Summary
An early granulomatous response, characterized by collections of white blood cells at foci surrounding pathogens, occurs after infection by many intracellular organisms, including Listeria, but how these clusters become organized and for what purpose remain poorly understood. Here, we show that dendritic cell (DC) activation by Listeria nucleates rapid clustering of innate cells, including granulocytes, NK cells and monocytes, to sites of bacteria propagation where IL-12 is expressed in the spleen. Clustered NK cells express IFNγ, which is necessary for the maturation and activation of co-localized monocytes to TNF- and iNOS-producing DCs (TipDCs). NK cell clustering is necessary for IFNγ production and requires pertussis toxin-sensitive recruitment, in part mediated by CCR5, and MyD88-mediated signaling. Thus, spatial organization of the immune response by DCs between 6 and 24 hr ensures functional activation of innate cells, which restricts pathogens before adaptive immunity is fully activated.
Introduction
The key role of DCs in ingesting, sensing, processing and presenting antigen to activate adaptive immunity has been amply documented (Banchereau et al., 2000). Although much is known about how adaptive immunity is regulated by DCs, little is known about their role in controlling innate immune cells at the early stage of infection. The capacity of DCs to capture antigens effectively renders these cells key sentinels in scanning for pathogens that have surmounted peripheral barriers. Similarly, DCs in the spleen respond rapidly to bloodborne infections and function to prime adaptive immunity (Jung et al., 2002). Increasing numbers of reports demonstrate cross-talk between DCs, NK cells and various myeloid cells, including granulocytes (Ludwig et al., 2006; Moretta et al., 2006; Newman and Riley, 2007), but little understanding exists regarding how these cells become positioned such that these cellular interactions can occur to control infection.
Listeria monocytogenes is a gram-positive bacterium with requirements from both innate and adaptive arms of immunity for its control by the host (Pamer, 2004). Innate cells such as macrophages, neutrophils and NK cells contribute to host survival by controlling infection at early times (Conlan and North, 1994; Czuprynski et al., 1994; Dunn and North, 1991; Rogers and Unanue, 1993; Unanue, 1997), but T cell-mediated adaptive immunity is necessary to achieve sterile eradication of the bacteria. Recently, subpopulations of CD11c+ DC-like cells have been implicated at various stages of Listeria immunity, including in splenic sequestration (Neuenhahn et al., 2006), in the early activation of CD8+ T cells (Jung et al., 2002) and in effector function through maturation to a specialized TNF- and iNOS-producing DC, the TipDC (Serbina et al., 2003a; Serbina et al., 2003b). Precisely how and which types of DCs interact with the previously identified components of innate immunity, including NK cells and neutrophils, is not entirely clear, but such insights may establish hierarchical ordering of the innate and adaptive immune responses that may reveal common underpinnings. With these questions in mind, we undertook a systematic flow cytometric- and immunohistochemistry-based approach to understand how the initial response to Listeria unfolds early after infection in vivo. The findings show a previously unappreciated role for DCs in the organization of innate immune cells into regional clusters where immune cell activation occurs prior to the induction of adaptive immunity.
Results
Clustering and activation of NK cells and myeloid cells after Listeria infection
In naive, uninfected mice, co-staining of spleen sections with Thy1.2 (T cells) and B220 (B cells) facilitated delineation of the T and B cell areas which comprise the white pulp surrounding the central arterioles, and the red pulp where cells remain transitorily before returning to the blood (Cyster, 2005) (Figure 1A). CD11b+ myeloid cells were individually dispersed within the red pulp. Due to difficulty in positioning endogenous NK cells in situ (Andrews et al., 2001; Bajenoff et al., 2006; Dokun et al., 2001a), the dynamic nature of NK cell activation has not been presented in histological studies. By using a perfusion-fixation method (Andrews et al., 2001), followed by tyramide amplification, we reliably visualized the C-type lectin receptor, NK1.1, which is highly expressed on NK cells. Although anti-NK1.1 antibody stains both NK and NKT cells as assessed by flow cytometric analysis of dispersed cells, expression of NK1.1 is lower on NKT cells (Figure 1C) and falls below the threshold of detection by immunohistochemistry, as confirmed by using mice deficient in NK cells (data not shown, and see below, Figure 4D, E). Thus, NK1.1 staining in tissues identifies NK1.1+ NK cells, and these are positioned in uninfected mice in the red pulp together with CD11b+ myeloid cells, consistent with prior reports (Andrews et al., 2001; Berg et al., 2005; Dokun et al., 2001a; Walzer et al., 2007). CD11b is expressed at lower levels on NK cells and is below the level of detection by immunohistochemistry using these methods (data not shown). After infection with Listeria, CD11b+ cells and NK cells underwent marked reorganization with the myeloid cells organized centrally and the NK cells arranged peripherally about the central myeloid cluster. The clusters of CD11b+ cells and NK cells were localized around the central arterioles in the T cell zones of the white pulp areas (Figure 1A, arrows).
Figure 1. Innate cell clustering and IFNγ production in response to Listeria.
(A) Positioning of myeloid (CD11b+), NK cells (NK1.1+) and B cells (B220+) in the spleens from mice either uninfected or infected with Listeria 24 hr previously. T and B areas labeled; RP, red pulp; arrows, central arteriole; 40X.
(B) Positioning of IFNγ-producing cells by immunohistochemistry. Conditions as in legend to (A).
(C) Gating strategy for NK cells, NKT cells and T cells in spleen before (−) and 24 hr after (+) Listeria infection. Intracellular IFNγ shown in right panels for cell types designated.
(D) Percentages of IFNγ-positive cells among designated cell types before (−) and 24 hr after (+) infection with Listeria. n = 3 for (−), 4 for (+); red bars, means.
(E) Total numbers of IFNγ-positive cells in the spleen before (−) and 24 hr after (+) infection with Listeria. n = 3 for (−), 4 for (+); red bars, means.
Figure 4. IFNγ from NK cells mediates the maturation of monocytes to TipDCs.
(A) Quantitation of numbers of respective cells after depletion and infection with 2000 Listeria shown. n = 6 for each group; **, p<0.01.
(B) Immunohistochemistry of mice treated with anti-Gr1 or isotype control antibody and infected 48 hrs later with Listeria. The spleens were examined 24 hr later. Myeloid cell depletion had no effect on NK cell clustering. 40X.
(C) Spleen immunohistochemistry of positioning of NK cells, myeloid cells and cells producing IFNγ 24 hr after Listeria infection in wild-type or CCR2-deficient (CCR2 KO) mice. 40X.
(D) Mice were treated with anti-asialoGM1 (anti-aGM1) or control Ab (rabbit IgG), and spleens analyzed 48 hr later for proportions of NK cells (uppermost gates) and NKT cells (right gates). Distribution of asialoGM1 staining on NK and NKT cells from spleen denoted in the right panel, with percentage of NKT cells that were stained with anti-aGM1 indicated.
(E) NK cell-independent clustering of CD11b+ myeloid cells. Mice were treated with control rabbit antibody (control) or anti-asialoGM1(aGM1) and infected 48 hr later with Listeria. The next day, spleens were harvested and imaged using immunohistochemistry. 40X.
(F) Monocytes in the spleen were gated as in Figure 3B and analyzed for expression of iNOS and MHC class II from the wild-type (WT), IFNγ-deficient (IFNγ KO) and RAG1-deficient (RAG KO) mice infected 48 hr earlier with Listeria. Representative flow cytometry plots left, summary of results right. Mean of WT arbitrarily set as 100%. MFI, Mean Fluorescence Intensity; n = 4 for each group; **, p<0.01.
(G) Analysis of TipDC-like monocytes in spleen from wild-type (WT) and γc-deficient ( γc KO) mice infected 48 hr earlier with Listeria. Summary with quantitation shown. Mean of WT arbitrarily set as 100%. n = 4 for WT, 3 for γc-deficient group; **, p<0.01.
(H) Analysis of TipDC-like monocytes in spleen from RAG1-deficient (RAG KO) mice treated with control rabbit antibody (control) or anti-asialoGM1 antibody (anti-aGM1) and infected with Listeria 48 hr before. Summary with quantitation shown. Mean of control Ab-treated mice arbitrarily set as 100%. n = 3 for control, 4 for anti-aGM1 group; *, p<0.05.
