Abstract
This study explores the dilemma in cellular signaling that triggering of CD95 (Fas/APO-1) in some situations results in cell death and in others leads to the activation of NF-κB. We established an integrated kinetic mathematical model for CD95-mediated apoptotic and NF-κB signaling. Systematic model reduction resulted in a surprisingly simple model well approximating experimentally observed dynamics. The model postulates a new link between c-FLIPL cleavage in the death-inducing signaling complex (DISC) and the NF-κB pathway. We validated experimentally that CD95 stimulation resulted in an interaction of p43-FLIP with the IKK complex followed by its activation. Furthermore, we showed that the apoptotic and NF-κB pathways diverge already at the DISC. Model and experimental analysis of DISC formation showed that a subtle balance of c-FLIPL and procaspase-8 determines life/death decisions in a nonlinear manner. We present an integrated model describing the complex dynamics of CD95-mediated apoptosis and NF-κB signaling.
Keywords: apoptosis, CD95 signaling, DISC, model reduction, NF-κB
Introduction
The CD95 protein (APO-1/Fas; Krammer et al, 2007) is a member of the death receptor (DR) family, a subfamily of the tumor necrosis factor receptor (TNF-R) superfamily (Ashkenazi and Dixit, 1998; Krammer, 2000). Cross linking of CD95 with its natural ligand, CD95L, or with agonistic anti-CD95 antibodies induces apoptosis in sensitive cells (Trauth et al, 1989). The signal transduction of CD95 starts with the formation of the death-inducing signaling complex (DISC) detectable within seconds after receptor stimulation (Kischkel et al, 1995). The DISC consists of CD95, the adaptor molecule FADD, procaspase-8a/b, procaspase-10 and c-FLIPL/S/R (Muzio et al, 1996; Scaffidi et al, 1999; Sprick et al, 2002; Golks et al, 2005; Krammer et al, 2007). Procaspase-8 is converted at the DISC, through a series of autoproteolytic cleavage steps, into p43/p41 and p18, which leads to the activation of effector caspase-3 (Lavrik et al, 2003). This can occur in two different ways in type I and type II cells (Scaffidi et al, 1998). Type I cells are characterized by high levels of CD95 DISC formation and increased amounts of active caspase-8, leading to the direct activation of downstream effector caspases-3 and -7 and subsequent apoptosis. Type II cells are characterized by lower levels of CD95 DISC formation, and caspase-3 activation requires additional amplification through the release of pro-apoptotic factors from mitochondria triggered by the caspase-8-mediated cleavage of the Bcl-2 family protein Bid.
Recently, experiments have demonstrated that CD95L is not only a potent apoptosis inducer but can also activate multiple nonapoptotic pathways, in particular induction of transcription factor NF-κB (Barnhart et al, 2004; Kreuz et al, 2004; Peter et al, 2007). The NF-κB family regulates the expression of genes crucial for innate and adaptive immune responses, cell growth and apoptosis. In most cells, the NF-κB dimer is sequestered in the cytosol by inhibitors of the κB protein (IκB), and its nuclear translocation can be induced by a wide variety of stimuli through activation of the IκB kinase (IKK) complex (Karin and Lin, 2002; Hayden and Ghosh, 2004). The IKK complex consists of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit IKKγ/NEMO. On activation of the IKK complex, IκB is phosphorylated and degraded in an ubiquitin-dependent manner. The NF-κB dimers can then translocate into the nucleus to induce transcription of target genes. However, the exact molecular mechanism of NF-κB activation through CD95 remains unclear (Peter et al, 2007).
It has been previously shown that DED-containing proteins, such as procaspase-8 and c-FLIP have a prominent role in NF-κB activation (Chaudhary et al, 2000; Hu et al, 2000; Kreuz et al, 2004; Dohrman et al, 2005; Su et al, 2005). c-FLIP N-terminal cleavage products p43-FLIP and p22-FLIP strongly induce NF-κB (Kataoka and Tschopp, 2004; Golks et al, 2006a). p43-FLIP is generated by procaspase-8 at the DISC on CD95 stimulation (Scaffidi et al, 1999; Krueger et al, 2001). It has been shown to interact with components of the TNFR-mediated NF-κB activation pathway, TNFR-associated factor 1 (TRAF1), TRAF2 and receptor-interacting protein (RIP), which together promote NF-κB activation (Kataoka and Tschopp, 2004; Dohrman et al, 2005). Interestingly, it has been shown that c-FLIPL/S/R form heterodimers with procaspase-8, resulting in the generation of the cleavage fragment p22-FLIP (Golks et al, 2006a). This protein mediates NF-κB activation by binding directly to the IKK complex. p22-FLIP differs from p43-FLIP in that it is generated in nonapoptotic cells without DR stimulation (Golks et al, 2006a). Furthermore, it was also reported that the binding of procaspase-8 in the MALT1–Bcl-10 adapter complex formed on TCR stimulation has a prominent role in NF-κB induction (Su et al, 2005). Thus, various molecules of the CD95 signaling machinery, such as procaspase-8 and c-FLIP have a complex role in CD95-mediated apoptosis and NF-κB pathways. These findings motivated our systems biology approach and prompted us to determine whether CD95-mediated signaling should be considered as a dynamic system resulting in life/death decisions. It has been shown repeatedly that decision making is often a process brought about by a reaction system rather than by the influence of a single molecule (Xiong and Ferrell, 2003; Legewie et al, 2006; Santos et al, 2007).
