Abstract
Inositol 1,4,5-trisphosphate (InsP3) receptors are calcium-release channels found in the endoplasmic/sarcoplasmic reticulum (ER/SR) membrane of diverse cell types. InsP3 receptors release Ca2+ from ER/SR lumenal stores in response to InsP3 generated from various stimuli. The complex spatial and temporal patterns of InsP3 receptor-mediated Ca2+ release regulate many cellular processes, ranging from gene transcription to memory. Ankyrins are adaptor proteins implicated in the targeting of ion channels and transporters to specialized membrane domains. Multiple independent studies have documented in vitro and in vivo interactions between ankyrin polypeptides and the InsP3 receptor. Moreover, loss of ankyrin-B leads to loss of InsP3 receptor membrane expression and stability in cardiomyocytes. Despite extensive biochemical and functional data, the validity of in vivo ankyrin-InsP3 receptor interactions remains controversial. This controversy is based on inconsistencies between a previously identified ankyrin-binding region on the InsP3 receptor and InsP3 receptor topology data that demonstrate the inaccessibility of this lumenal binding site on the InsP3 receptor to cytosolic ankyrin polypeptides. Here we use two methods to revisit the requirements on InsP3 receptor for ankyrin binding. We demonstrate that ankyrin-B interacts with the cytoplasmic N-terminal domain of InsP3 receptor. In summary, our findings demonstrate that the ankyrin-binding site is located on the cytoplasmic face of the InsP3 receptor, thus validating the feasibility of in vivo ankyrin-InsP3 receptor interactions.
Keywords: Trafficking, ankyrin, calcium, cytoskeleton, transport, InsP3 receptor
INTRODUCTION
Calcium release from intracellular endoplasmic/sarcoplasmic reticulum (ER/SR) stores is activated by inositol 1,4,5-trisphosphate (InsP3), a second messenger produced via the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2). Binding of InsP3 to membrane-associated InsP3 receptors increases InsP3 receptor calcium sensitivity and, in a biphasic manner, induces a stimulatory effect at low calcium concentrations and an inhibitory effect at higher calcium concentrations [Taylor and Laude, 2002]. InsP3 receptor-induced release of calcium from intracellular stores provides positive feedback whereby calcium amplifies its own release. This dynamic interplay allows complex spatial and temporal patterns of intracellular calcium signaling that are necessary for unique cellular functions.
The functional InsP3 receptor complex exists as a homo- or hetero-tetramer of 260 kD InsP3 receptor subunits. Distinct genes encode three InsP3 receptor isoforms in mammals (types I, II, and III), with splice variants identified for each type [Foskett et al., 2007]. Isoform type and expression levels vary among cell types: cerebellar Purkinje cells express InsP3 receptor type I almost exclusively, while pancreatic acinar cells express types II and III, and many epithelia express all three isoforms [Bush et al., 1994; Nathanson et al., 1994; Wojcikiewicz, 1995].
InsP3 receptors interact with a number of protein partners (recently reviewed by Foskett and colleagues) [Foskett et al., 2007]. In 1993, two independent groups identified an interaction between the InsP3 receptor and the adaptor protein ankyrin [Bourguignon et al., 1993a; Joseph and Samanta, 1993]. Specifically, Joseph and colleagues demonstrated that the InsP3 receptor co-immunoprecipitated with ankyrin-R from brain [Joseph and Samanta, 1993]. Moreover, Bourguignon and colleagues showed that ankyrin-R and InsP3 receptor co-purified from murine T-lymphoma cells [Bourguignon et al., 1995], interacted directly with high affinity, and that ankyrin-R inhibited InsP3-binding and InsP3-dependent radiolabeled calcium flux [Bourguignon et al., 1993a; Bourguignon et al., 1993b]. Over the past fifteen years, a number of groups have replicated ankyrin-InsP3 receptor interactions using purified proteins, co-immunoprecipitation assays, 3H-InsP3 binding experiments, and pulse-chase biosynthesis experiments [Feng and Kraus-Friedmann, 1993; Hayashi et al., 2000; Hayashi and Su, 2001; Liu et al., 2007; Mohler et al., 2005; Mohler et al., 2004a; Mohler et al., 2003; Mohler et al., 2004b; Tuvia et al., 1999]. Additional evidence for the relevance of the ankyrin-InsP3 receptor interaction comes from mouse models with reduced ankyrin-B expression that display significant defects in InsP3 receptor expression, subcellular localization, and decreased InsP3 receptor post-translational stability [Mohler et al., 2004a].
Despite overwhelming evidence supporting the ankyrin-InsP3 receptor interaction, questions remain as to the validity of this interaction in vivo. In 1995, an 11-residue motif in the InsP3 receptor (GGVGDVLRKPS corresponding to residues 2548–2558 on rat InsP3 receptor I, Figure 1A–B) was proposed to be the binding site for ankyrin based on its similarity to the ankyrin-binding site on CD44 [Bourguignon and Jin, 1995]. Furthermore, a peptide corresponding to this sequence competed ankyrin-InsP3 receptor interactions and blocked ankyrin-induced inhibitory effects on InsP3 binding and InsP3-dependent calcium release events [Bourguignon and Jin, 1995]. The ankyrin binding site was predicted to reside in the cytosol, allowing for the in vivo interaction of InsP3 receptor and cytosolic ankyrin [Bourguignon and Jin, 1995]. Subsequent studies that resolved InsP3 receptor topology placed the residues 2548–2558 on the lumenal side of the InsP3 receptor, which would effectively prohibit in vivo interactions between ankyrin and the receptor. To date, both ankyrin and InsP3 receptor fields remain unclear regarding the feasibility of in vivo ankyrin- InsP3 receptor interactions despite compelling biochemical and animal findings (see comments in [Foskett et al., 2007; Mohler et al., 2004a; Patel et al., 1999; Patterson et al., 2004; Roderick and Bootman, 2003; Vermassen et al., 2004]).