(I) Analysis of TipDC-like monocytes in spleen from RAG1-deficient mice and RAG1xIFNγ-deficient (RAG-IFNγ DKO mice infected with Listeria 48 hr before. Summary with quantitation shown. Mean of RAG1-deficient mice arbitrarily set as 100%. n = 3 for each group; **, p<0.01.
We investigated whether reorganization of these cells was accompanied by functional consequences. Since IFNγ is critical for resistance to Listeria and is produced by NK cells, we examined the location of IFNγ expression. The position of IFNγ-producing cells closely correlated with that of the NK cells collaring the myeloid clusters (Figure 1B). We also examined the sources of early IFNγ produced during the first 24 hr of infection by using quantitative flow cytometric analysis of isolated cells without further stimulation ex vivo. We observed three IFNγ-producing cells: NK (CD3−, NK1.1hi), NKT (CD3+, NK1.1lo), and CD3 T (CD3+, NK1.1−) cells (Figure 1C). Substantial percentages of NK and NKT cells produced IFNγ at this early time point (Figure 1C, D). Although the percentage of IFNγ-producing cells among CD3 T cells was much smaller than that of NK or NKT cells, IFNγ production by CD3 T cells is not trivial when measured by numbers of IFNγ-producing cells (Figure 1E).
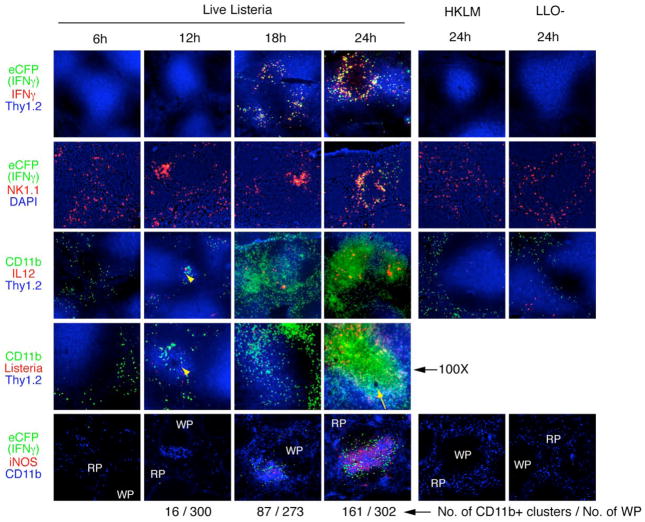
The resistance to Listeria infection requires IFNγ, IL-12, TNFα, iNOS, neutrophils, NK cells, and a recently identified cell type called TipDCs (Pamer, 2004; Serbina et al., 2003a; Serbina et al., 2003b; Unanue, 1997). However, the crosstalk between the innate cells and the regulation of their activation to produce cytokines and effector molecules have been little studied in situ. To understand better the organizational process, we carried out a kinetic analysis of the mobilization of NK and CD11b+ myeloid cells into clusters, the production of cytokines (IL-12, IFNγ) and the elaboration of the effector molecule, iNOS. To confirm the production of IFNγ by clustered NK cells, we used newly generated mice with a sensitive IFNγ reporter allele. These mice carry a targeted IFNγ allele with a gene for enhanced cyan fluorescent protein (eCFP) fused via a self-cleaving 2A sequence (de Felipe and Ryan, 2004) to the IFNγ start codon, thus generating an intracellular eCFP molecule each time that IFNγ mRNA is translated and thus reliably marking cells that have expressed, rather than taken up or bound, the IFNγ protein (Figure S2). The earliest IFNγ+ clusters were observed, albeit rarely, at 12 hr and this increased with each subsequent time point (Figure 2, first and second row). Importantly, co-staining with an NK cell marker and eCFP (IFNγ confirmed that the majority of NK cells in the clusters were producing IFNγ, whereas the unclustered NK cells that remained in the red pulp did not express IFNγ (Figure 2, second row; Figure S2). IFNγ production by NK cells in clusters suggested that NK cell activation might be spatially regulated. Clustering of NK cells and CD11b+ myeloid cells began at 12 hr around the terminal arterioles where IL-12 production and Listeria were first detected (Figure 2, third and fourth rows, arrowheads). The clusters expanded in size and increased in numbers around the central arterioles into the T cell zones (Figure 2, third and fourth row, arrow). iNOS was first detected in CD11b+ cells at 18 hr and increased over 48 hr, and was expressed in close association with IFNγ-producing cells (Figure 2, bottom row). The results of multiple histologic studies are summarized in Table S1.
Figure 2. Time course of innate immune response after Listeria infection in spleen.
Spleens from the IFNγ-reporter mice infected with Listeria were harvested at the indicated times. Molecules stained for are color-matched and shown to the left of each row: eCFP, enhanced cyan fluorescent protein as a marker for IFNγ, green; cytokines (IFNγ, IL-12), red; Thy1.2 staining marks T cells, blue; NK1.1, NK cells, red; CD11b, CD11b+ myeloid cells, green in the third and fourth rows, blue in the bottom row; Listeria, red; iNOS, inducible nitric oxide synthase, red; HKLM, heat-killed Listeria; LLO-, listeriolysin O-deficient Listeria; RP, red pulp; WP, white pulp; arrowheads, terminal arteriole; arrow, central arteriole; 40X except the penultimate row (100X).
Previous studies showed that when mice were challenged with either heat-killed Listeria (HKLM) or viable listeriolysin O (LLO)-deficient Listeria, which are unable to escape from intracellular vacuoles to propagate infection, IL-12, an important cytokine for IFNγ production, was not produced in vivo (Liu and Kurlander, 1995; Poston and Kurlander, 1992), and Mac-3+ myeloid cells were not recruited to the spleen (Serbina et al., 2003a) or to the infectious foci (Muraille et al., 2005). This raised the question whether invasive live Listeria, but not HKLM or LLO-deficient Listeria, could trigger spatial reorganization of innate cells for IFNγ production in addition to production of IL-12. Neither cell clusters nor immune cell activation occurred following injection of heat-killed Listeria or LLO-deficient Listeria, indicating that cytosolic invasion is critical for clustering of NK and CD11b+ cells, cytokine production and iNOS expression (Figure 2).
The identity and location of CD11b+ cells responding to Listeria infection
The synchronized clustering of CD11b+ cells and NK cells prompted us to investigate whether these processes are inter-dependent. To devise a way to deplete CD11b+ cells, we first characterized the types of CD11b+ myeloid cells in the spleen. Although previous reports have suggested that Listeria is trapped by macrophages in the marginal zone and carried into the white pulp by Mac-3+CD11bhi macrophage-like cells to activate DCs for T cell priming (Aichele et al., 2003; Conlan, 1996; Muraille et al., 2005), the characterization and functions of these cells are not clear. After excluding T, B and NK cells using standard markers, combinations of staining for PDCA-1, CD11c, CD11b, Ly6C and Ly6G were used to establish gating parameters for plasmacytoid DCs (pDCs), conventional DCs (cDCs), neutrophils (polymorphonuclear leukocytes, PMNs) and monocytes in resting, uninfected mice. Monocytes in resting spleen were defined by their CD11b+, Ly6C+, Ly6G−, MHC class IIlo phenotype (Figure 3A, B). These monocytes have a phenotype (Mac-3hi, CD11bint, Figure S3A) similar to that described on TipDC precursor monocytes identified in bone marrow (Serbina and Pamer, 2006), demonstrating that cells of this phenotype are present in uninfected spleen. After infection, the numbers of neutrophils and monocytes increased (data not shown), and some of the cells with a monocyte phenotype matured to take on the characteristic phenotype of TipDCs. Monocyte/TipDCs became MHC class IIhi and approximately 20 –30% of the cells were positive for iNOS (Figure 3B).
Figure 3. Characterization of CD11b+ myeloid cells in spleen.
(A) Gating scheme and markers for the delineation by flow cytometry of plasmacytoid DCs (pDC, orange), conventional DCs (cDC, green), polymorphonuclear leukocytes (PMNs, neutrophils, blue), and monocytes (mono, red) from spleens of uninfected mice. Expression of Ly6G on various myeloid populations which are color-coded as in the third panel and graphed at top right.
(B) Simplified scheme to separate neutrophils (PMNs) and monocytes (mono) in uninfected and infected mouse spleen and differentiation of monocytes to TipDCs. Mice were infected 48 hr previously with Listeria. Expression of iNOS and MHC class II from cells gated is depicted, with percentage of monocytes expressing iNOS shown. Gray histogram, isotype control; solid line, iNOS or class II.
