Abstract
Hepatic fibrosis results from excessive deposition of type I collagen. The roles of Smads in mediating the effect of transforming growth factor-β1 (TGFβ1) on activation of the α1(I) collagen promoter were determined. Smads bind in association with Sp1 to the CC(GG)-rich TGFβ1 responsive element of the promoter that lacks the classical Smad recognition element, and enhance binding of Sp1. In transfection experiments, TGFβ1 activated a proximal promoter, but not promoters mutated at sites that prevented Sp1 binding. Sp1 alone or the combination of Smad2 and Smad4 activated the promoter in transfected human LX-2 stellate cells. Sp1 or Smad2 knockdowns with siRNAs prevented the effect of TGFβ1 in enhancing the promoter. In conclusion, this study shows that Smads bind in association with Sp1 to the CC(GG)-rich TGFβ1 responsive element of the human α1(I) collagen promoter that lacks the classical Smad recognition element, thus enhancing the binding of Sp1 and in this manner activating the collagen promoter.
Introduction
Hepatic fibrosis and cirrhosis result from the excessive deposition of predominantly type I collagen, which is composed of two α1 and one α2 chains. Transforming growth factor-β1 (TGFβ1), a principal profibrogenic cytokine, activates the human α2(I) collagen promoter via Smads (Ghosh et al., 2001). TGFβ1 also activates the α1(I) human collagen promoter, and a TGFβ1 response element has been located between −164 and −142 in the proximal promoter (Jimenez et al., 1994; Tang et al., 2003). However, the role of Smads in the activation of the human α1(I) collagen promoter remains unknown. Smads recognize the CAGA nucleotide sequence (Smad recognition element) (Shi et al., 1998) that is present in the human α2(I) collagen promoter (Zhang et al., 2000). By contrast, no Smad recognition element has been demonstrated in the human α1(I) collagen promoter. Sp1, a ubiquitous zinc-finger family transcription factor, binds to numerous sites along the α1(I) collagen promoter (Gaidarova and Jimenez, 2002). Sp1 and Smad proteins cooperate in enhancing the TGFβ1-mediated expression of the human α2(I) collagen gene (Zhang et al., 2000) and other genes (Pardali et al., 2000). The aim of this study was to determine the roles of Smads and Sp1 in mediating the effect of TGFβ1 signaling on activation of the human α1(I) collagen promoter.
Materials and Methods
Materials
Dulbecco's modified Eagle's medium, and Schneider's Drosophila medium were purchased from Gibco-Invitrogen (Carlsbad, CA). Fetal bovine serum (FBS), dimethyl sulfoxide, acetyl coenzyme A sodium salt, and bovine serum albumin were obtained from Sigma (St. Louis, MO). Human TGFβ1 was purchased from R&D Systems (Minneapolis, MN). [14C] Chloramphenicol was from MP Biomedicals (Irvine, CA). Protease Inhibitor Cocktail was obtained from Roche (Indianapolis, IN). Poly(dIdC) was from GE Healthcare (Piscataway, NJ). [α-32P]dATP and [α-32P]dCTP were purchased from ICN Biochemicals (Irvine, CA).
Cell culture
LX-2, a human stellate cell line, was a gift from Dr. Scott L. Friedman from the Mount Sinai School of Medicine (New York). The LX-2 cells were cultured in 75-cm2 tissue culture flasks and maintained in Dulbecco's modified Eagle's medium containing 10% FBS, penicillin G (100 U/mL), streptomycin (100 mg/mL), and Fungizone (2.5 mg/mL) at 37°C with a humidified atmosphere of 5% CO2 and 95% air. Drosophila Schneider L2 cells were obtained from American Type Culture Collection (Manassas, VA). The Drosophila cells were maintained at room temperature in Schneider's Drosophila medium, supplemented with 10% FBS, penicillin G (100 U/mL), and streptomycin (100 mg/mL). Mouse embryonic fibroblast (MEF) cell lines, wild type (wt), and Smad3−/− were kindly provided by Dr. Kathleen C. Flanders from NIH. The MEF cells were cultured and maintained as described for the LX-2 cells.
Plasmids
The −2.3 kb to +42 CAT (p2.3k α1CAT) and the −174 to +42 CAT (p174 α1CAT) constructs of the human α1(I) collagen promoter (Jimenez et al., 1994) were provided by Dr. Sergio A. Jimenez from the Thomas Jefferson University (Philadelphia, PA). The luciferase construct of pGL3-2.3k α1 and the shorter pGL3-804 α1 were made as described previously (Tang et al., 2003). The pGL-174 α1 construct was made by cutting p174 α1CAT with HinfI and KpnI filling the 5′-end overhang caused by the HinfI cut and inserting it by blunt ligation into the KpnI site of the pGL3-Enhancer vector. pGL3-Enhancer, pGL3-Promoter, and phRL-CMV were purchased from Promega (Madison, WI). The expression vectors pPacSp1 and pPadh were obtained from Dr. Robert Tijan from the University of California (Berkeley, CA). The expression vectors pRK5F-Smad2, pRK5F-Smad3 and pRK-DPC4–Flag (Smad4), and the empty vector pRK5 were obtained form Dr. Rik Derynck from the University of California (San Francisco, CA). The p3TP-lux plasmid, a chimeric TGFβ inducible reporter containing multimerized TGFβ-binding elements inserted upstream of the human plasminogen activator one gene (Wrana et al., 1992), was a gift of Dr. Joan Massague from the Memorial Sloan-Kettering Cancer Center (New York).
Site-directed mutagenesis
Oligonucleotides containing 2 bp substitutions were designed for site-directed mutagenesis. The method for the mutagenesis of the α1(I) collagen promoter utilized the strategy of overlap extension using PCR described by Ho et al. (1989). Successful mutations were confirmed by the DNA sequencing facility of our Department of Biochemistry.
Nuclear protein extraction
LX-2 stellate cells were harvested for nuclear protein extraction as described previously (Potter et al., 1991). Protein concentration was determined by the method of Lowry et al. (1951). The nuclear protein extracts were aliquoted and stored at −80°C.
Electrophoretic mobility shift assay
The sequence of the α1(I) collagen promoter oligonucleotide used for electrophoretic mobility shift assay (EMSA) was Sp1.1 5′-CTTCCCTCCTCCTCCCCCTCTCC-3′ (−164 to −142) (Jimenez et al., 1994). Mutated oligonucleotides contained 2 bp nucleotide substitutions shown in Figure 1. Complimentary strands of oligonucleotide were annealed, and the double-stranded oligonucleotides were labeled with [α-32P]dATP and [α-32P]dCTP using Klenow enzyme according to the method of Feinberg and Vogelstein (1983). DNA–protein binding reactions were performed following the previously described EMSA procedure (Potter et al., 1991). For supershift EMSA experiments, rabbit polyclonal antibodies to Smad2/3, Smad4, and Sp1 (Santa Cruz Biotechnology, Santa Cruz, CA) and to cKrox (Rockland, Gilbertsville, PA) were used.
