Summary
Social interactions among microbes that engage in cooperative behaviours are well studied in laboratory contexts [1, 2], but little is known about the scales at which initially cooperative microbes diversify into socially conflicting genotypes in nature. The predatory soil bacterium Myxococcus xanthus responds to starvation by cooperatively forming multi-cellular fruiting bodies in which a portion of the population differentiates into stress-resistant spores [3, 4]. Natural M. xanthus populations are spatially structured [5] and genetically divergent isolates from distant origins exhibit striking developmental antagonisms that decrease spore production in chimaeric fruiting bodies [6]. Here we show that genetically similar isolates of M. xanthus from a centimeter-scale population [7] also exhibit strong and pervasive antagonisms when mixed in development. Negative responses to chimerism were less intense, on average, among local strains than among global isolates, although no significant correlation was found between genetic distance at multi-locus sequence typing (MLST) loci and the degree of social asymmetry between competitors. A test for self/non-self discrimination during vegetative swarming revealed a great diversity of distinct self-recognition types even among identical MLST genotypes. Such non-self exclusion may serve to direct the benefits of cooperation to close kin within diverse populations in which the probability of social conflict among neighbours is high.
Keywords: Myxococcus xanthus, social development, cooperation, kin discrimination
Results and Discussion
Biological incompatibilities take diverse forms: reproductive isolation across species [8], non-cooperation between social groups such as insect colonies both across and within species [9] and self/non-self recognition systems that discriminate between closely related individuals [10–12]. Some microorganisms engage in complex forms of social cooperation, such as fruiting body formation in the prokaryotic myxobacteria and eukaryotic slime molds, but do so preferentially with identical or highly similar genotypes and exhibit reduced cooperation with (or outright antagonism toward) divergent conspecifics [6, 13]. However, little is known about the spatial and genetic scales at which social incompatibilities evolve in natural populations of social microbes.
Myxococcus xanthus is a predatory bacterium that kills and eats other microbes (and also consumes organic polymers), by secreting toxic and lytic compounds [14]. Upon starvation, cells aggregate and exchange signals in the process of forming multi-cellular fruiting bodies [3]. Some cells within a fruiting body become stress-resistant spores, whereas others undergo autolysis or remain undifferentiated [4, 15]. This process of social development is likely maintained by kin selection because fruiting body construction can increase group-level fitness among close relatives [16–18].
Fiegna and Velicer (2005) [6] investigated whether cooperation extends across divergent M. xanthus genotypes. Nine isolates of M. xanthus that originated from distant global locations were mixed in all 36 possible pair-wise combinations and the effect of mixing on their spore production was quantified. Mixing was found to negatively affect the sporulation of one or both strains in most pairings, thus demonstrating that M. xanthus has diverged into many socially incompatible genotypes.
Natural populations of M. xanthus are spatially structured, with genetic variation within local (e.g. below meter-scale) populations greatly reduced relative to variation between populations separated by large distances (>102 − 103 km) [5]. Such spatial structuring of diversity in large sexual organisms is central to models of how reproductive isolation evolves (i.e. sympatric vs. allopatric modes of speciation [8]). Analogously, spatial structuring of social microbe populations raises the question of whether barriers to cooperation between distinct genotypes originate from divergence in sympatry, allopatry or both. We addressed this question by repeating the experiments of Fiegna & Velicer (2005) [6] with nine isolates collected from a 16 × 16 cm soil plot in Tübingen, Germany and comparing the results from these local cm-scale isolates to those from the global isolates (all strains listed in Fig. 1).
Figure 1.