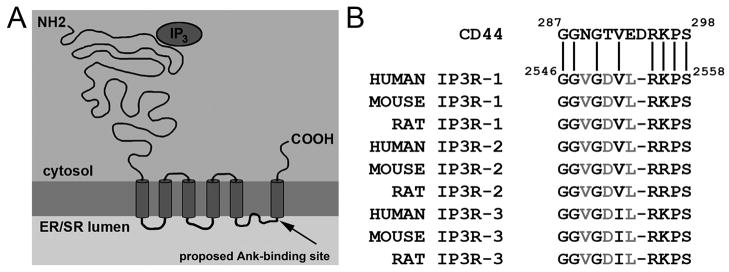
Figure 1. Domain organization of InsP3 receptor and InsP3 receptor intermolecular interactions.
A, Domain organization of the InsP3 receptor monomer, depicting membrane topology and cytoplasmic and lumenal protein orientation. Inositol 1,4,5-trisphosphate (InsP3; oval) activates Ca2+ release via binding to an N-terminal region. Arrow denotes location of previously proposed ankyrin-binding motif in C-terminal domain of InsP3 receptor (see Figure 1B). Note that these residues (rat 2546–2558) are located in the ER/SR lumen, inconsistent with an interaction with cytosolic ankyrin. B, Ankyrin-binding site in InsP3 receptor identified by sequence similarity with CD44 ankyrin-binding motif. Minimal ankyrin-binding residues on CD44 [Bourguignon and Jin, 1995]. Amino acid sequences below CD44 denote sequence homology of residues in InsP3 receptor C-terminus to the CD44 sequence.
Here, we use two approaches to revisit the structural requirements for ankyrin-binding on the InsP3 receptor. Using yeast two-hybrid and in vitro binding experiments, we map the ankyrin-binding region on the InsP3 receptor type I to residues 955–991. Based on the membrane topology of the InsP3 receptor, these data demonstrate that the ankyrin-binding region is located on the cytoplasmic face of the receptor, validating the feasibility of the in vivo ankyrin-InsP3 receptor interactions.
MATERIALS AND METHODS
InsP3 receptor/ankyrin-B constructs
InsP3 receptor constructs were engineered into pACT2 (Clontech) for yeast two-hybrid assays using standard molecular techniques and full-length rat InsP3 receptor type I as template. InsP3 receptor constructs were also engineered into pcDNA3.1(+) for in vitro translation experiments. Positive clones were analyzed by restriction digestion and subsequently sequenced. For in vitro translation constructs, an additional initiator methionine was introduced at the beginning of the coding sequence of each InsP3 receptor mutant. The membrane-binding domain (plus residues of the spectrin-binding domain) of human 220 kD ankyrin-B (residues 1–958) was inserted into pGEX-6P1 for expression as a GST-fusion protein. Protein was expressed in BL21(DE3)pLysS cells and purified using glutathione-sepharose.
Protein expression. A, In vitro transcription- translation
InsP3 receptor constructs were transcribed and translated using the TNT Coupled Reticulocyte Lysate System (Promega) with 20 μCi of Redivue L-[35S] methionine (GE Healthcare) and 0.75 μg of plasmid DNA. B, Full-length InsP3 receptor production. Baculovirus was used to generate full-length InsP3 receptor and a mutant InsP3 receptor lacking amino acids 924–991. Briefly, full-length wild-type or mutant cDNAs (generated from WT InsP3 receptor using standard molecular techniques) were co-expressed in a standard transfer vector (Clontech) with Bsu36I-digested BacPAK6 viral DNA into Spondoptera frugiperda cells (Sf21 cells, Clontech) using the BacPAK Baculovirus Expression System (Clontech). Sf21 cells were then infected as described [Mohler et al., 2005]. Recombinant full-length InsP3 receptor was solubilized from cell membranes as described [Mohler et al., 2005]. Full-length protein expression was confirmed by SDS-PAGE and immunoblot on cell lysates using an affinity-purified InsP3 receptor Ig generated against the distal C-terminus of the receptor[Mohler et al., 2004a]. For binding experiments, equal quantities of cell lysate were added to binding reactions.
In vitro binding
GST-ankyrin-B membrane-binding domain was expressed in BL21(DE3)pLysS bacteria, purified using glutathione sepharose (GE Healthcare), and eluted with glutathione. Twenty micrograms of purified GST or GST-ankyrin-B membrane-binding domain were coupled to glutathione sepharose for two hours at 4°C in binding buffer (50 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.1% Triton X-100). Following extensive washes in Buffer A (50 mM Tris pH 7.4, 1 mM EDTA, 1 mM EGTA, 500 mM NaCl, 1% Triton X-100), conjugated beads were incubated with InsP3 receptor in vitro translation products representing rat InsP3 receptor amino acids 924–1582, 924–1550, 924–1517, 924–1454, 924–1398, 924–1352, 924–1297, 924–1226, 924–1164, 924–1125, 924–991, 785–954, 955–1143, and 2403–2575, or full-length InsP3 receptor products generated by baculoviral expression for 4 hours at 4°C in binding buffer plus protease inhibitor cocktail (Sigma). Following incubations, binding reactions were extensively washed in Buffer A, eluted, and analyzed by SDS-PAGE. Radiolabeled proteins were detected by phosphorimaging. Full-length InsP3 receptor products were detected by immunoblot using affinity-purified InsP3 receptor Ig.