FIG. 1.
- WT: 5′-CTTCCCTCCTCCTCCCCCTCTCC-3′
- M1: 5′-CTTTTCTCCTCCTCCCCCTCTCC-3′
- M2: 5′-CTTCCTACCTCCTCCCCCTCTCC-3′
- M3: 5′-CTTCCCTTTTCCTCCCCCTCTCC-3′
- M4: 5′-CTTCCCTCCATCTCCCCCTCTCC-3′
- M5: 5′-CTTCCCTCCTTTACCCCCTCTCC-3′
- M6: 5′-CTTCCCTCCTCCTTTCCCTCTCC-3′
- M7: 5′-CTTCCCTCCTCCTCCTTCTCTCC-3′
- M8: 5′-CTTCCCTCCTCCTCCCCTACTCC-3′
- M9: 5′-CTTCCCTCCTCCTCCCCCTTACC-3′
Western blot analysis
Western blot was performed as described previously (Tang et al., 2003). The total protein from the nuclear extracts was resolved on a 4–15% gradient denaturing polyacrylamide gel. The Western blots were probed with antibodies to Smad2 (R&D Systems), Smad3, Smad4 (Rockland), phospho-Smad2 (Cell Signaling Technology, Danvers, MA), phospho-Smad3 (R&D Systems), Sp1 (Santa Cruz Biotechnology), cKrox (Rockland), or β-actin (Santa Cruz Biotechnology). Horseradish peroxidase–conjugated goat anti-rabbit IgG (GE Healthcare) was used as the secondary antibody. The blots were viewed by enhanced chemiluminescence reaction (ECL; GE Healthcare).
Transient transfections and luciferase assay
LX-2 stellate cells were grown as described until they reached 60% confluence. Each cell culture plate was transfected with 3 μg of the pGL3-α1 collagen promoter and 2 μg of pPacSp1, pRK5F-Smad2, pRK5F-Smad3, or pRK-DPC4-Flag (Smad4), and 3 μg of pGL3-Enhancer and pGL3-Promoter vectors as controls, using calcium phosphate precipitation (Di Nocera and David, 1983). Transfection efficiency was determined by cotransfection of 0.2 μg of the Renilla luciferase vector phRL-CMV (Promega). Four hours after transfecting, the cells were washed twice with phosphate-buffered saline (PBS) and then shocked with 10% dimethyl sulfoxide. A similar procedure (no shock) was followed for transfection in Drosophila cells. The cells were harvested 12–24 h after transfection. For siRNA experiments and experiment with MEFs, liposome-mediated transfection was employed using HiPerfect Transfection Reagent (Qiagen, Valencia, CA). Wild-type and Smad3−/− MEFs were transfected with 3 μg of pGL3-2.3k α1 or p3TP-lux expression vector and 0.3 μg of phRL-CMV. TGFβ1 (10 ng/mL) or reconstitution buffer (control) was added at 48 h, and the cells were harvested 24 h later. The harvested cells were exposed to two freeze–thaw cycle in Reporter Lysis Buffer (Promega). Firefly luciferase activity was determined using the Dual luciferase assay system (Promega) and normalized to total cell protein (Lowry et al., 1951).
CAT assay
CAT activity was determined as described previously (Potter et al., 1991). After autoradiography, the TLC plates were cut and counted by liquid scintillation to quantify the acetylated [14C]chloramphenicol. Percent acetylation was determined as the average quotient of acetylated products and total (unacetylated and acetylated chloramphenicol).
Ultraviolet cross-linking of nuclear protein to oligonucleotides
The binding reactions were carried out as in EMSA for 30 min at room temperature followed by 15 min on ice using nuclear extracts (16 μg of the protein content) and radioactively labeled oligonucleotides (50 fmol) specifying the Sp1.1-binding site. After completion of the reaction, the samples are kept on ice followed by exposure to UV radiation at 120,000 mJ for 10 min in a UV Crosslinker 1800 (Stratagene Cloning Systems, La Jolla, CA). The cross-linked protein–DNA complexes were resolved on 4–15% gradient denaturing SDS-polyacrylamide gel.
Chromatin immunoprecipitation assay
One- and two-step cross-link procedures were performed. For the two-step procedure (Nowak et al., 2005; Evans et al., 2007) protein–protein cross-linking, utilizing disuccinimidyl glutarate to facilitate n-hydroxysuccinimide–mediated protein–protein esterification, was done before protein–DNA cross-linking with formaldehyde. Briefly, T75 flasks containing LX2 cells in monolayer were washed three times with PBS, after which PBS containing 1 mM MgCl2, pH 8.0, to improve cell attachment, was added to each flask. Disuccinimidyl glutarate was then rapidly added, and mixed to a final concentration of 2 mM, and the cells were incubated at room temperature for 45 min. The cells were then washed three times with PBS, after which they were incubated at room temperature for 15 min in 10 mL of a fresh 1% formaldehyde solution in PBS containing 1 mM MgCl2, pH 8.0. The flasks were again washed three times with PBS, and the cells scraped and transferred to 2 mL microfuge tubes. Following the method outlined in the Magna chromatin immunoprecipitation (ChIP) procedure manual (Upstate, Charlottesville, VA), nuclei were prepared from the cells and lysed. The nuclear lysates were sonicated to produce soluble chromatin with an average length between 200 and 500 bp. The protein–DNA complexes were immunoprecipitated with the respective antibodies, the protein–DNA cross-links reversed and the DNA purified using spin columns (Qiagen). Anti-Acetyl Histone H3 was used for the positive control and normal rabbit IgG for the negative control. A one-step ChIP assay (Aparicio et al., 2005) was also done by washing the cultured LX-2 cells with PBS, then incubating them with 10 mL of fresh 1% formaldehyde solution in PBS for 15 min. The formaldehyde was then quenched by adding 1 M glycine to a final concentration of 145 mM, after which the flasks were incubated at room temperature for 5 min. The flasks were washed three times with PBS and scraped, and the DNA was recovered as described above (Nowak et al., 2005).
