Abstract
Design and synthesis of basic functional circuits are the fundamental tasks of synthetic biologists. Before it is possible to engineer higher-order genetic networks that can perform complex functions, a toolkit of basic devices must be developed. Among those devices, sequential logic circuits are expected to be the foundation of the genetic information-processing systems. In this study, we report the design and construction of a genetic sequential logic circuit in Escherichia coli. It can generate different outputs in response to the same input signal on the basis of its internal state, and ‘memorize’ the output. The circuit is composed of two parts: (1) a bistable switch memory module and (2) a double-repressed promoter NOR gate module. The two modules were individually rationally designed, and they were coupled together by fine-tuning the interconnecting parts through directed evolution. After fine-tuning, the circuit could be repeatedly, alternatively triggered by the same input signal; it functions as a push-on push-off switch.
Keywords: bistable switch, coupling modules, genetic sequential logic circuit, NOR gate, push-on push-off switch
Introduction
The first step of the bottom-up approach in synthetic biology is to create biological modules that can perform different basic functions, such as sensors to receive various environmental signals, actuators to implement physiological reactions, and processors to manipulate cellular information flow (Gardner et al, 2000; Park et al, 2003; Levskaya et al, 2005; Anderson et al, 2006, 2007; Skerker et al, 2008). Among these functions, information processing is essential for integrating and storing environmental inputs and producing appropriate outputs after in vivo computation. In this area, extensive studies on information processing in cells have been conducted at the transcriptional and translational level (McAdams and Shapiro, 1995; Ptashne and Gann, 1997; Guet et al, 2002; Shen-Orr et al, 2002; Buchler et al, 2003; Libby et al, 2007; Tkacik et al, 2008; Win and Smolke, 2008). This accumulated knowledge facilitates the design of artificial genetic circuits.
As in electronics, combinational and sequential logic circuits are two kinds of fundamental processors in cells. In a combinational logic circuit, the output depends only on the present inputs, whereas in a sequential logic circuit, the output also depends on the history of the input owing to its own memory. If we can successfully construct the two kinds of basic logic circuits in a cell, they can serve as building blocks to be assembled into high-order genetic circuits and implement more sophisticated computation.
Construction of genetic combinational logic circuits, such as AND, OR, and NOR gates, has been frequently reported in the last decade (Guet et al, 2002; Dueber et al, 2003; Anderson et al, 2007; Win and Smolke, 2008). Meanwhile, toggle switches, which can function as memory modules, have been implemented in prokaryotic and eukaryotic cells (Becskei et al, 2001; Kramer et al, 2004; Ajo-Franklin et al, 2007). Recently, researchers have designed, and in some cases constructed, sequential logic circuits. For example, Fritz et al (2007) designed a conditional memory in which a bistable switch is controlled by two external inputs. They demonstrated the conditional memory function in silico and believed that the corresponding in vivo construction of conditional memory circuits would be a milestone for synthetic biology. Ham et al (2008) constructed a sequential DNA-encoded memory using two inversion recombination systems, in which they intriguingly arranged the inverted sites of FimB and Hin recombinases. In this system, the heritable state is encoded into a DNA sequence, which can be manipulated by inducing the expression of the corresponding recombinase.
In this study, we designed and constructed a novel genetic sequential logic circuit (GSLC). It is composed of a bistable switch memory module, a double-repressed promoter NOR gate module, and two interconnecting parts connecting the two modules. GSLC can achieve a sequential logic function as follows: (1) it generates output based not only on the present input signal but also on its internal state and (2) the output feeds back to the memory module so that the history of the output signal can be registered. This enables the GSLC to function as a push-on push-off switch, which is essentially a binary counter. We used two strategies to construct the circuit. First, rational design was used to guide the construction of two basic modules (bistable switch and NOR gate). Second, directed evolution was used to fine-tune the interconnecting parts. We also constructed three circuits as controls to show that the system achieved the sequential logic function only because of our specific design.
Results
Overall structure of the genetic circuit
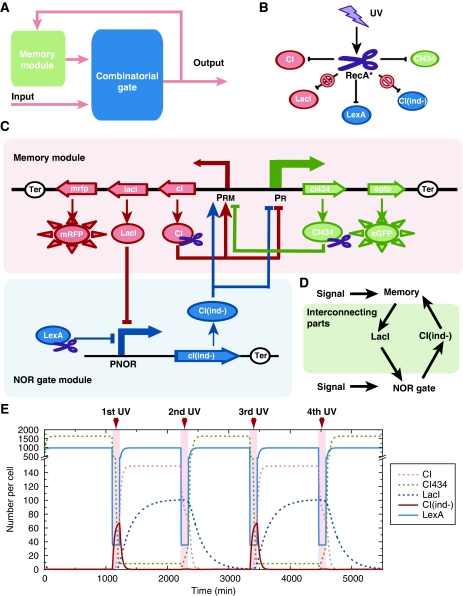
In principle, a sequential logic circuit can be composed of a combinatorial logic gate and a memory module (Figure 1A). In our design, the combinatorial logic gate is a NOR gate and the memory module is a clearable bistable switch. Two interconnecting parts are designed to connect the NOR gate and the bistable switch. The output signal generated by the NOR gate feeds back to the memory module and functions as the overall output of the circuit in the meantime. UV irradiation was used as both an external input signal and a reset signal for the clearable bistable switch. In the following sections, we describe all the parts one by one.
Figure 1.
The GSLC and the related properties of its elements. (A) A simplified diagram of a sequential logic circuit. It consists of a memory module and a combinatorial gate module. (B) The effect of UV input on all repressors. The scissor represents activated RecA*. Arrows represent UV irradiation activating RecA* to specifically degrade CI434, CI, and LexA, but with no effect on LacI and CIind−. (C) Detailed schematics of the GSLC. Boxes represent the memory module and the NOR gate module. The rectangles with arrow and the full oval circles, respectively, represent genes and their translated proteins. The scissors on proteins represent those proteins that can be cleaved by RecA*. Open circles represent terminators. Solid lines with arrow represent the regulating relations between genes. Arrows represent activation and blunt arrows represent repression. (D) The two parts interconnecting the memory and the NOR gate modules. LacI functions as an input of the NOR gate module and as an output of the memory module, whereas CIind− functions in the reverse manner. (E) Simulation results of the push-on push-off switch process. In simulation, the effect of UV irradiation persists for 120 min; the concentrations of repressors reach a stationary level before the next UV stimulation.