(C) Distinguishing monocytes and neutrophils by using 7/4, Ly6C and Ly6G. Mice were infected 48 hr previously with Listeria. Percentages of gated cell types among live cells shown. Expression of Ly6G (G1, red line; G2, blue line) and iNOS on gated populations graphed at right. Gray histogram, isotype control; solid line, iNOS.
(D) Positioning of monocytes and neutrophils in the spleen of mice before and 24 hr after Listeria infection. The inset areas in the leftmost panels shown at right. WP, white pulp; RP, red pulp; T, T cell area; B, B cell area; arrows, monocytes; arrowheads, neutrophils; 200X.
We next localized monocytes and neutrophils differentially in the spleen using immunohistochemistry. Based on recent reports showing that the combination of 7/4 and Ly6G could separate neutrophils (7/4hi, Ly6G+) from monocytes (7/4hi, Ly6G−) by flow cytometry (Taylor et al., 2003; Tsou et al., 2007), we determined whether gating by using 7/4 is comparable to our gating scheme for neutrophils and monocytes. The cells gated in G1 and G2 by using 7/4 and Ly6C (Figure 3C, second column) gave the same proportions as monocytes and neutrophils gated by using CD11b and Ly6C, respectively (Figure 3C, compare first with second column). Further, the cells in G1 and G2 are superimposed on monocytes and neurophils, respectively (Figure 3C, third column). Ly6G is expressed predominantly on G2 cells and iNOS by G1 cells (Figure 3C, histograms). Based on this knowledge, we could distinguish the location of monocytes (7/4+, Ly6G− : Figure 3D, arrows) and neutrophils (7/4+, Ly6G+ : Figure 3D, arrowheads) in spleens after co-staining for 7/4 and Ly6G. In resting, uninfected spleens, these cells are located in the red pulps (Figure 3D, upper panels). After infection, these cells were found in the white pulps (Figure 3D, lower panels). At high resolution, we observed that some CD11b+Ly6G− monocytes, but not the CD11b+Ly6G+ neutrophils, expressed iNOS, confirming their identification as TipDCs (Figure S3B). These cells were distinct from F4/80+ resident macrophages in the spleen (Figure S3C). Thus, CD11b+ cells revealed by immunohistochemistry represent monocytes and neutrophils, but not cDC or pDC.
The clustering of NK cells and CD11b+ cells are independent processes
Gr1 recognizes a common epitope shared by Ly6G expressed on neutrophils and Ly6C on neutrophils, monocytes, TipDCs and pDCs, and depletion of Gr1+ cells by using anti-Gr1 antibody, which efficiently removed neutrophils, TipDC-like monocytes and (less efficiently) pDCs from the spleen (Figure 4A, Figure S4A), had little impact on NK cell clustering (Figure 4B). In CCR2-deficient mice, where monocytes, but not neutrophils, are retained in the bone marrow and absent in the spleen ((Serbina and Pamer, 2006), data not shown), clustering of NK cells and CD11b+ cells was also little affected (Figure 4C). These CD11b+ cells are Ly6G+ neutrophils (data not shown), indicating that neutrophil clustering is also independent of monocytes. When NK cells were depleted using anti-asialoGM1, CD11b+ myeloid cells still clustered (Figure 4D, E; Figure S4B, C). Thus, clustering of NK cells is not required for CD11b+ cell clustering and vice versa.
Role of IFNγ produced by NK cells in TipDC differentiation
With our data showing the clustering and activation of NK cells surrounding the CD11b+ myeloid cells, we reasoned that IFNγ secretion from NK cells may be crucial for activation of monocytes to differentiate to TipDCs. The capacity to identify immature monocyte/TipDCs (Figure 3) allowed us to assess the role of IFNγ from NK cells in this maturation process by quantitating the expression of MHC class II and iNOS on these cells. Since NK cells, NKT cells and T cells contribute to early IFNγ after Listeria infection, we first compared TipDC maturation in IFNγ-deficient mice with WT or RAG1-deficient mice, which are deficient in NKT and T cells (and B cells), but not NK cells. Although TipDC maturation was unaffected in RAG1-deficient mice as compared to wild-type (WT) mice, mice deficient in IFNγ demonstrated a significant reduction in MHC classII+, iNOS+ TipDCs and in the degree of iNOS induction, as assessed by mean fluorescence intensity (Figure 4F). Further, TipDC maturation by these criteria was significantly decreased in γc-deficient mice, which lack NK cells and functional T cells, as well as in RAG1-deficient mice treated with anti-asialoGM1 to deplete NK cells (Figure 4G, H; Figure S4D, E). This was not due to impaired clustering of monocytes, however, since the clustering of myeloid cells, including monocytes, still occurred when NK cells were depleted (Figure 4E; Figure S4B, C). Finally, crossing RAG1-deficient mice onto the IFNγ-deficient background resulted in loss of TipDC maturation, consistent with a predominant and requisite role for IFNγ from NK cells in driving the maturation of monocytes to the TipDC phenotype characterized by high expression of MHC class II and iNOS (Figure 4I; Figure S4F). When monocytes isolated from bone marrow were cultured with IFNγ, they differentiated to iNOS+, MHC class II+ TipDCs (Figure S4G). Therefore, IFNγ is both necessary and sufficient for TipDC differentiation.
CD11c+ DCs are required for innate cell clustering and activation
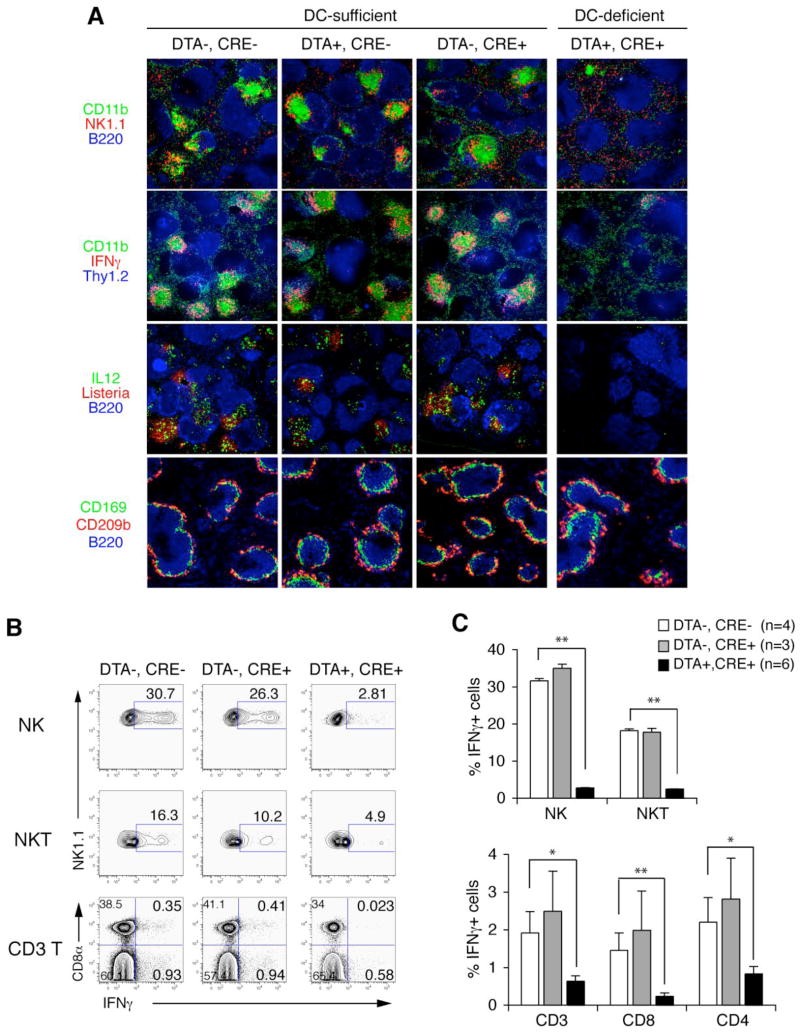
Although cross-talk between DCs and other innate cells has been shown by many in vitro studies (Ludwig et al., 2006; Moretta et al., 2006; Newman and Riley, 2007), little has been investigated in vivo. We hypothesized that CD11c+ DCs are required for innate cell clustering and thereby activation in the spleen in response to Listeria. We challenged mice expressing either a CD11c-cre transgene, which directs Cre recombinase expression to the CD11c-expressing population of cells, including DCs (Caton et al., 2007), and/or a diphtheria toxin A gene downstream of a flox-stop-flox site engineered into the ubiquitously expressed ROSA26 locus. As assessed by flow cytometric analysis, CD11c-cre-mediated depletion removed both conventional DCs and, although less completely, plasmacytoid DCs, but not NK cells, NKT cells or monocytes (Figure S5A, B and data not shown). After Listeria infection, innate cell clustering was abolished in CD11c-depleted mice but not in the controls (Figure 5A, first row). Depletion of the CD11c-expressing cells resulted in almost complete loss of IFNγ expression in NK cells, as well as in NKT and CD3 T cells, as assessed by immunohistochemistry and by quantitative flow cytometric analysis (Figure 5A, second row; Figure 5B, C); IL-12 expression was similarly lost (Figure 5A, third row). Consistent with the previous study showing the role of CD8α+ DC in bacterial entry and spreading in the spleen (Neuenhahn et al., 2006), infectious foci of propagating Listeria were diminished (although not absent) after CD11c-targeted cell depletion (Figure 5A, third row; Figure S5C). This is not due to absence of particle-trapping marginal zone macrophages and metallophilic macrophages, depletion of which was shown to impair the early control of Listeria infection (Aichele et al., 2003), because these cells are preserved in DC-deleter mice (Figure 5A, bottom row).
