephosphorylation. Mol Pharmacol. 2006;70:415–25. doi: 10.1124/mol.106.022954. [DOI] [PubMed] [Google Scholar]
- Lee C. H., Evans R. M. Peroxisome proliferator-activated receptor-γ in macrophage lipid homeostasis. Trends Endocrinol Metab. 2002;13:331–5. doi: 10.1016/s1043-2760(02)00668-9. [DOI] [PubMed] [Google Scholar]
- Lee K. S., Park S. J., Hwang P. H., Yi H. K., Song C. H., Chai O. H., Kim J. S., Lee M. K., Lee Y. C. PPAR-γ modulates allergic inflammation through up-regulation of PTEN. Faseb J. 2005;19:1033–5. doi: 10.1096/fj.04-3309fje. [DOI] [PubMed] [Google Scholar]
- Lee S. K., Jung S. Y., Kim Y. S., Na S. Y., Lee Y. C., Lee J. W. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol Endocrinol. 2001;15:241–54. doi: 10.1210/mend.15.2.0595. [DOI] [PubMed] [Google Scholar]
- Lehmann J. M., Moore L. B., Smith-Oliver T. A., Wilkison W. O., Willson T. M., Kliewer S. A. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPAR γ) J Biol Chem. 1995;270:12953–6. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- Lemon B., Inouye C., King D. S., Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–8. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Lennon A. M., Ramauge M., Dessouroux A., Pierre M. MAP kinase cascades are activated in astrocytes and preadipocytes by 15-deoxy-δ(12-14)-prostaglandin J(2) and the thiazolidinedione ciglitazone through peroxisome proliferator activator receptor γ-independent mechanisms involving reactive oxygenated species. J Biol Chem. 2002;277:29681–5. doi: 10.1074/jbc.M201517200. [DOI] [PubMed] [Google Scholar]
- Leuenberger N., Pradervand S., Wahli W. Sumoylated PPARalpha mediates sex-specific gene repression and protects the liver from estrogen-induced toxicity in mice. J Clin Invest. 2009;119:3138–48. doi: 10.1172/JCI39019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H., Takeuchi J. K., Von Both I., Walls J. R., McAuliffe F., Adamson S. L., Henkelman R. M., Wrana J. L., Rossant J., Bruneau B. G. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–12. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Lim H. J., Moon I., Han K. Transcriptional cofactors exhibit differential preference toward peroxisome proliferator-activated receptors α and δ in uterine cells. Endocrinology. 2004;145:2886–95. doi: 10.1210/en.2004-0011. [DOI] [PubMed] [Google Scholar]
- Lin C. Y., Duan H., Mazzone T. Apolipoprotein E-dependent cholesterol efflux from macrophages: kinetic study and divergent mechanisms for endogenous versus exogenous apolipoprotein E. J Lipid Res. 1999;40:1618–27. [PubMed] [Google Scholar]
- Ling Y., Sankpal U. T., Robertson A. K., McNally J. G., Karpova T., Robertson K. D. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32:598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Gomes P. J., Chen J. D. RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc Natl Acad Sci U S A. 1997;94:8479–84. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Chu M. J., Xu J. Tissue- and nuclear receptor-specific function of the C-terminal LXXLL motif of coactivator NCoA6/AIB3 in mice. Mol Cell Biol. 2007;27:8073–86. doi: 10.1128/MCB.00451-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Shuai K. Regulation of the sumoylation system in gene expression. Curr Opin Cell Biol. 2008;20:288–93. doi: 10.1016/j.ceb.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lordan S., Mackrill J. J., O'Brien N. M. Oxysterols and mechanisms of apoptotic signaling: implications in the pathology of degenerative diseases. J Nutr Biochem. 2009;20:321–36. doi: 10.1016/j.jnutbio.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Lorkowski S., Kratz M., Wenner C., Schmidt R., Weitkamp B., Fobker M., Reinhardt J., Rauterberg J., Galinski E. A., Cullen P. Expression of the ATP-binding cassette transporter gene ABCG1 (ABC8) in Tangier disease. Biochem Biophys Res Commun. 2001;283:821–30. doi: 10.1006/bbrc.2001.4863. [DOI] [PubMed] [Google Scholar]
- Louet J. F., Coste A., Amazit L., Tannour-Louet M., Wu R. C., Tsai S. Y., Tsai M. J., Auwerx J., O'Malley B. W. Oncogenic steroid receptor coactivator-3 is a key regulator of the white adipogenic program. Proc Natl Acad Sci U S A. 2006;103:17868–73. doi: 10.1073/pnas.0608711103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E. G., Peterson L. B., Adams A. D., Lam M. H., Burton C. A., Chin J., Guo Q., Huang S., Latham M., Lopez J. C., Menke J. G., Milot D. P., Mitnaul L. J., Rex-Rabe S. E., Rosa R. L., Tian J. Y., Wright S. D., Sparrow C. P. Different roles of liver X receptor α and β in lipid metabolism: effects of an α-selective and a dual agonist in mice deficient in each subtype. Biochem Pharmacol. 