Modeling in biology has a long tradition and can be dated back to Hodgkin and Huxley's model of a neuron (Hodgkin and Huxley, 1952). A wide gamut of modeling formalisms has been applied to biological systems ranging from qualitative approaches, such as Boolean and Bayesian networks (Sun and Zhao, 2004) to partial differential equations and stochastic simulations. A number of models typically based on ordinary differential equations (ODEs) have described apoptosis signaling. Fussenegger et al (2000) presented the first mathematical model encompassing both extrinsic and intrinsic apoptotic pathways. A more detailed model with parameters estimated from quantitative western blots verified a threshold for CD95 stimulation regulated by c-FLIP (Bentele et al, 2004). Advanced models of apoptosis have been used to discriminate between different hypotheses of apoptosis inhibition by Bcl-2 family members and XIAPs (Hua et al, 2005; Rehm et al, 2006). Others have addressed aspects, such as feedback and bistability (Eissing et al, 2004; Janes et al, 2005; Bagci et al, 2006; Legewie et al, 2006). A number of mathematical models for NF-κB activation have investigated the complex interplay of various IκB subunits in NF-κB oscillatory activity (Hoffmann et al, 2002; Nelson et al, 2004; Cheong et al, 2008). Furthermore, the signaling of the TNF pathway leading to NF-κB activation was modeled (Cheong et al, 2006; Park et al, 2006).
Despite much study on both apoptotic and NF-κB signaling pathways induced by CD95, there is a lack of a detailed mechanistic understanding of the divergence of these pathways. In the following text, we propose an integrated quantitative model supported by experimental data for CD95-mediated apoptosis and NF-κB activation. A direct interaction between p43-FLIP and the IKK complex is postulated by the model and validated experimentally. The model also predicts a divergence of the two pathways at the DISC and that the cellular decision depends on the balance between the levels of c-FLIPL and caspase-8. Experimental modulations of DISC protein amounts are all consistent with the model predictions. Our model gives mechanistic insights into the cellular response on CD95 activation and provides a way to predict cellular behavior.
Results
NF-κB and caspases show parallel activation
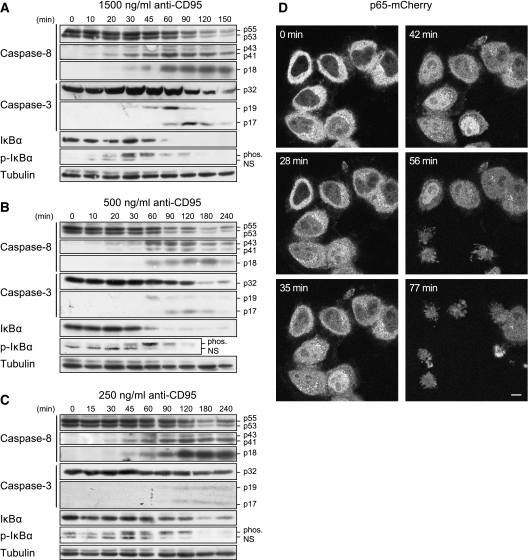
The CD95 protein is known to mediate the activation of caspases leading to apoptosis (Lavrik et al, 2005) and mediating the activation of NF-κB (Barnhart et al, 2004). However, it is not understood whether CD95 activates apoptotic and NF-κB pathways in a mutually exclusive manner, or whether the activation of both pathways takes place in parallel. To answer this question, we investigated CD95 signaling by western blot analysis of HeLa cells stably overexpressing CD95–GFP (HeLa-CD95) and treated the cells with agonistic anti-CD95 antibodies at three concentrations: 1500, 500 and 250 ng/ml (Figure 1A–C). Procaspase-8a/b was processed to p43/p41 and subsequently to p18. Procaspase-3 was also cleaved to the active subunit p17 showing the induction of the apoptotic pathway. In parallel, we observed the phosphorylation of IκBα followed by its degradation. Stimulation of CD95 caused apoptosis at all antibody concentrations (Supplementary Figure S1). The quantified blots demonstrate that apoptotic and NF-κB pathways were activated on a similar time scale (Figure 2A–G). Interestingly, IκBα phosphorylation was at its maximum (Figure 2F) roughly three times faster than active caspases (Figure 2B, D and G) at all antibody concentrations. Using live-cell images of HeLa-CD95 cells stably overexpressing p65–mCherry, we observed simultaneous apoptosis and nuclear translocation of p65–mCherry in each cell on induction of CD95 (Figure 1D). Thus, we could exclude the possibility that a subset of cells activated NF-κB, whereas another subset activated caspases. This demonstrates a parallel rather than a mutually exclusive activation of caspases and NF-κB. The simultaneous induction of apoptosis and NF-κB activation and their different dynamics led us to investigate the divergence point of the two pathways.
Figure 1.
NF-κB and caspases show parallel activation. (A–C) HeLa-CD95 cells were stimulated with (A)1500 ng/ml, (B) 500 ng/ml and (C) 250 ng/ml of agonistic anti-CD95 antibodies for the indicated periods of time. The cellular lysates were analyzed by western blotting using antibodies against caspases, IκBα and p-IκBα. Nonspecific bands of anti-p-IκBα are marked (NS). Results are representative of four different experiments. (D) To follow p65 localization, Hela-CD95 cells were stably transfected with p65–mCherry. Cells were stimulated with 1500 ng/ml anti-CD95 antibody and imaged using fluorescent microscopy over the respective time. Measurements of mCherry (depicted in gray) indicated translocation of p65 on CD95 activation. Induced apoptosis is observed by the appearance of apoptotic bodies. Scale bar: 10 μm.
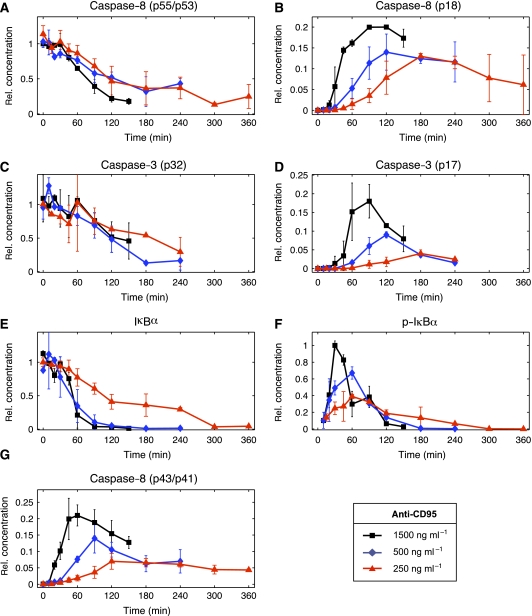
Figure 2.
