Abstract
Optimal sample handling techniques for tissue preparation and storage, RNA extraction and quantification, and target gene detection are crucial for reliable gene expression analysis. Methods for measuring low-expressing genes, such as interferons, in human cervical samples are not described in the scientific literature. To detect interferon mRNA in human cervical samples we obtained normal and dysplastic frozen and formalin-fixed cervical biopsies from colposcopy. Histopathological diagnosis was performed by one pathologist. Cervical keratinocytes were isolated using laser capture microdissection. Immortalized keratinocytes transduced with or devoid of an HPV oncogene were used for initial method development. RNA from samples was extracted and integrity tested to compare tissue storage and extraction methods. The expression of five housekeeping genes was analyzed in cell lines and patient tissue to permit normalization between samples using quantitative real-time polymerase chain reaction. The usefulness of cDNA amplification was assessed for the detection of low-expressing interferon κ in cervical tissue. Here we report optimal tissue storage conditions, RNA extraction, sample normalization, and transcript amplification, as well as the sensitivity of quantitative real-time polymerase chain reaction and laser capture microdissection, for interferon κ detection in cervical tissue. Without these optimized techniques, interferon κ detection would be unattainable in cervical samples.
Keywords: Cervical keratinocytes, Frozen and formalin-fixed cervical tissue, Housekeeping genes, Interferons, Interferon κ, Laser capture microdissection, Quantitative real-time polymerase chain reaction
Quantitative real-time polymerase chain reaction (qRT-PCR)1 for the molecular analysis of disease is a powerful and widely used tool. Extraction of high-quality RNA for use in gene expression analysis techniques such as reverse transcription (RT), qRT-PCR, and cDNA microarrays is of great importance. Although obtaining high-quality RNA from cell lines is relatively straightforward, the complexity and heterogeneity of human tissue pose considerable challenges for linking gene expression patterns with disease state. Nonetheless, ex vivo studies using human tissue samples are important for the validation of in vitro work and to closely mimic relevant biological processes. In recent years, the introduction of laser capture microdissection (LCM) has greatly enhanced the specificity of the molecular analysis of cell types within a sample. However, obtaining sufficient quantities of high-quality RNA from microdissected cells represents an additional obstacle. Performing reliable gene expression studies requires stringent monitoring of sample integrity during sample preparation, RNA extraction, and qRT-PCR steps.
Poor tissue storage conditions can degrade RNA [1]. Formalin-fixed, paraffin-embedded (FFPE) tissue produces optimal morphology for histological assessment, but this technique may not preserve RNA integrity [2]. The literature suggests that frozen (FR) tissue is optimal for the extraction of high-quality RNA [2,3]; however, this has not been shown for FR cervical tissue. Numerous techniques and commercially available kits exist for RNA extraction from FR tissue [4-7], and some have been used for cervical specimen extraction [8-10]. However, the integrity of the RNA and the reproducibility of the techniques are often overlooked. Current literature does not provide a consensus for the optimal protocol for high-quality RNA extraction, particularly from FR cervical specimens. In addition, the importance of LCM techniques for studying specific cell populations has resulted in the optimization of tissue preparation protocols specifically for LCM [11-13], but it has not been demonstrated that these work well for cells obtained from cervical tissue.
The quality of data obtained from qRT-PCR experiments is greatly influenced by RNA integrity, RT conditions, reference gene selection, and transcript abundance. The type of primers used for RT can also significantly alter the cDNA yield and specificity [14]. Although normalization of qRT-PCR-derived expression levels requires an appropriate reference or housekeeping gene (HKG) for compensating for differences between samples [15,16], HKG validation is required for each tissue type [17]. Furthermore, low transcript amounts represent an obstacle for simultaneous analysis of multiple targets or low-expressing genes, and thus, several methods exist for amplifying mRNA transcripts extracted from tissue samples [18-21]. However, none of these parameters have been described or optimized for gene expression analysis in cervical samples.
Human papillomavirus (HPV) infection is necessary for the formation of cervical premalignant lesions (i.e., cervical dysplasia), and chronic infection is required for progression to cervical cancer. We are interested in characterizing the mRNA transcript levels of low-expressing interferons in cervical tissue infected with HPV, with a particular emphasis on the novel interferon κ (IFN-κ), as its expression has never been established in ex vivo HPV-infected cervical tissue. The requisite methods to measure low-expressing cytokines in small amounts of cervical tissue samples are currently not described in the literature, which made it compulsory to develop and/or optimize these methods ourselves. Using stringent sample integrity monitoring, we describe the optimal methods for sample storage, RNA extraction, cDNA amplification, and qRT-PCR for the detection of low-abundant interferon genes in cervical biopsy tissue and laser capture microdissected cervical keratinocytes. Without this method optimization, IFN-κ, would have been undetectable in cervical samples, minimizing its role in cervical disease and, thus, profoundly affecting the outcome of this research.
Materials and methods
Sample preparation
Cell lines
Near-diploid immortalized foreskin keratinocytes (NIKS) [22] in the presence or absence of the human papillomavirus type 16 (HPV16) E6 oncogene [23] were grown in culture to 70% confluency. These cells were harvested and used to mimic normal and diseased cervical tissue in initial experiments to assess HKG suitability and cDNA amplification uniformity.