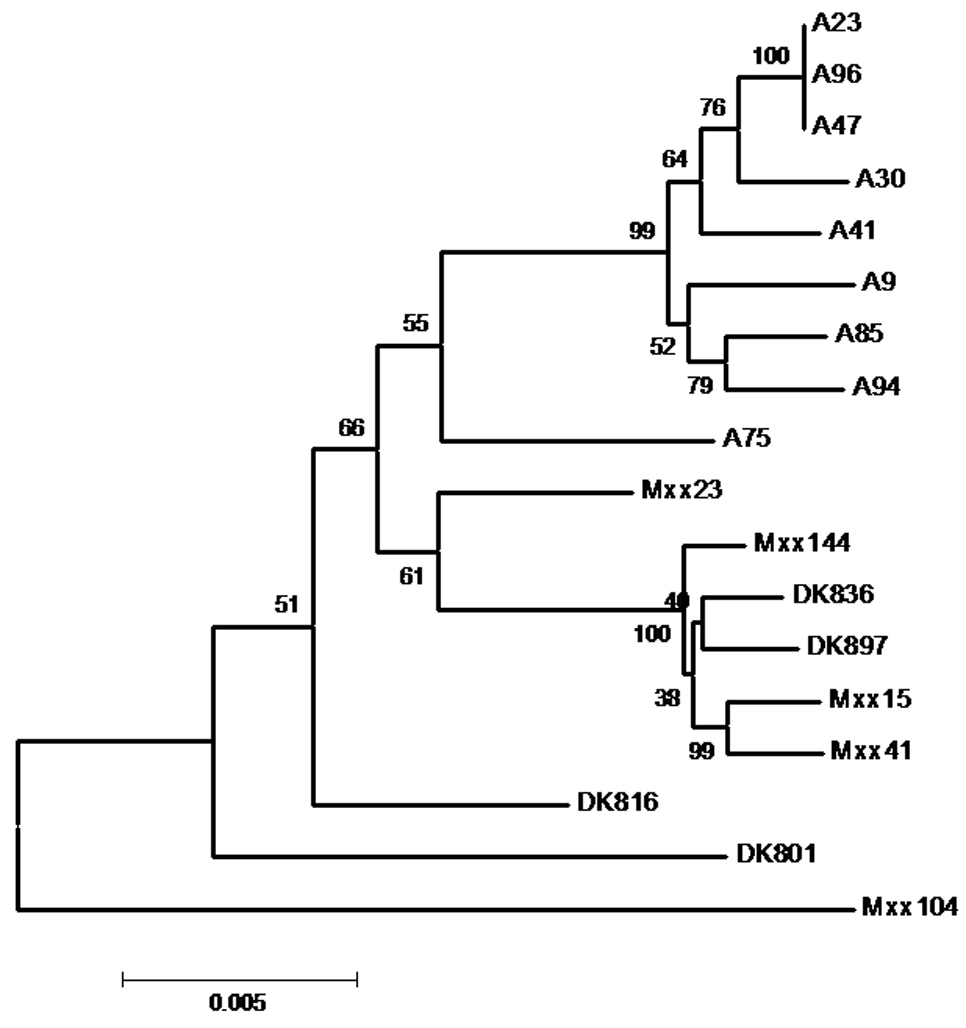
Neighbor-joining tree of nine local strains from Tübingen (prefix A) and nine global isolates (prefixs Mxx and DK). Bootstrap values (10,000 replicates) are indicated next to nodes. Number of base substitutions per site is indicated below the scale bar.
A previous study of 78 M. xanthus clones isolated from this cm-scale plot revealed 21 distinct genotypes based on sequences from three loci (csgA, fibA, and pilA) [7]. Seven of those 21 genotypes are represented among the nine isolates examined here, while three strains (A23, A47 and A96) share the same concatemer genotype. The genetic distances for all strain pairs within both the local and global isolate sets were calculated based on a concatemer of six MLST loci (Supplemental Experimental Procedures), which was also used to generate a tree of phylogenetic relationships among strains in which all of the Tübingen cm-scale isolates form a clade that does not include any of the global isolates (Fig. 1). The average pair-wise genetic distance among the local isolates (0.007) was significantly lower than that for global isolate set (0.012, p = 0.013 for global vs. local difference, two-tailed t-test, n = 35).
The co-development experiments performed here (see Supplemental Experimental Procedures) allowed us to test whether patterns of developmental incompatibility among relatively similar isolates at a local scale differ from those among relatively divergent isolates at the global scale. Our co-development assays allow rigorous quantification of social incompatibilities among strains, but were not designed to mimic how distinct genotypes actually interact in the soil. Because direct encounters between conspecifics are likely to occur during vegetative swarming via gliding motility, we also scored whether isolates distinguish self from non-self during active growth on nutrient agar.