Quantified response to anti-CD95 stimulation. Each of the panels (A–G) shows a quantification of western blots shown in Figure 1A–C. The symbols mark the average value and the lines show their linear interpolations. The s.d. was included when more than one value was obtained. Black, blue and red colors correspond to 1500, 500 and 250 ng/ml anti-CD95 antibodies, respectively, used to induce the cells at time t=0.
A mechanistic model assuming a direct interaction between p43-FLIP and the IKK complex is consistent with experimental observations
To understand the crosstalk between CD95-mediated apoptosis and NF-κB activation, we created a mathematical model of CD95 signaling. When we integrated known molecular interactions into a detailed model, we faced the problem that it is mechanistically not understood how signals from CD95 are transmitted to NF-κB. Taking into account earlier findings that the c-FLIPL cleavage products, p43-FLIP and p22-FLIP, are involved in NF-κB signaling (Kataoka and Tschopp, 2004; Golks et al, 2006a), and that p22-FLIP is not formed upon CD95 stimulation (Golks et al, 2006a), we hypothesized that p43-FLIP is the missing link between the DISC and the IKK complex. The kinetics of phosphorylated IκBα in Figure 2F indicate a temporal correlation between CD95 stimulation and IKK activity. The simplest scheme to explain this phenomenon would be through p43-FLIP generation at the DISC and subsequent interaction with the IKK complex. This led us to the mathematical model of CD95-mediated caspase and NF-κB activation shown in Figure 3.
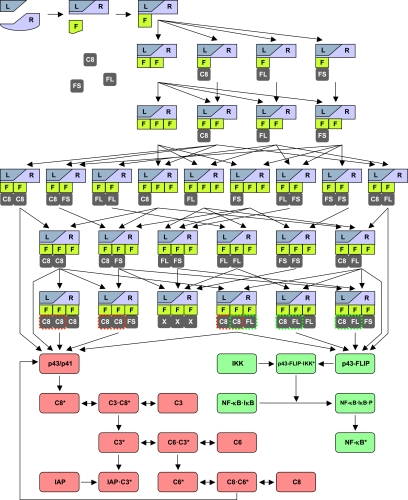
Figure 3.
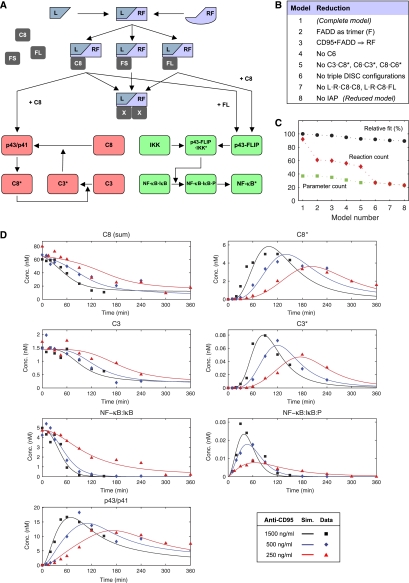
Model of CD95-mediated signaling. A graphic representation of the complete model illustrating the process of DISC formation and subsequent signaling of the apoptotic (depicted in red) and NF-κB pathway (depicted in green). Abbreviations: L, CD95 ligand; R, CD95 receptor; F, FADD; C8, procaspase-8 (p55/p53); C8*, active caspase-8; C3, procaspase-3; C3*, active caspase-3; C6, procaspase-6; C6*, active caspase-6; FL, c-FLIPL; FS, c-FLIPS; X, C8, FL or FS; p43/p41, 1st cleavage product of procaspase-8; and p43-FLIP, cleavage product of c-FLIPL.
Our model assumes a trimerized ligand (L) that binds to a trimerized CD95 receptor (R) that can recruit three copies of FADD (F) leading to the DISC formation. Subsequently, DED-containing proteins, such as procaspase-8 (C8), c-FLIPL (FL) and c-FLIPS (FS) can bind to FADD. The order of protein binding gives rise to a combinatorial variety of intermediates. Assembled DISCs can be categorized into three groups: the first group contains at least two copies of procaspase-8 and is further processed by p43/p41 into active protease subunits, p18 and p10, forming an active caspase-8 heterotetramer (C8*). This apoptotic branch of the model also includes procaspase-3 and procaspase-6 (C3 and C6), their active forms (C3* and C6*), the inhibitor IAP and a feedback loop from caspase-6 to caspase-8.
The second group of DISCs features at least one copy of procaspase-8 and one copy of c-FLIPL giving rise to p43-FLIP. The model postulates that p43-FLIP interacts with the IKK complex leading to phosphorylation of IκB (NF-κB·IκB·P), which entails its degradation and the translocation of p65 to the nucleus (NF-κB*). For simplicity, we assume that the entire pool of IκB is bound to NF-κB. In the third group of DISCs, we considered all remaining configurations. They do not participate in further signaling and are merged in a terminal state (L·R·F·F·F·X·X·X). The key features of the model are its partitioning into DISC formation, apoptotic and NF-κB signaling, and the confinement of all interactions between the two pathways to the DISC. In this model, the gene transcription induced by NF-κB was neglected, as we aimed to investigate early signaling events immediately after the stimulation of CD95.