Design of the memory module
The memory module is a toggle switch that consists of two mutually repressed repressors, CI and CI434. The corresponding cI and cI434 genes come from the lambda and 434 phages, respectively. The promoters PR and PRM control the transcription of cI434 and cI genes and they are in turn repressed by CI and CI434 (Figure 1C). RFP and GFP are expressed by promoters PRM and PR, respectively, to indicate promoter activity (Figure 1C). Note that promoter PR is about 30-fold stronger than PRM (Dodd et al, 2001, 2004).
The memory module is expected to function as follows: When CI is present, it can activate its transcription and repress the transcription of cI434, thus establishing a stable high CI/low CI434 state, which is defined as the ‘ON’ state of the memory module. In this case, RFP is expressed (Ptashne, 2004). Alternatively, when CI434 is present, it can repress the transcription of cI, thus maintaining the transcription of itself and establishing a stable low CI/high CI434 state, which is defined as the ‘OFF’ state. In this case, GFP is expressed. Thus, we expect each individual cell that carries the memory module to express RFP or GFP exclusively.
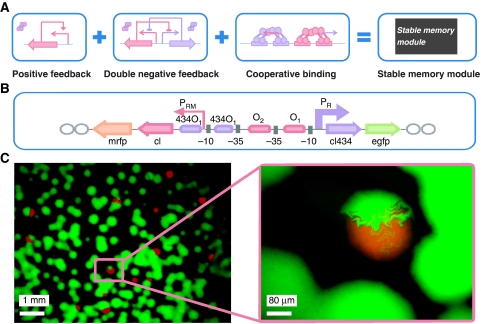
To design such a memory module, we took the OR promoter region of the wild-type lambda lysis/lysogeny switch (Ptashne, 2004) as the backbone of the promoter region, replaced the CI-binding site O3 with the CI434-binding site 434O1, and added another 434O1 downstream of the initiation site of promoter PRM (Figure 2B). The detailed sequence of the promoter region is shown in Supplementary Figure S3B. As a result, two CI dimers cooperatively bind O1 and O2, activate PRM, and meanwhile repress PR; two CI434 dimers also bind the two tandem 434O1 sites and repress the transcription of PRM. It is notable that our design incorporates a positive feedback loop, a double-negative feedback loop, and the cooperativity of the repressor dimers (Figure 2A). All three mechanisms have been proposed to cause or enhance the bistable behavior of a system (Ferrell, 2002). With a simple model, we theoretically showed that our designed circuit inhabits a much larger bistable parameter regime than a circuit with only one or two of these three mechanisms in the phase diagram (Supplementary Figure S2).
Figure 2.
Construction of the bistable memory module. (A) The memory module incorporates three mechanisms: positive feedback, double-negative feedback, and the repressor binding cooperativity. (B) The arrangement of the genes and promoter region in the memory module. Rectangles with arrow represent genes. Open ovals represent terminations. Lines with arrow represent the transcriptional strength and direction of the promoters. Rectangles with colors represent the binding sites of repressors. Gray squares represent the −10 and −35 regions of the promoters. (C) Images of cells carrying the memory module. Each dot represents a colony. The image in the right panel is an enlarged view of the image in the left panel and shows the colony with mixed colors.
Design of the NOR gate module
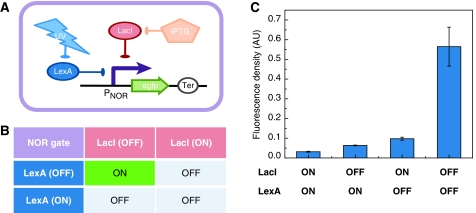
For the NOR gate module, the two immediate inputs (also called ‘internal inputs’) that were selected are LacI and LexA repressors; the output, which is also the output of the sequential logic circuit, is the transcriptional activity of a promoter (PNOR) (see the NOR gate module in Figure 1C). LacI is a repressor of the lac operon, whereas LexA is a repressor responsible for SOS response in Escherichia coli. Promoter PNOR is repressed by both LacI and LexA repressors, so that the corresponding output is ON only if both LacI and LexA are in the OFF state; promoter PNOR is OFF if any input is ON (Figure 3B).
Figure 3.
Construction of the NOR gate module. (A) The circuit for quantitative measurement of NOR gate. The promoter PNOR is suppressed by both LacI and LexA, which can be eliminated by IPTG and UV irradiation, respectively. GFP reports the promoter's activity. (B) The truth table of the NOR gate. The output of the NOR gate is ‘ON’ only when neither LacI nor LexA exists; else the output is ‘OFF’. (C) Experimental measurement of the NOR gate by flow cytometry. values and standard deviations were determined from four parallel experiments.
When designing the NOR gate, our objective was to maximize the difference in promoter activity between the ‘ON’ and ‘OFF’ states and eliminate intermediate activity. First, the consensus binding sites of LacI and LexA were used to suppress a strong promoter in which the consensus −10 and −35 regions come from the recA promoter. Second, we fixed the interoperator distance of the LacI-binding sites to 93 bp, with which the repression by LacI reaches a local maximum on the basis of previous quantitative experimental data (Müller et al, 1996). The detailed sequence of the NOR gate promoter is shown in Supplementary Figure S4B.
Interconnecting parts
Two interconnecting parts, LacI and CIind−, connect the memory module and the NOR gate module. The genes lacI and cIind− are transcribed by PRM and PNOR promoters, respectively (Figure 1C). Owing to its capability to repress the activity of PNOR, LacI serves as an internal input of the NOR gate (Figure 1D). CIind− is a mutant of CI that functions in the same manner as CI but cannot be degraded by the RecA* protease (Roberts and Roberts, 1975). Under the control of promoter PNOR, it can repress promoter PR and activate promoter PRM (Figure 1C), and thus functions as the feedback from the output of the NOR gate to the memory module (Figure 1D).
UV irradiation signal
As the external input, UV irradiation exerts an effect on the internal input LexA through the RecA* protease. UV irradiation can damage DNA and block the DNA replication process. The appearance of single-stranded DNA recruits RecA and converts it into a protease (RecA*). Subsequently, RecA* cleaves LexA, CI, and CI434 repressors specifically and rapidly (Figure 1B; Little, 1984). As LexA is an input of the NOR gate and CI/CI434 maintains the state of the bistable switch, UV irradiation not only works as an external input signal but also clears the memory of the bistable switch in order for it to accept a new state.