Sporulation mixing effects
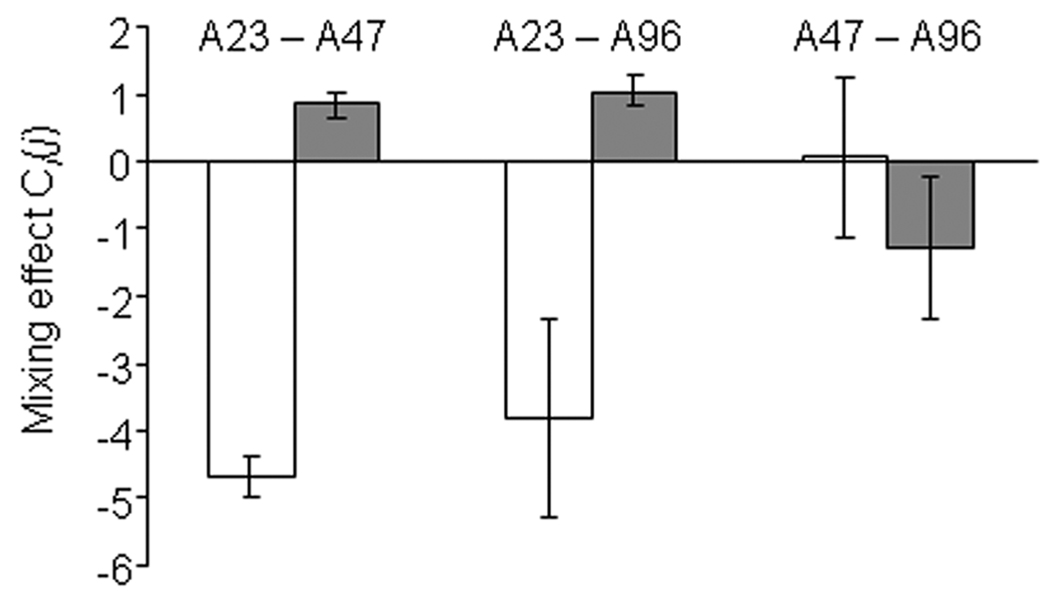
The effect of mixing two strains i and j on the sporulation efficiency of strain i was quantified by the one-way mixing effect parameter Ci(j) (Supplemental Experimental Procedures). A negative Ci(j) value indicates that strain i sporulates less efficiently in the presence of strain j than it does in pure culture, whereas positive values indicate that the presence of strain j increases the sporulation efficiency of strain i. In 11 of the 36 pair-wise mixes performed among the local Tübingen isolates, the reciprocal Ci(j) values for paired competitors were both negative, whereas in the remaining 25 mixes one clone responded negatively to mixing while the winning competitor responded positively. In contrast, only 13 of the 36 global isolate winners had positive Ci(j) values. Strikingly, some instances of strong antagonism associated with positive responses to mixing occurred between identical concatemer genotypes. For example, strains A23, A47 and A96 share a common MLST genotype, but both A47 and A96 strongly inhibit A23 sporulation while themselves sporulating more efficiently in the presence of A23 than in isolation (Fig. 2). On average, genetic chimaerism strongly reduced individual spore production (relative to clonal performance) among the centimetre scale isolates (average Ci(j) = −1.00, p < 0.001, one-sample t-test for difference from 0, n = 35) (Fig. 3).
Figure 2.
Co-development interactions among the three most closely related local isolates (A23, A47 and A96). The effect of mixing two clones on the (log-transformed) sporulation efficiency each is given as Ci(j) (with ‘i’ referring to whichever strain is being considered). Open bars show the mixing effect on the first clone in each pair and grey bars show the effect on the second clone. Error bars represent 95% confidence intervals.