Yeast two-hybrid
Following one round of AH109 transformation to ensure that none of the plasmids induced autoactivation, 0.05 μg of pACT2 DNA carrying the InsP3 receptor insert and 0.05 μg of pAS2-1 DNA carrying ankyrin-B membrane-binding domain were co-transformed into AH109 yeast (in the presence of herring sperm DNA) using a standard lithium acetate protocol and cultured at 30° C on YPD media lacking leucine (-L), lacking tryptophan (-T), and lacking both leucine and tryptophan (-LT). Double transformants (identified as colonies capable of growth on media lacking both leucine and tryptophan (-LT)), were further selected on media lacking adenine, histidine, leucine, and tryptophan (-AHLT). The positive control was a co-transformation of 0.05 μg of TD1-1 and 0.05 μg of pVA3 and the negative control was a co-transformation of TD1-1 and pLAM5. Transformants exhibiting growth on –AHLT media following three to five days of 30° C incubation were considered a positive interactions.
RESULTS
Ankyrin-B interacts with the InsP3 receptor cytoplasmic N-terminal domain
We used the yeast two-hybrid system to revisit the structural requirements on the InsP3 receptor for ankyrin-binding. We previously demonstrated ankyrin-B associates with the InsP3 receptor via its membrane-binding domain [Mohler et al., 2004a]. We expressed the ankyrin-B membrane-binding domain fused to GAL4 DNA-binding domain (yeast two-hybrid “bait”). InsP3 receptor constructs generated from rat InsP3 receptor I (identical cDNA used as in [Bourguignon and Jin, 1995]) were fused to GAL4 activation domain (yeast two-hybrid “prey”). InsP3 receptor constructs were designed based on prior limited trypsin proteolysis mapping [Yoshikawa et al., 1999] to preserve the major folding domains of the InsP3 receptor in our binding assays (Figure 2). Due to the large size of the InsP3 receptor cytoplasmic N-terminus, this region was subdivided into seven fragments (see Figure 2B).
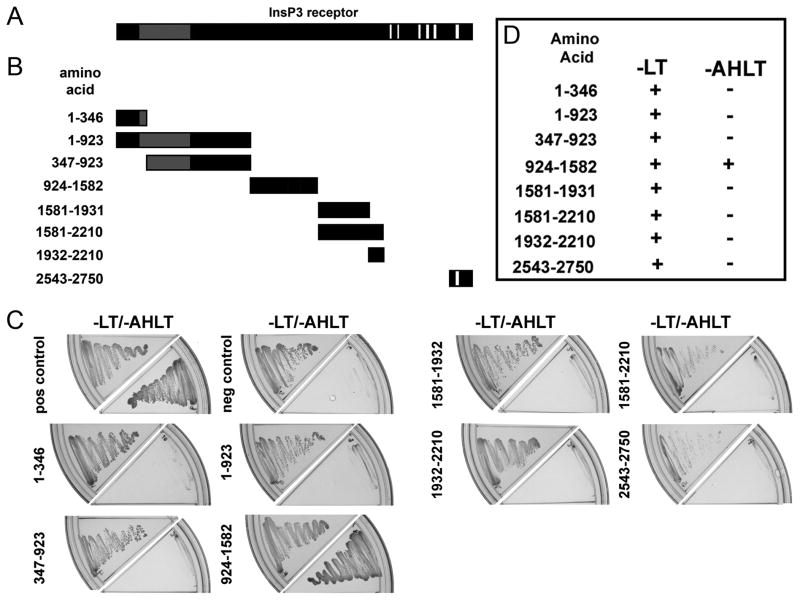
Figure 2. Identification of requirements on InsP3 receptor for ankyrin-B association.
A, Diagram of the InsP3 receptor monomer [da Fonseca et al., 2003]. Gray box represents InsP3 binding domain and white boxes represent transmembrane domains. B, InsP3 receptor yeast two-hybrid constructs. Fragments of the cytoplasmic regions of the InsP3 receptor monomer used in the initial assessment of the structural requirements of InsP3 receptor for interaction with ankyrin-B. InsP3 receptor construct 2543–2750 contains the previously predicted ankyrin-binding domain [Bourguignon and Jin, 1995]. Constructs were designed based on identified structural folding boundaries defined by limited trypsin digestion [Yoshikawa et al., 1999]. C, AH109 yeast were co-transformed with ankyrin-B membrane-binding domain (bait) and a unique InsP3 receptor construct (prey). Co-expression of bait and prey proteins was confirmed by inoculating onto -LT media. Positive interactions were determined by growth on -AHLT media after five days incubation. TD1-1 served as bait for both the positive and negative controls, while pVA3 and pLAM5 were used as prey for the positive and negative controls, respectively. D, Summary data for ankyrin-B membrane-binding domain-InsP3 receptor interactions.