For model simulations, the molecular interactions depicted in Figure 3 were translated into a system of coupled ODEs (for details on reaction equations and parameters, see Supplementary Tables S1 and S2). To reduce the large number of free parameters, we simplified the reactions to be irreversible as previously described with the exception of a reversible enzymatic scheme used for caspase activation (Bentele et al, 2004). The model contains unknown reaction constants and concentrations that we estimated from our data by least-squares optimization. Remarkably, the model was able to reproduce the experimental data set (Supplementary Figure S2) despite the almost exclusive use of an irreversible reaction scheme. Interestingly, it shows that the activity of effector caspases is not required for reproducing the dynamics of CD95-mediated NF-κB activation. In addition, the model postulates a direct interaction between p43-FLIP and the IKK complex.
p43-FLIP interacts with the IKK complex upon CD95 stimulation
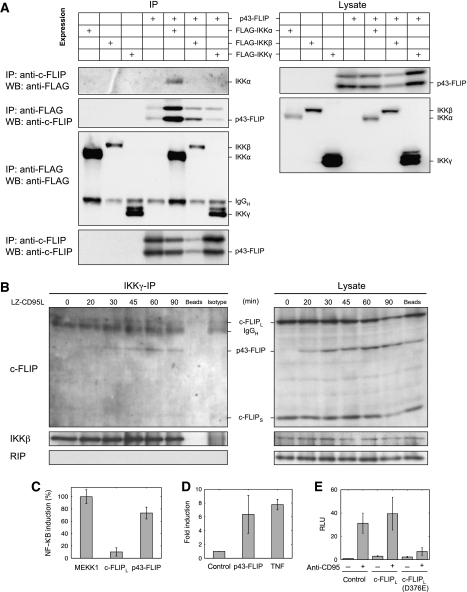
To test our model hypothesis that p43-FLIP interacts with the IKK complex, we transiently co-transfected p43-FLIP with FLAG-tagged IKKα, IKKβ and IKKγ into 293T cells, and then performed an immunoprecipitation (IP) using anti-FLAG (FLAG-IP) and anti-FLIP antibodies (FLIP-IP; Figure 4A). We observed IKKα in the FLIP-IP and p43-FLIP mostly in the FLAG-IKKα-IP. Thus, we established that p43-FLIP can interact with IKKα. Next, we tested whether this interaction also occurs under endogenous conditions on CD95 stimulation in Hela-CD95 cells. An IP of the IKK complex using anti-IKKγ antibodies showed a recruitment of p43-FLIP to the IKK complex at 30 min after the stimulation of CD95 (Figure 4B). To avoid co-IP of the CD95 DISC with the IKK complex, we used LZ-CD95L and not anti-CD95 antibodies for stimulation in this experiment. The RIP protein, invoked in CD95-mediated NF-κB activation (Kreuz et al, 2004), was not detectable at the IKK complex under our experimental conditions. In the IKKγ-IP, we also detected a band corresponding to the size of c-FLIPL, however, the band was also detected in the isotype control IP and, hence, is considered unspecific. In a CD95 DISC-IP (CD95-IP), we could not detect IKK-α/β, suggesting that the IKK complex is not associated to the DISC (Supplementary Figure S3). Positive controls for the CD95 DISC-IP showed that the DISC was formed efficiently, for example, procaspase-8 and its cleavage product p43/p41 were present at the DISC.
Figure 4.
p43-FLIP interacts with the IKK complex on CD95 stimulation. (A) The association of p43-FLIP with components of the IKK complex was determined using co-immunoprecipitations (IPs). 293T cells were co-transfected with FLAG-tagged IKKα, IKKβ or IKKγ with or without p43-FLIP. The lysates were immunoprecipitated using antibodies against the FLAG tag or against c-FLIP and analyzed by western blotting using antibodies against the FLAG tag or c-FLIP (left side). In parallel, corresponding lysates were also analyzed using western blot analysis with the same antibodies as the immunoprecipitation (right side). (B) The association of p43-FLIP with the IKK complex under endogenous conditions was determined using co-immunoprecipitations of IKKγ (IKKγ-IP). HeLa-CD95 cells were stimulated with 500 ng/ml of LZ-CD95L for the indicated time intervals and immunoprecipitated using an antibody against IKKγ. All subunits of the IKK complex were immunoprecipitated in this procedure due to IKK complex stability. The immunoprecipitated proteins (IKKγ-IP, left side) and the corresponding lysates (lysate, right side) were analyzed by western blotting using antibodies against c-FLIP, IKKβ and RIP. IKKβ served as a loading control. (C) 0.5 × 105 293T cells were co-transfected with MEKK1, p43-FLIP and c-FLIPL, and the luciferase reporter plasmid. NF-κB–luciferase activity was determined at 16 h after transfection. The results represent the mean±s.d. values of quadruplet cultures. GFP transfection was performed to control 100% transfection efficiency. (D) 1 × 105 HeLa-CD95 cells were transfected with a NF-κB-dependent luciferase reporter plasmid (1 μg per well) and a Renilla reporter plasmid (100 ng per well). Cells were either untreated (control), co-transfected with p43-FLIP (2 μg per well) or stimulated with TNF (500 ng/ml). NF-κB–luciferase activity was determined at 16 h after transfection. Renilla transfection was performed to normalize the transfection efficiency. (E) 1 × 105 HeLa-CD95 cells were co-transfected with a luciferase (1 μg per well) and a Renilla reporter plasmid (100 ng per well), as well as pcDNA3 as a control plasmid (control), WT c-FLIPL or c-FLIPL (D376E; 2 μg per well). At 24 h after transfection, cells were left untreated or were stimulated with 1000 ng/ml of anti-CD95. To prevent apoptosis cells were pretreated with 20 μM zVAD-fmk at 30 min before CD95 stimulation. NF-κB–luciferase activity was determined at 16 h after CD95 stimulation. Renilla transfection was performed to normalize the transfection efficiency.