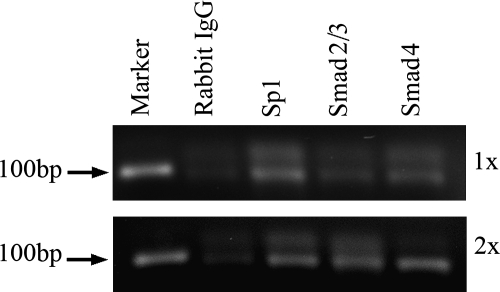
The purified DNA was used as a template for PCR amplification. Primers specific for the detection of the region −199 to −93 were as follows: 5′-AGAGCTGCGAAGAGGGGAGAT-3′ and 5′CAGCCAATCAGAGCTGCCT-3′. Forward and reverse primers of E-cadherin (R&D Systems), which is known to associate with Smad2/3, were used as positive controls for the PCR detection of the E-cadherin promoter. Owing to the high GC content of the region of the collagen gene to be amplified, the PCR was optimized by titrating enhancer solution (Gibco-Invitrogen) in the reaction and performing 30 cycles of PCR amplification. A 1× enhancer solution concentration proved optimal.
siRNA suppression of Smad2, Smad4, or Sp1 in LX-2 cells
Four siRNA sequences against different regions of the human Smad2, Smad3, Smad4, or Sp1 genes together with negative and positive control siRNA obtained from Qiagen were tested. The siRNA with the highest silencing efficiency was selected for the subsequent experiments. The siRNA target sequences were 5′-ATGGTGCGAGAAGGCGGTCAA-3′ (for Smad3 silencing), 5′-AGCAAGGTTGCACATAGGCAA-3′ (for Smad4 silencing), and 5′-CAGCAAGTTCTGACAGGACTA-3′ (for Sp1 silencing). The Smad2-validated siRNA target sequence was not disclosed by the manufacturer. LX-2 cells were transfected with the siRNA using HiPerFect transfection reagent (Qiagen). Silencing of Smad2, Smad3, Smad4, or Sp1 expression was confirmed by RT-qPCR and Western blot.
Statistical analysis
Data were analyzed with Student's t-test when appropriate or by two-way analysis of variance when comparing means of more than two groups.
Results
Effect of the α1(I) collagen promoter mutations on binding of nuclear proteins
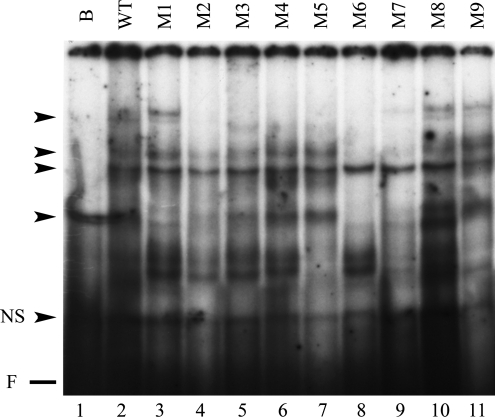
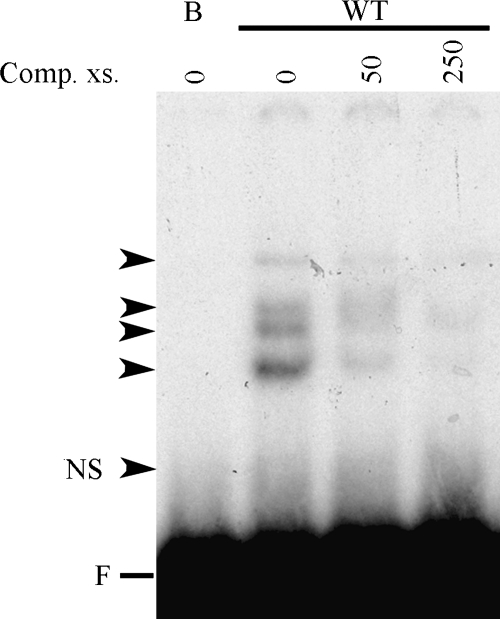
A mutational analysis of the TGFβ1-binding responsive sequence of the α1(I) collagen promoter was performed by EMSA of protein–DNA complexes formed with the labeled Sp1.1 oligonucleotide that was sequentially mutated by two base pairs between −161 and −144 (Fig. 1). This analysis reveals four major complexes with the wt oligonucleotide. The distal mutation of the oligonucleotide (M2) eliminates the uppermost and lowest complexes, while a more proximal mutation of the oligonucleotide (M6) eliminates the upper two complexes and the lowest complex. None of the mutations eliminate the third complex from the top. All four complexes formed with the wt oligonucleotide are competed away in the presence of the unlabeled oligonucleotide (Fig. 2).
FIG. 2.
EMSA showing binding of nuclear extracts form LX-2 stellate cells to the wild-type (wt) Sp1.1 oligonucleotide in the presence of excess competing unlabeled oligonucleotide. The complexes formed are indicated by the arrowheads. NS indicates nonspecific band; F indicates the position of the free probe.
The role of TGFβ1 in activation of the α1(I) collagen promoter
In transient transfection experiments in LX-2 cells, the addition of TGFβ1 (10 ng/mL) activated the wt p174 α1CATwt promoter (p < 0.05), but not the mutated p174 α1CATmut2 or p174 α1CATmut6 promoters (Table 1). Of note is that p174 α1CATmut2 had an enhanced basal activity as compared with the p174 α1CATwt promoter (p < 0.01), which was suppressed by the TGFβ1.
Table 1.
Effects of Transforming Growth Factor β1 on the Activation of the Wild-Type and Mutated α1(I) Collagen Promoter
| |
CAT activity expressed as a percentage of the wild promoter |
|
|---|---|---|
| Promoter | Control | TGFβ1 |
| p174 α1CAT wt | 100 ± 16 | 141 ± 2.3a |
| p174 α1CATmut2 | 198 ± 26b | 90 ± 6.4c |
| p174 α1CATmut6 | 92 ± 15 | 56 ± 1.7 |
The promoters were transfected into LX-2 cells using the calcium precipitation method.
The cells were treated with TGFβ1 (10 ng/mL) or reconstitution buffer (control) for 24 h.
Data are expressed as means ± SE of six determinations.
p < 0.05 versus respective control.
p < 0.01 versus respective control.
p < 0.05 versus p174 α1CATmut2.