We detected no interaction between ankyrin-B membrane-binding domain and an InsP3 receptor fusion protein containing the proposed ankyrin binding site at residues 2548–2558 (InsP3 receptor construct 2543–2750; see Figure 2B-D;[Bourguignon and Jin, 1995]). Instead ankyrin-B membrane-binding domain interacted with an InsP3 receptor fusion protein containing residues 924–1582 (Figure 2B-D, note significant growth on –AHLT plate in 2C). No interactions were detected between ankyrin-B membrane-binding domain and InsP3 receptor fusion proteins using InsP3 receptor 1–346, 1–923, 347–923, 1581–1931, 1581–2210, and 1932–2210, even though all constructs were expressed in yeast (note –LT plates in Figure 2C). These data demonstrate that a large N-terminal region of rat InsP3 receptor I contains the ankyrin-B binding motif. Moreover, these data demonstrate that the ankyrin-binding region resides on the cytoplasmic face of the InsP3 receptor.
Defining the minimal binding region on InsP3 receptor for ankyrin-B
Our initial binding assays identified a domain within the N-terminal region of the InsP3 receptor (residues 924–1582) that is required for ankyrin-binding (see Figure 2). However, due to the large size of the InsP3 receptor N-terminus, we were interested in further narrowing the ankyrin-binding region. We used standard molecular biology techniques to create additional InsP3 receptor prey plasmids based on residues 924–1582 (Figure 3). Ten additional mutants were engineered and screened for interaction in yeast against the ankyrin-B membrane-binding domain (Figure 3). As shown in Figure 4A, all ten InsP3 receptor truncated constructs bound to the ankyrin-B membrane-binding domain. Even the smallest construct, corresponding to InsP3 receptor I residues 924–991, displayed significant ankyrin-binding activity (Figure 4A).
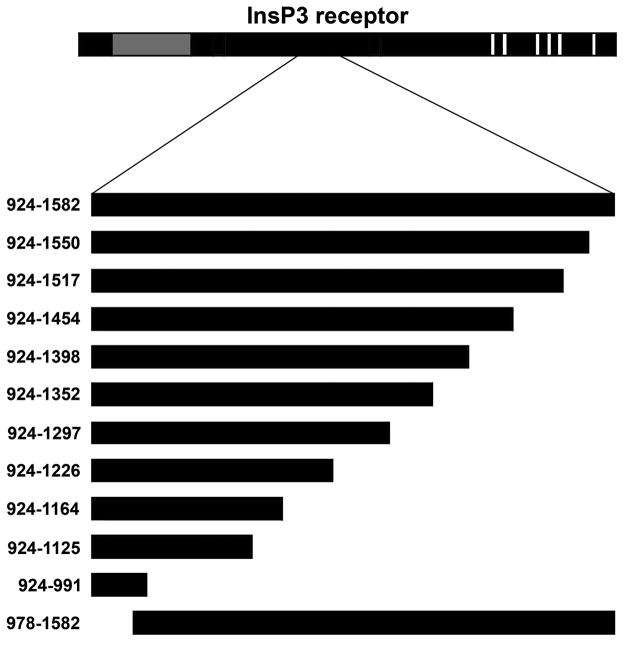
Figure 3. Identification of minimal requirements on InsP3 receptor for ankyrin-binding.
Eleven InsP3 receptor truncation constructs were generated to define the minimal structural requirements on InsP3 receptor for ankyrin-binding. Constructs represent truncations based on rat InsP3 receptor I residues 924–1582.
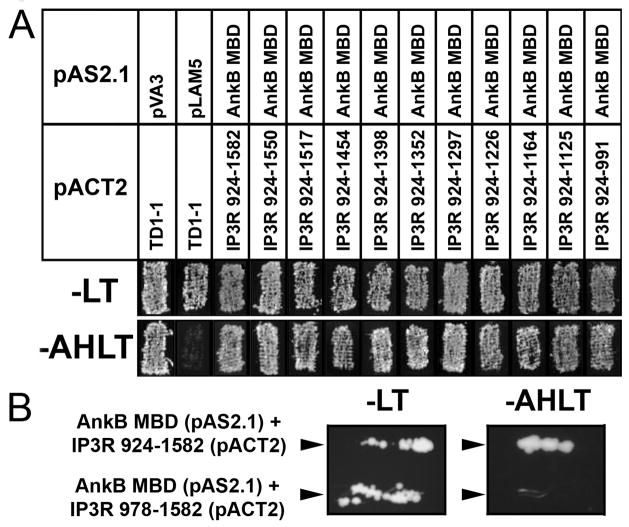
Figure 4. Defining the minimal ankyrin-binding site on InsP3 receptor N-terminus.
A, Ankyrin-B membrane binding domain (bait) and a series of InsP3 receptor constructs (prey, see Figure 3) were co-transformed into AH109 yeast to identify a minimal region of the InsP3 receptor responsible for interaction with ankyrin-B. Co-transformants were selected on –LT media and analyzed for HIS3 reporter gene activation by evaluating growth on –AHLT after five days incubation. Positive interaction is seen with all constructs, with the smallest interacting region of the InsP3 receptor corresponding to amino acids 924–991. Note that this region is located on the cytoplasmic N-terminus of the InsP3 receptor. B, InsP3 receptor lacking residues 978–1582 fails to interact with ankyrin-B membrane-binding domain in yeast. Positive control for experiments was InsP3 receptor 924–1582. Positive interaction was assessed after 5 days on AHLT medium.
We performed yeast two-hybrid analyses with an additional InsP3 receptor construct that lacked residues 924–977 (Figure 3, construct 978–1582) to determine whether these residues were necessary for the ankyrin-B - InsP3 receptor interaction. While we observed co-expression of InsP3 receptor construct 978–1582 with ankyrin-B in AH109 cells (see –LT plate, Figure 4B), we were unable to detect binding of this GAL4-fusion protein with the ankyrin-B membrane-binding domain (note –AHLT plate, Figure 4B). Our combined data demonstrate that InsP3 receptor residues within 924–991 are necessary, and sufficient, for interaction with ankyrin-B in the context of yeast.