2006;71:453–63. doi: 10.1016/j.bcp.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Mabb A. M., Miyamoto S. SUMO and NF-kappaB ties. Cell Mol Life Sci. 2007;64:1979–96. doi: 10.1007/s00018-007-7005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A., Horikoshi S., Gohda T., Tsuge T., Maeda K., Tomino Y. Pioglitazone attenuates TGF-β(1)-induction of fibronectin synthesis and its splicing variant in human mesangial cells via activation of peroxisome proliferator-activated receptor (PPAR)γ. Cell Biol Int. 2005;29:422–8. doi: 10.1016/j.cellbi.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Maekawa M., Kikuchi J., Kotani K., Nagao K., Odgerel T., Ueda K., Kawano M., Furukawa Y., Sakurabayashi I. A novel missense mutation of ABCA1 in transmembrane α-helix in a Japanese patient with Tangier disease. Atherosclerosis. 2009;206:216–22. doi: 10.1016/j.atherosclerosis.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Majdalawieh A., Zhang L., Fuki I. V., Rader D. J., Ro H. S. Adipocyte enhancer-binding protein 1 is a potential novel atherogenic factor involved in macrophage cholesterol homeostasis and inflammation. Proc Natl Acad Sci U S A. 2006;103:2346–51. doi: 10.1073/pnas.0508139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalawieh A., Zhang L., Ro H. S. Adipocyte enhancer-binding protein-1 promotes macrophage inflammatory responsiveness by up-regulating NF-kappaB via IkappaBalpha negative regulation. Mol Biol Cell. 2007;18:930–42. doi: 10.1091/mbc.E06-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalawieh A., Ro H. S. LPS-induced suppression of macrophage cholesterol efflux is mediated by adipocyte enhancer-binding protein 1. Int J Biochem Cell Biol. 2009;41:1518–25. doi: 10.1016/j.biocel.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Mak P. A., Laffitte B. A., Desrumaux C., Joseph S. B., Curtiss L. K., Mangelsdorf D. J., Tontonoz P., Edwards P. A. Regulated expression of the apolipoprotein E/C-I/C-IV/C-II gene cluster in murine and human macrophages. A critical role for nuclear liver X receptors α and β. J Biol Chem. 2002;277:31900–8. doi: 10.1074/jbc.M202993200. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf D. J., Borgmeyer U., Heyman R. A., Zhou J. Y., Ong E. S., Oro A. E., Kakizuka A., Evans R. M. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 1992;6:329–44. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- Marx N., Sukhova G., Murphy C., Libby P., Plutzky J. Macrophages in human atheroma contain PPARgamma: differentiation-dependent peroxisomal proliferator-activated receptor γ(PPARgamma) expression and reduction of MMP-9 activity through PPARgamma activation in mononuclear phagocytes in vitro. Am J Pathol. 1998;153:17–23. doi: 10.1016/s0002-9440(10)65540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauldin J. P., Nagelin M. H., Wojcik A. J., Srinivasan S., Skaflen M. D., Ayers C. R., McNamara C. A., Hedrick C. C. Reduced expression of ATP-binding cassette transporter G1 increases cholesterol accumulation in macrophages of patients with type 2 diabetes mellitus. Circulation. 2008;117:2785–92. doi: 10.1161/CIRCULATIONAHA.107.741314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzone T. Apolipoprotein E secretion by macrophages: its potential physiological functions. Curr Opin Lipidol. 1996;7:303–7. doi: 10.1097/00041433-199610000-00008. [DOI] [PubMed] [Google Scholar]
- Mazzone T., Reardon C. Expression of heterologous human apolipoprotein E by J774 macrophages enhances cholesterol efflux to HDL3. J Lipid Res. 1994;35:1345–53. [PubMed] [Google Scholar]
- McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–68. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–74. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Miao B., Zondlo S., Gibbs S., Cromley D., Hosagrahara V. P., Kirchgessner T. G., Billheimer J., Mukherjee R. Raising HDL cholesterol without inducing hepatic steatosis and hypertriglyceridemia by a selective LXR modulator. J Lipid Res. 2004;45:1410–7. doi: 10.1194/jlr.M300450-JLR200. [DOI] [PubMed] [Google Scholar]
- Miksa M., Wu R., Cui X., Dong W., Das P., Simms H. H., Ravikumar T. S., Wang P. Vasoactive hormone adrenomedullin and its binding protein: anti-inflammatory effects by up-regulating peroxisome proliferator-activated receptor-γ. J Immunol. 2007;179:6263–72. doi: 10.4049/jimmunol.179.9.6263. [DOI] [PubMed] [Google Scholar]
- Miller A. R., Etgen G. J. Novel peroxisome proliferator-activated receptor ligands for Type 2 diabetes and the metabolic syndrome. Expert Opin Investig Drugs. 2003;12:1489–500. doi: 10.1517/13543784.12.9.1489. [DOI] [PubMed] [Google Scholar]