Computational simulation and the function of the genetic circuit
After defining all elements and interconnections of the genetic circuit, we used a set of ordinary differential equations to simulate the dynamic process. With a set of reasonable parameters (see Supplementary information), simulation results showed that the GSLC could function as a push-on push-off switch (Figure 1E). When the stimulus of UV is present repeatedly, the circuit's output, CIind−, alternatively generates ‘OFF’ and ‘ON’ pulse signals. The output signal can be registered into the memory module; thus, the memory module will be alternatively in the ‘ON’ or ‘OFF’ state after the effect of UV irradiation ends. A detailed description is as follows:
As the initial state of the bistable module could be in either of the two stable states, we assume the initial state of the bistable switch (memory module) to be in the high CI/low CI434 state (defined as the ‘ON’ state), wherein CI represses promoter PR and activates promoter PRM (Figure 1C and E). As LexA is constitutively expressed in the cell and LacI is cotranscribed with repressor CI, promoter PNOR, and thus transcription of CIind−, is repressed by the presence of both LexA and LacI (Figure 1C and E). After exposure to an appropriate dose of UV irradiation, both LexA and CI are degraded rapidly by the RecA* protease (Figure 1B, C, and E). As promoter PR is much stronger than PRM as mentioned above, activity of PR will increase and activity of PRM will decrease because of the absence of CI. As a result, the transcripts of gene cI434 predominantly accumulate. Meanwhile, PNOR is still repressed by LacI, and hence the output of promoter PNOR, CIind−, is still in the ‘OFF’ state (Figure 1C and E).
After the effect of UV irradiation ends, LexA reappears and replaces the gradually diluted LacI to continuously suppress PNOR; therefore, CIind− is maintained in the ‘OFF’ state. Meanwhile, the CI434 repressor is rapidly generated from the accumulated cI434 transcripts and represses the transcription of its antagonist CI. Thus, the low-CI and high-CI434 state (defined as the ‘OFF’ state) becomes a stable state, which gives a high GFP expression (Figure 1C and E).
When the memory module is in the low CI/high CI434 state (the ‘OFF’ state), CI434 represses the expression of CI and LacI, and hence LexA, but not LacI, is present to repress promoter PNOR. Once the cell is exposed to UV light, both LexA and CI434 are degraded rapidly (Figure 1B), and PNOR is released because of the absence of LacI and LexA. As a result, gene cIind− is transcribed from promoter PNOR, followed by translation of the CIind− repressor. With neither repressor present, the output of promoter PNOR is now in the ‘ON’ state (Figure 1C and E).
After the UV effect ends again, the accumulated CIind− will activate promoter PRM to express CI and LacI, and repress PR (Figure 1C). Promoter PNOR is suppressed by both recovered LexA and LacI, whereas the newly expressed CI replaces the gradually diluted CIind−. The memory module thus returns to the ‘ON’ state with a high RFP expression (Figure 1C and E).
In summary, when the internal state of the memory is in the ‘ON’ state, the external UV input makes the circuit's output promoter PNOR generate an ‘OFF’ pulse signal and register the ‘OFF’ state into the memory; when the internal state is in ‘OFF’ state, the same external UV input induces the circuit's output promoter PNOR to generate an ‘ON’ pulse signal and register the ‘ON’ state into the memory. Thus, if the switch can be flipped efficiently, we should observe that the memory of the GSLC is repeatedly flipped between the ‘ON’ and ‘OFF’ state by the same UV stimulus, which resembles a push-on push-off switch. The function is demonstrated by the state transitions of Boolean logic (Supplementary Figure S1B), which is generated by an analogous digital circuit (Supplementary Figure S1A).
Experimental realizations and tests
Construction of the bistable memory and the NOR gate module
Following the rational design described above, we constructed the bistable memory module on a 20- to 30-copy plasmid (pSB3K3). We transformed the plasmid into E. coli and observed that the red and green colonies coexisted on agar plates after overnight growth in a 37°C incubator (Figure 2C). Interestingly, we observed that a few colonies were of both red and green color, and the patterns of colors were not overlapping but sectored. These sectored colorful colonies indicated that during growth to form colonies, some competent cells with the newly transformed plasmid did not establish a stable inheritable state before bearing two new cells, and each of the newborn cells entered different stable states, which were inherited to their offspring. Therefore, the individuals in the colony exclusively emit red or green fluorescence, and these states are stable and heritable (Figure 2C). These results prove that cells carrying the memory module indeed have bistable behavior. Moreover, the bistability of the module is quite robust, as the memory module still shows bistable behaviors when it is transferred into a 3- to 4-copy plasmid with pSC101* origin (data are not shown, but this low-copy plasmid was used in the following experiments).
Similarly, we constructed the NOR gate module on the basis of the above rational design. When measuring the property of the NOR gate, we inserted a GFP under the control of promoter PNOR (Figure 3A). The quantitative data generated by flow cytometry indicate that the NOR gate works as expected. Expression of GFP is high only if both LacI and LexA are absent; otherwise GFP expression is low (Figure 3C). When coupled with other parts in our circuits, the NOR gate module was located on a biobrick's standard plasmid (pSB3K3).
Construction of the mutation library
After constructing the memory and the NOR gate module, we coupled the two modules together to obtain a GSLC. Directed evolution was used to select proper coupling strengths between the two modules. More specifically, we fine-tuned the expression of the two interconnecting parts LacI and CIind− by mutating their ribosome-binding sites (RBSs). When constructing mutation libraries of these RBSs, we adopted a detuned strategy that destabilized the function of some elements (Haseltine and Arnold, 2007). We took two of the strongest RBSs, SDY and SDA, as backbones and varied their translational activity by mutating their key points (as shown in Table I). To avoid unnecessary mutation on the memory module, the lacI gene and its promoter PRM in the circuit were not localized on the same plasmid of the memory module but on another plasmid, pLXcm (see Supplementary Table S2), whereas the PNOR promoter and the cIind− gene remained on the pSB3K3 plasmid. The saturation mutation library of the RBSs of lacI and cIind− were constructed using a mutagenesis kit (Toyobo, Japan), as described under Materials and methods. The two libraries were simultaneously transformed into E. coli cells harboring the memory module plasmid. After growth on agar plates with appropriate antibiotics, colonies containing all three plasmids were selected. Among the colonies, the intensity of green and red fluorescence varied significantly, which indicated the efficiency of the mutation libraries (Supplementary Figure S5).