Binding of nuclear proteins to the α1(I) collagen promoter
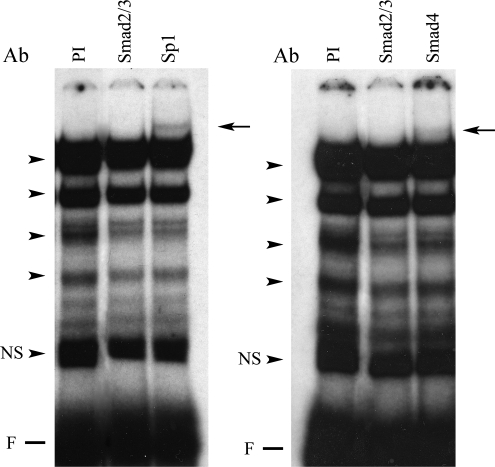
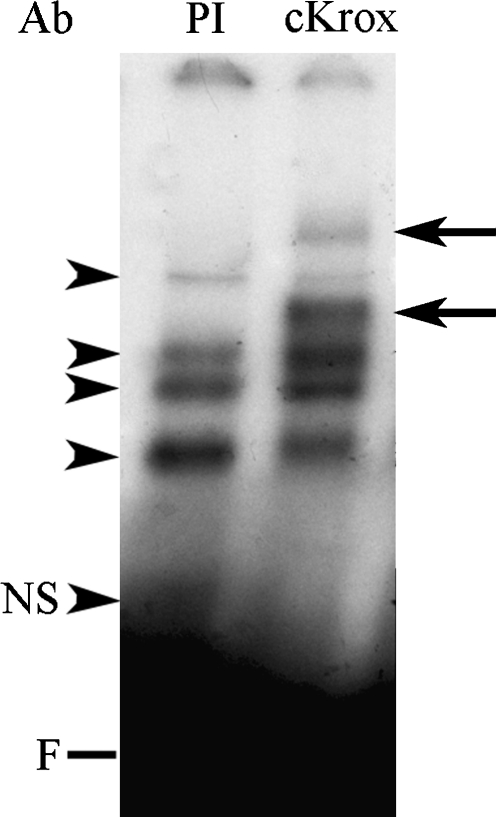
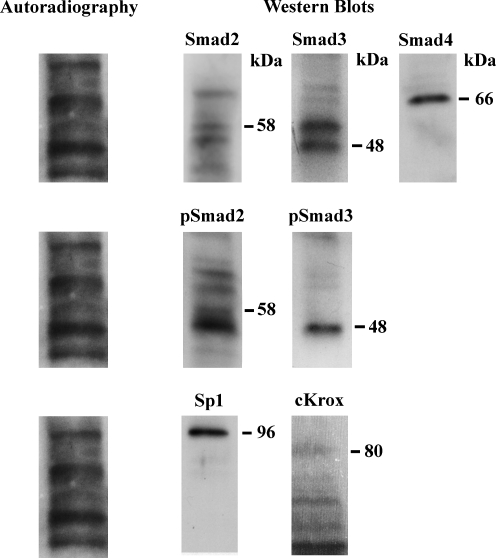
EMSA of the Sp1.1 oligonucleotide with nuclear protein extracts from the LX-2 cells shows four DNA–protein complexes (Fig. 3). Antibodies to Sp1 and to Smad4 (Fig. 3) did not decrease or eliminate any of the DNA–protein complexes but resulted in the appearance of supershifts. Antibody to Smad2/3 (Santa Cruz Biotechnology) did not change the complexes or result in the appearance of a supershift. Antibody to cKrox resulted in a decrease in the uppermost and lowest complexes and the appearance of two supershifted bands (arrows) (Fig. 4). To further evaluate the binding of the Sp1 and Smads to the α1(1) collagen promoter, nuclear extracts from LX-2 cells treated with TGFβ1 (10 ng/mL) were UV crosslinked with the Sp1.1 oligonucleotide specifying the region −164 to −142. Figure 5 shows that Smad2 (pSmad2), Smad3 (pSmad3), and Smad4 with a kDa of 58, 48, and 66, respectively, bind to the Sp1 oligonucleotide (kDa of 17.8). The binding is stronger for pSmad3 than for pSmad2. Sp1 with a kDa of 96 and cKrox with a kDa of 80 also bind to the Sp1 oligonucleotide, and in this case the bound cKrox is weaker than the bound Sp1.
FIG. 3.
EMSA with supershift showing binding of nuclear extracts form LX-2 stellate cells to the Sp1.1 oligonucleotide. The EMSAs were performed with 10 μg of nuclear extract and the labeled oligonucleotide. For supershifts the antibodies (Ab) to Smad2/3, Smad4, and Sp1 (1:200 dilution) or preimmune serum (PI) were added to the reaction mixture. The arrowheads indicate the protein–DNA complexes formed. The arrows indicate the location of the supershifted complexes. NS indicates a nonspecific band; F indicates the position of the free probe.
FIG. 4.
EMSA with supershift showing binding of nuclear extracts form LX-2 stellate cells to the Sp1.1 oligonucleotide. The EMSAs were performed with 10 μg of nuclear extract and the labeled oligonucleotide. For supershifts the antibody (Ab) to cKrox (1:200 dilution) or preimmune serum (PI) was added to the reaction mixture. The arrowheads indicate the protein–DNA complexes formed. The arrows indicate the location of the supershifted complexes. NS indicates a nonspecific band; F indicates the position of the free probe.
FIG. 5.
Autoradiography and Western blot showing the presence of Smad2, Smad3, Smad4, pSmad2, pSmad3, Sp1, and cKrox in nuclear extract from TGFβ1-treated LX-2 cells UV crosslinked to the labeled Sp1.1 oligonucleotide. The cells were exposed to TGFβ1 (10 ng/mL) for 24 h. The blots were exposed to film to determine the UV binding of the proteins and incubated with antibodies to the Smads, Sp1, and cKrox. The location of the bound proteins and their molecular mass (kDa) obtained from protein standards is shown.
The binding of Sp1 and Smads was further determined in vivo by ChIP assays after 24 h exposure of the LX-2 cells in culture to TGFβ1 (10 ng/mL). The ChIP assays demonstrate binding of Sp1 and Smads to the promoter region between −199 and −93 (Fig. 6). The 2× ChIP assay demonstrates that the binding of Smad 2/3 and Smad4 occurs in association with Sp1, and their combination results in enhanced binding of Sp1.
FIG. 6.
Chromatin immunoprecipitation (ChIP) assay of Sp1 and Smad binding. One- (1×) and two-step (2×) cross-linked DNA–protein complexes were immunoprecipitated with antibodies to Sp1, Smad 2/3, and Smad4. Negative control was performed with purified rabbit IgG. The covalent linkage was reversed, and the precipitated double-stranded DNA was amplified to the region −199 to −93 (106 bp) of the α1(I) collagen promoter.