- Mizukami J., Taniguchi T. The antidiabetic agent thiazolidinedione stimulates the interaction between PPAR γ and CBP. Biochem Biophys Res Commun. 1997;240:61–4. doi: 10.1006/bbrc.1997.7602. [DOI] [PubMed] [Google Scholar]
- Nagy L., Tontonoz P., Alvarez J. G., Chen H., Evans R. M. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–40. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- Niino M., Iwabuchi K., Kikuchi S., Ato M., Morohashi T., Ogata A., Tashiro K., Onoe K. Amelioration of experimental autoimmune encephalomyelitis in C57BL/6 mice by an agonist of peroxisome proliferator-activated receptor-γ. J Neuroimmunol. 2001;116:40–8. doi: 10.1016/s0165-5728(01)00285-5. [DOI] [PubMed] [Google Scholar]
- Nishida T., Yasuda H. PIAS1 and PIASxalpha function as SUMO-E3 ligases toward androgen receptor and repress androgen receptor-dependent transcription. J Biol Chem. 2002;277:41311–7. doi: 10.1074/jbc.M206741200. [DOI] [PubMed] [Google Scholar]
- Nishizawa H., Yamagata K., Shimomura I., Takahashi M., Kuriyama H., Kishida K., Hotta K., Nagaretani H., Maeda N., Matsuda M., Kihara S., Nakamura T., Nishigori H., Tomura H., Moore D. D., Takeda J., Funahashi T., Matsuzawa Y. Small heterodimer partner, an orphan nuclear receptor, augments peroxisome proliferator-activated receptor γ transactivation. J Biol Chem. 2002;277:1586–92. doi: 10.1074/jbc.M104301200. [DOI] [PubMed] [Google Scholar]
- Oberfield J. L., Collins J. L., Holmes C. P., Goreham D. M., Cooper J. P., Cobb J. E., Lenhard J. M., Hull-Ryde E. A., Mohr C. P., Blanchard S. G., Parks D. J., Moore L. B., Lehmann J. M., Plunket K., Miller A. B., Milburn M. V., Kliewer S. A., Willson T. M. A peroxisome proliferator-activated receptor γ ligand inhibits adipocyte differentiation. Proc Natl Acad Sci U S A. 1999;96:6102–6. doi: 10.1073/pnas.96.11.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D., Stone J. F., Takata Y., Blaschke F., Chu V. H., Towler D. A., Law R. E., Hsueh W. A., Bruemmer D. Liver x receptor agonists inhibit cytokine-induced osteopontin expression in macrophages through interference with activator protein-1 signaling pathways. Circ Res. 2005;96:e59–67. doi: 10.1161/01.RES.0000163630.86796.17. [DOI] [PubMed] [Google Scholar]
- Ohshima T., Koga H., Shimotohno K. Transcriptional activity of peroxisome proliferator-activated receptor γ is modulated by SUMO-1 modification. J Biol Chem. 2004;279:29551–7. doi: 10.1074/jbc.M403866200. [DOI] [PubMed] [Google Scholar]
- Oiknine J., Aviram M. Increased susceptibility to activation and increased uptake of low density lipoprotein by cholesterol-loaded macrophages. Arterioscler Thromb. 1992;12:745–53. doi: 10.1161/01.atv.12.6.745. [DOI] [PubMed] [Google Scholar]
- Panini S. R., Sinensky M. S. Mechanisms of oxysterol-induced apoptosis. Curr Opin Lipidol. 2001;12:529–33. doi: 10.1097/00041433-200110000-00008. [DOI] [PubMed] [Google Scholar]
- Park S. J., Lee K. S., Kim S. R., Min K. H., Choe Y. H., Moon H., Chae H. J., Yoo W. H., Lee Y. C. Peroxisome proliferator-activated receptor γ agonist down-regulates IL-17 expression in a murine model of allergic airway inflammation. J Immunol. 2009;183:3259–67. doi: 10.4049/jimmunol.0900231. [DOI] [PubMed] [Google Scholar]
- Park J. G., Muise A., He G. P., Kim S. W., Ro H. S. Transcriptional regulation by the gamma5 subunit of a heterotrimeric G protein during adipogenesis. Embo J. 1999;18:4004–12. doi: 10.1093/emboj/18.14.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual G., Fong A. L., Ogawa S., Gamliel A., Li A. C., Perissi V., Rose D. W., Willson T. M., Rosenfeld M. G., Glass C. K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-γ. Nature. 2005;437:759–63. doi: 10.1038/nature03988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet D. J., Turley S. D., Ma W., Janowski B. A., Lobaccaro J. M., Hammer R. E., Mangelsdorf D. J. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR α. Cell. 1998b;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- Peet D. J., Janowski B. A., Mangelsdorf D. J. The LXRs: a new class of oxysterol receptors. Curr Opin Genet Dev. 1998a;8:571–5. doi: 10.1016/s0959-437x(98)80013-0. [DOI] [PubMed] [Google Scholar]