Table 1. The sequences of the RBS of the wild type and three functional mutants.
| Wild type or mutant of SDA (the RBS of the cIind− gene) | Wild type or mutant of SDY (the RBS of the lacI gene) | |
|---|---|---|
| RBS, ribosome-binding site. | ||
| The mutated nucleotides and start codons are in bold. N=A, T, C, or G. | ||
| Wild type | 5′-AGGAGGAAAAAAATG-3′ | 5′-AGAGGAGAAATTAAGCATG-3′ |
| Mutation library | 5′-ANNNNNAAAAAAATG-3′ | 5′-AGNNNNNAAATTAAGCATG-3′ |
| 8A | 5′-ATGTCGAAAAAAATG-3′ | 5′-AGTGTGTAAATTAAGCATG-3′ |
| 11E | 5′-ATGTCGAAAAAAATG-3′ | 5′-AGGTTTTAAATTAAGCATG-3′ |
| 11H | 5′-ATGTGTAAAAAAATG-3′ | 5′-AGTGAGTAAATTAAGCATG-3′ |
The screening process and results
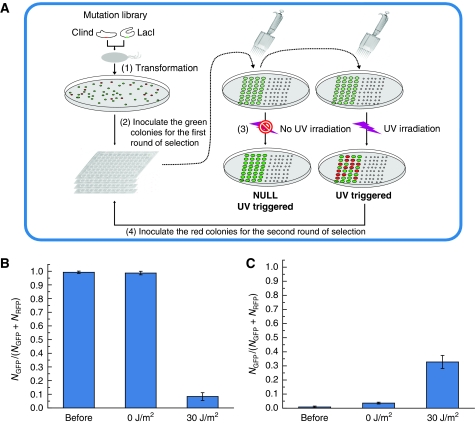
Using efficient mutation libraries, we developed a new screening method to select functional GSLCs. The experimental process is described under Materials and methods (also see Figure 4). We performed two rounds of selection. In the first round of selection, about 300 mutants out of 1000 were chosen. In the second round, however, only three mutants, 8A, 11E, and 11H, were selected; the names refer to their positions on the 96-well plate. Their corresponding RBS sequences are listed in Table I.
Figure 4.
The screening process and the switching efficiency of a functional mutant. (A) Schematics of the screening process to obtain the functional sequential logic circuits. The process is as follows: (1) The two RBS libraries for CIind− and LacI were transformed simultaneously into cells harboring a switch module, and the colors of mutant colonies were identified with a fluorescence stereomicroscope. (2) Green colonies were inoculated into 96-well plates, and the cultures were incubated and diluted appropriately. (3) The cultures were transferred onto two of the same type of agar plates with appropriate dilution, one of which was exposed to 25 J/m2 of UV irradiation and the other was not. (4) After overnight incubation at 37°C, the colors of each mutant on the two plates were compared using the fluorescence stereomicroscope. The mutants that changed color from green to red after UV irradiation were selected for the next round of screening. The second round of selection was similar as the first one, except that colonies that changed color from red to green after UV stimulus were now selected. (B) The switching efficiency of a functional mutant from the green state (OFF) to the red state (ON). (C) The switching efficiency of the same mutant from the red state to the green state. The mark ‘before’ means samples were taken from the culture before UV irradiation; ‘30 J/m2’ and ‘0 J/m2’ represent cells exposed to 30 J/m2 or unexposed to UV irradiation, respectively. Each bar represents the fraction of green cells in the population. Mean values and standard deviations were derived from eight parallel experiments.
After obtaining the three selected mutants, we isolated green and red cells and measured their respective switching efficiencies. It is noteworthy that all isolated cells came from replicated plates that had never been exposed to UV irradiation. By using a similar method as the screening process (see Materials and methods), we quantitatively measured switching efficiency by flow cytometry. As shown in Figure 4B, if the initial state was ‘OFF’ with green color, the fraction of green cells in the population was near 100% before UV stimulus, whereas less than 10% of cells remained in the green ‘OFF’ state after UV stimulus (Figure 4B). This result indicates that the switch from ‘OFF’ to ‘ON’ is quite complete. Unfortunately, the switch from ‘ON’ to ‘OFF’ was not as efficient: only about one-third of the population switched to the ‘OFF’ state after UV stimulation (Figure 4C). Nonetheless, the switch is still significant, compared with the population not exposed to UV irradiation (Figure 4B and C). These results show that the fine-tuned GSLC can generate different output signals under the same input on the basis of the internal state of its memory, and register the output signal into its memory as the new internal state.
Control circuits
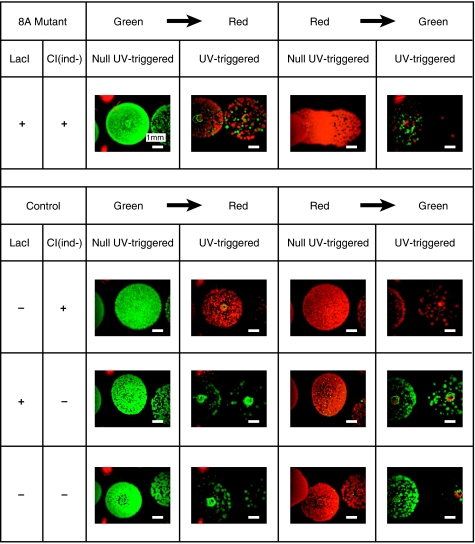
We also constructed three control circuits to show that decoupled circuits cannot achieve the sequential logic function. The memory module and the NOR gate module were decoupled by removing the two interconnecting parts. In the first control circuit, LacI was removed. Without LacI, LexA becomes the only effective input for the NOR gate. As a consequence, on UV stimulus, promoter PNOR always generates a high output signal, and the ‘ON’ state (high CI and low CI434) is latched in the memory with the help of CIind−. Correspondingly, the color of the cells will change to red. Our experimental results are consistent with this expectation: irrespective of the initial states, the cells always changed to the red state after UV stimulation (Figure 5). In the other two control circuits, CIind−, or both LacI and CIind−, was removed. Owing to the lack of the feedback part CIind−, when the output of promoter PNOR is ‘ON’, no output signal can be registered into the memory. In this case, the memory module will spontaneously enter the low CI/high CI434 state after UV stimulus. The experimental results of both cases confirmed our expectation: irrespective of the initial states, the final states of cells were always green (Figure 5). In summary, these control experiments show that only the GSLC designed by us can achieve the defined functions: (1) its output is determined by both UV input and its memory's state and (2) the output signal can be registered into the memory module as the internal state.
Figure 5.
A comparison of the switching process between a functional GSLC and control circuits. Indicated are colonies of cells that were exposed to 25 J/m2 (labeled as UV triggered) or not exposed to UV irradiation (labeled as NULL UV triggered), respectively. Row-1 was taken from a functional GSLC 8A, whereas Row-2 to Row-4 were taken from the circuit with removed lacI gene, the circuit with removed cIind− gene, and the circuit with both lacI and cIind− genes removed, respectively.