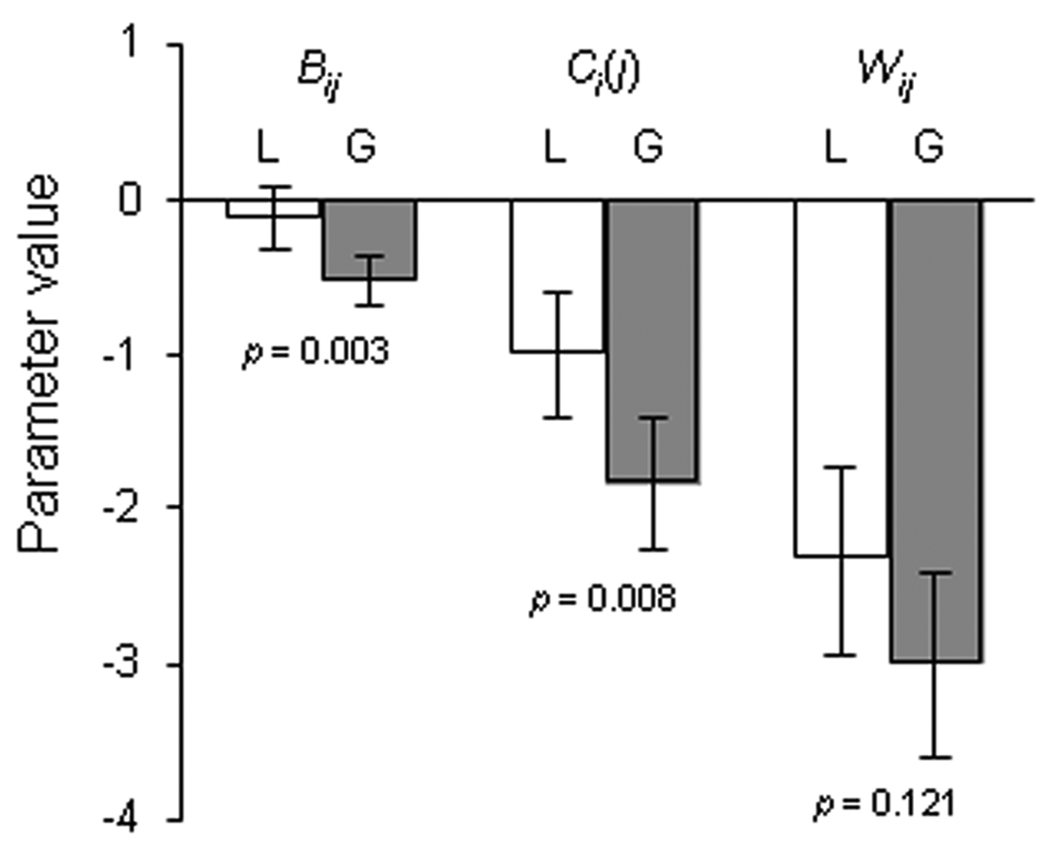
Figure 3.
A comparison of average one-way (Ci[j]) mixing effect, bidirectional Bij mixing effectand relative sporulation efficiency (Wij) parameters between the local (L) isolates and global (G) isolates. Error bars represent 95% confidence intervals.
The effect of mixing strains on total group spore production is represented by the bidirectional mixing effect parameter Bij (Supplemental Experimental Procedures). Among the local isolates, Bij was neither significantly negative for any individual strain (Table S1) nor on average across all nine strains (mean Bij local = −0.13), indicating that total group productivity was not severely affected by mixing of distinct clones. (For many local isolate pairings, the Ci(j) value for the losing competitor was strongly negative, whereas the corresponding value for the paired winning competitor was very small and/or positive, with the result that total group productivity was largely unaffected. Thus, the average Ci(j) for local strains can be significantly negative while the average of Bij is not (Fig. 3) because the former includes the individual responses from both the winner and loser in each pair.)
Relative sporulation
The fitness parameter Wij quantifies the sporulation efficiency of strain i relative to that of j during co-development (Supplemental Experimental Procedures). It thus reflects any asymmetry in spore production by two strains when they undergo development together. The average Wij of developmental competition losers among the local isolates was found to be very large (−2.33, Fig. 3) revealing stark fitness differences (>100-fold on average) among these neighboring strains during codevelopment. Wij correlates strongly with the difference between paired Ci(j) and Cj(i) values (parameter Cij in [6], r2 = 0.913, p < 0.001), but not with differences between pure-culture sporulation measures (log(Di/Dj), Pearsons r = 0.041, p = 0.813), indicating that fitness asymmetries in mixture are caused primarily by chimerism per se rather than by pure-culture sporulation abilities. As for the global isolates [6], the hierarchy of developmental fitness ranks among local isolates (Table S2) was significantly linear (Kendall’s technique [19], K = 0.57, p = 0.045). This result reflects a prevalence of transitive (A beats B and C and B beats C) rather than non-transitive (A beats B and B beats C, but C beats A) fitness relationships among all possible 3-way strain comparisons.