- Petrova T. V., Akama K. T., Van Eldik L. J. Cyclopentenone prostaglandins suppress activation of microglia: down-regulation of inducible nitric-oxide synthase by 15-deoxy-Delta12,14-prostaglandin J2. Proc Natl Acad Sci U S A. 1999;96:4668–73. doi: 10.1073/pnas.96.8.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan C. A., Weaver J. M., Steger D. J., Joshi S., Maslany J. T., Collins J. L., Zuercher W. J., Willson T. M., Walker M., Jaye M., Lazar M. A. Selective partial agonism of liver X receptor α is related to differential corepressor recruitment. Mol Endocrinol. 2008;22:2241–9. doi: 10.1210/me.2008-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature. 2004;429:771–6. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P., Wu Z., Park C. W., Graves R., Wright M., Spiegelman B. M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Spiegelman B. M. Peroxisome proliferator-activated receptor-γ coactivator 1 α (PGC-1 α): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- Qi C., Zhu Y., Pan J., Yeldandi A. V., Rao M. S., Maeda N., Subbarao V., Pulikuri S., Hashimoto T., Reddy J. K. Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor α-regulated gene expression. Proc Natl Acad Sci U S A. 1999;96:1585–90. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet E. M., Savio D. A., Halpern A. R., Chen L., Miller C. P., Nambi P. Gene-selective modulation by a synthetic oxysterol ligand of the liver X receptor. J Lipid Res. 2004;45:1929–42. doi: 10.1194/jlr.M400257-JLR200. [DOI] [PubMed] [Google Scholar]
- Quinet E. M., Savio D. A., Halpern A. R., Chen L., Schuster G. U., Gustafsson J. A., Basso M. D., Nambi P. Liver X receptor (LXR)-β regulation in LXRalpha-deficient mice: implications for therapeutic targeting. Mol Pharmacol. 2006;70:1340–9. doi: 10.1124/mol.106.022608. [DOI] [PubMed] [Google Scholar]
- Rangwala S. M., Lazar M. A. The dawn of the SPPARMs? Sci STKE. 2002;2002:pe9. doi: 10.1126/stke.2002.121.pe9. [DOI] [PubMed] [Google Scholar]
- Reilly C. M., Oates J. C., Cook J. A., Morrow J. D., Halushka P. V., Gilkeson G. S. Inhibition of mesangial cell nitric oxide in MRL/lpr mice by prostaglandin J2 and proliferator activation receptor-γ agonists. J Immunol. 2000;164:1498–504. doi: 10.4049/jimmunol.164.3.1498. [DOI] [PubMed] [Google Scholar]
- Renaud J. P., Rochel N., Ruff M., Vivat V., Chambon P., Gronemeyer H., Moras D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–9. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- Repa J. J., Turley S. D., Lobaccaro J. A., Medina J., Li L., Lustig K., Shan B., Heyman R. A., Dietschy J. M., Mangelsdorf D. J. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000a;289:1524–9. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000b;14:2819–30. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa J. J., Mangelsdorf D. J. The liver X receptor gene team: potential new players in atherosclerosis. Nat Med. 2002;8:1243–8. doi: 10.1038/nm1102-1243. [DOI] [PubMed] [Google Scholar]
- Ricote M., Huang J., Fajas L., Li A., Welch J., Najib J., Witztum J. L., Auwerx J., Palinski W., Glass C. K. Expression of the peroxisome proliferator-activated receptor γ (PPARgamma) in human atherosclerosis and regulation in macrophages by colony stimulating factors and oxidized low density lipoprotein. Proc Natl Acad Sci U S A. 1998a;95:7614–9. doi: 10.1073/pnas.95.13.7614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricote M., Li A. C., Willson T. M., Kelly C. J., Glass C. K. The peroxisome proliferator-activated receptor-γ is a negative regulator of macrophage activation. Nature. 1998b;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- Rizzo G., Fiorucci S. PPARs and other nuclear receptors in inflammation. Curr Opin Pharmacol. 2006;6:421–7. doi: 10.1016/j.coph.2006.03.012. [DOI] [PubMed] [Google Scholar]
- Ro H. S., Zhang L., Majdalawieh A., Kim S. W., Wu X., Lyons P. J., Webber C., Ma H., Reidy S. P., Boudreau A., Miller J. R., Mitchell P., McLeod R. S. Adipocyte enhancer-binding protein 1 modulates adiposity and energy homeostasis. Obesity (Silver Spring) 2007;15:288–302. doi: 10.1038/oby.2007.569. [DOI] [PubMed] [Google Scholar]
- Ro H. S., Kim S. W., Wu D., Webber C., Nicholson T. E. Gene structure and expression of the mouse adipocyte enhancer-binding protein. Gene. 2001;280:123–33. doi: 10.1016/s0378-1119(01)00771-5. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Glass C. K. Coregulator codes of transcriptional regulation by nuclear receptors. J Biol Chem. 2001;276:36865–8. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- Rosen E. D., Spiegelman B. M. PPARgamma : a nuclear regulator of metabolism, differentiation, and cell growth. J Biol Chem. 2001;276:37731–4. doi: 10.1074/jbc.R100034200. [DOI] [PubMed] [Google Scholar]
- Ruan H., Pownall H. J., Lodish H. F. Troglitazone antagonizes tumor necrosis factor-α-induced reprogramming of adipocyte gene expression by inhibiting the transcriptional regulatory functions of NF-kappaB. J Biol Chem. 2003;278:28181–92. doi: 10.1074/jbc.M303141200. [DOI] [PubMed] [Google Scholar]