Biopsy material
FR and FFPE cervical biopsies were obtained with written consent from women attending the Colposcopy Clinic at the Thunder Bay Regional Health Sciences Centre between November 2005 and November 2006. Biopsies were either snap-frozen in liquid nitrogen and transferred to −80 °C or immediately submerged in 10% (v/v) buffered formaldehyde solution for 3 h at room temperature. The tissue was then dehydrated by submerging it first into ascending grades of ethanol (70, 80, 95, and 100% v/v in water) and then into 100% (v/v) xylene, followed by wax impregnation with paraffin [24]. FR cervical tissue was sectioned on a cryostat (Leica CM1850, Leica Microsystems, Richmond Hill, ON, Canada), maintaining tissue temperature at −20 °C using Tissue Tek embedding medium (O.C.T. Compound, Sakura Finetek, Torrance, CA, USA), while FFPE cervical tissue was sectioned using a microtome (Leica 1720 digital microtome, Leica Microsystems). Tweezers, brushes, and surfaces were cleaned with diethylpyrocarbonate-treated 70% (v/v) ethanol between specimens to reduce RNase activity and RNA cross-contamination. Ten-micrometer-thick tissue sections were processed for hematoxylin and eosin staining [24] and subsequent histopathological diagnosis by the same pathologist (NE). Tissue samples were diagnosed as morphologically normal tissue or cervical dysplasia.
RNA extraction
RNA was extracted from 10 × 10-μm-thick FFPE and FR cervical tissue sections as well as from 1 × 106 NIKS using three different methods: Sigma TriReagent (Sigma–Aldrich), following the recommended protocol for extraction using 1 ml of TriReagent with the addition of 20 μg of RNase-free glycogen carrier (Fermentas Lifesciences, Burlington, ON, Canada) and with 2 U of terminal DNase treatment (Ambion); the Ambion RNAqueous-4PCR (Ambion Inc., Austin, TX, USA), following the described method for extraction from FR tissue/cell pellets; and the Arcturus PicoPure RNA Isolation Kit, following the protocol described for isolation from CapSure Macro LCM Caps (Arcturus) with the addition of 13.6 Kunitz units of RNase-free DNase treatment (Qiagen, Catalog No. 79254) and slight modifications when using tissue sections [25]. Briefly, 100 μl of extraction buffer (XB) was added to samples and incubated at 42 °C for 30 min. The addition of an extra 50 μl of XB made tissue homogenization more manageable when working with tissue sections. One hundred microliters of 70% (v/v) ethanol was then added (1:1 ratio with XB) to the cell extract, and RNA isolation was performed as described in the protocol. The Arcturus kit was also used for RNA extraction from LCM-derived cervical keratinocytes from 8-μm-thick FR tissue following the recommended protocol for use with CapSure Macro LCM caps (Arcturus).
RNA integrity assessment
The quality and quantity of RNA extracted from cell lines and cervical tissue were assessed using the Bio-Rad Experion Automated Electrophoresis System. RNA integrity from samples was assessed by visual inspection of the electropherograms [26] as well as the digital gel images. Samples possessing distinct 18S and 28S ribosomal peaks in the electropherogram [26], as well as sharp, abundant 18S and 28S ribosomal RNA (rRNA) bands in the gel images, are indicative of high RNA integrity. Calculated rRNA 28S/18S ratios did not always correlate with superior images [27] and are therefore reported here but were not used for sample integrity assessment.
High-sensitivity chips (Experion RNA HighSens Analysis Kit, Bio-Rad) were used to analyze RNA from LCM-obtained samples and from FFPE and FR cervical biopsy sections, whereas standard-sensitivity chips (Experion RNA StdSens Analysis Kit, Bio-Rad) were used to assess RNA from cell lines. When specified, initial cell line cDNA was quantified using a Nanodrop spectrophotometer (ND-1000, NanoDrop Technologies, Thermo Fisher Scientific, Wilmington, DE, USA), and RNA integrity was later confirmed using the Experion (data not shown).
Laser capture microdissection
The Arcturus Histogene Frozen Section Staining Kit was used to prepare samples for microdissection. FR biopsy sections were fixed, stained, and dehydrated following the Arcturus method with slight modifications. Briefly, 1× ProtectRNA (ProtectRNA RNase Inhibitor 500× concentrate, Catalog No. R7397, Sigma-Aldrich) was added to the staining solution [25], and tissue was dehydrated as normal with the addition of an extra 100% (v/v) ethanol step to acquire intact RNA [11]. Tissue sections were allowed to dry for up to 3 h prior to capturing 5000–25,000 cells in 1000–5000 captures using CapSure Macro LCM caps. Typically, one tissue section was adequate to obtain 1000 LCM captures of cervical keratinocytes, and four or five sections were required to obtain 5000 LCM captures. Macro caps were cleaned of unwanted debris using CapSure pads (Arcturus) prior to being deposited into 0.5-ml microfuge tubes for RNA extraction. The laser spot size was consistently 15 μm in diameter, whereas the laser power and duration varied slightly between sections (ranging from 80 to 95 mW and 0.65 to 0.8 ms in duration). LCM images were taken at 10× magnification.
Reverse transcription and transcript amplification
RNA isolated from samples was reverse transcribed to cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems) according to the manufacturer’s directions with random hexamer primers. All RT reactions were performed in 18-μl volume. RNA from cell lines whose cDNA was not amplified was reverse transcribed at 20 ng/μl, whereas RNA from cell lines requiring cDNA amplification and cervical biopsy tissue was reverse transcribed at 4 ng/μl. RNA isolated from microdissected cervical samples was reverse transcribed at 0.06-0.6 ng/μl, depending on acquired LCM sample RNA concentration.
Fifty nanograms of cDNA from NIKS and whole biopsy sections, as well as 0.75 or 7.5 ng of cDNA from 1000 and 5000 LCM captures, respectively, was amplified using the TaqMan PreAmp Master Mix Kit (Applied Biosystems), unless otherwise specified. For amplification uniformity assessment, 7.5 ng of cDNA from NIKS and biopsy sections, as well as 2.5 ng of cDNA from a laser capture microdissected sample, was amplified and compared with 7.5 ng of unamplified cDNA from NIKS and biopsy material and 1.8 ng of unamplified cDNA from the laser capture microdissected sample. NIKS were used in initial experiments to determine the optimal amplification conditions. Uniform transcript amplification was assessed within each sample by comparing unamplified versus amplified gene expression between two genes using the ΔΔCt method, as described (Applied Biosystems TaqMan PreAmp Master Mix Kit Protocol, Appendix A: Checking Preamplification Uniformity). A ΔΔCt value of 0 ± 1.5 indicated uniform amplification.