We used in vitro binding assays to confirm the yeast two-hybrid mapping data. Briefly, InsP3 receptor truncation constructs were sub-cloned into an expression plasmid containing a T7 promoter for in vitro transcription/translation (in the presence of 35S-methionine). Following the generation of radiolabeled InsP3 receptor products, the reactions were purified and incubated with either GST or GST-ankyrin-B membrane-binding domain (generated and purified from bacteria). Following the binding reaction and extensive washes, bound radiolabeled InsP3 receptor products were eluted and analyzed by SDS-PAGE and phosphorimaging (see Experimental Procedures). Consistent with prior experiments performed in yeast, in vitro binding experiments demonstrated binding of all InsP3 receptor truncation constructs (including InsP3 receptor 924–991 see Figure 5A–B) with GST-ankyrin-B membrane-binding domain. We observed no binding of control GST with any InsP3 receptor product (Figure 5A–B).
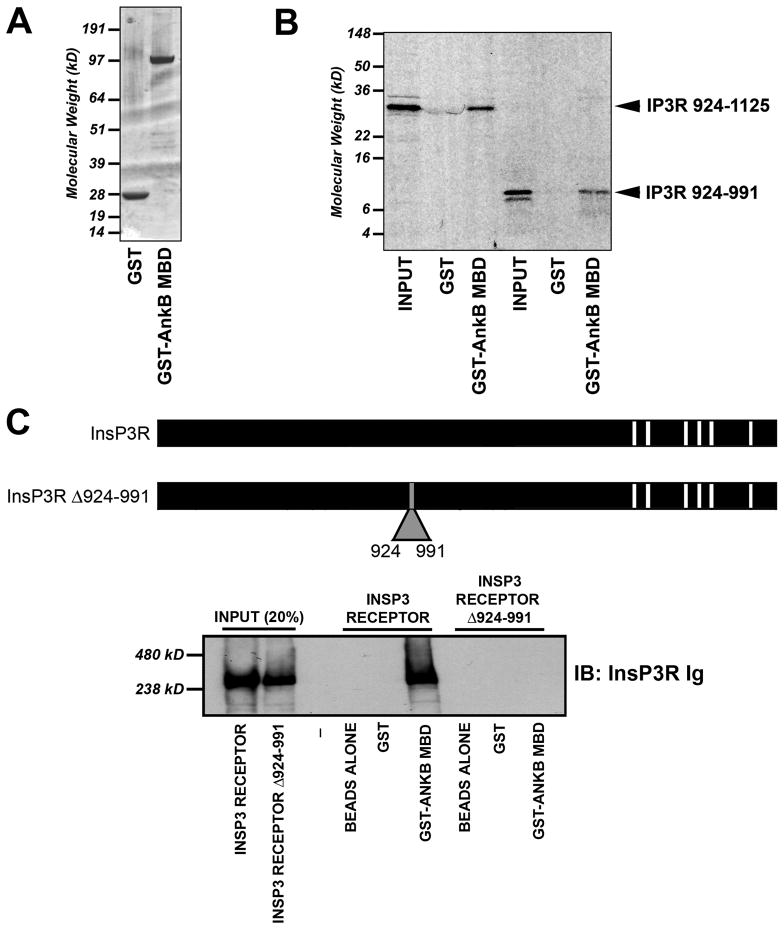
Figure 5. InsP3 receptor residues 924–991 are required and sufficient for ankyrin-B association.
A, Coomassie Blue stained gel depicting purified GST and GST-ankyrin-B (10 μg of each protein) membrane-binding domain (InsP3 receptor binding domain on ankyrin-B). B, InsP3 receptor residues 924–991 are sufficient to mediate interaction of InsP3 receptor with ankyrin-B membrane-binding domain. Constructs shown in Figure 3 and 5 were inserted into pcDNA3.1(+) and in vitro transcribed/translated in the presence of 35S-methionine. Products were used in binding experiments with purified GST or GST-ankyrin-B membrane-binding domain. Shown are representative experiments from two radiolabeled N-terminal samples including 924–991 and 924–1125. We observed no binding of InsP3 receptor constructs with GST alone. C, Full-length InsP3 receptor Δ924–991 lacks ankyrin-B-binding activity. Full-length InsP3 receptor and InsP3 receptor Δ924–991 were generated using baculovirus (Sf21 cells) and individually incubated with GST-ankyrin-B membrane-binding domain immobilized on glutathione sepharose. Bound protein was eluted and analyzed by immunoblot using affinity-purified InsP3 receptor Ig (generated against InsP3 receptor C-terminal domain). Input equals 20%.
To validate our binding results in the context of a full-length InsP3 receptor we used baculoviral expression (in Sf21 cells) to generate full-length InsP3 receptor [Cardy et al., 1997] and an InsP3 receptor mutant lacking residues 924–991 (InsP3 receptor Δ924–991, Figure 5C). Approximately equal amounts of InsP3 receptor proteins were incubated to immobilized GST or GST-ankyrin-B membrane-binding domain. Following extensive high salt washes, bound protein was eluted and analyzed by SDS-PAGE and immunoblot using affinity-purified InsP3 receptor Ig. While we observed association of wild-type full-length InsP3 receptor with ankyrin-B membrane-binding domain, no association was detected between ankyrin and InsP3 receptor Δ924–991 (Figure 5C). Together, our binding data clearly demonstrate that InsP3 receptor residues 924–991 are necessary and sufficient for ankyrin-B- InsP3 receptor interaction.