- Rust S., Walter M., Funke H., von Eckardstein A., Cullen P., Kroes H. Y., Hordijk R., Geisel J., Kastelein J., Molhuizen H. O., Schreiner M., Mischke A., Hahmann H. W., Assmann G. Assignment of Tangier disease to chromosome 9q31 by a graphical linkage exclusion strategy. Nat Genet. 1998;20:96–8. doi: 10.1038/1770. [DOI] [PubMed] [Google Scholar]
- Rytinki M. M., Palvimo J. J. SUMOylation attenuates the function of PGC-1alpha. J Biol Chem. 2009;284:26184–93. doi: 10.1074/jbc.M109.038943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salma N., Xiao H., Mueller E., Imbalzano A. N. Temporal recruitment of transcription factors and SWI/SNF chromatin-remodeling enzymes during adipogenic induction of the peroxisome proliferator-activated receptor γ nuclear hormone receptor. Mol Cell Biol. 2004;24:4651–63. doi: 10.1128/MCB.24.11.4651-4663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schissel S. L., Dunsmore S. E., Liu X., Shine R. W., Perrella M. A., Layne M. D. Aortic carboxypeptidase-like protein is expressed in fibrotic human lung and its absence protects against bleomycin-induced lung fibrosis. Am J Pathol. 2009;174:818–28. doi: 10.2353/ajpath.2009.080856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R. J., Ficorilli J. V., Zhang Y., Bramlett K. S., Beyer T. P., Borchert K., Dowless M. S., Houck K. A., Burris T. P., Eacho P. I., Liang G., Guo L. W., Wilson W. K., Michael L. F., Cao G. A 15-ketosterol is a liver X receptor ligand that suppresses sterol-responsive element binding protein-2 activity. J Lipid Res. 2006;47:1037–44. doi: 10.1194/jlr.M500526-JLR200. [DOI] [PubMed] [Google Scholar]
- Schoonjans K., Peinado-Onsurbe J., Lefebvre A. M., Heyman R. A., Briggs M., Deeb S., Staels B., Auwerx J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. Embo J. 1996;15:5336–48. [PMC free article] [PubMed] [Google Scholar]
- Schuster G. U., Parini P., Wang L., Alberti S., Steffensen K. R., Hansson G. K., Angelin B., Gustafsson J. A. Accumulation of foam cells in liver X receptor-deficient mice. Circulation. 2002;106:1147–53. doi: 10.1161/01.cir.0000026802.79202.96. [DOI] [PubMed] [Google Scholar]
- Schwartz K., Lawn R. M., Wade D. P. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem Biophys Res Commun. 2000;274:794–802. doi: 10.1006/bbrc.2000.3243. [DOI] [PubMed] [Google Scholar]
- Seo J. B., Moon H. M., Kim W. S., Lee Y. S., Jeong H. W., Yoo E. J., Ham J., Kang H., Park M. G., Steffensen K. R., Stulnig T. M., Gustafsson J. A., Park S. D., Kim J. B. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor γ expression. Mol Cell Biol. 2004;24:3430–44. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serfaty-Lacrosniere C., Civeira F., Lanzberg A., Isaia P., Berg J., Janus E. D., Smith M. P., Jr., Pritchard P. H., Frohlich J., Lees R. S. Homozygous Tangier disease and cardiovascular disease. Atherosclerosis. 1994;107:85–98. doi: 10.1016/0021-9150(94)90144-9. [DOI] [PubMed] [Google Scholar]
- Sfeir Z., Ibrahimi A., Amri E., Grimaldi P., Abumrad N. Regulation of FAT/CD36 gene expression: further evidence in support of a role of the protein in fatty acid binding/transport. Prostaglandins Leukot Essent Fatty Acids. 1997;57:17–21. doi: 10.1016/s0952-3278(97)90487-7. [DOI] [PubMed] [Google Scholar]
- Shang Y., Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–8. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Shin D. J., Osborne T. F. Peroxisome proliferator-activated receptor-γ coactivator-1alpha activation of CYP7A1 during food restriction and diabetes is still inhibited by small heterodimer partner. J Biol Chem. 2008;283:15089–96. doi: 10.1074/jbc.M710452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singaraja R. R., Visscher H., James E. R., Chroni A., Coutinho J. M., Brunham L. R., Kang M. H., Zannis V. I., Chimini G., Hayden M. R. Specific mutations in ABCA1 have discrete effects on ABCA1 function and lipid phenotypes both in vivo and in vitro. Circ Res. 2006;99:389–97. doi: 10.1161/01.RES.0000237920.70451.ad. [DOI] [PubMed] [Google Scholar]