Quantitative real-time polymerase chain reaction
Reactions were carried out according to the qRT-PCR protocol specified in the TaqMan PreAmp Master Mix Kit. Amplified or unamplified cDNA (6.25 μl) was added to each reaction. Triplicate reactions of 25-μl volume were added to a 96-optical well plate (Applied Biosystems) and incubated under standard qRT-PCR conditions (50 °C for 5 min, 95 °C for 10 min, and then cycled at 95 °C for 15 s, and 60 °C for 1 min, for 40 cycles). Candidate HKGs were chosen based on an extensive literature review. The optimal HKG for normalization in cervical samples was determined using the relative expression software tool (REST) [28-30] with reaction efficiencies set to one and target gene values set to zero. P values ≤0.05 obtained with REST indicate significant differences in expression. TaqMan gene expression assays for hypoxanthine phosphoribosyltransferase 1 (HPRT1), actin, phospholipase A (PLA), 18S ribosomal subunit (18S), β2-microglobulin (B2M), IFN-γ (79-bp amplicon), IFN-β (73-bp amplicon), and IFN-κ (108-bp amplicon) were used. Negative controls in which cDNA was omitted or the enzyme was omitted from the RT reaction were run to monitor for contamination or nonspecific primer binding. A positive control was included on every plate to control for variation between runs. After demonstrating that its expression was unaffected by HPV infection, target Ct values were normalized to HPRT1. Relative quantification of target genes was performed using auto Ct and baseline settings and a threshold of 0.20 (Applied Biosystems 7300/7500/7500 Fast Real-Time PCR System Software).
Results
Sample storage, RNA extraction, and RNA integrity
RNA degradation, resulting from inadequate tissue storage or extraction techniques, could significantly alter the resulting transcript profile within a sample. Several methods exist to assess RNA integrity, including denaturing gel electrophoresis, UV spectrophotometry, and microfluidics. Traditional gel electrophoresis requires large amounts of RNA to visualize ribosomal bands, whereas UV spectrophotometry assesses sample purity not RNA integrity. Microfluidic gel electrophoresis, such as the Bio-Rad Experion, permits integrity assessment of small amounts of RNA. This system was therefore used to assess RNA integrity in cervical biopsies stored under different conditions and extracted using various techniques, as well as in laser capture microdissected cervical epithelium.
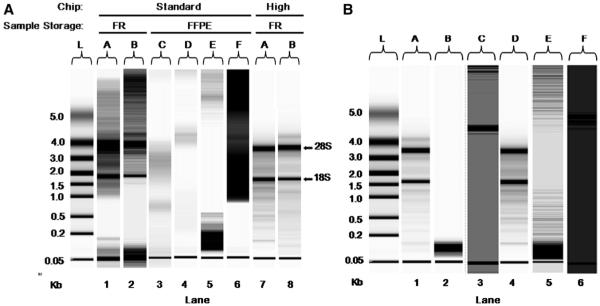
RNA extracted from FR cervical biopsy tissue showed higher integrity than RNA extracted from FFPE cervical tissue (Fig. 1A). With standard-sensitivity Experion chips (lanes 1–6), RNA integrity was assessed in two FR samples (lanes 1 and 2) and four FFPE samples (lanes 3–6). No visible rRNA bands were found in the FFPE samples. However, as seen in lanes 7 and 8, the accuracy of the integrity assessment was greatly improved for the intact FR samples when high-sensitivity Experion chips were used, as demonstrated by the more pronounced rRNA bands shown. These data demonstrate that FR tissue specimens preserved the integrity of RNA, whereas formalin fixation procedures degrade it. However, of all FR biopsy specimens tested, only 74% yielded high-quality RNA (data not shown), indicating that RNA integrity is not equal between samples despite identical extraction and storage conditions.
Fig. 1.
RNA integrity of cervical tissue using different sample storage and extraction methods. (A) Integrity of RNA in FR versus FFPE cervical tissue. RNA was extracted from FR cervical biopsy sections (lanes 1, 2, 7, 8) and FFPE cervical sections (lanes 3–6) using the Arcturus PicoPure RNA Isolation Kit. RNA in lanes 1 through 6 was measured using standard-sensitivity Experion chips. RNA from FR samples A and B was remeasured using an Experion high-sensitivity chip (lanes 7 and 8) resulting in 28S/18S rRNA ratios of 0.83 and 1.49, respectively. (B) Integrity of RNA from six frozen cervical biopsies extracted using three different extraction kits. The Arcturus (lanes 1 and 4), Sigma (lanes 2 and 5), and Ambion (lanes 3 and 6) extraction kits were tested. Experion standard-sensitivity chips (lanes 3 and 6) and high-sensitivity chips (lanes 1, 2, 4, 5) were used. “L” represents the RNA ladder (kb). Representative gel images are shown. The 28S/18S rRNA ratios for samples A and D were 1.49 and 0.83, respectively. Two bands represent 28S (top) and 18S (bottom) rRNA.
The Ambion extraction method is based on guanidinium thiocyanate cell disruption and silica-based filtration, whereas the TriReagent method involves phenol and guanidinium thiocyanate RNA extraction and simultaneous preservation, chloroform isolation, and isopropanol precipitation. The Arcturus isolation kit components were not revealed by the manufacturer; however, RNA extracted from FR cervical biopsy sections was of highest quality when using this method (Fig. 1B). No visible rRNA bands were present when the Ambion or Sigma method was used for extraction. Thus, the Arcturus method was optimal for the extraction of high-quality RNA from small amounts of FR cervical biopsy tissue. Conversely, RNA yield and quality from NIKS were highest when the Ambion method was used (data not shown).