Figure 5. CD11c+ cells are required for innate cell clustering and activation in response to Listeria.
(A) Spleen immunohistochemistry for indicated markers 24 hr after (upper panels) and before (bottom panels) infection with Listeria. DTA, diphtheria toxin A; Cre, CD11c-cre recombinase; CD169 (sialoadhesin, MOMA-1), a marker for metallophilic macrophages; CD209b (SIGNR1, ER-TR9), a marker for marginal zone macrophages; 40X.
(B) Representative flow cytometric analysis of NK cells, NKT cells, and T cells that express intracellular IFNγ when isolated from the indicated mice 24 hr after infection with Listeria. Percentages of IFNγ-positive cells among designated cell types shown.
(C) Quantitation of flow cytometric analysis from multiple experiments. *, p<0.05; **, p<0.01.
Clustering of NK cells is required for IFNγ production
Our data showing that only the clustered NK cells produce IFNγ led us to the hypothesis that NK cell clustering is a pre-requisite for IFNγ production. To test this, we used pertussis toxin (PTX), which inhibits Gαi-protein-coupled receptors including chemokine receptors, to attempt to block the mobilization of NK cells after infection. Treatment of mice with pertussis toxin (PTX) resulted in loss of clustering by NK cells as well as the almost complete abrogation of IFNγ production from NK cells and other cells at 24 hr as confirmed by both immunohistochemistry and flow cytometric analysis (Figure 6A, top and middle rows; Figure 6B). PTX treatment neither affected the entry of Listeria into the spleen nor the appearance of early IL-12 in association with bacteria (Figure 6A, bottom row), which were impaired by the absence of DCs (Figure 5). Thus, a PTX-sensitive pathway acts downstream of sensing of bacteria by DCs. PTX did not decrease the total numbers of NK cells in the spleen (data not shown). However, the ratio between the numbers of monocytes and neutrophils changed with PTX treatment, consistent with some differential effects of PTX under these conditions (Figure S6B). To confirm whether the effect of PTX was intrinsic to NK cells, we isolated NK cells from RAG1-deficient mice, labeled them with CFSE and treated the cells with either PTX or vehicle control before adoptively transferring the cells intravenously to recipient wild-type mice. On the next day, mice were infected with Listeria and the activation of host (CFSE-negative) and donor (CFSE-positive) NK cells was assayed by immunohistochemistry to assess cell positioning and IFNγ production, and by flow cytometry for quantitation of IFNγ production. The association of NK cells with clusters was completely inhibited by PTX pre-treatment, and the capacity to generate IFNγ was PTX-sensitive and entirely cluster-associated (Figure 6C, D; Figure S6A).
Figure 6. NK cell clustering and activation requires a pertussis-toxin sensitive receptor.
(A) Spleen immunohistochemistry for indicated markers 24 hr after Listeria infection of mice pre-treated with PBS or pertussis toxin (PTX). 40X.
(B) Representative (upper panels, percentages of IFNγ-positive cells among designated cell types shown) and quantitative summary (lower panels) of intracellular IFNγ detection from the indicated spleen cells 24 hr after Listeria infection. n= 4 for each group; **, p<0.01.
(C) NK cells purified from RAG1-deficient mice were labeled with CFSE and treated with PBS (vehicle control) or PTX before transfer to recipient WT mice. Spleen sections were stained for CFSE and IFNγ 24 hr after infection with Listeria. Quantitation of movement of donor (CFSE+), as compared to host (CFSE−) NK cells into clusters, as defined by association with an IFNγ-producing cluster, from multiple experiments is shown below. n = 3 for PBS, 4 for PTX group; **, p<0.01.
(D) NK cells prepared as in (C) were isolated from infected mice 24 hr after Listeria infection and stained for IFNγ Loss of IFNγ production in PTX-treated NK cells shown in graph. Mean of IFNγ-producing host cells was set arbitrarily as 100%. n = 4 for each group; **, p<0.01.
(E) Spleen immunohistochemistry of positioning of NK cells, myeloid cells, and cells producing IFNγ 24 hr after Listeria infection in wild-type or CCR5-deficient (CCR5 KO) mice. 40X.
(F) Quantitation of flow cytometric analysis from multiple experiments of spleen cells staining for intracellular IFNγ 24 hr after Listeria infection from the designated mice. Mean of WT was set arbitrarily as 100%; n = 9 for WT, 8 for CCR5 KO group; *, p<0.05, **, p<0.01.
The capacity of PTX to inhibit Gαi protein-coupled receptors suggested that chemokine and chemokine receptor interactions might underlie the nucleation of NK cells in clusters in the spleen. We extensively screened a number of chemokine receptors expressed on NK cells (Gregoire et al., 2007) in Listeria-infected mice for effects on NK cell clustering and IFNγ expression. Many, including CCR2, CCL2, CCL7, CCR1, CXCR3, CCL3, CCL5 and CX3CR1, showed no deficit in genetically deficient animals (Figure 4C, Figure S7). In contrast, mice deficient in CCR5 had a consistent but partial defect in NK cell clustering; myeloid cell clustering was little effected. Although fewer in number, clustered NK cells appeared to express IFNγ, suggesting that CCR5 ligand(s) is required for the nucleation of NK cells but not for their activation once clustered. Altogether, CCR5 deficiency caused a significant reduction in the numbers of IFNγ-secreting cells by approximately 50% in the various cell populations (Figure 6E, F; Figure S6C). Altogether, these results support the notion that activation of innate cells is spatially controlled and regulated by their attraction into clusters.