To test whether c-FLIPL or its cleavage product p43-FLIP can induce NF-κB, we performed NF-κB luciferase assays (Figure 4C–E). c-FLIPL and p43-FLIP were overexpressed in 293T cells, which was followed by a luciferase-based NF-κB activity assay. Noticeably, we found that p43-FLIP is a strong inducer of NF-κB activity, whereas c-FLIPL exhibited low induction efficiency compared with MEKK1 used as a positive control (Figure 4C). In addition, HeLa-CD95 cells transfected with p43-FLIP showed an induction of NF-κB similar to the one observed after TNF induction (Figure 4D). To confirm that p43-FLIP, not c-FLIPL, for example, in the absence of CD95 activation mediates NF-κB activation, we generated a mutant form of c-FLIPL, which cannot be cleaved to p43-FLIP due to a substitution at D376 (c-FLIPL (D376E)). To avoid unspecific effects by the cleavage of c-FLIPL to p22-FLIP, constructs of c-FLIP were mutated in the p22 cleavage site (D196E). c-FLIPL, c-FLIPL (D376E) or an empty pcDNA3 vector, as a control, was transfected into HeLa-CD95 cells along with a NF-κB reporter construct and a Renilla reporter construct for normalization. HeLa-CD95 cells transfected with c-FLIPL (D376E) showed decreased CD95-induced NF-κB activation, as compared with HeLa-CD95 cells transfected with WT c-FLIPL (Figure 4E). Taken together, these data confirm our model postulation that cleavage of c-FLIPL to p43-FLIP is required for the CD95-mediated NF-κB activation through interaction of p43-FLIP with the IKK complex.
Apoptotic and NF-κB signalings diverge at the DISC
When establishing the model structure, we encountered the problem in deciding whether the reported cleavage of NF-κB pathway members by active caspase also has a role in our experiments (Fischer et al, 2003). We decided to omit these interactions as long as the model was in agreement with our experimental observations. As a consequence, our model postulates that the apoptotic and NF-κB pathways diverge already at the DISC. During model fitting, we found that this was sufficient to reproduce the dynamics of IκB phosphorylation and degradation. To test our model hypothesis, we blocked the apoptotic pathway with the pan-caspase inhibitor, zVAD-fmk, and investigated CD95 signaling. It was shown previously that zVAD-fmk blocks caspase activation downstream from the DISC, but does not affect the processing of procaspase-8 to the catalytically active subunit p43/p41 at the DISC, which mediates the processing of c-FLIP to p43-FLIP (Golks et al, 2006b; Hughes et al, 2009). The inhibitor may bind caspase-8 only after the so-called substrate switch that is necessary for caspase-3 and Bid cleavage but not for p43/p41 and p43-FLIP formation (Hughes et al, 2009). Therefore, application of zVAD-fmk provides a unique experimental tool that allows to switch off the apoptotic signaling of CD95 and follow only the nonapoptotic events mediated by p43-FLIP. We stimulated HeLa-CD95 cells, pre-treated with zVAD-fmk, with anti-CD95 and analyzed NF-κB activation and cell death signaling (Supplementary Figure S4). Using live-cell imaging of HeLa-CD95 cells stably overexpressing p65–mCherry, we observed nuclear translocation of p65–mCherry on induction of CD95 (Supplementary Figure S4A). Apoptosis, however, was not detected. Thus, we confirmed that pre-treatment with zVAD-fmk blocks CD95-induced apoptosis but does not inhibit NF-κB activation. Western blot analysis shows that cleavage of c-FLIPL to p43-FLIP and phosphorylation of IκB were affected only slightly by zVAD-fmk (Supplementary Figure S4B). Thus, we could validate the model prediction that processed caspases are not required for NF-κB activation and that the apoptotic and NF-κB pathways diverge already at the DISC. These findings underline the important role of the DISC in distributing signals between caspases and NF-κB.
Reduced model is sufficient to explain dynamics of life/death signaling
The model of CD95-mediated signaling depicted in Figure 3 is a comprehensive integration of data for CD95 signaling pathways. However, its complexity might hide key features of the mechanism of pathway divergence. Furthermore, an overly complex model is prone to overfitting. To simplify the model without compromising its predictive power, we tested which of the proteins and reactions of the complete model are required to reproduce the observed dynamics. A clear advantage of a smaller model is that it yields much more reliable parameter estimates. To determine the most important interactions, we simplified the complete model in a step-wise manner, obtaining a model of considerably lower complexity (Figure 5A). The simplification steps are listed in Figure 5B and the corresponding intermediate models can be found in the Supplementary information (Supplementary Figure S5A–G). We repeated the parameter estimation after each step of model reduction. The fit of the complete model was set to 100% and the performance of the reduced models was normalized against it. Figure 5C shows that with an increasing number of reduction steps the goodness of fit decreased only slightly. Further model reduction led to a marked decrease in model fit (data not shown); hence we decided to consider model 8 as the final reduced model. This reduced model 8 (Figure 5A, Supplementary Figure S5G) still approximated well with the experimental data set (Figure 5D), however, it should be noted that a permutation of the simplification steps might lead to other equally well-fitting models. The number of reactions decreased from 92 in the complete to 23 in the reduced model (Figure 5C). For parameter estimation, the search space of the optimization method was reduced by 17 dimensions because of the lower number of unknown reaction constants and concentrations. Thus, the reduced model yields a much higher confidence in the parameter estimates and is more suitable for computer-based predictions (Supplementary Figure S6). Throughout the remainder of this study, we used the reduced model 8 for prediction and comparison to experimental results.
Figure 5.
Reduced model is sufficient to explain dynamics of life/death signaling. (A) Reduced model of CD95-mediated caspase and NF-κB activation. CD95 receptor and FADD are merged into one entity named RF. (B) The table lists the consecutive reduction steps performed from models 1 to 8. (C) Model comparison using the relative goodness of fit, in which the χ2 measure of model 1 is set to 100%. Red diamonds indicate the number of reactions of each model and squares show the corresponding number of parameters. (D) Simulated concentrations of proteins in HeLa-CD95 cells based on the reduced model after receptor stimulation. Black, blue and red colors correspond to 1500, 500 and 250 ng/ml anti-CD95, respectively, used to induce the cells at time t=0. C8 (sum) is observable, which sums all concentrations of proteins containing one copy of procaspase-8, for example, L·RF·C8·FS. Squares, diamonds and triangles indicate experimental data obtained by western blot (cf. Figure 2).