Function as a push-on push-off switch
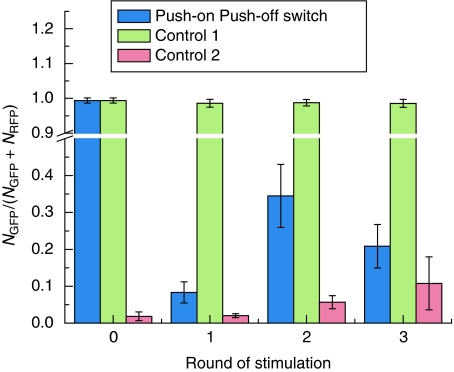
In the above experiments, we showed that GSLC could be flipped once by UV stimulus. But the circuit, in principle, should be flipped back and forth multiple times by the same UV signal. Our deterministic model also indicated that multiple switching is feasible (see Figure 1E). Thus, we sequentially stimulated a homogeneous population of cells with the same dose of UV signal multiple times. The results are summarized in Figure 6. The first UV stimulus caused the fraction of green cells in the population to decrease from 99.3% to 8.4% (Figure 6), so that more than 90% of the population switched from the ‘OFF’ to the ‘ON’ state. The second UV stimulus resulted in the fraction of green cells increasing from 8.4% to 34.5%. Therefore, only 26.1% of the population switched back to the ‘OFF’ state. These results are comparable with the measurement results of switching efficiency shown in Figure 4B and C. With repeated exposure to UV irradiation, the population increasingly started to appear like a mixture of the two states, the ratio of which gradually reached a steady state. The push-on–push-off function of the circuit was thus lost at the population level. In contrast, we also measured the spontaneous switching rate for the green or red initial population (Figure 6). For the green population, more than 98% of individuals were still in the green state after three rounds of UV stimulation. At the same time, about 90% of the population, whose initial state was red, remained in the red state (Figure 6). In our experiments, the time span of three rounds of stimulation was more than 60 h. It means that the spontaneous switching rates between the two stable states are extremely low.
Figure 6.
Multiple sequentially switching events of the push-on push-off switch and the spontaneous switching rates of the initially green and red population. Each bar represents the fraction of green cells in the population. Mean values and standard deviations were derived from eight parallel experiments. Blue bars represent the cells that were repeatedly stimulated by 30 J/m2 UV irradiation. Green bars (control-1) and red bars (control-2) represent, respectively, the initially green and red populations that were never exposed to UV irradiation. Mean values and standard deviations were derived from eight parallel experiments.
Discussion
In this work, we show the design and synthesis of a genetically encoded device that can perform sequential logic. The sequential logic circuit is composed of a bistable memory module coupled to a NOR gate. We show both theoretically and experimentally that a single input, UV radiation, will lead to distinct outputs depending on the initial state of the system. With the ability to genetically encode both combinatorial and sequential logic circuits, synthetic biologists now possess the necessary repertoire to engineer biological machines to perform elaborate computations.
The crux of our system is that we patch into the endogenous E. coli SOS response to switch between states by inducing rapid proteolytic degradation of the state-defining repressor proteins. Although rapid proteolytic degradation is an important design feature and provides many benefits to our system, the manner in which we implement it has some associated caveats that should be noted. To activate the RecA* protease, we used UV irradiation to trigger the SOS response. This may have several unintended effects in our experimental system. First, UV irradiation may result in nontargeted mutations in the E. coli genome, in addition to the specific RBS region in which we performed targeted mutagenesis. Such mutations could create E. coli variants that affect the switching function. To rule out such mutational effects, we purified plasmids from the successfully screened GSLC cells and retransformed them into fresh E. coli cells. When we checked the switching behavior, we found that the fresh retransformed cells retained the same switching behavior as the cells from which the plasmids were isolated. Furthermore, control circuits were also constructed on the basis of purified mutated plasmids. Such constructions ensured that the GSLC and control circuits differed solely in the region encoding lacI and cIind−.
Second, UV irradiation is toxic for cell growth. In our experiments, 25–30 J/m2 UV irradiation was used to induce the switch. This strong irradiation caused the death of more than 90% of the individuals in the population (see the Supplementary Table S3). Even worse, when the cells were repeatedly exposed to UV irradiation more than three times, they began to grow ectopically. After three exposures to UV irradiation, the population grew significantly slower than the initial population under normal growth conditions (data not shown).
To overcome such disadvantages of the UV input signal, we could use constitutively active RecA mutants, for example RecA730 (Lavery and Kowalczykowski, 1992), instead of the wild type. If such a constitutive mutant is controlled by an inducible promoter, the GSLC would become a new module that could be used to assemble more complex genetic circuits, such as an N-bit counter. Supplementary Figure S6A provides an example of 3-bit ripple carry binary counter, which requires three distinct proteases, each with unique specificity. The corresponding state transitions are shown in Supplementary Figure S6B.
Switching efficiency is another issue that merits discussion. Although the switch from the ‘OFF’ to the ‘ON’ state is nearly complete, the reverse switch from ‘ON’ to ‘OFF’ is not as efficient. The possible reasons include (1) incomplete degradation of the CI repressor, (2) leaky expression of CIind−, and (3) insufficient CI434 repressor accumulation when cells were triggered by UV irradiation. Several strategies could be adopted to improve the efficiency of the reverse switch. First, we could further optimize the present circuit by fine-tuning the expression of each regulator through more rounds of directed evolution. Second, we could add accessory modules (e.g. a second NOR gate and a ‘ghost’ memory of CI434) into the present circuit (Supplementary Figure S7). The accessory modules could help our circuit switch from the ‘ON’ to the ‘OFF’ state by expressing more CI434 repressor molecules. Nonetheless, the switching process will remain probabilistic rather than deterministic.
In summary, we successfully assembled a memory module and combinatorial NOR gate module into a functional sequential logic circuit. We combined rational design with directed evolution to generate the push-on push-off switch. In this work, we demonstrated that directed evolution is a powerful tool to search the in vivo parameter space to generate functional circuits from multiple rationally designed synthetic device modules. We anticipate that this approach will lend itself well to the next step in synthetic biology, combining multiple circuits, each composed of several device modules, to create useful synthetic systems that perform sophisticated computation.
Materials and methods
Media, chemical, and other reagents
All E. coli strains were grown in LB liquid media or agar plates supplemented with appropriate antibiotics or inducers. Reagents were purchased from MERCK or Sigma-Aldrich. The working concentrations were 50 mg/ml for ampicillin (MERCK), 10 mg/ml for kanamycin (MERCK), 25 mg/ml for chloramphenicol (MERCK), 10 mg/ml for cycloheximide (Sigma-Aldrich), 1 mM for L-arabinose (Sigma-Aldrich), and 1 mM for IPTG (Sigma-Aldrich). Enzymes were from Takara unless otherwise indicated. Taq and KOD polymerase were purchased from TRANStar and Toyobo, respectively.