Signaling pathways involved in the clustering and activation of innate cells
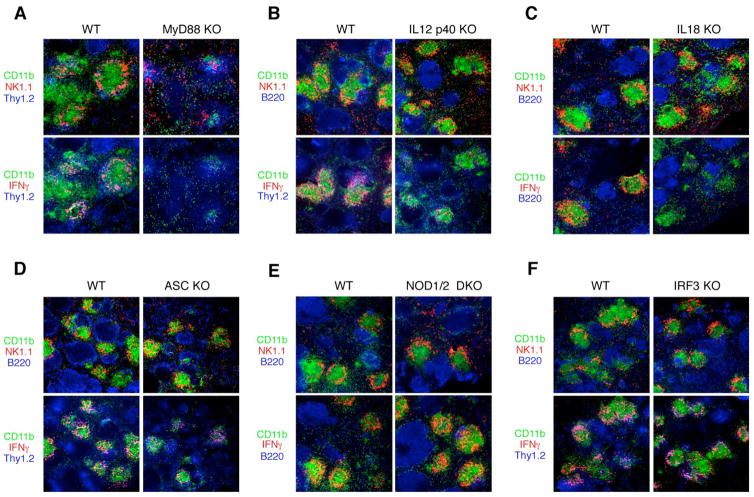
Next, we investigated the signaling pathways responsible for the clustering of innate cells and their subsequent production of IFNγ. Clustering was substantially, but not completely, attenuated and IFNγ production was almost completely attenuated in the absence of MyD88 (Figure 7A), presumably due to the absence of signaling through Toll-like receptors and IL-1-family members that use receptors coupled with this adaptor, such as IL-1 and IL-18, which have been implicated in early innate defense against Listeria (Seki et al., 2002; Unanue, 1997). This defect is not due to lesser numbers of Listeria present in the spleen (Figure S8A). Both IL-18 and IL-12 were required for IFNγ production by clustered NK cells but not for clustering itself, which was intact in mice lacking IL-12p40 or IL-18 (Figure 7B, C). Thus, these data differentiate the role of chemokines required to cluster innate cells from the role of effector cytokines required to activate them for IFNγ production. In contrast, a defect in IL-1R signaling due to lack of IL-1R1 chain has no effect on clustering or IFNγ production (Figure S8B). Since our data showed that cytosolic invasion is critical for clustering and IFNγ production (Figure 2), we reasoned that cytosolic sensor(s) might initiate or cooperate with MyD88-mediated signaling pathways. A critical component of a major cytosolic sensing program is ASC (apoptosis-associated speck-like protein containing a CARD), a key part of the inflammasome used to activate caspase-1, which is required for the post-translational processing of IL-1 and IL-18 to functional proteins. WT Listeria, but not HKLM or LLO-deficient Listeria, have been demonstrated to activate the inflammasome pathway (Mariathasan et al., 2004; Mariathasan et al., 2006; Ozoren et al., 2006). Further, bacterial RNA or transfected cytoplasmic DNA induced activation of caspase-1 through the ASC-inflammasome (Kanneganti et al., 2006; Muruve et al., 2008). Another candidate is cytosolic NOD (nucleotide-binding oligomerization domain) molecules which are activated by peptidoglycans, a major cell wall component of bacteria (Kanneganti et al., 2007). However, ASC-deficiency or combined NOD1/NOD2 deficiency had negligible impact on the clustering of NK cells or their activation to produce IFNγ (Figure 7D, E). Intracellular bacterial DNA also triggers type I IFN production (Stetson and Medzhitov, 2006) via IRF3. However, IRF3 (Figure 7F) or type I IFN receptor, IFNAR1 (data not shown) were not required as assessed by the normal phenotype for these cellular activities in genetically deficient mice.
Figure 7. Signaling pathways involved in innate immune cell clustering and IFNγ production.
(A)–(F), Immunohistochemistry of co-infected matched WT or designated knockout (KO) mice to assess NK cell clustering (upper panels) and IFNγ production (lower panels) 24 hr after Listeria infection.
Discussion
These experiments establish a hierarchical program for the activation of innate immune cells that is orchestrated by CD11c+ DCs in the spleen. By a process dependent on live Listeria that are competent to reach the cytosol, but independent of the cytosolic ASC-associated inflammasome or NOD1/2 proteins, IL-12 and IL-18 are induced at foci of bacterial infection within rapidly forming clusters of neutrophils and monocytes. CCR5 ligands and as-yet-unidentified other chemoattractants recruit and localize NK cells to the outside edges of the myeloid cell clusters that surround infectious foci. Clustering of NK cells at sites of IL-12 and IL-18 production results in IFNγ expression, which in turn is necessary for the maturation of monocytes to TipDCs which express iNOS and TNFα, and which are required to restrict early infection (Serbina et al., 2003a; Serbina et al., 2003b). Although NKT cells and T cells contribute to IFNγ production, NK cells are the predominant source of IFNγ necessary for TipDC maturation. Thus, DCs, which have a well-described role in activating adaptive immunity by facilitating the clustering of T cells, are also crucial for the recruitment and activation of innate cells into clusters that surround pathogens before adaptive responses can be activated. Clustering was required for the activation of NK cells, exposing a novel paradigm by which innate cells become recruited into ‘fields’ of cytokine-expressing cells in order to spatially regulate their activation.
The findings presented here (Figure 5), consistent with a previous report (Neuenhahn et al., 2006), suggest a model wherein bloodborne bacteria are trapped and sensed by either resident or blood immigrant DCs in the spleen near the terminal arterioles where clustering initiates (Figure 2). An alternative possibility, which is not mutually exclusive, is that DCs might sense the presence of Listeria indirectly and initiate an inflammatory response, which traps the organisms but thereby enhances their local proliferation and eventual spread in the absence of adaptive immune responses. Previous studies showed that marginal zone macrophages and metallophilic macrophages trap Listeria (Aichele et al., 2003; Conlan, 1996) and that when they were depleted, Listeria spread to the red pulp and other organs at high doses of infectious bacteria (Aichele et al., 2003). However, in the mice deleted of DCs as used in our study, these macrophage populations are left intact (Figure 5A) and Listeria was not found in organs other than the spleen and liver (data not shown). Therefore, DCs facilitate the entry and early expansion of bacteria within clusters of innate immune cells in the spleen, thus localizing the infection to sites amenable to later control by the innate and adaptive immune system. Further study of the precise role of DCs in coordinating these activities between these two arms of the immune system are needed. Very recently, Aoshi et al visualized the redistribution of Listeria to T cell areas and the subsequent clustering of Listeria-specific T cells, a process which was mediated by DCs at early times after infection (Aoshi et al., 2008), providing a link to the adaptive limb of immunity. On a technical note, their use of PTX 24 hr earlier than as used in our experimental protocol blocked the translocation of Listeria to the white pulps, probably due to impaired mobilization of Listeria-trapping cells.
Our study demonstrates a novel regulatory mechanism used to activate early IFNγ production in response to bacterial infection. Although it is known that IFNγ is produced by NK cells and bystander memory CD8 T cells taken from the spleens of Listeria-infected mice as measured ex vivo (Berg et al., 2003, 2005; Lertmemongkolchai et al., 2001), the activation of cells to produce IFNγ in vivo through a spatially regulated process has not been previously noted. Here, we show the causal link between the mobilization of NK cells and their activation to produce IFNγ. From the experiments performed in the various genetically deficient mice we report, it appears that NK cells are recruited to areas where the cytokines IL-12 and IL-18 can activate IFNγ production. Although it is likely that contact-dependent events additionally regulate this process, inhibition of the activating receptor NKG2D on NK cells with a blocking antibody did not impede NK cell clustering or IFNγ secretion (data not shown). Although clustering proceeded independently of Gr1-positive myeloid cells, NK cells take a strategic position around the centrally clustered monocytes and provide the predominant source of early IFNγ required for differentiation of monocytes to TipDCs, as shown through induction of iNOS expression. Thus, in contrast to a previous study showing a lack of mobilization of adoptively transferred NK cells in Listeria infection (Berg et al., 2005), our study reveals a mechanistic explanation of how NK cells can activate monocytes to mature into TipDCs. A subset of NK cells were localized in marginal zone and lymphoid follicles when mice were infected with viruses (Dokun et al., 2001a) and it will be interesting to see whether this clustering is also associated with functional activation of NK cells and other innate cells interacting with NK cells. Several studies have examined the differential expression of IFNγ among NK cell subsets in viral infection (Daniels et al., 2001; Dokun et al., 2001b; Tay et al., 1999), but such studies have not been done in bacterial infection. Since it has been shown that NK cell subsets can be defined by the differential expression of Ly49 molecules, we used an array of antibodies against different Ly49 molecules. The overall contribution for IFNγ-producing NK cells by each subset was proportional to the frequency of each subset in NK cells, indicating a widespread response among NK cell subsets after Listeria infection (Figure S9A). Recent studies suggested that a subset of NK cells expressing B220 and CD11c produced more IFNγ than B220− and/or CD11clow/− NK cells when cultured in vitro with various stimuli (Blasius et al., 2007; Vosshenrich et al., 2007). However, we did not detect any biased expression of these molecules among IFNγ-producing NK cells in our studies (Figure S9B).
We tested a number of chemokines and chemokine receptors that have been reported to be important for immunity mediated by NK cells. Although not entirely quantitative, the ability to screen many clusters in many genotypes of mice was sufficient to detect a partial defect in NK cell clustering and IFNγ production in CCR5-deficient mice that was verified by flow cytometric studies. This partial inhibition, together with our observation that some iNOS is still expressed in the absence of IFNγ, may reconcile the difference with the previous report that CCR5-deficient mice have minimal differences in total spleen IFNγ, resistance to Listeria, or TipDC differentiation, which were measured by different experimental schemes (Zhong et al., 2004). The partial phenotype may reflect redundancy in the chemokine ligands or the functional redundancy of CCR5 with other receptors. For example, some of the ligands of CCR5 are shared by CCR1 (Cyster, 2005), which may mask the contribution of each receptor. However, testing this through the use of doubly-deficient mice will be challenging because the ccr5 gene is located close to the ccr1 gene. Similarly, a partial phenotype due to multiple ligands for a chemokine receptor was observed for the CCR2 ligands, CCL2 (MCP-1) and CCL7 (MCP-3) (Jia et al., 2008; Tsou et al., 2007).