Life/death threshold concentration is independent of the amount of CD95
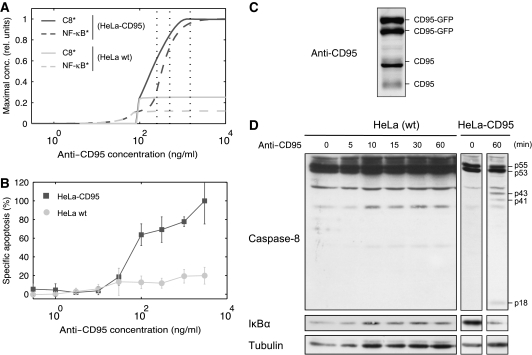
Previous study on CD95-induced apoptosis demonstrated the existence of an apoptotic threshold (Bentele et al, 2004). To test whether our model is consistent with this finding, we analyzed the model for such a threshold behavior using a simulated dose–response experiment. We varied the amount of anti-CD95 antibodies and recorded the maximum level of active caspase-8 and NF-κB as readouts for apoptotic and NF-κB signaling, respectively, (Figure 6A). The model predicts a threshold of approximately 100 ng/ml required to trigger the apoptotic pathway. To validate this prediction, we performed a titration study by measuring cell death by flow cytometry and found the threshold to be in the range of 30–100 ng/ml (Figure 6B).
Figure 6.
Life/death threshold concentration is independent of the amount of CD95. (A) The simulated dose–response curves display active NF-κB and caspase-8 as a function of the simulation strength for HeLa cells (light gray) and HeLa-CD95 cells (dark gray). The highest concentrations attained by NF-κB* and C8* in a simulation of 60 h for a given stimulus are shown. Concentrations were scaled to unity for comparison. Vertical dotted black lines indicate the used experimental concentrations of anti-CD95 antibody (1500/500/250 ng/ml; cf. Figure 1). (B) HeLa-CD95 and WT HeLa cells were stimulated with various amounts of anti-CD95 antibodies for 18 h. Cell death was determined by flow cytometry. (C) Western blot analysis of CD95 expression in HeLa-CD95 cells. GFP-tagged CD95 and endogenous CD95 are indicated. Glycosylation of CD95 gave rise to multiple bands in the range of 42–52 kDa for endogenous CD95 and 59–69 kDa for GFP-tagged CD95. (D) HeLa and HeLa-CD95 cells were stimulated using 500 ng/ml of anti-CD95 antibodies for the indicated time intervals. Total cellular lysates were analyzed by western blotting using antibodies against caspase-8, IκBα and tubulin (loading control).
Next, we sought to determine how the amount of CD95 influences signal transduction of the two pathways in our cell lines. We compared CD95-overexpressing HeLa cells with WT HeLa cells. The endogenous amount of CD95 was estimated to be approximately 12 times less than GFP-tagged CD95 (Figure 6C). We repeated the simulated dose–response experiment with a decreased number of CD95 receptors. This gave rise to two predictions: we expected a greatly reduced activity of both pathways and, more interestingly, that the same amount of anti-CD95 (∼100 ng/ml) would be sufficient to trigger apoptosis despite the lower death rate (Figure 6A). Experimentally, we confirmed that the apoptotic threshold of HeLa WT cells was identical to that of HeLa-CD95 cells (between 30 and 100 ng/ml). However, HeLa-CD95 cells exhibited significantly higher rates of apoptosis (Figure 6B). Moreover, we observe a much higher level of caspase-8 cleavage and IκBα degradation by western blot analysis in HeLa-CD95 cells stimulated with 500 ng/ml anti-CD95 (Figure 6D) than in WT HeLa cells. In summary, the increased receptor number in HeLa-CD95 cells resulted in the amplification of both CD95-mediated apoptosis and NF-κB signaling. Furthermore, we observed that the amount of CD95 receptors determines the death rate, in contrast to the apoptotic threshold concentration that remained constant.