Local vs. global comparisons
The local and global strain sets differed significantly in two of the three parameters measured here (Ci(j) and Bij). While average Ci(j) was significantly negative within both the local and global [6] data sets, the negative response of the global strains was significantly greater than that for the local isolates (Fig. 3, p = 0.008 for difference, 2-tailed t-test, n = 35). Moreover, the effect of mixing on total group spore production was significantly negative among the global isolates [6] but not among the local strains (Fig. 3, p < 0.001). The average Wij of developmental competition losers was slightly lower among global isolates than among local strains, but not significantly so (Fig. 3, p = 0.12). The greater negativity of mixing effects on individual and group spore production among the global isolates examined here suggests that social incompatibilities are generally more severe among isolates from allopatric origins than among sympatric strains.
Developmental incompatibility vs. genetic distance
The high degree of antagonism in the local population revealed by the Ci(j) and Wij parameters (Fig. 3) suggests that social incompatibilities often evolve among recently diverged lineages. Indeed, some of the largest competitive asymmetries documented here occur among the least divergent strains examined (A23, A47 and A96, Fig. 1, Fig 2), suggesting that overall genomic divergence (as represented by MLST loci) fails to predict the magnitude of fitness asymmetries among strains. Although average genetic diversity and the magnitude of mixing parameters Ci(j) and Bij were greater among the global isolates (see above and Fig. 3), neither of these parameters nor the fitness asymmetry parameter Wij were found to correlate significantly with pair-wise genetic distance at MLST loci, either for the local and global data sets combined (Ci(j) r2 = 0.00, p = 0.36; Wij r2 = 0.01, p = 0.69; Bij r2 = 0.03, p = 0.13) or within either data set analyzed individually (results not shown).
Kin discrimination during vegetative growth
Different M. xanthus genotypes are likely to encounter one another during active swarming through soil. To test whether swarms of different clones merge upon encounter or remain separate through some mechanism of kin discrimination (or ‘allorecognition’), we scored the presence or absence of swarm boundaries between paired clones swarming on nutrient agar (Supplemental Experimental Procedures). This assay is a modified form of the Dienes test, which is a highly discriminatory typing method originally developed to identify swarming Proteus mirabilis strains [20, 21]. In this assay, distinct isolates were spotted next to each other on an agar surface and the interface of their respective growing swarms was examined to determine whether they merged into a single continuous swarm or if a line of demarcation formed between them.
Initial experiments indicated that all 21 csgA/fibA/pilA concatemer genotypes among the 78 Tübingen cm-scale isolates were distinct allorecognition types (i.e. swarms of distinct genotypes never freely merged). Subsequently, all isolates with identical concatemer genotypes were tested for allorecognition (Table S3). Fourteen genotypes were each represented by multiple (2–15) isolates, whereas seven genotypes were represented by a single clone (A2, A75, A79, A81, A82, A83, A98) and thus could not be staged with a different isolate of the same genotype. A total of 260 such within-genotype comparisons revealed 38 distinct incompatible types among the 71 clones that share a common genotype with at least one other isolate (e.g. between A23 and A47, Fig. 4). Thus, a total of 45 unique allorecognition types appear to be present in our collection of 78 cm-scale isolates (including the seven genotypes unique to single isolates).
Figure 4.