As assessed using a combined histologic and flow cytometric approach, we established the identities of the diverse types of CD11b+ myeloid cells in the spleen. As we demonstrated, treatment with anti-Gr1 antibody depletes neutrophils, monocytes and pDCs, suggesting that care be taken in interpreting individual roles for neutrophils or plasmacytoid DCs (Conlan and North, 1994; Czuprynski et al., 1994; Rogers and Unanue, 1993; Yoneyama et al., 2005) by using anti-Gr1 antibody. We also confirmed that MyD88 is essential in organizing innate immune cells within infectious foci in the spleen, in support of prior findings that the recruitment of neutrophils to infectious foci in the liver was impaired in MyD88-deficient mice (Seki et al., 2002). Studies of the inflammasome have been made in a number of infection models (Mariathasan and Monack, 2007), including Listeria, in which cytosolic invasion and nucleic acids of bacterial origin were shown to activate caspase-1 via the ASC-inflammasome, leading to production of active IL-1 and IL-18 in vitro (Kanneganti et al., 2006; Mariathasan et al., 2004; Mariathasan et al., 2006; Muruve et al., 2008; Ozoren et al., 2006). However, our data show little contribution of the ASC-inflammasome in vivo in either the clustering of innate immune cells or in the activation of IFNγ production. Indeed, studies in caspase-1-deficient mice showing weak susceptibility (Tsuji et al., 2004) and no defect in the differentiation of TipDCs (Serbina et al., 2003a). The difference in the defect of IFNγ production between IL-18-deficient mice and ASC-deficient mice, as shown in our studies, indicates that ASC-mediated maturation of IL-18 is not critical for IFNγ production in vivo under these conditions. Whether a recently identified sensor for cytosolic DNA, DAI (DNA-dependent activator of IFN-regulatory factors, DLM-1/ZBP1) (Takaoka et al., 2007) plays a role in these processes awaits further investigation. A previous study described the capacity of DCs to prime NK cells through type I IFNs and IL-15 in the draining lymph nodes by an IL-12-independent pathway after challenge of mice with TLR-ligands or Listeria (Lucas et al., 2007). However, our results show that type I IFNs and IRF3 were not necessary for the recruitment and activation of NK cells after systemic Listeria challenge, perhaps favoring the concept of a permissive role for the type I IFN pathway in this infection (Auerbuch et al., 2004; Carrero et al., 2004; O’Connell et al., 2004). Conversely, IL-12, along with IL-18, was required for optimal IFNγ production by NK cells in our study, suggesting that NK cells priming is a pathway distinct from the processes we describe. We speculate that innate cell clustering may underlie a common function used by DCs to link the detection of a variety of pathogens with the early immune response, and that further understanding of the pathways involved will discover new strategies for optimizing crosstalk between innate and adaptive immunity.
Experimental Procedures
Mice
C57BL/6, CCR2 knockout (KO), CCR5 KO, CX3CR1 KO, CCL3 KO, CCL2 KO, CCL5 KO, IL-12p40 KO, IL-18 KO, IL-1R1 KO, common γc KO and RAG1 KO were purchased from Jackson Laboratories. CCR1 KO mice were from Taconic. CCR2 KO and CCL7 KO mice were kindly provided by Dr. I. Charo (UCSF), CXCR3 KO and CCR5 KO by Dr. A. Luster (Harvard), CD11c-CRE by Dr. B. Reizis (Columbia), MyD88 KO by Dr. A. DeFranco (UCSF), IFNAR1 KO by Dr. M. Matloubian (UCSF), ASC KO by Dr. V. Dixit (Genentech), IRF3 KO by Dr. G. Cheng (UCLA), NOD1/2 double KO by Dr. D. Portnoy (UC Berkeley). Mice were C57BL/6 background, and age-and gender-matched for all of the studies. The generation of IFNγ reporter mice (DTAG) is described in Figure S2. Heterozygote DTAG mice were used for all studies. ROSA-DTA mice were previously described. Mice used in this study are referenced in Supplemental data. Mice were kept under specific pathogen-free conditions in the laboratory animal resource center and experimental procedures were performed under approval from the Institutional Animal Care and Use Committee of the University of California, San Francisco.
Antibodies
Antibodies for flow cytometry and immunohistochemistry were from BD Biosciences, Invitrogen, eBioscience, or Serotec, unless otherwise indicated. The antibodies used were: CD3ε (clone 145-2C11 or 17A2), CD4 (RM4–5), CD8 (53–6.7), CD11b (M1/70), CD11c (N418, HL3), CD19 (1D3), CD45R (B220, RA3-6B2), CD49b (DX5), CD90.2 (Thy1.2, 53-2.1), IFNγ (XMG1.2), IL-12 (C17.8), NK1.1 (PK136), PDCA-1 (eBio927), Ly6C (AL-21), Ly6G (1A8), Gr1 (RB6-8C5), class II (M5/114.15.2), F4/80 (BM8) and Mac-3 (M3/84), CD209b (eBio22D1), CD169 (MOMA-1).
Infection
Mice were infected with 2000 live L. monocytogenes (strain: 10403S) per mouse for FACS analysis and with 104 live Listeria, 109 heat-killed Listeria (HKLM) or 108 LLO-deficient Listeria for immunohistochemistry. There was no difference in the pattern of immune responses determined by immunohistochemistry between infections with 2000 or 104 Listeria.
Treatment of mice
For NK cell depletion, mice were injected i.p. with control antibody (Chromopure rabbit IgG, Jackson Immunoresearch) or anti-asialoGM1 (Dako), 50 μl/mouse, at day -2 and day 0. For Gr1+ cell depletion, mice were injected i.v. with purified rat anti-Gr1 antibody or isotype-matched control Ig (rat IgG2b) for flow cytometry, 250 μg/mouse, or PBS for immunohistochemistry as control 2 days before infection. For PTX treatment, mice were injected i.p. with PBS (control) or 1 μg/mouse PTX 30 min – 1 hr before infection.
Flow cytometry
Spleens were collected and dispersed into cells by passing through a mesh (BD Falcon). To isolate DCs, spleens were pre-treated with Liberase 3 and DNase I (Roche). For unfixed cell staining, cells were blocked with 10% normal mouse serum (Sigma) and/or anti-CD16 + CD32, and then stained for the surface molecules. DAPI (4′,6′-diamidine-2′-phenylindole dihydrochloride, Roche) was used to exclude dead cells. For fixed cell staining, dead cells were excluded by using Live/Dead® fixable violet dead-cell stain kit (Invitrogen) following the manufacturer’s recommendations after staining the surface molecules. Surface-stained cells were fixed in 2% paraformaldehyde, permeabilized in 0.5% saponin including 10% normal goat serum and 1% rat serum, and stained with anti-IFNγ antibody or rabbit anti-iNOS antibody (Upstate) followed by A647-goat anti-rabbit antibody (Invitrogen). In intracellular staining of IFNγ, surface-bound IFNγ was excluded by pre-blocking with purified anti-IFNγ antibody during surface staining. Live cells were counted with counting beads (Invitrogen). Data were acquired on a LSRII (BD Biosciences) and analyzed using FlowJo (Tree Star) software.