c-FLIPL recruitment kinetics explains the differential dynamics of the two pathways
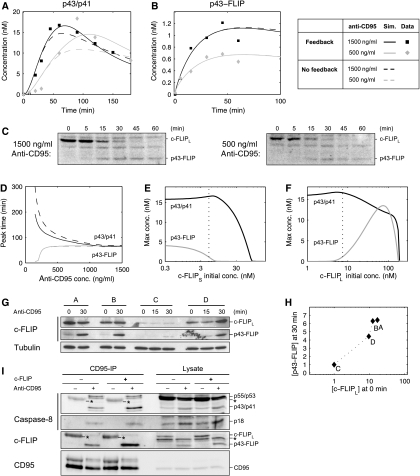
The quantified western blots of the apoptotic and NF-κB pathways revealed an interesting feature of differential dynamics (Figure 2). The peak of phosphorylated IκBα was reached within 1h and was rather invariant to the stimulus intensity (Figure 2F). On the contrary, the dynamics of caspase-8 and caspase-3 activation showed a marked delay with decreasing stimulus strength (Figure 2B, D and G). The initial hypothesis was that the feedback loop in the apoptotic pathway between caspase-8, -3 and -6 is the cause for a gradual build-up of active caspases, timing of which is strongly dependent on the stimulus concentration. Alternatively, the differential dynamics might reflect an inherent feature of the DISC formation process, in which different reaction constants create a kinetic asymmetry for the signaling of the apoptotic and NF-κB pathways. To discriminate between these two hypotheses, we analyzed the reaction speeds of DISC assembly in our model. The result of parameter estimation that procaspase-8 had a lower affinity for the DISC than c-FLIPL is consistent with previous studies (Chang et al, 2002; Lavrik et al, 2007). Model simulations demonstrated that this leads to qualitatively different dynamics of p43/p41 and p43-FLIP generation: with increasing stimulus strength, p43/p41 peaked earlier, whereas there was almost no difference when p43-FLIP reached its maximum (Figure 7A and B). To validate these predictions, we analyzed c-FLIPL cleavage by western blotting and confirmed that while the amplitude of p43-FLIP generation varied between stimulations with 1500 and 500 ng/ml antibody, the timing of the curves was identical (Figure 7C). The predicted dynamics of p43-FLIP were matched well by the quantified blots despite a slight disagreement at the latest time point, in which a decrease was observed experimentally (Figure 7B). The qualitative difference of timing of procaspase-8 and c-FLIPL cleavage becomes evident in Figure 7D, in which we examined when p43/p41 and p43-FLIP arrived at their maximal values as a function of stimulus strength. With decreasing stimulus strength, p43/p41 reaches its peak level increasingly late, whereas p43-FLIP attains the maximum level earlier. Further model analysis revealed that this was the cause of the different timing of caspase-3 and NF-κB activation observed experimentally (Figure 2D and F). To confirm that the apoptotic feedback loop is not the reason for the differential dynamics, we simulated the model after disrupting the loop by removing the activation of procaspase-8 by caspase-3. We observed that the disruption influenced p43/p41 processing only weakly and mainly with respect to its amplitude (Figure 7A, B and D; dashed line). This argues against our initial hypothesis that the feedback loop caused the delayed caspase-3 activation at low doses of anti-CD95. Instead, these results show that a kinetic asymmetry in the process of DISC assembly was likely the reason for the differential dynamics of apoptotic and NF-κB signaling.
Figure 7.
c-FLIPL recruitment kinetics explains the differential dynamics of the two pathways. (A) Simulation and experimental data of p43/p41 dynamics for 1500 and 500 ng/ml of stimulating anti-CD95 antibody. (B) Same as (A) for p43-FLIP. The data points were obtained by quantifying the blots shown in (C). (C) HeLa-CD95 cells stimulated with 1500/500 ng/ml anti-CD95 for the indicated time points. Lysates were used for western blots analysis using an antibody against c-FLIP. The full-length form of c-FLIPL and its cleavage product p43-FLIP are indicated. (D) The curves show the time when p43/p41 and p43-FLIP reached their peak level in the simulation as a function of the antibody concentration. (E) Model prediction of the maximal concentration of p43/p41 and p43-FLIP depending on the initial concentration of c-FLIPS. The dotted line indicates the estimated level of c-FLIPS in our setting. Stimulation intensity corresponds to 1000 ng/ml of anti-CD95 antibodies. (F) Same as in (E) but with c-FLIPL. (G) Western blotting of four different clones of HeLa-CD95 cells stably overexpressing c-FLIPL in different amounts. Western blots show the full-length form of c-FLIPL and its cleavage product p43-FLIP after induction with 1000 ng/ml of anti-CD95 for the indicated time points. Tubulin serves as a loading control. (H) Quantification of (G) by plotting the intensity of c-FLIPL at 0 min against p43-FLIP at 30 min. For normalization, the amounts of c-FLIPL and p43-FLIP in clone C were set to 1, as they exhibited the lowest levels. (I) HeLa-CD95 cells with a stable c-FLIP downregulation (c-FLIP−) and control-transfected HeLa-CD95 cells (c-FLIP+) were generated by RNA interference. The cells were kept under nonstimulated conditions for control (anti-CD95−) or stimulated with 500 ng/ml anti-CD95 for 30 min (anti-CD95+). The CD95-IP was performed using anti-CD95 antibodies. The immunoprecipitated proteins (CD95-IP) and the lysates were analyzed by western blot using antibodies against caspase-8, c-FLIP and CD95. Secondary antibodies recognize the heavy chain (50 kDa) of the antibody used for the IPs. Thus, the heavy chain of the antibody used for immunoprecipitation is marked in the western blot with an asteric (*=IgGH). Importantly, the amount of antibody in the unstimulated lane was always higher as it was added after cell lysis.
The generation of p43/p41 is a hallmark of caspase activation and apoptosis, whereas p43-FLIP engages in NF-κB signaling in our model. Hence, we consider these proteins as end points of signal processing at the DISC. They are the result of the dynamic process of DISC assembly, which is mainly determined by the levels of c-FLIP and procaspase-8. To understand the role of c-FLIP isoforms in determining the balance between apoptotic and NF-κB signaling, we altered their concentrations in the simulation. We charted the maximum levels of p43/p41 and p43-FLIP as a function of the initial concentration of c-FLIPS after a stimulus corresponding to 1000 ng/ml anti-CD95 antibody (Figure 7E). As expected from the model structure, c-FLIPS inhibited both pathways, although p43-FLIP generation was inhibited at a lower threshold than p43/p41 generation. We repeated the virtual experiment with c-FLIPL with a different result (Figure 7F). Low and high levels of c-FLIPL both inhibited p43-FLIP generation, giving rise to a bell-shaped activation profile. This could readily be explained by the fact that cells expressing significantly more c-FLIPL than procaspase-8 would predominantly feature c-FLIPL trimers at the DISC, effectively blocking downstream signaling. The vertical dotted line in Figure 7F indicates the estimated level of c-FLIPL in our cell line. Our model predicts that increasing the concentration of c-FLIPL by one order of magnitude would lead to a steep increase in p43-FLIP generation until it reaches a maximum, beyond which the curve drops. To test this prediction, we generated HeLa cells stably co-overexpressing CD95 and c-FLIPL. Four clones that exhibited different levels of c-FLIPL were selected. The CD95/c-FLIPL-overexpressing cells were induced with 1000 ng/ml of anti-CD95 antibody and analyzed by western blotting (Figure 7G). A quantification of c-FLIPL before stimulation and p43-FLIP after 30 min of stimulation is shown in Figure 7H. The measured activation profile is in good agreement with the predicted curve of p43-FLIP generation in Figure 7F. Cell line C exhibited the lowest level of c-FLIPL overexpression and correspondingly less p43-FLIP generation. With increasing levels of c-FLIPL we also found an increase in p43-FLIP. The model predicted that at very high concentrations no p43-FLIP is generated. This drop-off was not found experimentally, as our cell lines probably do not overexpress c-FLIPL at sufficiently high levels. As the processing of caspase-8 and c-FLIP can be different in the cytosolic pool as compared to processing at the CD95 DISC, we performed a CD95-IP after stably reducing the amount of c-FLIP by RNA interference (Figure 7I). We observed that lowering the amount of c-FLIP leads to less p43-FLIP generation at the DISC and marginally decreased levels of p43/p41. These two observations confirm our model prediction in which lowered levels of c-FLIPL result in very little p43-FLIP but almost unchanged levels of p43/p41 (Figure 7F), further validating our model of signal processing at the CD95 DISC. Thus, the analysis of the kinetic model elucidated the complex signal processing of c-FLIPL and procaspase-8 to p43-FLIP and p43/p41, respectively, and explained how this leads to differential dynamics of caspase-3 and NF-κB activation.