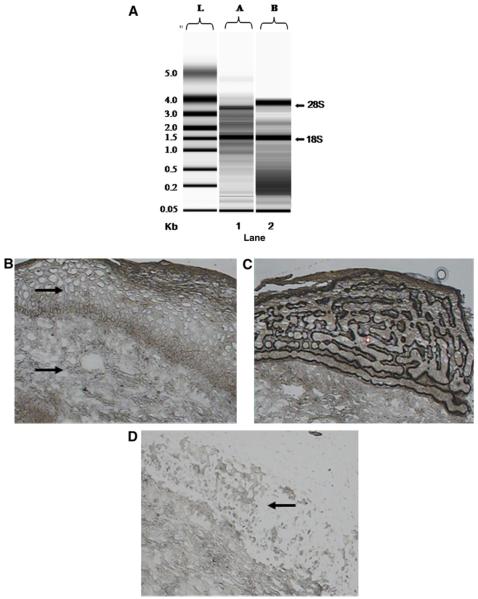
LCM selects for specific cells within a heterogenous cell population. When target genes of interest are cell type specific, such as IFN-κ, which is expressed mainly in epithelial cells [31], it is crucial to isolate only those cells of interest to gain an accurate assessment of the gene expression profile within a tissue microecology. Obtaining intact RNA from microdissected samples is equally as important for reliable mRNA analysis. Cervical biopsy tissue prepared for LCM using the Arcturus method allowed isolation of high-quality RNA from cervical keratinocytes using 1000 or 5000 LCM captures (Fig. 2A), as distinct 28S and 18S rRNA bands can be seen in both laser capture microdissected samples. Furthermore, the morphology of the tissue was adequate for differentiation between different cell types within the tissue (Fig. 2B–D), as cervical keratinocytes were easily differentiated from surrounding stroma. These data demonstrate that preparation of laser capture microdissected tissue using the reported method provides suitable tissue morphology and high-quality RNA for use in downstream applications.
Fig. 2.
RNA integrity and tissue morphology of laser capture microdissected cervical tissue. (A) Intact RNA obtained from 1000 (lane 2: 190 pg/μl RNA, 28S/18S = 0.38) and 5000 (lane 3: 630 pg/μl, 28S/18S = 1.50) laser capture microdissected cervical keratinocytes. Two bands represent 28S (top) and 18S (bottom) rRNA. (B) Cervical biopsy section illustrating cervical keratinocytes (top arrow) and stroma (bottom arrow). (C) One thousand LCM captures before film was lifted from slide. (D) Excised epithelial region after film was lifted from slide (black arrow). Representative gel and histological images are shown.
Reference gene suitability for qRT-PCR
The use of HKGs for normalization between samples is critical, especially when using heterogenous tissue samples. Although all qRT-PCR studies use HKGs for normalization between samples, few studies determine optimal reference genes for specific tissues. Expression of the HKGs HPRT1, β-actin, 18S, B2M, and PLA was analyzed in NIKS in the presence and absence of an HPV16 oncogene (Fig. 3A). HPRT1, B2M, and PLA mRNA levels were statistically similar (P ≥ 0.05) in the two cell lines. By contrast, β-actin and 18S genes differed significantly in expression (P < 0.05). The similarity of HPRT1 and PLA gene expression in normal (n = 4) and dysplastic (n = 12) cervical biopsy tissue (Fig. 3B) demonstrates their suitability as reference genes in cervical tissue. However, similar to the cell line data, HPRT1 (P = 0.796) had a higher P value than PLA (P = 0.728) and, thus, represented a better choice to use for expression normalization when comparing normal and diseased cervical tissue.
Fig. 3.
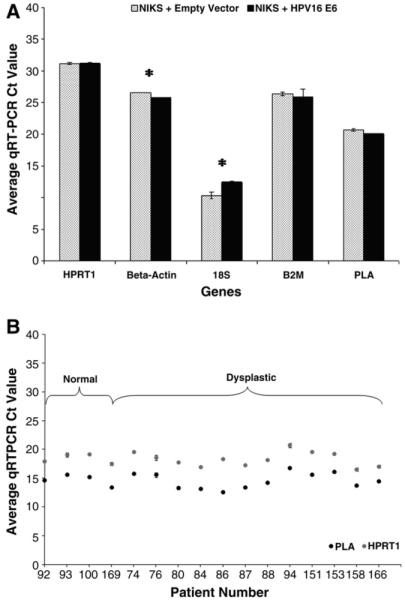
Housekeeping gene suitability for normalization across cervical disease states. (A) HPRT1, β-actin, 18S, B2M, and PLA housekeeping gene expression in NIKS ± HPV16 E6 measured using qRT-PCR and unamplified cDNA. HPRT1 (P = 0.955), B2M (P = 0.367), and PLA (P = 0.065) were expressed the same in the cell lines. cDNA was quantified using a Nanodrop. (B) HPRT1 and PLA housekeeping gene expression in normal (n = 4: patients 92, 93, 100, 169) and dysplastic (n = 12: patients 74–166) cervical tissue. HPRT1 (P = 0.796) and PLA (P = 0.728) are expressed the same in the control and dysplastic groups. Values represent raw average qRT-PCR Ct value ± SD using amplified cDNA. Asterisks indicate significant difference in expression (P ≤ 0.05) between the cell lines. cDNA input concentrations were determined using the Experion and are not always equal between genes.
Uniform transcript amplification
Complementary DNA amplification involves the uniform amplification of specific target genes of interest within a sample and increases the ability to detect low-expressing gene transcripts within a suitable range using qRT-PCR. The Taqman PreAmp Master Mix Kit (Applied Biosystems) uniformly amplified HKG cDNA transcripts within NIKS, whole cervical biopsy sections, and microdissected cervical epithelium (Fig. 4), as all ΔΔCt values were less than 1.0. Target IFNs also show uniform amplification in NIKS (data not shown). With the use of NIKS, the optimal cDNA amplification conditions were determined to be 14 cycles using 50 ng of unamplified cDNA, as a negative affect on the ΔΔC t value was not observed when the cDNA concentration was lowered and the cycle number was increased (data not shown).