- Smoak K., Madenspacher J., Jeyaseelan S., Williams B., Dixon D., Poch K. R., Nick J. A., Worthen G. S., Fessler M. B. Effects of liver X receptor agonist treatment on pulmonary inflammation and host defense. J Immunol. 2008;180:3305–12. doi: 10.4049/jimmunol.180.5.3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C., Kokontis J. M., Hiipakka R. A., Liao S. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc Natl Acad Sci U S A. 1994;91:10809–13. doi: 10.1073/pnas.91.23.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straus D. S., Pascual G., Li M., Welch J. S., Ricote M., Hsiang C. H., Sengchanthalangsy L. L., Ghosh G., Glass C. K. 15-deoxy-δ 12,14-prostaglandin J2 inhibits multiple steps in the NF-κ B signaling pathway. Proc Natl Acad Sci U S A. 2000;97:4844–9. doi: 10.1073/pnas.97.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surapureddi S., Yu S., Bu H., Hashimoto T., Yeldandi A. V., Kashireddy P., Cherkaoui-Malki M., Qi C., Zhu Y. J., Rao M. S., Reddy J. K. Identification of a transcriptionally active peroxisome proliferator-activated receptor α -interacting cofactor complex in rat liver and characterization of PRIC285 as a coactivator. Proc Natl Acad Sci U S A. 2002;99:11836–41. doi: 10.1073/pnas.182426699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzawa M., Takada I., Yanagisawa J., Ohtake F., Ogawa S., Yamauchi T., Kadowaki T., Takeuchi Y., Shibuya H., Gotoh Y., Matsumoto K., Kato S. Cytokines suppress adipogenesis and PPAR-γ function through the TAK1/TAB1/NIK cascade. Nat Cell Biol. 2003;5:224–30. doi: 10.1038/ncb942. [DOI] [PubMed] [Google Scholar]
- Szewczyk J. W., Huang S., Chin J., Tian J., Mitnaul L., Rosa R. L., Peterson L., Sparrow C. P., Adams A. D. SAR studies: designing potent and selective LXR agonists. Bioorg Med Chem Lett. 2006;16:3055–60. doi: 10.1016/j.bmcl.2006.02.050. [DOI] [PubMed] [Google Scholar]
- Tabas I. Consequences of cellular cholesterol accumulation: basic concepts and physiological implications. J Clin Invest. 2002;110:905–11. doi: 10.1172/JCI16452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangirala R. K., Bischoff E. D., Joseph S. B., Wagner B. L., Walczak R., Laffitte B. A., Daige C. L., Thomas D., Heyman R. A., Mangelsdorf D. J., Wang X., Lusis A. J., Tontonoz P., Schulman I. G. Identification of macrophage liver X receptors as inhibitors of atherosclerosis. Proc Natl Acad Sci U S A. 2002;99:11896–901. doi: 10.1073/pnas.182199799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaka N., Hiroshima A., Ariga A., Honzumi S., Koieyama T., Inaba T., Fujiwara T. Liver X receptor agonists inhibit tissue factor expression in macrophages. Febs J. 2005;272:1546–56. doi: 10.1111/j.1742-4658.2005.04599.x. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Graves R. A., Budavari A. I., Spiegelman B. M. mPPAR γ 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994a;8:1224–34. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Devine J., Beale E. G., Spiegelman B. M. PPAR γ 2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol Cell Biol. 1995;15:351–7. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P., Nagy L., Alvarez J. G., Thomazy V. A., Evans R. M. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–52. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- Tontonoz P., Hu E., Spiegelman B. M. Stimulation of adipogenesis in fibroblasts by PPAR γ 2, a lipid-activated transcription factor. Cell. 1994b;79:1147–56. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- Torchia J., Rose D. W., Inostroza J., Kamei Y., Westin S., Glass C. K., Rosenfeld M. G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–84. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Torti F. M., Torti S. V., Larrick J. W., Ringold G. M. Modulation of adipocyte differentiation by tumor necrosis factor and transforming growth factor β. J Cell Biol. 1989;108:1105–13. doi: 10.1083/jcb.108.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traves P. G., Hortelano S., Zeini M., Chao T. H., Lam T., Neuteboom S. T., Theodorakis E. A., Palladino M. A., Castrillo A., Bosca L. Selective activation of liver X receptors by acanthoic acid-related diterpenes. Mol Pharmacol. 2007;71:1545–53. doi: 10.1124/mol.106.031906. [DOI] [PubMed] [Google Scholar]
- Tsubouchi Y., Kawahito Y., Kohno M., Inoue K., Hla T., Sano H. Feedback control of the arachidonate cascade in rheumatoid synoviocytes by 15-deoxy-δ(12,14)-prostaglandin J2. Biochem Biophys Res Commun. 2001;283:750–5. doi: 10.1006/bbrc.2001.4847. [DOI] [PubMed] [Google Scholar]
- Unno A., Takada I., Takezawa S., Oishi H., Baba A., Shimizu T., Tokita A., Yanagisawa J., Kato S. TRRAP as a hepatic coactivator of LXR and FXR function. Biochem Biophys Res Commun. 2005;327:933–8. doi: 10.1016/j.bbrc.2004.12.095. [DOI] [PubMed] [Google Scholar]
- Van Eck M., Herijgers N., Vidgeon-Hart M., Pearce N. J., Hoogerbrugge P. M., Groot P. H., Van Berkel T. J. Accelerated atherosclerosis in C57Bl/6 mice transplanted with ApoE-deficient bone marrow. Atherosclerosis. 2000;150:71–80. doi: 10.1016/s0021-9150(99)00372-x. [DOI] [PubMed] [Google Scholar]