Combination of Smad expression vectors activates the α1(I) collagen promoter
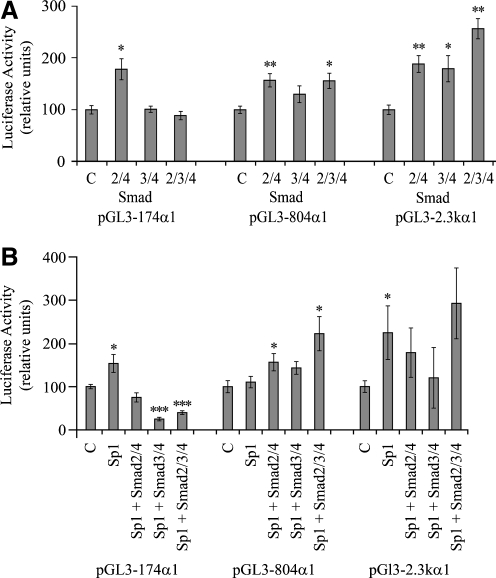
In cotransfection experiments the activating effect by the combined Smad2 and Smad4 expression vectors was of similar magnitude in the short (pGL3-174 α1wt), intermediate length (pGL3-804 α1 wt), and long (pGL3-2.3 k α1 wt) promoter (Fig. 7A), indicating that the site of action of Smad2 in combination with Smad4 is located proximally in the promoter. The combinations of Smad3 and Smad4 resulted in activation of pGL3-2.3 k α1 wt, indicating a possible site of action of Smad3 between −804 bp and −2.3 k in the α1(I) collagen promoter.
FIG. 7.
Effect of Smad expression vectors (pRK5F-Smad2, pRK5F-Smad3, and pRK-DPC4–Flag [Smad4]) on the activity of α1(I) collagen promoters in transfected LX-2 cells in (A) the absence and (B) the presence of the Sp1 expression vector (pPacSp1). Luciferase activity is expressed as the percentages of the control empty vectors, pRK5 (for Smads), pPadh (for Sp1), or their combination. The data are expressed as means ± SE of six determinations. *p < 0.05 versus control; **p < 0.01 versus control; ***p < 0.001 versus control.
The Sp1 expression vector activates the α1(I) collagen promoters
The Sp1 expression vector activated pGL3-174 α1 wt and pGL3-2.3 k α1 wt in transfected LX-2 cells (Fig. 7B). Sp1 in the presence of cotransfected Smad2 and Smad4 did not activate pGL3-174 α1 wt, while the combination of Sp1 and Smad3 and Smad4 inhibited the activity of pGL3-174 α1 wt, but did not inhibit the activities of pGL3-804 α1 wt and pGL3-2.3 k α1 wt.
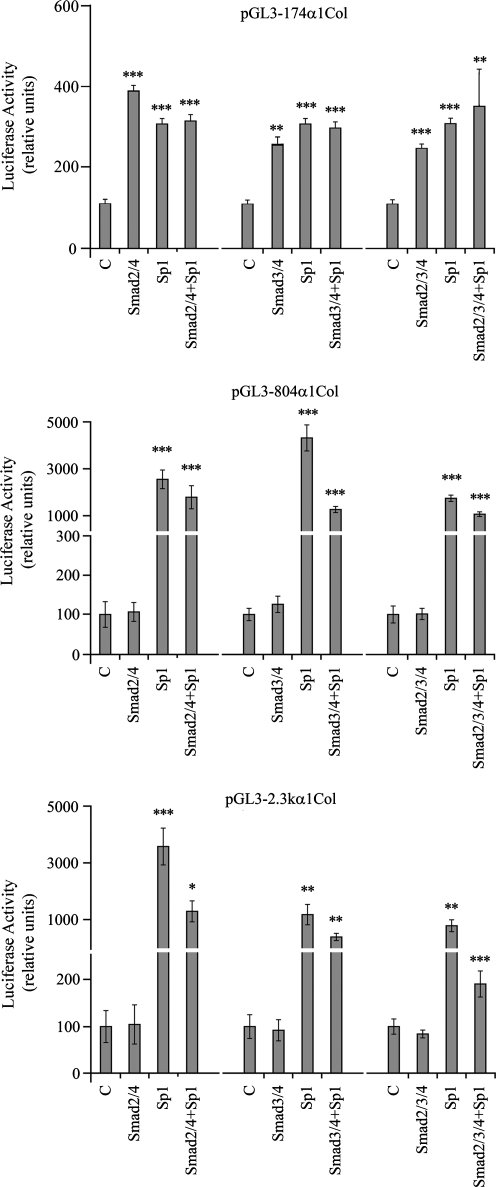
To eliminate the possible interference of endogenous Sp1, the cotransfections were repeated in Drosophila Schneider cells, which lack endogenous Sp1. The Smad expression vectors and Sp1 each independently enhanced pGL3-174 α1 wt (Fig. 8). There was no additional activation of 174 α1 wt with the combination of Smads and Sp1 than seen with either alone. The Smad expression vectors had no effect on the activity of the longer promoters, while Sp1 increased the activity of the longer promoters in the absence or presence of the cotransfected Smad expression vectors.
FIG. 8.
Effect of Smad expression vectors (pRK5F-Smad2, pRK5F-Smad3, and pRK-DPC4–Flag [Smad4]) on the activity of α1(I) collagen promoters in transfected Drosophila Schneider cells in the absence and the presence of the Sp1 expression vector (pPacSp1). Luciferase activity is expressed as the percentages of the control empty vectors, pRK5 (for Smads), pPadh (for Sp1), or their combination. The data are expressed as means ± SE of six determinations. *p < 0.05 versus control; **p < 0.01 versus control; ***p < 0.001 versus control.
Effects of Smad2, Smad3, Smad4, and Sp1 siRNAs on activation of the α1(I) collagen promoter
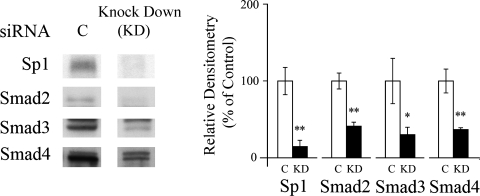
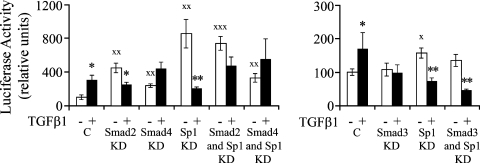
Transfection of LX-2 cells with siRNAs for Sp1, Smad2, Smad3, and Smad4 in the presence of TGFβ1 (10 ng/mL) selectively decreased their protein expression determined by Western blot (Fig. 9). The mean percent decreases in protein were 86%, 59%, 71%, and 64% for Sp1, Smad2, Smad3, and Smad4, respectively. In cotransfection experiments siRNA-mediated silencing of Smad2 expression abrogated the enhancing effect of TGFβ1 on the activity of the α1(1) collagen promoter (Fig. 10). Smad3 and Smad4 siRNAs had no effect on activation of the promoter, while Sp1 siRNA in the absence or presence of Smad2 or Smad3, but not Smad4 siRNA, prevented the effect of TGFβ1 in enhancing the promoter activity.