Strains and plasmids
E. coli strain JM109 was used to amplify all the plasmids. The NOR gate was tested in strain JM110 (a kind gift from Yanhui Xian, CAS), for the strain is SOS-response positive and carries lacIq (a mutant of wild-type lacI but with a stronger binding affinity) on the F′ plasmid. Testing of the bistable switch and screening of saturation mutation libraries were performed using the strain AAEClac− (MG1655 Δfim ΔlacI), which was constructed from AAEC072 (MG1655 Δfim, a kind gift from IC, Blomfield, UK) by knocking out lacI using a standard lambda red method. The wild-type strain MG1655 was purchased from the Coli Genetic Stock Center. All information regarding the strains is provided in Supplementary Table S2.
The plasmids used for this study were constructed by the iGEM standard BioBrick assembly unless noted otherwise (http://parts.mit.edu/wiki/index.php/Standard_Assembly) and the flow of construction is shown in Supplementary Figure S8. All information on plasmids is listed in Supplementary Table S2 as well. To construct p15Aswitch (p15A origin, Kanr), we sequentially inserted parts into plasmid pSB3K3 (iGEM biobrick's) by several steps. First, the modified OR promoter region was amplified by PCR from wild-type lambda phage (a kind gift from L Huang, Institute of Microbiology, CAS) and inserted into plasmid pSB3K3 to generate plasmid pSB3K3-OR; the egfp gene was amplified by PCR (the forward primer carries SDY RBS) from BBa_I13522 (iGEM biobrick) and inserted into plasmid pSB3K3 to generate plasmid pSB3K3-egfp. Second, the cI gene was amplified by PCR (the forward primer carries SDA RBS) from the lambda phage and inserted into plasmid pSB3K3-OR to generate plasmid pSB3K3-cI-OR; meanwhile, cI434 was amplified by PCR (the forward primer carries SDcro RBS) from BBa_C0056 (iGEM biobrick) and inserted into plasmid pSB3K3-egfp to generate plasmid pSB3K3-cI434-egfp. Next, the cI434-egfp cassette was cleaved from plasmid pSB3K3-cI434-egfp by XbaI and PstI and inserted between the SpeI and PstI sites on plasmid pSB3K3-cI-OR to generate plasmid pSB3K3-cI-OR-cI434-egfp. Finally, the mrfp gene was amplified by PCR (the forward primer carries SDY RBS) from BBa_I13521 (iGEM biobrick) and inserted into plasmid PSB3K3-cI-OR-cI434-egfp to construct the switch plasmid, p15Aswitch (Supplementary Figure S8A). All sequences were verified by sequencing.
To decrease the copy number of the bistable switch module, we transferred the whole switch cassette into plasmid pZS*A to generate plasmid pZS*Aswitch (pSC101* origin, Ampr). The construction was accomplished in two steps. First, the switch cassette was cleaved from plasmid p15Aswitch by AatII and XhoI and inserted into the AatII and XhoI sites of plasmid pZS*24MCS (bought from Expressys, Germany; Lutz and Bujard, 1997) to generate plasmid pZS*switch. Second, the ampr gene with its promoter was PCR-amplified from plasmid pAB1A3 (iGEM biobrick) and inserted into plasmid pZS*switch between the AatII and SacI sites to generate plasmid pZS*Aswitch. All sequences were verified by sequencing.
The testing plasmid pLXnor and the NOR gate plasmid p15AcIindnor were constructed as follows: To construct plasmid pLXnor, the pNOR promoter was amplified by overlapping PCR from the promoter region of recA on the MG1655 genome, and was inserted into plasmid pLX012 (colE origin, Ampr; constructed by Q Ouyang's laboratory) between the XhoI and XbaI sites, upstream of the reporter gene egfp. To construct plasmid p15AcIindnor, first, the cIind− gene was subcloned from plasmid pLC002 (constructed by Q Ouyang's laboratory) and inserted into plasmid pSB3K3 to generate plasmid p15AcIind. Second, the pNOR promoter was subcloned from plasmid pLXnor and inserted into plasmid p15AcIind to finally generate plasmid p15AcIindnor (Supplementary Figure S8C). All sequences were verified by sequencing.
Plasmid pLXcmlacIOR (pMB origin, Cmr) was constructed in three steps. First, the lacI gene with SDY RBS was PCR-amplified from E. coli strain MG1655 by iGEM standard primers, digested by EcoRI and SpeI, and inserted between the EcoRI and XbaI sites of plasmid pSB3K3-OR to generate plasmid pSB3K3-lacI-OR (Supplementary Figure S8B). Second, the cat gene with its constitutive promoter was PCR-amplified from plasmid PKD3 (Datsenko and Wanner, 2000), digested by AatII and PstI, and inserted into plasmid pLX002 (constructed by Q Ouyang's laboratory) in place of the ampr gene to generate plasmid pLXcm. Finally, the lacI-OR cassette was cleaved from plasmid pSB3K3-lacI-OR by AatII and XhoI, and inserted into plasmid pLXcm to finally generate plasmid pLXcmlacIOR. All sequences were verified by sequencing.
Plasmids of the control circuits were constructed by deleting lacI from plasmid PLXcmLacIOR and deleting cIind− from p15AcIindnor. With the selected functional circuit (8A) as template, we designed a pair of back-to-back primers outside of the lacI or cIind− genes to amplify all other sequences on the mutated PLXcmLacIOR and p15AcIindnor plasmids. These amplifications were performed with KOD-plus DNA polymerase (Toyobo). The PCR products were purified by gel shift and directly ligated by T4 ligase (Takara).
Saturation mutagenesis
The RBS library for CIind− or LacI was generated using the KOD-plus mutagenesis kit (code no. SMK-101; Toyobo) with a few modifications. First, we designed a pair of 5′-phosphated primers for each RBS library on the basis of the template plasmid p15AcIindnor or pLXcmlacIOR, respectively. Second, with the 5′-phosphated primers, we PCR-amplified the entire template plasmid, digested the methylated DNA templates by DpnI, and ligated the PCR products themselves to finally generate the saturation mutation library.