Fig. 4.
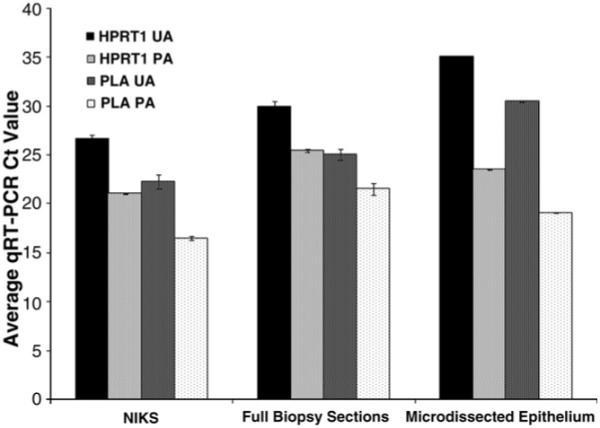
Uniform cDNA amplification in cervical samples. HPRT1 and PLA gene expression in NIKS, cervical biopsy sections, and laser capture microdissected cervical keratinocytes was analyzed before (UA) and after (PA) cDNA amplification using qRT-PCR. Amplification uniformity was assessed using the ΔΔCt method. HPRT1 and PLA in NIKS (ΔΔCt = 0.03), cervical sections (ΔΔCt = 0.93), and microdissected cervical keratinocytes (ΔΔCt = 0.14), respectively, were amplified uniformly. Representative cases are shown for tissue samples. Five thousand LCM captures were taken for microdissected cervical samples. Values represent raw qRT-PCR Ct value ± SD. cDNA input concentrations were determined using the Experion.
Sensitivity of qRT-PCR and LCM
Quantitative RT-PCR is a method that allows for the simultaneous detection, real-time amplification, and quantification of RNA transcripts. With the TaqMan (Applied Biosystems) methodology and reagents for gene expression analysis, this method was used to analyze the gene expression of HKGs and target genes in samples to determine transcript input and LCM sensitivity for the detection of low-expressing IFNs. Whole biopsy sections from three cases of cervical dysplasia were analyzed by qRT-PCR (Fig. 5A). With the methods described, all IFNs were within a detectable range for qRT-PCR analysis. In general, the expression of IFNs followed the order IFN-κ < IFN-β < IFN-γ. One thousand LCM captures (~1 ng of total RNA) were sufficient to measure HKGs and target genes present at only a moderate level, such as IFN-γ, in cervical epithelium (Fig. 5B). However, for the detection of low-expressing genes, such as IFN-β and IFN-κ, 5000 LCM captures were required (~10 ng of total RNA).
Fig. 5.
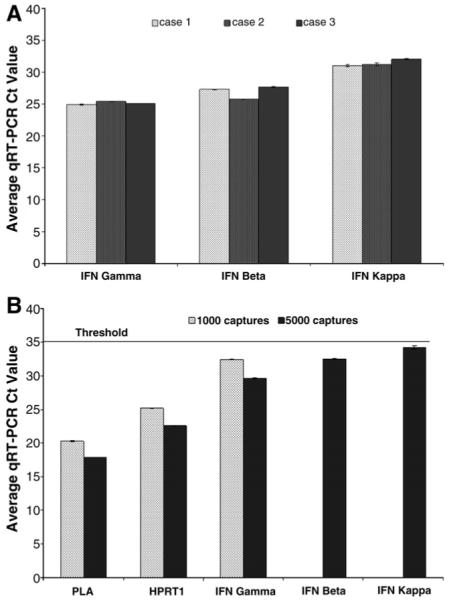
Sensitivity of qRT-PCR and LCM for the detection of interferon gene expression in dysplastic cervical tissue. (A) Three cases of cervical dysplasia were analyzed for IFN gene expression using qRT-PCR and 50 ng of RNA from whole biopsy sections. cDNA input values were determined from RNA values measured using the Experion. (B) Housekeeping and target IFN mRNA detection using qRT-PCR and 1000 and 5000 LCM captures of cervical keratinocytes yielding 1 and 10 ng of RNA. Values represent raw qRT-PCR Ct value ± SD. cDNA input concentrations were determined using the Experion. The horizontal dashed line represents the upper limit for qRT-PCR detection. A representative case is shown.
Discussion
In this study, using cell lines, cervical tissue, and microdissected cervical keratinocytes, we assessed various sample preparation and analysis techniques that can influence gene expression studies to devise optimal methods for analyzing low-abundant genes in cervical samples. First, the method of tissue storage was assessed for the preservation of high-quality RNA in cervical biopsy specimens. In agreement with previous studies [2,3,32], our data strongly suggested that FR cervical tissue was optimal for obtaining a high quality and quantity of RNA (Fig. 1A), whereas FFPE tissue processing caused considerable degradation and low yield, possibly as a result of nucleic acid crosslinking with proteins, covalent modifications, and strand breaks [33]. RNA from FFPE tissue was not suitable for gene analysis. For the detection of quality and quantity of RNA in the range 20–40 ng/μl, the Experion high-sensitivity kit produced more reliable results than the standard kit.