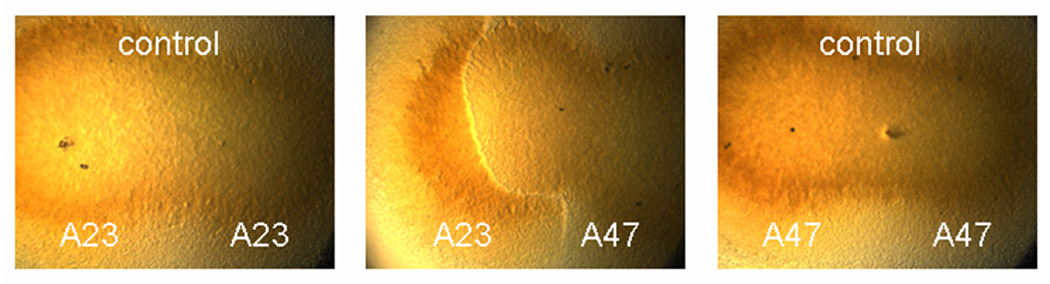
Kin recognition between vegetatively growing swarms. A clear line of demarcation is evident between swarms of isolates A23 and A47. The control treatments show two swarms of a single isolate that have merged into a continuous swarm with no line of demarcation.
Discussion
This study reveals that evolutionary diversification of a highly cooperative bacterium into many antagonistic genotypes has occurred among genetically highly similar individuals living in sympatry. Closely related clones of M. xanthus clones occupying the same centimeter-scale soil patch exhibit high levels of conflict when undergoing mixed social development (Fig. 2, Fig 3) and kin discrimination while swarming during vegetative growth (Fig. 4). Such discrimination of self from non-self has also been observed between genetically distinct swarms of the γ-proteobacteria Proteus mirabilis [20, 21] and Pseudomonas aeruginosa [22].
Although interactions among local strains were strongly antagonistic, our data suggest that divergence in allopatry augments incompatibilities that first evolve in sympatry (Fig. 3). Spatially isolated populations may have undergone unique patterns of antagonistic social coevolution and/or some form of local adaptation (e.g. adaptation to distinct prey communities). Distinct evolutionary effects of different local selective conditions may contribute to increased incompatibility across vs. within local populations.
Chimaeric organisms can arise through grafting in vertebrates [23], somatic fusion in invertebrates [24], vegetative fusion in fungi [25, 10] or aggregation in slime molds [11, 13]. In systems where this process occurs, non-self recognition systems are often in place to limit fusion to genetically identical or closely related individuals. The great diversity of self-recognition types in M. xanthus appears to be analogous to these eukaryotic systems.
We propose that non-self exclusion between distinct swarms of M. xanthus may be a selected trait that benefits genotypes that would be inferior competitors under conditions of direct cell-cell contact within genetically heterogeneous groups. Our experiments show that strains co-existing in the soil often have dramatically asymmetric fitness values under conditions of direct cell-cell competition during development and that fitness ranks among these strains are strongly hierarchical under our laboratory conditions. Were distinct clonal groups to merge freely in the soil, our data suggest that a small number of competitively superior genotypes would come to dominate within local areas and thus reduce local diversity relative to a scenario in which different clonal groups are able to maintain swarm integrity upon encounter. The ability to prevent territorial invasion by antagonistic genotypes could serve as a general xenophobic strategy [16] to prevent susceptible genotypes from being overtaken by superior competitors that engage in antagonistic and/or exploitative behaviors [6, 26].
In a manner analogous to M. xanthus, starvation induces individual amoebae of the predatory slime mold Dictyostelium discoideum to aggregate and form multicellular fruiting bodies. Aggregating populations first form migrating slugs, which then transform into stalked, spore-bearing fruiting bodies [28, 27]. Despite their similar responses to starvation, M. xanthus and D. discoideum differ in their predatory phase. M. xanthus cells usually remain in dense swarms bound together by extracellular polysaccharides [29] and individuals rarely leave established groups to move alone [30]. Such social cohesion promotes high relatedness when cells deplete local resources and initiate development. In contrast, D. discoideum predation is individualistic [31], making genotypic interspersion during predation and the probability of chimeric coaggregation at the onset of development more likely than in Myxococcus. Even with solitary predation, only a minority of Dictyostelium fruiting bodies in nature appear to be chimeric [17]. When D. discoideum genotypes do coaggregate in nature, the degree of cooperation between them has been found to decrease with increasing genomic divergence [32]. This result stands in stark contrast to our findings with Myxococcus, in which mixing of swarms is strongly binary, with even very closely related strains being able to discriminate and exclude each other.