- Venkateswaran A., Laffitte B. A., Joseph S. B., Mak P. A., Wilpitz D. C., Edwards P. A., Tontonoz P. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR α. Proc Natl Acad Sci U S A. 2000b;97:12097–102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran A., Repa J. J., Lobaccaro J. M., Bronson A., Mangelsdorf D. J., Edwards P. A. Human white/murine ABC8 mRNA levels are highly induced in lipid-loaded macrophages. A transcriptional role for specific oxysterols. J Biol Chem. 2000a;275:14700–7. doi: 10.1074/jbc.275.19.14700. [DOI] [PubMed] [Google Scholar]
- von Knethen A., Soller M., Tzieply N., Weigert A., Johann A. M., Jennewein C., Kohl R., Brune B. PPARgamma1 attenuates cytosol to membrane translocation of PKCalpha to desensitize monocytes/macrophages. J Cell Biol. 2007;176:681–94. doi: 10.1083/jcb.200605038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B. L., Valledor A. F., Shao G., Daige C. L., Bischoff E. D., Petrowski M., Jepsen K., Baek S. H., Heyman R. A., Rosenfeld M. G., Schulman I. G., Glass C. K. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23:5780–9. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak R., Tontonoz P. PPARadigms and PPARadoxes: expanding roles for PPARgamma in the control of lipid metabolism. J Lipid Res. 2002;43:177–86. [PubMed] [Google Scholar]
- Walczak R., Joseph S. B., Laffitte B. A., Castrillo A., Pei L., Tontonoz P. Transcription of the vascular endothelial growth factor gene in macrophages is regulated by liver X receptors. J Biol Chem. 2004;279:9905–11. doi: 10.1074/jbc.M310587200. [DOI] [PubMed] [Google Scholar]
- Wang N., Lan D., Chen W., Matsuura F., Tall A. R. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc Natl Acad Sci U S A. 2004;101:9774–9. doi: 10.1073/pnas.0403506101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Yang X. Y., Zhang X., Farrar W. L. Inhibition of adhesive interaction between multiple myeloma and bone marrow stromal cells by PPARgamma cross talk with NF-kappaB and C/EBP. Blood. 2007;110:4373–84. doi: 10.1182/blood-2006-07-038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J. S., Ricote M., Akiyama T. E., Gonzalez F. J., Glass C. K. PPARgamma and PPARdelta negatively regulate specific subsets of lipopolysaccharide and IFN-γ target genes in macrophages. Proc Natl Acad Sci U S A. 2003;100:6712–7. doi: 10.1073/pnas.1031789100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney K. D., Watson M. A., Goodwin B., Galardi C. M., Maglich J. M., Wilson J. G., Willson T. M., Collins J. L., Kliewer S. A. Liver X receptor (LXR) regulation of the LXRalpha gene in human macrophages. J Biol Chem. 2001;276:43509–15. doi: 10.1074/jbc.M106155200. [DOI] [PubMed] [Google Scholar]
- Wigren J., Surapureddi S., Olsson A. G., Glass C. K., Hammarstrom S., Soderstrom M. Differential recruitment of the coactivator proteins CREB-binding protein and steroid receptor coactivator-1 to peroxisome proliferator-activated receptor γ/9-cis-retinoic acid receptor heterodimers by ligands present in oxidized low-density lipoprotein. J Endocrinol. 2003;177:207–14. doi: 10.1677/joe.0.1770207. [DOI] [PubMed] [Google Scholar]
- Williams S., Bledsoe R. K., Collins J. L., Boggs S., Lambert M. H., Miller A. B., Moore J., McKee D. D., Moore L., Nichols J., Parks D., Watson M., Wisely B., Willson T. M. X-ray crystal structure of the liver X receptor β ligand binding domain: regulation by a histidine-tryptophan switch. J Biol Chem. 2003;278:27138–43. doi: 10.1074/jbc.M302260200. [DOI] [PubMed] [Google Scholar]
- Willson T. M., Brown P. J., Sternbach D. D., Henke B. R. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–50. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- Willson T. M., Cobb J. E., Cowan D. J., Wiethe R. W., Correa I. D., Prakash S. R., Beck K. D., Moore L. B., Kliewer S. A., Lehmann J. M. The structure-activity relationship between peroxisome proliferator-activated receptor γ agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem. 1996;39:665–8. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- Willy P. J., Umesono K., Ong E. S., Evans R. M., Heyman R. A., Mangelsdorf D. J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–45. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Xu H. E., Lambert M. H., Montana V. G., Parks D. J., Blanchard S. G., Brown P. J., Sternbach D. D., Lehmann J. M., Wisely G. B., Willson T. M., Kliewer S. A., Milburn M. V. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- Yamagata K., Yoshimochi K., Daitoku H., Hirota K., Fukamizu A. Bile acid represses the peroxisome proliferator-activated receptor-γ coactivator-1 promoter activity in a small heterodimer partner-dependent manner. Int J Mol Med. 2007;19:751–6. [PubMed] [Google Scholar]