Methods for extracting RNA differ with respect to reagents, filter columns, and the amount of sample required. Furthermore, many published techniques claiming high-quality RNA isolation from FR tissue either relied only on expression of HKGs [2,3] and A260/A280 ratios [34,35] or did not perform RNA quality assessment at all [8,10,36]. Here we show that contrary to previous results with FR tissue [4,37-39], the modified Arcturus RNA extraction method was optimal for the extraction of high-quality RNA compared with the Ambion and Sigma methods for cervical biopsy sections (Fig. 1B). Although limited material prohibited a direct comparison of all techniques with the same sample, representative images are shown for each extraction method (Fig. 1B). The Trizol extraction method had previously been shown to produce poorer-quality RNA when using small amounts of cervical cells [9]. Small amounts of sample material may limit RNA yield for some extraction methods. Although the amount of tissue used was in the range specified by the Ambion and Sigma kits, high-quality RNA was only obtained using the Arcturus kit, which is designed for extraction from microdissected samples. This indicates that for extraction from small amounts of cervical tissue, as seen previously in prostate tissue [25], methods optimized for extraction from microdissected samples produced favorable results.
Tissue preparation protocols for LCM and FR tissue often differ in many respects, including the dehydration procedure and the staining solutions. Recent evidence suggested that inclusion of an RNase inhibitor [25], as well as a prolonged 100% (v/v) ethanol incubation [11,25], generated higher-quality RNA from microdissected samples. It was previously argued that the protocol described in the Arcturus Histogene Frozen Section Staining Kit is not adequate for obtaining high-quality RNA from FR laser capture microdissected samples [11]. This could be due to active RNases present in the tissue and/or improper dehydration leading to RNA degradation. Together with recent results using similar methodology and FR specimens [12,25], our data demonstrate that a modified version of the Arcturus staining method combined with the Arcturus extraction method is highly suitable for morphological assessment and high-quality RNA extraction from LCM-obtained cervical keratinocytes (Fig. 2). This finely tuned technique is a significant advancement, as obtaining intact RNA from laser capture microdissected samples is often challenging, and our modification may be extended for the preparation of other FR tissues for LCM. RNAlater-ICE (Ambion), tested for its impact on RNA integrity preservation, rendered the biopsy specimen difficult to section and had no impact on sample RNA integrity (data not shown). In addition, cervical tissue scraped off the slide displayed similar HKG expression compared with LCM-obtained cervical material from the same specimen (data not shown), indicating that the microdissection process did not compromise RNA integrity.
HPRT is a commonly used reference gene, previously used for the study of low-level cytokine expression [17] and cervical cancer [40]. HPRT has been verified as an optimal normalizer between samples in colon, prostate, breast, skin, and bladder tissue [41], but had not been established as a suitable normalizer between normal and diseased cervical tissue. We demonstrated that HPRT1 is the optimal HKG for normalization compared with β-actin, B2M, 18S, and PLA using REST software, which employs a Pair Wise Fixed Reallocation Randomization Test and is recommended for experiments demanding highly sensitive, specific, and reproducible mRNA quantification [28]. This was shown in NIKS transduced or devoid of the HPV16 E6 oncogene (Fig. 3A), as well as in cervical tissue in the presence or absence of HPV (Fig. 3B). Relatively constant expression of HKGs in specimens is also indicative of sample integrity at this stage of sample processing. We found that PLA was also a suitable HKG in cervical tissue, though its expression may vary in the presence of IFN [42] and, therefore, may change in virus-infected tissue. HPRT was reported to represent the mean expression of many commonly used normalizing genes [41], eliminating the need to use multiple genes for normalization. It is suitable for use with sensitive detection methods [43], and, similar to our data, HPRT has been shown to exhibit a low expression level suitable for the measurement of low-expressing genes [41], such as IFN-κ. Random hexamer primers were chosen for RT of RNA to cDNA, as these primers resulted in minimal differences in HKG expression between samples and slightly higher mRNA levels compared with RT using oligo(dT) primers (data not shown). Random hexamer primers are thus ideal when preparing samples for the comparison of low-expressing gene levels between pathological conditions.
Although several methods exist for transcript amplification, they are often tedious and time consuming. Complementary DNA amplification is comparatively simple, but its utility has never been reported in cervical samples for the analysis of multiple low-expressing target genes. Our data demonstrated that cDNA amplification significantly increased the expression level of target genes within NIKS, whole biopsy specimens, and laser capture microdissected samples (Fig. 4). Furthermore, determining the optimal amplification conditions (i.e., the number of cycles and the least amount of input cDNA required for target mRNA detection using qRT-PCR) greatly increased the ability of our techniques to measure target genes in limited amounts of cervical samples. We have shown that a reasonable amount of RNA isolated from cervical specimens, with the aid of cDNA amplification, permitted detection of all IFNs within a suitable qRT-PCR level (≤35Ct) in both whole cervical sections (Fig. 5A) and microdissected cervical keratinocytes (Fig. 5B). IFN-κ gene transcripts were not detectable in cervical biopsy tissue or microdissected keratinocytes using unamplified cDNA (data not shown) and, along with IFN-β, were not detected in microdissected cervical keratinocytes using amplified cDNA from 1000 LCM captures (Fig. 5B). This is a significant sensitivity advancement, and to our knowledge, this is the first time IFN-κ detection has been demonstrated in cervical keratinocytes from human cervical biopsy specimens. Additionally, the relationship between IFN-κ and other prominent IFN gene expression in ex vivo cervical material has not previously been shown. IFN-κ mRNA was expressed at a lower level with respect to IFN-β and IFN-γ (Fig. 5A and B).
Although present at low levels within the cervical microenvironment, IFN-κ gene expression is profoundly influenced by HPV infection compared with other IFNs (DeCarlo et al., unpublished data), demonstrating the biological relevance of detecting this IFN at low levels. By use of the methods described, preliminary investigations involving microdissected cervical tissue revealed a link between cervical disease and IFN-κ upregulation (DeCarlo et al., unpublished data). Along with an upregulation of IFN-κ expression, this technique allows for the discrimination of specific cell populations involved in the observed change in expression. LCM revealed a correlation between gene expression of IFN-κ in stromal cells and cervical disease progression (data not shown). Further investigations are required to establish the link between HPV and IFN-κ; however, based on initial data, toll-like receptor pathways represent an intriguing bridge between HPV infection and innate immunity mediated by IFN-κ (DeCarlo et al., unpublished data).