The molecular basis of the developmental antagonisms and territorial exclusion among M. xanthus clones documented here remains to be characterized, but might involve differential production of diffusible secondary metabolites [33], altered cell-surface chemistry [21] or both. [34]. The strains examined here exhibit a wide variety of secondary metabolite production profiles, and even the genetically most similar strains (A23, A47 and A96) produce different metabolite combinations [34]. Consistent with their higher level of genetic diversity, the global isolates from Fiegna & Velicer (2005) [6] exhibit a greater diversity of metabolite profiles than do the local Tübingen isolates [30], which may contribute to the larger reduction in total spore production observed among mixtures of the global isolates.
This study, as well as previous work on P. mirabilis [20 and 21 and P. aeruginosa [22], suggests that intra-specific allorecognition is commonplace in natural populations of swarming bacteria. Moreover, our results show that such non-self exclusion, as well as antagonism during multicellular development, evolves among very closely related, sympatric genotypes. This finding is consistent with the discovery of single mutations that strongly affect social interactions between mutants and their parents in both M. xanthus [26, 35] and P. mirabilis [21]. Exclusion of genetically distinct strains promotes high relatedness within local groups, which in turn favors the evolution and maintenance of social cooperation and multicellular complexity [1, 12].
Supplementary Material
Acknowledgments
We thank Heike Keller for assistance with gene sequencing and Duur Aanen, Angus Buckling, Kevin Foster, Andy Gardner and three anonymous reviewers for helpful discussion and/or comments on the manuscript. This work was supported by the Max-Planck Society, the Deutsche Forschungsgemeinschaft, the National Institutes of Health (NIH) and a Netherlands Organisation for Scientific Research (NWO) Rubicon grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu. Rev. Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 2.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The social lives of microbes. Annu. Rev. Ecol. Evol. Syst. 2007;38:53–77. [Google Scholar]
- 3.Kaiser D. Signaling in myxobacteria. Annu. Rev. Microbiol. 2004;58:75–98. doi: 10.1146/annurev.micro.58.030603.123620. [DOI] [PubMed] [Google Scholar]
- 4.Nariya H, Inouye M. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell. 2008;132:55–66. doi: 10.1016/j.cell.2007.11.044. [DOI] [PubMed] [Google Scholar]
- 5.Vos M, Velicer GJ. Isolation by distance in the spore-forming soil bacterium Myxococcus xanthus. Curr. Biol. 2008;18:386–391. doi: 10.1016/j.cub.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 6.Fiegna F, Velicer GJ. Exploitative and hierarchical antagonism in a cooperative bacterium. PLoS Biol. 2005;3:e370. doi: 10.1371/journal.pbio.0030370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vos M, Velicer GJ. Genetic population structure of the soil bacterium Myxococcus xanthus at the centimeter scale. Appl. Environ. Microbiol. 2006;72:3615–3625. doi: 10.1128/AEM.72.5.3615-3625.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coyne JA, Orr HA. Speciation. Sunderland: Sinauer Associates; 2004. [Google Scholar]
- 9.Bourke AFG, Franks NR. Social Evolution in Ants. Princeton: Princeton University Press; 1995. [Google Scholar]
- 10.Aanen DK, Debets AJM, de Visser JAGM, Hoekstra RF. The social evolution of somatic fusion. Bioessays. 2008;30:1193–1203. doi: 10.1002/bies.20840. [DOI] [PubMed] [Google Scholar]
- 11.Buss LW. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl Acad. Sci. USA. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grosberg RK, Strathmann RR. The evolution of multicellularity: a minor major transition? Annu. Rev. Ecol. Evol. 2007;38:621–654. [Google Scholar]
- 13.Mehdiabadi NJ, Jack CN, Farnham TT, Platt TG, Kalla SE, Shaulsky G, Queller DC, Strassmann JE. Social evolution: kin preference in a social microbe. Nature. 2006;442:881–882. doi: 10.1038/442881a. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg E, Varon M. Antibiotics and lytic enzymes. In: Rosenberg E, editor. Myxobacteria: Development and Cell Interactions. New York: Springer-Verlag; 1984. pp. 109–125. [Google Scholar]