Screening processes and control experiments
The two RBS libraries for CIind− and LacI were simultaneously transformed into AAEClac− cells harboring pZS*Aswitch by electroporation and plated on LB agar media containing an appropriate concentration of the three antibiotics (Cm, Kan, Amp). We then screened for the functional GSLC mutants. The screening process of the two interconnecting parts was as follows: The colors of the mutant colonies were identified using a fluorescence stereomicroscope. Green colonies were inoculated into 96-well plates. The cultures were incubated in 37°C with shaking at 200 r.p.m., and diluted appropriately. The cultures were then transferred onto two of the same type of agar plates with appropriate dilution, one of which was exposed to 25 J/m2 UV irradiation and the other was not. After overnight incubation at 37°C, the colors of each mutant on the two plates were compared using a fluorescence stereomicroscope. The mutants that changed color from green to red after UV irradiation were selected for the next round of screening. The second round of selection was similar to the first one, except with regard to selection of colonies that changed color from red to green after UV irradiation. After obtaining the functional GSLC mutants, we isolated their green and red colonies from the replicated plates, which had not been exposed to UV irradiation, and stimulated these green and red cell cultures by 25 J/m2 UV irradiation in the same way as in the above screening process. Images of the resulting colonies were taken using a fluorescence stereomicroscope (Figure 5, row 1).
Similarly, we also isolated the green and red colonies of cells that carried control circuits, and inoculated them into LB medium. The cultures were manipulated as above. Images of the resulting colonies were taken using a fluorescence stereomicroscope (Figure 5, rows 2–4).
Fluorescence stereomicroscopy
Images of E. coli colonies were taken using a steREO lumar.12 fluorescence stereomicroscope and Axivision 2.0 software (Carl Zeiss). Two images were taken for each view with appropriate exposure time. One is in the GFP channel (filter set number 13, blue/green filter), whereas the other one is in the RFP channel (filter set number 20, green/red filter). The two pictures were directly merged using Adobe Photoshop CS2 9.0 (Adobe). The fluorescence illuminator was a Halogen Lamp (15 V, 150 W).
Quantitative measurement of the NOR gate
To measure the property of the NOR gate promoter, we transformed plasmid pLXnor into the testing strain JM110. The promoter's activity was measured under four conditions: with both LacI and LexA, without LacI but with LexA, without LexA but with LacI, and with neither LacI nor LexA. Repression of LacI was eliminated by adding 1 mM isopropyl-β-D-galactopyranoside (IPTG) into the medium, and repression of LexA was eliminated by exposing cells to 30 J/m2 UV irradiation. For the condition with neither LacI nor LexA, we maintained cells in the exponential growth phase in 2 ml of LB medium (50 μg/ml ampicillin, 1 mM IPTG) for 6 h (37°C and 250 r.p.m.). Each culture (OD600 around 0.4) was precipitated, washed, and resuspended in 4 ml of 10 mM MgSO4 (50 μg/ml ampicillin, 1 mM IPTG). The clear culture was spread out on the surface of a sterilized dish (Fisher Scientific, Φ=15 mm) and exposed to 30 J/m2 UV irradiation. The same volume of 2 × LB medium (50 μg/ml ampicillin, 1 mM IPTG) was then added to the culture. The culture was transferred into a flask and left to grow in the dark for 2 h until the SOS response reached the maximum (Ni et al, 2008). A 1 ml sample was taken from the culture, precipitated, and resuspended in 1 ml PBS with 10 μg/ml cycloheximide. For the other conditions, cells were manipulated in the same way, except without IPTG for the LacI-positive conditions and without UV exposure for the LexA-positive conditions. Four parallel experiments were performed for each condition. The eGFP expression levels for all samples were measured by flow cytometry (LSRII; BD). A total of 100 000 events for each sample were acquired. The concentration of eGFP in each cell was estimated by FITC-A over FSC-A. As it was log-normally distributed in population, the geometric mean was used to represent the promoter's activity of the sample. Results are shown in Figure 3C.
Quantitative measurement of the GSLC switching efficiency and behavior of the push-on push-off switch
To measure switching efficiency, we triggered the switch of the selected GSLC by a method similar to that used for the screening process. We inoculated eight pure green and eight pure red colonies each into 2 ml of LB medium with appropriate concentrations of the three antibiotics. The cultures were maintained in the exponential growth phase for 6 h (37°C and 250 r.p.m.), and then triggered by UV irradiation at OD600 value of approximately 0.4. Each culture was diluted 1:10 and 1:1000 in 1 ml of fresh LB medium with antibiotics. The remaining volume of the culture was precipitated and resuspended in 1 ml of PBS with 10 μg/ml cycloheximide, as the sample ‘Before UV irradiation’. A volume of 15 μl of the 1:10 diluted culture was placed on the LB agar plates with antibiotics and exposed to 30 J/m2 UV irradiation, whereas 15 μl of the 1:1000 diluted culture was placed on another plate with antibiotics but not exposed to UV irradiation. After overnight incubation at 37°C, several hundred colonies emerged on each plate. The colonies on both plates were scraped and resuspended in 1 ml of PBS with 10 μg/ml cycloheximide and quantitatively measured as sample ‘30 J/m2’ and ‘0 J/m2’, respectively.
The fraction of green cells in each sample was measured by flow cytometry (LSRII; BD). The laser/detector setting for GFP and RFP was 488/530 and 561/610 nm, respectively. A total of 100 000 events were collected for each sample. The fluorescence intensity of both colors for each cell was plotted as a dot plot with FITC-A on the x-axis and mCherry-A on the y-axis. The whole area was divided into four regions: high green and high red, high green but low red, low green but high red, and low green and low red. Note that only a few cells fell in the high green and high red or low green and low red region. The fraction of green cells was calculated by the cell number in the high green but low red region over that in the low green but high red region and the high green but low red region. The Mean mean value and standard deviation were derived from all the eight samples. Results are shown in Figure 4B and C.
For measuring the behavior of the push-on push-off switch, we adopted a similar strategy. Pure green cultures were maintained in the exponential growth phase for 6 h, diluted 1:10, and exposed to 30 J/m2 UV irradiation, as carried out in the switching efficiency measurements. The plates were incubated overnight; all colonies were scraped and resuspended in 1 ml of PBS. Before adding cycloheximide, 100 μl was taken and diluted in 2 ml of fresh LB medium with antibiotics and incubated for the next round of stimulus. The whole cycle was repeated three times until the population tended to reach a steady state. All the samples from the plates exposed to 30 J/m2 UV irradiation were measured by flow cytometry, as described in the preceding paragraph, to indicate the fraction of green cells in the population in each round of UV stimulus. The spontaneous switching rate from the initially green and red population was measured at the same time. The pure green and pure red cultures were manipulated in the same way, except without exposure to UV irradiation. Mean values and standard deviations were calculated from eight parallel experiments. Results are shown in Figure 6.