The development of precise methodology for measurement of cell-specific targets is instrumental for experimental pathologists to investigate the molecular profile of disease. The methods described herein were crucial for the comprehensive analysis of interferons in cervical tissue and likely could be extended to other keratinocyte-based investigations. Combined with high-throughput gene expression technologies, such as cDNA microarrays, these methods could lead to improvements in immunological profiling and disease diagnosis.
Acknowledgments
We thank the Northern Cancer Research Foundation and the Ontario Graduate Scholarship Program for generously funding our work. The assistance of the TBRHSC central laboratory staff was greatly appreciated.
Footnotes
- B2M
- β2-microglobulin
- FFPE
- formalin-fixed paraffin-embedded
- FR
- frozen
- HKG
- housekeeping gene
- HPRT1
- hypoxanthine phosphoribosyltransferase 1
- HPV
- human papillomavirus
- HPV16
- human papillomavirus type 16
- IFN
- interferon
- LCM
- laser capture microdissection
- NIKS
- near-diploid immortalized keratinocytes
- PLA
- phospholipase A
- qRT-PCR
- quantitative real-time polymerase chain reaction
- REST
- relative expression software tool
- RT
- reverse transcription
- XB
- extraction buffer
References
- [1].Holland NT, Smith MT, Eskenazi B, Bastaki M. Biological sample collection and processing for molecular epidemiological studies. Mutat. Res. 2003;543:217–234. doi: 10.1016/s1383-5742(02)00090-x. [DOI] [PubMed] [Google Scholar]
- [2].Karsten SL, Van Deerlin VMD, Sabatti C, Gill LH, Geschwind DH. An evaluation of tyramide signal amplification and archived fixed and frozen tissue in microarray gene expression analysis. Nucleic Acids Res. 2002;30:2e4. doi: 10.1093/nar/30.2.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hiller T, Snell L, Watson PH. Microdissection RT-PCR analysis of gene expression in pathologically defined frozen tissue sections. Biotechniques. 1996;21:38–44. doi: 10.2144/96211bm07. [DOI] [PubMed] [Google Scholar]
- [4].Wallard MJ, Pennington CJ, Veerakumarasivam A, Burtt G, Mills IG, Warren A, et al. Comprehensive profiling and localization of the matrix metalloproteinases in urothelial carcinoma. Br. J. Cancer. 2006;94:569–577. doi: 10.1038/sj.bjc.6602931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Strand AD, Aragaki AK, Baquet ZC, Hodges A, Cullingham P, Holmans P. Conservation of regional gene expression in mouse and human brain. PLoS Genet. 2007;3:e59. doi: 10.1371/journal.pgen.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Krizman DB, Chuaqui RF, Meltzer PS, Trent JM, Duray PH, Linehan WM. Construction of a representative cDNA library from prostatic intraepithelial neoplasia. Cancer Res. 1996;56:5380–5383. [PubMed] [Google Scholar]
- [7].Wang YX, Martin-McNulty B, Freay AD, Sukovich DA, Halks-Miller M, Li WW. Angiotensin II increases urokinase-type plasminogen activator expression and induces aneurysm in the abdominal aorta of apolipoprotein E-deficient mice. Am. J. Pathol. 2001;159:1455–1464. doi: 10.1016/S0002-9440(10)62532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kendrick JE, Conner MG, Huh WK. Gene expression profiling of women with varying degrees of cervical intraepithelial neoplasia. J. Low. Genit. Tract Dis. 2007;11:25–28. doi: 10.1097/01.lgt.0000230124.68996.38. [DOI] [PubMed] [Google Scholar]
- [9].Lamarcq L, Deeds J, Ginzinger D, Perry J, Padmanabha S, Smith-McCune K. Measurements of human papillomavirus transcripts by real-time quantitative reverse transcription-polymerase chain reaction in samples collected for cervical cancer screening. J. Mol. Diagn. 2002;4:97–102. doi: 10.1016/S1525-1578(10)60687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Futakuchi H, Ueda M, Kanda K, Fujino K, Yamaguchi H, Noda S. Transcriptional expression of survivin and its splice variants in cervical carcinomas. Int. J. Gynecol. Cancer. 2007;17:1092–1098. doi: 10.1111/j.1525-1438.2007.00833.x. [DOI] [PubMed] [Google Scholar]
- [11].Morrogh M, Olvera N, Bogomolniy F, Borgen PI, King TA. Tissue preparation for laser capture microdissection and RNA extraction from fresh frozen breast tissue. Biotechniques. 2007;43:41–48. doi: 10.2144/000112497. [DOI] [PubMed] [Google Scholar]
- [12].Wang H, Owens JD, Shih JH, Li MC, Bonnerm RF, Mushinski JF. Histological staining methods preparatory to laser capture microdissection significantly affect the integrity of the cellular RNA. BMC Genomics. 2006;7:97–104. doi: 10.1186/1471-2164-7-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kihara AH, Moriscot AS, Ferreira PJ, Hamassaki DE. Protecting RNA in fixed tissue: an alternative method for LCM users. J. Neurosci. Methods. 2005;148:103–107. doi: 10.1016/j.jneumeth.2005.04.019. [DOI] [PubMed] [Google Scholar]
- [14].Zhang J, Byrne CD. Differential priming of RNA templates during cDNA synthesis markedly affects both accuracy and reproducibility of quantitative competitive reverse-transcriptase PCR. Biochem. J. 1999;337:231–241. [PMC free article] [PubMed] [Google Scholar]
- [15].Karge WH, 3rd, Schaefer EJ, Ordovas JM. Quantification of mRNA by polymerase chain reaction (PCR) using an internal standard and a nonradioactive detection method. Methods Mol. Biol. 1998;110:43–61. doi: 10.1385/1-59259-582-0:43. [DOI] [PubMed] [Google Scholar]
- [16].Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–337. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- [17].Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- [18].Ginsberg SD, Che S. RNA amplification in brain tissues. Neurochem. Res. 2002;27:981–992. doi: 10.1023/a:1020944502581. [DOI] [PubMed] [Google Scholar]