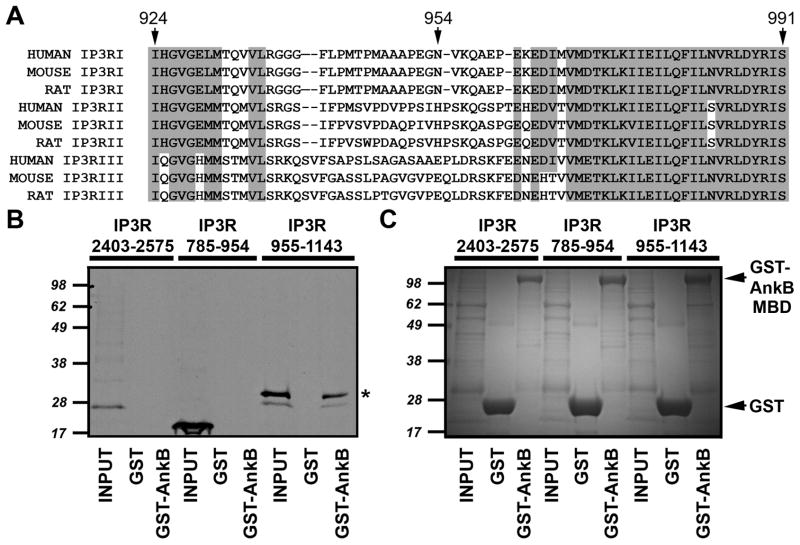
Finally, to narrow the ankyrin-binding domain on the InsP3 receptor, we performed in vitro binding reactions using GST-ankyrin-B membrane-binding domain and in vitro translation products of each half of the identified ankyrin-binding domain (InsP3 receptor residues 924–991). Specifically, we generated in vitro translation products for InsP3 receptor residues 785–954 (contains InsP3 receptor 924–954) and 955–1143 (contains InsP3 receptor residues 955–991, Figure 6A). Larger products were used to incorporate additional methionine residues for in vitro translation as well as increase polypeptide size for SDS-PAGE analysis. InsP3 receptor 955–1143 associated with ankyrin-B membrane-binding domain (Figure 6B and C). In contrast, InsP3 receptor residues 785–954 lacked ankyrin-binding activity (Figure 6B and C). Consistent with our yeast two-hybrid data (Figure 2), an InsP3 receptor C-terminal construct (2403–2575) containing the previously predicted ankyrin-binding motif [Bourguignon and Jin, 1995] also lacked ankyrin-binding activity. Our new data clearly demonstrate that the minimal structural requirements for InsP3 receptor binding of ankyrin-B (InsP3 receptor residues 955–991) reside in the cytoplasmic N-terminal domain of InsP3 receptor.
Figure 6. Ankyrin-B associates with InsP3 receptor residues 955–991.
A, Alignment of ankyrin-binding domain of InsP3 receptor. Sequences of human, mouse, and rat InsP3 receptor I, II, and III. Note high sequence conservation in N- and C-terminal regions of the domain. Residue numbers are based on rat InsP3 receptor amino acid sequence. B, InsP3 receptor ankyrin-binding domain (rat InsP3 receptor residues 924–991) was split into two constructs and in vitro translated in the presence of 35S-methionine. An InsP3 receptor C-terminal domain construct (residues 2403–2575) containing the previously predicted ankyrin-binding domain was also in vitro translated. Radiolabeled proteins were incubated with purified GST or GST-ankyrin-B membrane binding domain (GST-AnkB). Bound proteins were eluted and analyzed by SDS-PAGE and phosphorimaging. C, Gel used in binding studies was stained with Coomassie Blue to demonstrate protein purity and equal protein concentrations.
DISCUSSION
The current study was designed to evaluate the amino acid requirements on InsP3 receptor for ankyrin-B interaction. Consistent with prior studies, we observed interaction between ankyrin-B and the InsP3 receptor [Bourguignon et al., 1993a; Bourguignon et al., 1993b; Feng and Kraus-Friedmann, 1993; Hayashi et al., 2000; Hayashi and Su, 2001; Joseph and Samanta, 1993; Liu et al., 2007; Mohler et al., 2005; Mohler et al., 2004a; Mohler et al., 2003; Mohler et al., 2004b; Tuvia et al., 1999]. However, our data establish that ankyrin interacts with the cytoplasmic and not the lumenal face of the InsP3 receptor. Specifically, ankyrin-B interacts with the cytoplasmic N-terminal domain of the InsP3 receptor within residues 955–991. The new InsP3 receptor cytosolic ankyrin-binding region is consistent with in vivo ankyrin - InsP3 receptor interactions.
All three major InsP3 receptor isoforms associate with ankyrin-B [Mohler et al., 2004a]. Furthermore, the expression of the three InsP3 receptor isoforms is reduced in tissues derived from ankyrin-B+/− or ankyrin-B−/− (null) mice [Mohler et al., 2004a]. Therefore, the precise binding site on the InsP3 receptor is likely a conserved motif within the receptor subtypes (Figure 6A). While a previous study proposed that ankyrin directly associated with InsP3 receptor at a site based on limited sequence similarity to an ankyrin-binding region of CD44 [Bourguignon and Jin, 1995], it is now clear that most ankyrin-binding proteins do not display conserved sequence binding motifs (reviewed in [Bennett and Baines, 2001]). Instead, ankyrin-binding proteins associate with unstructured and non-conserved protein motifs (reviewed in [Bennett and Baines, 2001]). We previously identified a series of positively-charged residues on the ankyrin- B membrane-binding domain that are essential for InsP3 receptor binding. Based on these data, we predict that ankyrin-B will likely interact with a conserved, negatively-charged site within the 955–991 region of the InsP3 receptor. Future goals will be to further refine the binding site for ankyrin and to analyze the cellular targeting of an InsP3 receptor that lacks ankyrin-binding. Unfortunately, due to the homo- and hetero-trimeric organization of the InsP3 receptor protein complex, these experiments will unlikely be straightforward. Additionally, using current viral technologies, it is difficult to express full-length InsP3 receptor mutants in primary cells.