FIG. 9.
Effects of Sp1 and Smad siRNAs on protein expression of Sp1 and Smads determined by Western blot in the presence of TGFβ1 (10 ng/mL). C, negative control siRNA. KD, knock down. The values are expressed as means ± SE of relative densitometry of six measurements per group *p < 0.05 versus control; **p < 0.01 versus control.
FIG. 10.
Effects of selective inhibition of Sp1, Smad2, Smad3, and Smad4 by siRNAs on the activity of the cotransfected pGL3-2.3k α1 wt collagen promoter in LX-2 cells treated or not treated with TGFβ1 (10 ng/mL). C, negative control siRNA. KD, knock down. The data are expressed as means ± SE of six determinations. *p < 0.05 versus respective group not treated with TGFβ1; **p < 0.01 versus respective group not treated with TGFβ1; Xp < 0.05 versus control not treated with TGFβ1; XXp < 0.01 versus control not treated with TGFβ1; XXXp < 0.01versus control not treated with TGFβ1.
Effect of TGFβ1 on the α1(I) collagen promoter in Smad3 −/− MEFs
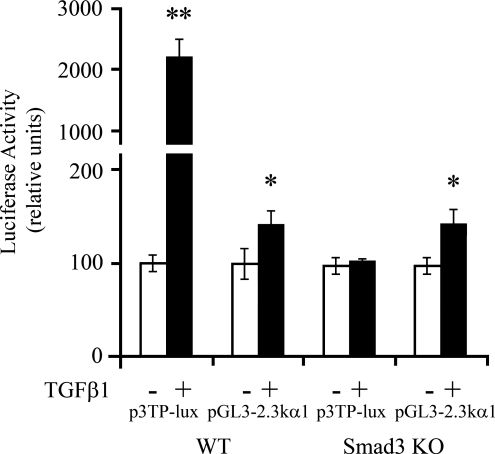
To determine whether the enhancing effect of TGFβ1 on the α1(I) collagen promoter was mediated by Smad3, the pGL3-2.3 k α1 wt promoter and the p3TP-LUX reporter were transfected into wt and Smad3−/− MEFs. As shown in Figure 11, TGFβ1 resulted in similar activation of the pGL3-2.3 k α1 wt in wt and Smad3−/− cells, while the effect of TGFβ1 on the activation of p3TP-lux was present in the wt cells but absent in the Smad3−/− cells. This indicates that Smad3 does not mediate the enhancing effect of TGFβ1 on the α1(I) collagen promoter.
FIG. 11.
Effect of TGFβ1 on the α1(I) collagen promoter in Smad3−/− (Smad3 KO) mouse embryonic fibroblasts (MEF). The pGL3-2.3k α1 wt promoter and the p3TP-lux reporter, a TGFβ1 inducible reporter containing three TGFβ1-binding elements, were transfected into wt and Smad3−/− MEF. The cells were exposed or not exposed to TGFβ1 (10 ng/mL) for 24 h. Luciferase activity is expressed as a percentage of control. Data are presented as means ± SE of six determinations. *p < 0.05 versus control without TGFβ1; **p < 0.001 versus control without TGFβ1.
Discussion
TGFβ1 is known to stimulate the human α1(I) collagen promoter and previous studies showed that the TGFβ1 responsive sequences are located between −164 and −142 bp of the promoter (Jimenez et al., 1994). Sp1 binds to this responsive element and is known to activate the collagen promoter. TGFβ1 is known to regulate the human α2(I) collagen promoter principally through the cellular Smad signal transduction pathway (Ghosh et al., 2001). TGFβ1 signaling is initiated by binding to type I and II receptors, which activates phosphorylation of Smad2 and Smad3, followed by recruitment of Smad4 forming a complex that migrates to the nucleus where it binds directly to DNA or associates with other transcription factors (Attisano and Wrana, 2002). Smad2 and Smad3 bind to a Smad recognition element that has the CAGA sequence. This sequence is present in many TGFβ1 responsive genes, including the human α2(I) collagen promoter (Chen et al., 2000), but was not found on our search of the human α1(I) collagen promoter. Our in vitro DNA binding study with EMSA and UV crosslinking indicated that in addition to Sp1, Smads may bind to the CC(GG)-rich −164 to −142 bp sequence of the α1(I) collagen promoter. The binding of Smads to the −164 to −142 sequence was demonstrated by UV crosslinking, while the binding of Smad4 was additionally demonstrated by detection of a supershifted complex on EMSA with the Smad4 antibody. The antibodies to Smad4 and to pSmad3 show single bands on Western blot in the crosslink experiments, while antibodies to Smad2 and Smad3 are less specific for the individual Smads and cross react showing two bands, namely, the 58 and 48 kDa bands of Smad2 and Smad3, respectively. In addition, we show that by EMSA with supershift and by UV crosslinking that the transcription factor, cKrox also binds to the −164 to −142 sequence. Previous studies had shown that cKrox binds to the −224 to −199 (Widom et al., 1997) and −61 to −122 sequences of the human α1(I) collagen promoter (Kypriotou et al., 2007) and to other GC-rich sequences in the murine α1(I) collagen promoter (Widom et al., 1997). The binding of Sp1 corresponds on EMSA and UV crosslinking to the upper DNA-binding complex, while the binding of Smads corresponds to a lower complex. The binding of cKrox by UV crosslinking corresponds to a small middle complex. In our in vivo study of DNA binding with ChIP assay shows that Smad proteins bind in association with Sp1 to the CC(GG)-rich −164 and −142 bp sequence of the α1(I) collagen promoter, thus enhancing the binding of Sp1 and in this manner activating the collagen promoter. Smads and Sp1 interact by forming complexes that mediate the effects of TGFβ in activating transcription of a variety of genes (Docagne et al., 2004) This is demonstrated with the α1(I) collagen promoter in this study by the 2 × crosslink experiment in which the associated proteins (Sp1 and Smads) are crosslinked before the protein–DNA crosslinking. It is also known that various transcription factors can recruit Smads to promoters without the need for a specific Smad DNA-binding site (Grinberg and Kerppola, 2003). As in our study, Smads were found to interact with Sp1 and activate the promoter of the human p21Waf1/Cip1 gene that has Sp1-binding sites but no known Smad recognition sites (Moustakas and Kardassis, 1998; Pardali et al., 2000). However, in a study of TGFβ1-dependent transcription of an activin target gene, the mammalian forkhead domain protein, FAST2, was shown to recruit complexes of Smad2 and Smad4 to bind to a GC-rich site of the promoter and activate it (Labbe et al., 1998). For the human α2(I) collagen promoter, a physical interaction between Sp1 and Smad was demonstrated, but in this case there are also separate Sp1 and Smad recognition elements (Poncelet et al., 2001; Zhang et al., 2005). The mutated p174 α1CAT promoters, in which the Sp1- and Smad-binding sites were eliminated, are not responsive to the enhancing effect of TGFβ1, indicating that both Sp1 and Smads are important in mediating the effect of TGFβ1.