- [19].Day RC, McNoe L, McKnight RC. Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser microdissected endosperm. Int. J. Plant Genomics. 2007:61028. doi: 10.1155/2007/61028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kurimoto K, Yabuta Y, Ohinata Y, Saitou M. Global single-cell cDNA amplification to provide a template for representative high-density oligonucleotide microarray analysis. Nat. Protoc. 2007;2:739–752. doi: 10.1038/nprot.2007.79. [DOI] [PubMed] [Google Scholar]
- [21].Noutsias M, Rohde M, Block A, Klippert K, Lettau O, Blunert K. Preamplification techniques for real-time RT-PCR analyses of endomyocardial biopsies. BMC Mol. Biol. 2008;9(2):3. doi: 10.1186/1471-2199-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Allen-Hoffmann BL, Schlosser SJ, Ivarie CAR, Sattler CA, Meisner LF, O’Connor SL. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line. NIKS, J. Invest. Dermatol. 2000;114:444–445. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- [23].Lichtiq H, Alqrisi M, Botzer LE, Abadi T, Verbitzky Y, Jackman A. HPV16 E6 natural variants exhibit different activities in functional assays relevant to the carcinogenic potential of E6. Virology. 2006;350:216–227. doi: 10.1016/j.virol.2006.01.038. [DOI] [PubMed] [Google Scholar]
- [24].Culling CFA, Allison T, Barr WT, editors. Cellular Pathology Technique. fourth ed. Butterworths; London: 1984. pp. 55–59. [Google Scholar]
- [25].Kube DM, Savci-Heijink CD, Lamblin AF, Kosari F, Vasmatzis G, Cheville JC. Optimization of laser capture microdissection and RNA amplification for gene expression profiling of prostate cancer. BMC Mol. Biol. 2007;8:25–38. doi: 10.1186/1471-2199-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schroeder A, Mueller O, Stocker S, Salowsky R, Leiber M, Gassmann M. The RIN: an RNA integrity number for assigning integrity calues to RNA measurements. BMC Mol. Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Marx V. RNA quality: defining the good, the bad and the ugly. Genomics Proteomics. 2004;4:14–21. [Google Scholar]
- [28].Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;9:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pfaffle MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: bestkeeper-excel based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- [31].Lafleur DW, Nardelli B, Tsareva T, Mather D, Feng P, Semenuk M. Interferon-κ, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 2001;276:39765–39771. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- [32].Qin Y, Heine VM, Karst H, Lucassen PJ, Joels M. Gene expression patterns in rat dentate granule cells: comparison between fresh and fixed tissue. J. Neurosci. Methods. 2003;131:205–211. doi: 10.1016/j.jneumeth.2003.08.010. [DOI] [PubMed] [Google Scholar]
- [33].Inoue T, Nabeshima K, Kataoka H, Koono M. Feasibility of archival non-buffered formalin-fixed and paraffin-embedded tissues for PCR amplification: an analysis of resected gastric carcinoma. Pathol. Int. 1996;46:997–1004. doi: 10.1111/j.1440-1827.1996.tb03580.x. [DOI] [PubMed] [Google Scholar]
- [34].Madabusi LV, Latham GT, Andruss BF. RNA extraction for arrays. Methods Enzymol. 2006;411:1–13. doi: 10.1016/S0076-6879(06)11001-0. [DOI] [PubMed] [Google Scholar]
- [35].Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- [36].Moreau ME, Dubreuil P, Molinaro G, Chagnon M, Muller-Esterl W, Lepage Y. Expression of metallopeptidases and kinin receptors in swine oropharyngeal tissues: effects of angiotensis I-converting enzyme inhibition and inflammation. J. Pharmacol. Exp. Ther. 2005;315:1065–1074. doi: 10.1124/jpet.105.088005. [DOI] [PubMed] [Google Scholar]
- [37].Gaffney DK, Winter K, Fuhrman C, Flinner R, Greven K, Ryu J. Feasibility of RNA collection for micro-array gene expression analysis in the treatment of cervical carcinoma: a scientific correlate of RTOG C-0128. Gynecol. Oncol. 2005;97:607–611. doi: 10.1016/j.ygyno.2005.01.014. [DOI] [PubMed] [Google Scholar]
- [38].Micke P, Ohshima M, Tahmasebpoor S, Ren ZP, Ostman A, Ponten F. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab. Invest. 2006;86:202–211. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- [39].Akilesh S, Petkova S, Sproule TJ, Shaffer DJ, Christianson GJ, Roopenian D. The MHC class I-like Fc receptor promotes humorally mediated autoimmune disease. J. Clin. Invest. 2004;113:1328–1333. doi: 10.1172/JCI18838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL. Gene expression analysis in preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67:10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- [41].de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab. Invest. 2005;85:154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- [42].Lindbom J, Ljungman AG, Lindahl M, Tagesson C. Increased gene expression of novel cytosolic and secretory phospholipase A(2) types in human airway epithelial cells induced by tumor necrosis factor-alpha and IFN-gamma. J. Interferon Cytokine Res. 2002;22:947–955. doi: 10.1089/10799900260286650. [DOI] [PubMed] [Google Scholar]
- [43].Foss DL, Baarsch MJ, Murtaugh MP. Regulation of hypoxanthine phosphoribosyltransferase glyceraldehyde-3-phosphate dehydrogenase and beta-actin mRNA expression in porcine immune cells and tissues. Anim. Biotechnol. 1998;9:67–78. doi: 10.1080/10495399809525893. [DOI] [PubMed] [Google Scholar]