Supplementary Material
Supplementary tables S1–3, Supplementary figures S1–8
In the version of this supplementary file originally posted online on 9 March 2010, the authors used [number of molecules/cell] as variable's units and [mol/L] as unit for binding affinities. The authors have now converted [mol/L] units into [number of molecules/cell]. In addition, the parameter rmRNA was replaced in equations 1-5 and in Table S1 by five mRNA-specific decay rates rcImRNA, rcI434mRNA, rlacImRNA, rcIindmRNA, rlexAmRNA. These errors have been corrected in this file as of 17 May 2010.
Acknowledgments
This work is a part of the project for the Peking University team in the International Genetically Engineered Machine (iGEM) competition. The team was awarded the Grand Prize in the competition in 2007. We thank Chao Tang, Luhua Lai, Francois Taddei, Shumo Liu, Yiping Wang, Fangting Li, and Xiaojing Yang for discussion and suggestions; Qi Liu, David Pincus, Christopher McClendon, Karsten Temme, and Alvin Tamsir for critically reading the paper; Justin Lui, Anting Xu, Tao Yu, Yifan Yang, Mingzhi Qu, Qinsi Zheng, Tao Ma, Ting Liu, and Zhe Ren for participating in the related experiments; Liying Du and Wendell Lim for helping with the flow cytometer; Jingjing Wang, Junli Teng, and Yingying Liu for technical help. This work was supported by The Natural Science Foundation of China (10721403) and by the Department of Science and Technology of China (2009CB918503). Author contributions CBL conceived the project; CBL, XLL, and QOY designed the experiments; CBL and MN performed the simulation; XLL, DL, LLJ, MCW, CL, DZC, CYC, XYC, LY, and HSM constructed the NOR gate and quantitatively measured the NOR gate and the GSLC; CBL, MN, YQH, QSH, and LWH constructed the memory module and the mutation libraries, and screened for the functional GSLC; JGC and QOY contributed new reagents/analytic tools; CBL, XLL, MN, and QOY wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EPS, Landgraf D, Phillips I Silver PA (2007) Rational design of memory in eukaryotic cells. Genes Dev 21: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA (2006) Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol 355: 619–627 [DOI] [PubMed] [Google Scholar]
- Anderson JC, Voigt CA, Arkin AP (2007) Environmental signal integration by a modular AND gate. Mol Syst Biol 3: 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becskei A, Séraphin B, Serrano L (2001) Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J 20: 2528–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchler NE, Gerland U, Hwa T (2003) On schemes of combinatorial transcription logic. Proc Natl Acad Sci USA 100: 5136–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB (2001) Octamerization of ? CI repressor is needed for effective repression of P RM and efficient switching from lysogeny. Genes Dev 15: 3013–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB (2004) Cooperativity in long-range gene regulation by the ? CI repressor. Genes Dev 18: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber JE, Yeh BJ, Chak K, Lim WA (2003) Reprogramming control of an allosteric signaling switch through modular recombination. Science 301: 1904–1908 [DOI] [PubMed] [Google Scholar]
- Ferrell JE (2002) Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr Opin Cell Biol 14: 140–148 [DOI] [PubMed] [Google Scholar]
- Fritz G, Buchler N, Hwa T, Gerland U (2007) Designing sequential transcription logic: a simple genetic circuit for conditional memory. Syst Synth Biol 1: 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner TS, Cantor CR, Collins JJ (2000) Construction of a genetic toggle switch in Escherichia coli. Nature 403: 339–342 [DOI] [PubMed] [Google Scholar]
- Guet CC, Elowitz MB, Hsing W, Leibler S (2002) Combinatorial synthesis of genetic networks. Science 296: 1466–1470 [DOI] [PubMed] [Google Scholar]
- Ham TS, Lee SK, Keasling JD, Arkin AP (2008) Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3: e2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseltine EL, Arnold FH (2007) Synthetic gene circuits: design with directed evolution. Annu Rev Biophys Biomol Struct 36: 1–19 [DOI] [PubMed] [Google Scholar]
- Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M (2004) An engineered epigenetic transgene switch in mammalian cells. Nat Biotechnol 22: 867–870 [DOI] [PubMed] [Google Scholar]
- Lavery PE, Kowalczykowski SC (1992) Biochemical basis of the constitutive repressor cleavage activity of recA730 protein. A comparison to recA441 and recA803 proteins. J Biol Chem 267: 20648–20658 [PubMed] [Google Scholar]
- Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, Voigt CA (2005) Engineering Escherichia coli to see light. Nature 438: 441–442 [DOI] [PubMed] [Google Scholar]
- Libby E, Perkins TJ, Swain PS (2007) Noisy information processing through transcriptional regulation. Proc Natl Acad Sci USA 104: 7151–7156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW (1984) Autodigestion of lexA and phage lambda repressors. Proc Natl Acad Sci USA 81: 1375–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucl Acids Res 25: 1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams H, Shapiro L (1995) Circuit simulation of genetic networks. Science 269: 650–656 [DOI] [PubMed] [Google Scholar]
- Müller J, Oehler S, Müller-Hill B (1996) Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol 257: 21–29 [DOI] [PubMed] [Google Scholar]
- Ni M, Yang L, Liu X, Qi O (2008) Fluence-response dynamics of the UV-induced SOS response in Escherichia coli. Curr Microbiol 57: 521–526 [DOI] [PubMed] [Google Scholar]
- Park S, Zarrinpar A, Lim WA (2003) Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299: 1061–1064 [DOI] [PubMed] [Google Scholar]
- Ptashne M (2004) A Genetic Switch: Phage Lambda Revisited. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Ptashne M, Gann A (1997) Transcriptional activation by recruitment. Nature 386: 569–577 [DOI] [PubMed] [Google Scholar]
- Roberts JW, Roberts CW (1975) Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci USA 72: 147–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U (2002) Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet 31: 64–68 [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT (2008) Rewiring the specificity of two-component signal transduction systems. Cell 133: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkacik G, Callan CG, Bialek W (2008) Information flow and optimization in transcriptional regulation. Proc Natl Acad Sci USA 105: 12265–12270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win MN, Smolke CD (2008) Higher-order cellular information processing with synthetic RNA devices. Science 322: 456–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables S1–3, Supplementary figures S1–8
In the version of this supplementary file originally posted online on 9 March 2010, the authors used [number of molecules/cell] as variable's units and [mol/L] as unit for binding affinities. The authors have now converted [mol/L] units into [number of molecules/cell]. In addition, the parameter rmRNA was replaced in equations 1-5 and in Table S1 by five mRNA-specific decay rates rcImRNA, rcI434mRNA, rlacImRNA, rcIindmRNA, rlexAmRNA. These errors have been corrected in this file as of 17 May 2010.