Ankyrins likely play multiple roles in normal InsP3 receptor function. Based on work from multiple labs, ankyrin has a central role in the targeting of InsP3 receptor to endoplasmic/sarcoplasmic reticulum membrane domains in both non-excitable and excitable tissues [Bourguignon et al., 1993a; Bourguignon et al., 1993b; Feng and Kraus-Friedmann, 1993; Hayashi et al., 2000; Hayashi and Su, 2001; Joseph and Samanta, 1993; Liu et al., 2007; Mohler et al., 2005; Mohler et al., 2004a; Mohler et al., 2003; Mohler et al., 2004b; Singleton and Bourguignon, 2004; Tuvia et al., 1999]. Moreover, ankyrin isoforms presumably stabilize the receptor in close proximity with effector and regulatory proteins [Bhasin et al., 2007; Lencesova et al., 2004; Liu et al., 2007; Mohler et al., 2005].
In summary, our new data identifies the ankyrin binding region on the endoplasmic/sarcoplasmic reticulum InsP3 receptor. Consistent with an in vivo association of cytosolic ankyrin and the InsP3 receptor, our data demonstrate that ankyrin-B directly associates with residues 955–991 in the cytoplasmic N-terminal domain of InsP3 receptor.
Acknowledgments
This work is supported by the Pew Scholars Trust and by NIH R01HL084583 and R01HL083422 (PJM). CFK is supported by a Graduate Research Fellowship from the National Science Foundation (NSF). This work was supported in part by the Cardiovascular Interdisciplinary Research Fellowship to TJH (5 T32 HL007131). We acknowledge Sonja Smith, Brandon Bates, and Patrick Wright for excellent technical assistance.
Grant sponsor: NIH; Grant numbers: R01HL084583 and R01HL083422 (PJM); Pew Scholars Trust (PJM), and NSF Graduate Research Fellowship (CFK).
References
- Bennett V, Baines AJ. Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol Rev. 2001;81:1353–92. doi: 10.1152/physrev.2001.81.3.1353. [DOI] [PubMed] [Google Scholar]
- Bhasin N, Cunha SR, Mudannayake M, Gigena MS, Rogers TB, Mohler PJ. Molecular basis for PP2A regulatory subunit B56{alpha} targeting in cardiomyocytes. Am J Physiol Heart Circ Physiol. 2007;293:H109–19. doi: 10.1152/ajpheart.00059.2007. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Iida N, Jin H. The involvement of the cytoskeleton in regulating IP3 receptor-mediated internal Ca2+ release in human blood platelets. Cell Biol Int. 1993a;17:751–8. doi: 10.1006/cbir.1993.1136. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Jin H. Identification of the ankyrin-binding domain of the mouse T-lymphoma cell inositol 1,4,5-trisphosphate (IP3) receptor and its role in the regulation of IP3-mediated internal Ca2+ release. J Biol Chem. 1995;270:7257–60. doi: 10.1074/jbc.270.13.7257. [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Jin H, Iida N, Brandt NR, Zhang SH. The involvement of ankyrin in the regulation of inositol 1,4,5- trisphosphate receptor-mediated internal Ca2+ release from Ca2+ storage vesicles in mouse T-lymphoma cells. J Biol Chem. 1993b;268:7290–7. [PubMed] [Google Scholar]
- Bush KT, Stuart RO, Li SH, Moura LA, Sharp AH, Ross CA, Nigam SK. Epithelial inositol 1,4,5-trisphosphate receptors. Multiplicity of localization, solubility, and isoforms. J Biol Chem. 1994;269:23694–9. [PubMed] [Google Scholar]
- Cardy TJ, Traynor D, Taylor CW. Differential regulation of types-1 and -3 inositol trisphosphate receptors by cytosolic Ca2+ Biochem J. 1997;328 (Pt 3):785–93. doi: 10.1042/bj3280785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Fonseca PC, Morris SA, Nerou EP, Taylor CW, Morris EP. Domain organization of the type 1 inositol 1,4,5-trisphosphate receptor as revealed by single-particle analysis. Proc Natl Acad Sci U S A. 2003;100:3936–41. doi: 10.1073/pnas.0536251100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Kraus-Friedmann N. Association of the hepatic IP3 receptor with the plasma membrane: relevance to mode of action. Am J Physiol. 1993;265:C1588–96. doi: 10.1152/ajpcell.1993.265.6.C1588. [DOI] [PubMed] [Google Scholar]
- Foskett JK, White C, Cheung KH, Mak DO. Inositol trisphosphate receptor Ca2+ release channels. Physiol Rev. 2007;87:593–658. doi: 10.1152/physrev.00035.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Maurice T, Su TP. Ca(2+) signaling via sigma(1)-receptors: novel regulatory mechanism affecting intracellular Ca(2+) concentration. J Pharmacol Exp Ther. 2000;293:788–98. [PubMed] [Google Scholar]