Immunohistochemistry
Mice were perfused at the indicated time with 2% paraformaldehyde/PBS through the left ventricle of the heart. Spleens were isolated, washed overnight with PBS, soaked in 30% sucrose and frozen in OCT embedding compound (Sakura Finetek). Frozen spleens were cut into 6 m sections with a Leica cryomicrotome. For staining for 7/4, CD209b and CD169, spleens were isolated, frozen, cut and dehydrated with acetone. Endogenous peroxidase was quenched using 1% H2O2 and 0.1% sodium azide in PBS for 1 hr and sections were blocked with PBS containing 5–10% normal goat and/or 10% mouse serum and/or 5% rat serum, followed with avidin/biotin (Vector laboratories). Sections were stained as follows: 1) eCFP: FITC-conjugated goat-anti-GFP (Abcam) followed by Alexa488-conjugated goat-anti-fluorescein/Oregon green (Invitrogen); 2) CD11b, Ly6G: FITC-conjugated anti-CD11b or anti-Ly6G followed by Alexa488-conjugated goat-anti-fluorescein/Oregon green, or biotin-conjugated anti-CD11b followed by Alexa647 or Alexa555-conjugated streptavidin (SA, Invitrogen); 3) Thy1.2: APC-conjugated anti-Thy1.2 followed by rabbit anti-APC antibody (Biomeda), and then by Alexa647-conjugated goat anti-rabbit; 4) B220: APC-conjugated anti-B220 alone or APC-conjugated anti-B220 followed by rabbit anti-APC antibody, and then by Alexa647-conjugtaed goat anti-rabbit; 5) 7/4: biotin-conjugated 7/4 antibody (Serotec) followed by Alexa555-SA; 6) Cytokines (IFNγ, IL-12), NK1.1, NKp46: biotin-conjugated rat anti-cytokine, or biotin-conjugated mouse anti-NK1.1 was detected by horseradish peroxidase (HRP)-SA which was amplified by biotin-tyramide (Perkin Elmer) and Alexa555-conjugated SA (Invitrogen). Goat anti-mouse NKp46 (R&D systems) followed by biotin-conjugated donkey anti-goat was detected by HRP-SA and Alexa555-tyramide (Invitrogen). Tyramide signal amplification (TSA) was done according to the manufacturer’s instructions with some modifications. When sequential TSA was done, HRP was quenched by incubating with 3% H2O2 and 0.1% sodium azide in PBS for 30 min; 7) Gr1: FITC-conjugated anti-Gr1 followed by HRP-conjugated sheep anti-FITC antibody (Perkin Elmer) was detected by FITC-tyramide (Perkin Elmer) or by Alexa488-conjugated goat-anti-fluorescein/Oregon green; 8) iNOS: rabbit anti-iNOS followed by Alexa555-conjugated goat anti-rabbit; 9) Listeria: rabbit anti-Listeria (Abcam) followed by Alexa555-conjugated goat anti-rabbit; 10) CFSE was amplified with Alexa488-conjugated goat-anti-fluorescein/Oregon green. Nuclei were stained with DAPI. The micrographs were captured by a Nikon Eclipse e800 microscope equipped with SimplePCI software (Compix Inc). Immunohistochemistry represents comparable data from at least two independent experiments with three or more age-matched cohorts, or, in the case of negative data, at least two age-matched cohorts.
NK cell transfer
NK cells from RAG1-deficient mice were enriched to 90% purity using DX5 mAb-coupled beads (Miltenyi). Cells were incubated with 2 M CFSE for flow cytometry or 5 μM CFSE for immunohistochemistry and washed thoroughly before adoptive transfer. Where indicated, cells were treated with 200 ng/ml PTX (Sigma or Calbiochem) or PBS (control)-containing RPMI-1640 media for 2 hr. Cells were washed three times, resuspended in PBS and 1–1.5 million cells were transferred intravenously to each mouse. The next day, mice were infected with Listeria and the spleens were harvested for analysis 24 hr later. The cluster association index was calculated as 1000 X [No. of green cells associating with clusters/No. of total green cells/No. of clusters] to normalize the infectivity among mice. Four different areas of spleens per mouse, which cover almost the whole spleen area, were used for quantitation.
Statistical analysis
The statistical analysis was performed by unpaired, two-tailed, Student’s t-test. Each symbol (circle) indicates individual mouse and the red bar indicates the mean value of the group. Due to the experimental variance, the data points were sometimes normalized to the mean of WT or control for inter-comparisons. Error bars represent standard deviations.
Supplementary Material
Acknowledgments
We thank I. Charo (UCSF), G. Cheng (UCLA), A. DeFranco (UCSF), V. Dixit (Genentech), A. Luster (Harvard), M. Matloubian (UCSF) and D. Portnoy (UC Berkeley) who generously provided mice. We thank J. Leber (UC Berkeley), S. Mariathasan (Genentech), N. Meyer-Morse (UC Berkeley) and C-L. Tsou (UCSF) for help with animals, N. Flores, C. McArthur and L. Stowring for technical assistance, and L. Lanier and W. Seaman for critical reading of the manuscript. This work was supported by the NIH AI026918 and AI078869, the HHMI and the Sandler Asthma Basic Research Center at UCSF. S-J. Kang was supported by a Cancer Research Institute Postdoctoral Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aichele P, Zinke J, Grode L, Schwendener RA, Kaufmann SH, Seiler P. Macrophages of the splenic marginal zone are essential for trapping of blood-borne particulate antigen but dispensable for induction of specific T cell responses. J Immunol. 2003;171:1148–1155. doi: 10.4049/jimmunol.171.3.1148. [DOI] [PubMed] [Google Scholar]
- Andrews DM, Farrell HE, Densley EH, Scalzo AA, Shellam GR, Degli-Esposti MA. NK1.1+ cells and murine cytomegalovirus infection: what happens in situ? J Immunol. 2001;166:1796–1802. doi: 10.4049/jimmunol.166.3.1796. [DOI] [PubMed] [Google Scholar]
- Aoshi T, Zinselmeyer BH, Konjufca V, Lynch JN, Zhang X, Koide Y, Miller MJ. Bacterial Entry to the Splenic White Pulp Initiates Antigen Presentation to CD8(+) T Cells. Immunity. 2008 doi: 10.1016/j.immuni.2008.06.013. [DOI] [PubMed] [Google Scholar]
- Auerbuch V, Brockstedt DG, Meyer-Morse N, O’Riordan M, Portnoy DA. Mice lacking the type I interferon receptor are resistant to Listeria monocytogenes. J Exp Med. 2004;200:527–533. doi: 10.1084/jem.20040976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajenoff M, Breart B, Huang AY, Qi H, Cazareth J, Braud VM, Germain RN, Glaichenhaus N. Natural killer cell behavior in lymph nodes revealed by static and real-time imaging. J Exp Med. 2006;203:619–631. doi: 10.1084/jem.20051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu YJ, Pulendran B, Palucka K. Immunobiology of dendritic cells. Annu Rev Immunol. 2000;18:767–811. doi: 10.1146/annurev.immunol.18.1.767. [DOI] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, Forman J. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J Exp Med. 2003;198:1583–1593. doi: 10.1084/jem.20031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg RE, Crossley E, Murray S, Forman J. Relative contributions of NK and CD8 T cells to IFN-gamma mediated innate immune protection against Listeria monocytogenes. J Immunol. 2005;175:1751–1757. doi: 10.4049/jimmunol.175.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasius AL, Barchet W, Cella M, Colonna M. Development and function of murine B220+CD11c+NK1.1+ cells identify them as a subset of NK cells. J Exp Med. 2007;204:2561–2568. doi: 10.1084/jem.20070991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrero JA, Calderon B, Unanue ER. Type I interferon sensitizes lymphocytes to apoptosis and reduces resistance to Listeria infection. J Exp Med. 2004;200:535–540. doi: 10.1084/jem.20040769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J Exp Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan JW. Early pathogenesis of Listeria monocytogenes infection in the mouse spleen. J Med Microbiol. 1996;44:295–302. doi: 10.1099/00222615-44-4-295. [DOI] [PubMed] [Google Scholar]
- Conlan JW, North RJ. Neutrophils are essential for early anti-Listeria defense in the liver, but not in the spleen or peritoneal cavity, as revealed by a granulocyte-depleting monoclonal antibody. J Exp Med. 1994;179:259–268. doi: 10.1084/jem.179.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyster JG. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- Czuprynski CJ, Brown JF, Maroushek N, Wagner RD, Steinberg H. Administration of anti-granulocyte mAb RB6-8C5 impairs the resistance of mice to Listeria monocytogenes infection. J Immunol. 1994;152:1836–1846. [PubMed] [Google Scholar]