- Yamashita D., Yamaguchi T., Shimizu M., Nakata N., Hirose F., Osumi T. The transactivating function of peroxisome proliferator-activated receptor γ is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells. 2004;9:1017–29. doi: 10.1111/j.1365-2443.2004.00786.x. [DOI] [PubMed] [Google Scholar]
- Yao T. P., Oh S. P., Fuchs M., Zhou N. D., Ch'ng L. E., Newsome D., Bronson R. T., Li E., Livingston D. M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–72. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- Yu K., Bayona W., Kallen C. B., Harding H. P., Ravera C. P., McMahon G., Brown M., Lazar M. A. Differential activation of peroxisome proliferator-activated receptors by eicosanoids. J Biol Chem. 1995;270:23975–83. doi: 10.1074/jbc.270.41.23975. [DOI] [PubMed] [Google Scholar]
- Yu C., Markan K., Temple K. A., Deplewski D., Brady M. J., Cohen R. N. The nuclear receptor corepressors NCoR and SMRT decrease peroxisome proliferator-activated receptor γ transcriptional activity and repress 3T3-L1 adipogenesis. J Biol Chem. 2005;280:13600–5. doi: 10.1074/jbc.M409468200. [DOI] [PubMed] [Google Scholar]
- Zelcer N., Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–14. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Y., Gaynor P. M., Kruth H. S. Apolipoprotein E produced by human monocyte-derived macrophages mediates cholesterol efflux that occurs in the absence of added cholesterol acceptors. J Biol Chem. 1996a;271:28641–6. doi: 10.1074/jbc.271.45.28641. [DOI] [PubMed] [Google Scholar]
- Zhang B., Berger J., Hu E., Szalkowski D., White-Carrington S., Spiegelman B. M., Moller D. E. Negative regulation of peroxisome proliferator-activated receptor-γ gene expression contributes to the antiadipogenic effects of tumor necrosis factor-α. Mol Endocrinol. 1996b;10:1457–66. doi: 10.1210/mend.10.11.8923470. [DOI] [PubMed] [Google Scholar]
- Zhang F., Lavan B. E., Gregoire F. M. Selective Modulators of PPAR-γ Activity: Molecular Aspects Related to Obesity and Side-Effects. PPAR Res. 2007;2007:32696. doi: 10.1155/2007/32696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Reidy S. P., Nicholson T. E., Lee H. J., Majdalawieh A., Webber C., Stewart B. R., Dolphin P., Ro H. S. The role of AEBP1 in sex-specific diet-induced obesity. Mol Med. 2005;11:39–47. doi: 10.2119/2005-00021.Ro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Chen W., Yang L., Chen L., Stimpson S. A., Diehl A. M. PPARgamma agonists prevent TGFbeta1/Smad3-signaling in human hepatic stellate cells. Biochem Biophys Res Commun. 2006;350:385–91. doi: 10.1016/j.bbrc.2006.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y. C., Waxman D. J. Cross-talk between janus kinase-signal transducer and activator of transcription (JAK-STAT) and peroxisome proliferator-activated receptor-α (PPARalpha) signaling pathways. Growth hormone inhibition of pparalpha transcriptional activity mediated by stat5b. J Biol Chem. 1999;274:2672–81. doi: 10.1074/jbc.274.5.2672. [DOI] [PubMed] [Google Scholar]
- Zhou M., Wu R., Dong W., Jacob A., Wang P. Endotoxin downregulates peroxisome proliferator-activated receptor-γ via the increase in TNF-α release. Am J Physiol Regul Integr Comp Physiol. 2008;294:R84–92. doi: 10.1152/ajpregu.00340.2007. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Calandra C., Rao M. S., Reddy J. K. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor γ. Gene Expr. 1996;6:185–95. [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Gong B., Bisgaier C. L., Aviram M., Newton R. S. Induction of PPARgamma1 expression in human THP-1 monocytic leukemia cells by 9-cis-retinoic acid is associated with cellular growth suppression. Biochem Biophys Res Commun. 1998;251:842–8. doi: 10.1006/bbrc.1998.9567. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Jain S., Rao M. S., Reddy J. K. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J Biol Chem. 1997;272:25500–6. doi: 10.1074/jbc.272.41.25500. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Qi C., Korenberg J. R., Chen X. N., Noya D., Rao M. S., Reddy J. K. Structural organization of mouse peroxisome proliferator-activated receptor γ (mPPAR γ) gene: alternative promoter use and different splicing yield two mPPAR γ isoforms. Proc Natl Acad Sci U S A. 1995;92:7921–5. doi: 10.1073/pnas.92.17.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingarelli B., Sheehan M., Hake P. W., O'Connor M., Denenberg A., Cook J. A. Peroxisome proliferator activator receptor-γ ligands, 15-deoxy-δ(12,14)-prostaglandin J2 and ciglitazone, reduce systemic inflammation in polymicrobial sepsis by modulation of signal transduction pathways. J Immunol. 2003;171:6827–37. doi: 10.4049/jimmunol.171.12.6827. [DOI] [PubMed] [Google Scholar]