- Hayashi T, Su TP. Regulating ankyrin dynamics: Roles of sigma-1 receptors. Proc Natl Acad Sci U S A. 2001;98:491–496. doi: 10.1073/pnas.98.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SK, Samanta S. Detergent solubility of the inositol trisphosphate receptor in rat brain membranes. Evidence for association of the receptor with ankyrin. J Biol Chem. 1993;268:6477–86. [PubMed] [Google Scholar]
- Lencesova L, O’Neill A, Resneck WG, Bloch RJ, Blaustein MP. Plasma membrane-cytoskeleton-endoplasmic reticulum complexes in neurons and astrocytes. J Biol Chem. 2004;279:2885–93. doi: 10.1074/jbc.M310365200. [DOI] [PubMed] [Google Scholar]
- Liu XL, Miyakawa A, Aperia A, Krieger P. Na,K-ATPase generates calcium oscillations in hippocampal astrocytes. Neuroreport. 2007;18:597–600. doi: 10.1097/WNR.0b013e3280b07bc9. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Davis JQ, Bennett V. Ankyrin-B Coordinates the Na/K ATPase, Na/Ca Exchanger, and InsP(3) Receptor in a Cardiac T-Tubule/SR Microdomain. PLoS Biol. 2005;3:e423. doi: 10.1371/journal.pbio.0030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler PJ, Davis JQ, Davis LH, Hoffman JA, Michaely P, Bennett V. Inositol 1,4,5-trisphosphate receptor localization and stability in neonatal cardiomyocytes requires interaction with ankyrin-B. J Biol Chem. 2004a;279:12980–7. doi: 10.1074/jbc.M313979200. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Schott JJ, Gramolini AO, Dilly KW, Guatimosim S, duBell WH, Song LS, Haurogne K, Kyndt F, Ali ME, Rogers TB, Lederer WJ, Escande D, Le Marec H, Bennett V. Ankyrin-B mutation causes type 4 long-QT cardiac arrhythmia and sudden cardiac death. Nature. 2003;421:634–9. doi: 10.1038/nature01335. [DOI] [PubMed] [Google Scholar]
- Mohler PJ, Splawski I, Napolitano C, Bottelli G, Sharpe L, Timothy K, Priori SG, Keating MT, Bennett V. A cardiac arrhythmia syndrome caused by loss of ankyrin-B function. Proc Natl Acad Sci U S A. 2004b;101:9137–42. doi: 10.1073/pnas.0402546101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson MH, Fallon MB, Padfield PJ, Maranto AR. Localization of the type 3 inositol 1,4,5-trisphosphate receptor in the Ca2+ wave trigger zone of pancreatic acinar cells. J Biol Chem. 1994;269:4693–6. [PubMed] [Google Scholar]
- Patel S, Joseph SK, Thomas AP. Molecular properties of inositol 1,4,5-trisphosphate receptors. Cell Calcium. 1999;25:247–64. doi: 10.1054/ceca.1999.0021. [DOI] [PubMed] [Google Scholar]
- Patterson RL, Boehning D, Snyder SH. Inositol 1,4,5-trisphosphate receptors as signal integrators. Annu Rev Biochem. 2004;73:437–65. doi: 10.1146/annurev.biochem.73.071403.161303. [DOI] [PubMed] [Google Scholar]
- Roderick HL, Bootman MD. Bi-directional signalling from the InsP3 receptor: regulation by calcium and accessory factors. Biochem Soc Trans. 2003;31:950–3. doi: 10.1042/bst0310950. [DOI] [PubMed] [Google Scholar]
- Singleton PA, Bourguignon LY. CD44 interaction with ankyrin and IP3 receptor in lipid rafts promotes hyaluronan-mediated Ca2+ signaling leading to nitric oxide production and endothelial cell adhesion and proliferation. Exp Cell Res. 2004;295:102–18. doi: 10.1016/j.yexcr.2003.12.025. [DOI] [PubMed] [Google Scholar]
- Taylor CW, Laude AJ. IP3 receptors and their regulation by calmodulin and cytosolic Ca2+ Cell Calcium. 2002;32:321–34. doi: 10.1016/s0143416002001859. [DOI] [PubMed] [Google Scholar]
- Tuvia S, Buhusi M, Davis L, Reedy M, Bennett V. Ankyrin-B is required for intracellular sorting of structurally diverse Ca2+ homeostasis proteins. J Cell Biol. 1999;147:995–1008. doi: 10.1083/jcb.147.5.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermassen E, Parys JB, Mauger JP. Subcellular distribution of the inositol 1,4,5-trisphosphate receptors: functional relevance and molecular determinants. Biol Cell. 2004;96:3–17. doi: 10.1016/j.biolcel.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Wojcikiewicz RJ. Type I, II, and III inositol 1,4,5-trisphosphate receptors are unequally susceptible to down-regulation and are expressed in markedly different proportions in different cell types. J Biol Chem. 1995;270:11678–83. doi: 10.1074/jbc.270.19.11678. [DOI] [PubMed] [Google Scholar]
- Yoshikawa F, Iwasaki H, Michikawa T, Furuichi T, Mikoshiba K. Trypsinized cerebellar inositol 1,4,5-trisphosphate receptor. Structural and functional coupling of cleaved ligand binding and channel domains. J Biol Chem. 1999;274:316–27. doi: 10.1074/jbc.274.1.316. [DOI] [PubMed] [Google Scholar]