- 15.Wireman JW, Dworkin M. Developmentally induced autolysis during fruiting body formation in Myxococcus xanthus. J. Bacteriol. 1977;129:798–802. doi: 10.1128/jb.129.2.798-802.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Travisano M, Velicer GJ. Strategies of microbial cheater control. Trends Microbiol. 2004;12:72–78. doi: 10.1016/j.tim.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 17.Gilbert OM, Foster KR, Mehdiabadi NJ, Strassmann JE, Queller DC. High relatedness maintains multicellular cooperation in a social amoeba by controlling cheater mutants. Proc. Natl Acad. Sci. USA. 2007;104:8913–8917. doi: 10.1073/pnas.0702723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Queller DC. Relatedness and the fraternal major transitions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000;355:1647–1655. doi: 10.1098/rstb.2000.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Appleby MC. The probability of linearity in hierarchies. Anim. Behav. 1983;31:600–608. [Google Scholar]
- 20.Pfaller MA, Mujeeb I, Hollis RJ, Jones RN, Doern GV. Evaluation of the discriminatory powers of the Dienes test and ribotyping as typing methods for Proteus mirabilis. J. Clin. Microbiol. 2000;38:1077–1080. doi: 10.1128/jcm.38.3.1077-1080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munson EL, Pfaller MA, Doern GV. Modification of dienes mutual inhibition test for epidemiological characterization of Pseudomonas aeruginosa isolates. J. Clin. Microbiol. 2002;40:4285–4288. doi: 10.1128/JCM.40.11.4285-4288.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buss LW, Green DR. Histoincompatibility in vertebrates: the relict hypothesis. Dev. Comp. Immunol. 1985;9:191–201. doi: 10.1016/0145-305x(85)90110-7. [DOI] [PubMed] [Google Scholar]
- 24.Grosberg RK. The evolution of allorecognition specificity in clonal invertebrates. Q. Rev. Biol. 1988;63:377–412. [Google Scholar]
- 25.Nauta MJ, Hoekstra RF. Evolution of vegetative incompatibility in filamentous ascomycetes. I. Deterministic models. Evolution. 1994;48:979–995. doi: 10.1111/j.1558-5646.1994.tb05287.x. [DOI] [PubMed] [Google Scholar]
- 26.Velicer GJ, Kroos L, Lenski RE. Developmental cheating in the social bacterium Myxococcus xanthus. Nature. 2000;404:598–601. doi: 10.1038/35007066. [DOI] [PubMed] [Google Scholar]
- 27.Bonner JT. The Social Amoebae: The Biology of Cellular Slime Moulds. Princeton: Princeton University Press; 2009. [Google Scholar]
- 28.Strassmann JE, Zhu Y, Queller DC. Altruism and social cheating in the social amoeba Dictyostelium discoideum. Nature. 2000;408:965–967. doi: 10.1038/35050087. [DOI] [PubMed] [Google Scholar]
- 29.Kearns DB, Bonner PJ, Smith DR, Shimkets LJ. An extracellular matrix-associated zinc metalloprotease is required for dilauroyl phosphatidylethanolamine chemotactic excitation in Myxococcus xanthus. J. Bacteriol. 2002;184:1678–1684. doi: 10.1128/JB.184.6.1678-1684.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus (Myxobacterales): Two gene systems control movement. Mol. Gen. Genet. 1979;117:177–191. [Google Scholar]
- 31.Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge: Cambridge University Press; 2001. [Google Scholar]
- 32.Ostrowski EA, Katoh M, Shaulsky G, Queller DC, Strassmann JE. Kin discrimination increases with genetic distance in a social amoeba. PLoS Biol. 2008;6:e287. doi: 10.1371/journal.pbio.0060287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichenbach H. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotech. 2001;27:149–156. doi: 10.1038/sj.jim.7000025. [DOI] [PubMed] [Google Scholar]
- 34.Krug D, Zurek G, Revermann O, Vos M, Velicer GJ, Muller R. Discovering the hidden secondary metabolome of Myxococcus xanthus: A study of intraspecific diversity. Appl. Environ. Microbiol. 2008;74:3058–3068. doi: 10.1128/AEM.02863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiegna F, Yu YT, Kadam SV, Velicer GJ. Evolution of an obligate social cheater to a superior cooperator. Nature. 2006;441:310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.