In the present study, the effect of TGFβ1 in activating the α1(I) collagen promoter was mediated principally by Smad2/Smad4, and by Sp1. The site of action of the combination of Smad2 and Smad4 was located to the proximal pGL3-174 α1wt promoter, since the activation was similar to that obtained with the longer pGL3-2.3k α1wt promoter. By contrast, the site of action of the combination of Smad3 and Smad4 in the transfected LX-2 cells was only apparent in the GL3-2.3k α1wt promoter. These findings differ from the effects of TGFβ1 on activation of the human α2 (I) collagen transcription (Ghosh et al., 2001) as well as on the transcription of many other profibrotic genes (Panish et al., 2006) that are principally mediated by Smad3 signaling. However, it has become apparent that the activation of other genes such as matrix-metalloproteinase-2 is mediated by Smad2 (Panish et al., 2006).
Sp1 activated the short pGL3-174 α1wt promoter and longer α1(I) collagen promoter in LX-2 cells, in agreement with prior studies (Chen et al., 1998). In the Sp1-deficient Drosophila Schneider cells, Sp1 also resulted in activation of the proximal pGL3-174 α1 wt promoter, and marked activation of the longer promoters, suggesting that Sp1 acts at various sites in the promoter. The human α1(I) collagen promoter has multiple GC box elements, of which three have been located in the proximal pGL3-174 α1 wt promoter at − 77 to −93, −121 to −112, and −164 to −142 (Chen et al., 1998), and others are located more distally (Jimenez et al., 1994). Sp1 in combination with Smads2 and 4 failed to activate the pGL3-174 α1 wt promoter in the LX-2, while the combination of Sp1 and Smads 3, and 4 resulted in inhibition of promoter activity. This however was not the case in the Sp1-deficient Drosophila cell transfections, where the activation of the pGL3-174 α1 wt promoter was similar with either the Sp1 or Smads alone or their combinations. The results with transfection in the LX2 cells are hard to interpret since endogenous Sp1 proteins in these cells could alter the binding and or interact with other transcription factors such as Smads. Other investigators have shown differences in results of the effects of Sp1 on the effects and other transcription factors on activation of the murine α1(I) collagen promoter depending on whether the transfected cells are NIH 3T3 fibroblasts or Drosophila cells. In those studies Sp1 and nuclear protein I inhibited the promoter in the NIH 3T3 fibroblasts, while enhancing it in the Drosophila cells (Nehls et al., 1992).
The importance of Sp1 in mediating activation of the α1(I) collagen promoter was further demonstrated by showing that Sp1 knockdown with Sp1 siRNA prevented the effect of TGFβ1 in enhancing promoter activity. As regard Smads, knockdown of Smad2 by siRNA also prevented the effect of TGFβ1 in enhancing the promoter, while knockdown of Smad3 by siRNA had no effect. Knockdowns are rarely complete. The percent knockdown of Smad2 by Smad2 siRNA was 59% and clearly abrogated the enhancement by TGFβ of the α1(I) collagen promoter activity in the transfection experiments. By contrast, while the percent knockdown of Smad3 by Smad3 siRNA was higher, at 71%, it had no effect on the activation of the promoter. Further, activation by TGFβ1 of the α1(I) collagen promoter was similar in transfected wt and Smad3−/− MEFs, indicating that Smad3 does not mediate the enhancing effect of TGFβ1 on the α1(I) collagen promoter. Other signaling pathways and transcription factors were found to act as mediators of the effect of TGFβ1 (Panish et al., 2006) on gene regulation, but their role relating to the up-regulation of the human α1(I) collagen gene remains unknown. These alternative pathways include the phosphoinositol-3-kinase pathway and MAPK signaling, which interacts with the Smad pathway (Roberts, 2002).
In conclusion, this study shows that Smads bind in association with Sp1 to the CC(GG)-rich TGFβ1 responsive element of the human α1(I) collagen promoter that lacks the classical Smad recognition element, thus enhancing the binding of Sp1 and in this manner activating the collagen promoter.
Acknowledgments
This work was supported by Grant AA000626 from the United States Public Health Service. P.S. was a postdoctoral fellow on National Research Service Award 2 T32 AA07467 from the National Institute of Alcohol Abuse and Alcoholism (NIH). The authors acknowledge assistance from The Hopkins Digestive Diseases Basic Research Development Center (NIH Grant 2464388) in the performance of this study.
Disclosure Statement
No competing financial interests exist.