- Daniels KA, Devora G, Lai WC, O’Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe P, Ryan MD. Targeting of proteins derived from self-processing polyproteins containing multiple signal sequences. Traffic. 2004;5:616–626. doi: 10.1111/j.1398-9219.2004.00205.x. [DOI] [PubMed] [Google Scholar]
- Dokun AO, Chu DT, Yang L, Bendelac AS, Yokoyama WM. Analysis of in situ NK cell responses during viral infection. J Immunol. 2001a;167:5286–5293. doi: 10.4049/jimmunol.167.9.5286. [DOI] [PubMed] [Google Scholar]
- Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001b;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- Dunn PL, North RJ. Early gamma interferon production by natural killer cells is important in defense against murine listeriosis. Infect Immun. 1991;59:2892–2900. doi: 10.1128/iai.59.9.2892-2900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire C, Chasson L, Luci C, Tomasello E, Geissmann F, Vivier E, Walzer T. The trafficking of natural killer cells. Immunol Rev. 2007;220:169–182. doi: 10.1111/j.1600-065X.2007.00563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia T, Serbina NV, Brandl K, Zhong MX, Leiner IM, Charo IF, Pamer EG. Additive roles for MCP-1 and MCP-3 in CCR2-mediated recruitment of inflammatory monocytes during Listeria monocytogenes infection. J Immunol. 2008;180:6846–6853. doi: 10.4049/jimmunol.180.10.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S, Unutmaz D, Wong P, Sano G, De los Santos K, Sparwasser T, Wu S, Vuthoori S, Ko K, Zavala F, et al. In vivo depletion of CD11c(+) dendritic cells abrogates priming of CD8(+) T cells by exogenous cell-associated antigens. Immunity. 2002;17:211–220. doi: 10.1016/s1074-7613(02)00365-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanneganti TD, Lamkanfi M, Nunez G. Intracellular NOD-like receptors in host defense and disease. Immunity. 2007;27:549–559. doi: 10.1016/j.immuni.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Ozoren N, Body-Malapel M, Amer A, Park JH, Franchi L, Whitfield J, Barchet W, Colonna M, Vandenabeele P, et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature. 2006;440:233–236. doi: 10.1038/nature04517. [DOI] [PubMed] [Google Scholar]
- Lertmemongkolchai G, Cai G, Hunter CA, Bancroft GJ. Bystander activation of CD8+ T cells contributes to the rapid production of IFN-gamma in response to bacterial pathogens. J Immunol. 2001;166:1097–1105. doi: 10.4049/jimmunol.166.2.1097. [DOI] [PubMed] [Google Scholar]
- Liu W, Kurlander RJ. Analysis of the interrelationship between IL-12, TNF-alpha, and IFN-gamma production during murine listeriosis. Cell Immunol. 1995;163:260–267. doi: 10.1006/cimm.1995.1125. [DOI] [PubMed] [Google Scholar]
- Lucas M, Schachterle W, Oberle K, Aichele P, Diefenbach A. Dendritic cells prime natural killer cells by trans-presenting interleukin 15. Immunity. 2007;26:503–517. doi: 10.1016/j.immuni.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig IS, Geijtenbeek TB, van Kooyk Y. Two way communication between neutrophils and dendritic cells. Curr Opin Pharmacol. 2006;6:408–413. doi: 10.1016/j.coph.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O’Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Moretta L, Ferlazzo G, Bottino C, Vitale M, Pende D, Mingari MC, Moretta A. Effector and regulatory events during natural killer-dendritic cell interactions. Immunol Rev. 2006;214:219–228. doi: 10.1111/j.1600-065X.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- Muraille E, Giannino R, Guirnalda P, Leiner I, Jung S, Pamer EG, Lauvau G. Distinct in vivo dendritic cell activation by live versus killed Listeria monocytogenes. Eur J Immunol. 2005;35:1463–1471. doi: 10.1002/eji.200526024. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Neuenhahn M, Kerksiek KM, Nauerth M, Suhre MH, Schiemann M, Gebhardt FE, Stemberger C, Panthel K, Schroder S, Chakraborty T, et al. CD8alpha+ dendritic cells are required for efficient entry of Listeria monocytogenes into the spleen. Immunity. 2006;25:619–630. doi: 10.1016/j.immuni.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Newman KC, Riley EM. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat Rev Immunol. 2007;7:279–291. doi: 10.1038/nri2057. [DOI] [PubMed] [Google Scholar]
- O’Connell RM, Saha SK, Vaidya SA, Bruhn KW, Miranda GA, Zarnegar B, Perry AK, Nguyen BO, Lane TF, Taniguchi T, et al. Type I interferon production enhances susceptibility to Listeria monocytogenes infection. J Exp Med. 2004;200:437–445. doi: 10.1084/jem.20040712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozoren N, Masumoto J, Franchi L, Kanneganti TD, Body-Malapel M, Erturk I, Jagirdar R, Zhu L, Inohara N, Bertin J, et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J Immunol. 2006;176:4337–4342. doi: 10.4049/jimmunol.176.7.4337. [DOI] [PubMed] [Google Scholar]
- Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- Poston RM, Kurlander RJ. Cytokine expression in vivo during murine listeriosis. Infection with live, virulent bacteria is required for monokine and lymphokine messenger RNA accumulation in the spleen. J Immunol. 1992;149:3040–3044. [PubMed] [Google Scholar]
- Rogers HW, Unanue ER. Neutrophils are involved in acute, nonspecific resistance to Listeria monocytogenes in mice. Infect Immun. 1993;61:5090–5096. doi: 10.1128/iai.61.12.5090-5096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki E, Tsutsui H, Tsuji NM, Hayashi N, Adachi K, Nakano H, Futatsugi-Yumikura S, Takeuchi O, Hoshino K, Akira S, et al. Critical roles of myeloid differentiation factor 88-dependent proinflammatory cytokine release in early phase clearance of Listeria monocytogenes in mice. J Immunol. 2002;169:3863–3868. doi: 10.4049/jimmunol.169.7.3863. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Kuziel W, Flavell R, Akira S, Rollins B, Pamer EG. Sequential MyD88-independent and -dependent activation of innate immune responses to intracellular bacterial infection. Immunity. 2003a;19:891–901. doi: 10.1016/s1074-7613(03)00330-3. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003b;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Tay CH, Yu LY, Kumar V, Mason L, Ortaldo JR, Welsh RM. The role of LY49 NK cell subsets in the regulation of murine cytomegalovirus infections. J Immunol. 1999;162:718–726. [PubMed] [Google Scholar]
- Taylor PR, Brown GD, Geldhof AB, Martinez-Pomares L, Gordon S. Pattern recognition receptors and differentiation antigens define murine myeloid cell heterogeneity ex vivo. Eur J Immunol. 2003;33:2090–2097. doi: 10.1002/eji.200324003. [DOI] [PubMed] [Google Scholar]
- Tsou CL, Peters W, Si Y, Slaymaker S, Aslanian AM, Weisberg SP, Mack M, Charo IF. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. J Clin Invest. 2007;117:902–909. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji NM, Tsutsui H, Seki E, Kuida K, Okamura H, Nakanishi K, Flavell RA. Roles of caspase-1 in Listeria infection in mice. Int Immunol. 2004;16:335–343. doi: 10.1093/intimm/dxh041. [DOI] [PubMed] [Google Scholar]
- Unanue ER. Inter-relationship among macrophages, natural killer cells and neutrophils in early stages of Listeria resistance. Curr Opin Immunol. 1997;9:35–43. doi: 10.1016/s0952-7915(97)80156-2. [DOI] [PubMed] [Google Scholar]
- Vosshenrich CA, Lesjean-Pottier S, Hasan M, Richard-Le Goff O, Corcuff E, Mandelboim O, Di Santo JP. CD11cloB220+ interferon-producing killer dendritic cells are activated natural killer cells. J Exp Med. 2007;204:2569–2578. doi: 10.1084/jem.20071451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T, Blery M, Chaix J, Fuseri N, Chasson L, Robbins SH, Jaeger S, Andre P, Gauthier L, Daniel L, et al. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc Natl Acad Sci U S A. 2007;104:3384–3389. doi: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama H, Matsuno K, Toda E, Nishiwaki T, Matsuo N, Nakano A, Narumi S, Lu B, Gerard C, Ishikawa S, Matsushima K. Plasmacytoid DCs help lymph node DCs to induce anti-HSV CTLs. J Exp Med. 2005;202:425–435. doi: 10.1084/jem.20041961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong MX, Kuziel WA, Pamer EG, Serbina NV. Chemokine receptor 5 is dispensable for innate and adaptive immune responses to Listeria monocytogenes infection. Infect Immun. 2004;72:1057–1064. doi: 10.1128/IAI.72.2.1057-1064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.