References
- Aparicio O. Geisberg J.V. Sekinger E. Yang A. Moqtaderi Z. Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005;Chapter 21:21.3.1–21.3.33. doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- Attisano L. Wrana J.L. Signal transduction by the TGF-β superfamilty. Science. 2002;296:1646–1647. doi: 10.1126/science.1071809. [DOI] [PubMed] [Google Scholar]
- Chen S.J. Artlett C.M. Jimenez A. Varga J. Modulation of human α1(I) procollagen gene activity by interaction with Sp1 and Sp3 transcription factors in vitro. Gene. 1998;215:101–110. doi: 10.1016/s0378-1119(98)00268-6. [DOI] [PubMed] [Google Scholar]
- Chen S.J. Yuan W. Lo S. Trojanowska M. Varga J. Interaction of Smad3 with a proximal Smad-binding element of the human α2 (I) procollagen gene promoter required for transcriptional activation of TGF-β. J Cell Physiol. 2000;183:381–392. doi: 10.1002/(SICI)1097-4652(200006)183:3<381::AID-JCP11>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Di Nocera P.P. David I.B. Transient expression of genes introduced into cultured cells in Drosophila. Proc Natl Acad Sci USA. 1983;80:7095–7098. doi: 10.1073/pnas.80.23.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docagne F. Gabriel C. Lebeurrier N. Lesne S. Hommet Y. Plawinski L. Mackenzie E.T. Vivien D. Sp1 and Smad transcription factors cooperate to mediate TGF-β-dependent activation of amyloid-β precursor protein gene transcription. Biochem J. 2004;383:383–399. doi: 10.1042/BJ20040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P.M. Zhang W. Chen X. Yang J. Bhakat K. Liu C. Kruppel-like factor 4 is acetylated by p300 and regulates gene transcription via modulation of histone acetylation. J Biol Chem. 2007;282:33994–34002. doi: 10.1074/jbc.M701847200. [DOI] [PubMed] [Google Scholar]
- Feinberg A.P. Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gaidarova S. Jimenez S.A. Inhibition of basal and transforming growth factor–β-induced stimulation of COL1A1 transcription by the DNA intercalators, mitoxantrone and WP631, in cultured human dermal fibroblasts. J Biol Chem. 2002;277:38737–38745. doi: 10.1074/jbc.M201742200. [DOI] [PubMed] [Google Scholar]
- Ghosh A.K. Yuan W. Mori Y. Chen S.J. Varga J. Antagonistic regulation of type I collagen gene expression by interferon-γ and transforming growth factor-β. Integration at the level of p300/CP transcriptional coactivators. J Biol Chem. 2001;276:11041–11048. doi: 10.1074/jbc.M004709200. [DOI] [PubMed] [Google Scholar]
- Grinberg A.V. Kerppola T. Both Max and TFB3 cooperate with Smad proteins to bind the plasminogen activator inhibitor-1 promoter but they have opposite effects on trasncriptional activity. J Biol Chem. 2003;278:11227–11236. doi: 10.1074/jbc.M211734200. [DOI] [PubMed] [Google Scholar]
- Ho S.N. Hunt H.D. Horton R.M. Pullen J.K. Pease L.R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- Jimenez S.A. Varga J. Olsen A. Li L. Diaz A. Herhal J. Koch J. Functional analysis of human α1(I) procollagen gene promoter. Differential activity in collagen-producing and–nonproducing cells and response to transforming growth factor β1. J Biol Chem. 1994;269:12684–12691. [PubMed] [Google Scholar]
- Kypriotou M. Beauchef G. Chadjichristos C. Widom R. Renard E. Jimenez S.A. Korn J. Maquart F.X. Oddos T. Von Stetten O. Pujol J.P. Galera P. Human collagen Krox up-regulates type I collagen expression in normal and scleroderma fibroblasts through interaction with Sp1 and Sp3 transcription factors. J Biol Chem. 2007;282:32000–32014. doi: 10.1074/jbc.M705197200. [DOI] [PubMed] [Google Scholar]
- Labbe E. Silvestri C. Hoodless P.A. Wrana J.L. Attisano L. Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol Cell. 1988;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- Lowry O.H. Rosebrough N.J. Farr A.L. Randall R.J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Moustakas A. Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci USA. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls M.C. Grapilon M.L. Brenner D.A. NF-I/Sp1 switch elements regulate collagen α1(I) gene expression. DNA Cell Biol. 1992;11:443–452. doi: 10.1089/dna.1992.11.443. [DOI] [PubMed] [Google Scholar]
- Nowak D.E. Tian B. Brasier A.R. Two-step cross-linking method for identification of NF-KappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39:715–724. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- Panish M.K. Wahab N.A. Colville-Nash P. Hendry B.M. Dockrell M.E.C. The differential role of Smad2 and Smad3 in the regulation of profibrotic TGFβ1 responses in human proximal-tubule epithelial cells. Biochem J. 2006;393:601–607. doi: 10.1042/BJ20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali K. Kurisaki A. Moren A. ten Dijke P. Kardassis D. Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21Waf1/Cip1 regulation by transforming growth factor-β. J Biol Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- Poncelet A.C. Schnaper H.W. Sp1 and Smad proteins cooperate to mediate transforming growth factor-β1-induced α2(I) collagen expression in human glomerular mesagial cells. J Biol Chem. 2001;276:6983–6992. doi: 10.1074/jbc.M006442200. [DOI] [PubMed] [Google Scholar]
- Potter J.J. Mezey E. Christy R.J. Crabb D.W. Stein P.M. Yang V.W. CCAAT/enhancer binding protein binds and activates the promoter to the rat class I alcohol dehydrogenase gene. Arch Biochem Biophys. 1991;285:246–251. doi: 10.1016/0003-9861(91)90356-n. [DOI] [PubMed] [Google Scholar]
- Roberts A.B. The ever-increasing complexity of TGF-β signaling. Cytokine Growth Factor Rev. 2002;13:3–5. doi: 10.1016/s1359-6101(01)00027-2. [DOI] [PubMed] [Google Scholar]
- Shi Y. Wang Y.F. Jayaraman L. Yang H. Massague J. Pavletich N.P. Crystal structure of a SmadMH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Tang M. Potter J.J. Mezey E. Activation of the human α2(I) collagen promoter by leptin in not mediated by transforming growth factor β responsive elements. Biochem Biophys Res Commun. 2003;31:629–633. doi: 10.1016/j.bbrc.2003.10.167. [DOI] [PubMed] [Google Scholar]
- Widom R.L. Culic I. Lee J.Y. Korn J.H. Cloning and characterization of hcKrox, a transcriptional regulator of extracellular matrix expression. Gene. 1997;198:407–420. doi: 10.1016/s0378-1119(97)00360-0. [DOI] [PubMed] [Google Scholar]
- Wrana J.L. Attisano L. Carcamo J. Zentella A. Doody J. Laiho M. Wang X.F. Massague J. TGFβ signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Zhang W. Ou J. Inagaki Y. Greenwel P. Ramirez F. Synergistic cooperation between Sp1 and Smad3/Smad4 mediates transforming growth factor β1 stimulation of the α2(I)-collagen (COL1A2) transcription. J Biol Chem. 2000;275:39237–39245. doi: 10.1074/jbc.M003339200. [DOI] [PubMed] [Google Scholar]
- Zhang X. Yang J. Li Y. Liu Y. Both Sp1 and Smad participate in mediating TGF-β1-induced HGF receptor expression in renal epithelial cells. Am J Physiol Renal Physiol. 2005;288:F16–F26. doi: 10.1152/ajprenal.00318.2003. [DOI] [PubMed] [Google Scholar]