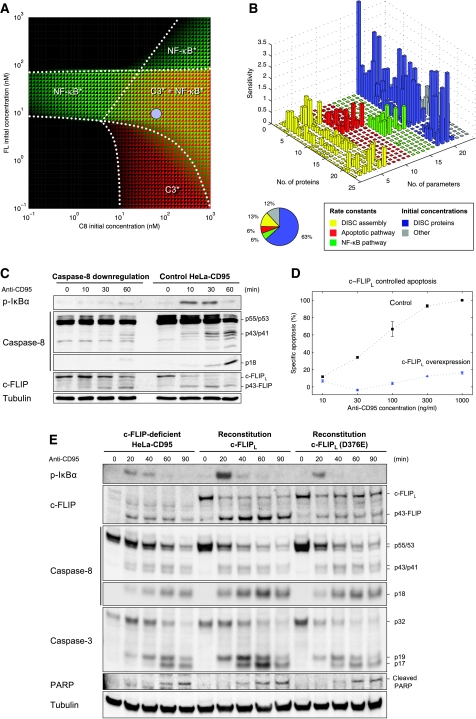
DISC protein stoichiometry is determinant of cell fate
To better understand the nonlinear interplay of DISC proteins in the determination of cell fate, we analyzed the activity of caspase-3 and NF-κB as a function of procaspase-8 and c-FLIPL levels (Figure 8A). We found in our simulations that the decision of apoptosis and NF-κB activation is controlled by both proteins. Different scenarios occurred, which show combination or absence of either caspase-3 activity or NF-κB activity. The estimated levels of procaspase-8 and c-FLIPL in HeLa-CD95 cells are located in the central area of the phase-like diagram. Small changes in concentration can trigger different scenarios, helping to reconcile earlier conflicting observations (Kataoka et al, 2000; Chang et al, 2002; Micheau et al, 2002; Budd et al, 2006) regarding the role of c-FLIPL in cell-fate decision. Our model predicts that c-FLIPL disables, promotes and inhibits NF-κB activation, respectively, depending on whether the level of c-FLIPL is low, in an intermediary range or high, respectively. To further corroborate the importance of DISC protein levels we performed a sensitivity analysis (Figure 8B). The initial concentrations of DISC proteins—especially of c-FLIP isoforms—were found to be the most sensitive parameters, summing up to 63% of the total sensitivities. The remaining 20 parameters contributed only to 37% of total sensitivities. This result explains how relatively small changes in the abundance of DISC proteins might lead to drastic changes in downstream signaling as already observed for the amount of CD95 (Figure 6).
Figure 8.
DISC protein stoichiometry is determinant of cell fate. (A) This model-based prediction illustrates how different levels of procaspase-8 and c-FLIPL may lead to different phenotypes. The logarithm of the maximum concentration of active NF-κB (green) and caspase-3 (red) in a simulation of 6 h after CD95 stimulation is shown as a function of the initial concentration of procaspase-8 and c-FLIPL. Dotted lines were drawn manually to delineate different regimes of activation. The blue circle indicates the estimated level of c-FLIPL and procaspase-8 in HeLa-CD95 cells. (B) Sensitivity analysis. Absolute values of relative sensitivities were averaged over a time interval of 360 min. The pie chart shows the contribution of different parameter categories to the total sensitivity. For variable and parameter numbers see Supplementary Tables S1 and S2. (C) HeLa-CD95 cells stably downregulating procaspase-8 were generated by RNA interference and compared against control HeLa-CD95 cells. Both cell lines were stimulated with 500 ng/ml anti-CD95 for the indicated time intervals. The lysates were used for western blot analysis against caspase-8, p-IκBα, c-FLIP and tubulin. (D) Cell death assay of HeLa-CD95 cells with c-FLIPL overexpression. Specific apoptosis normalized to 100% for HeLa-CD95 cells (control) is shown. Cells were treated with the depicted amount of anti-CD95 for 18 h. Cell death was determined with PI staining and analyzed with flow cytometry. Specific cell death was calculated as described in Materials and methods section. (E) c-FLIP-deficient HeLa-CD95 cells were transiently transfected with WT c-FLIPL (middle) or c-FLIPL (D376E; right; 15 ng per well). At 24 h after transfection, cells were stimulated with 1000 ng/ml anti-CD95 for the indicated time intervals. The lysates were used for western blot analysis against p-IκBα, c-FLIP, caspase-8, caspase-3, PARP and tubulin.
To determine the role of procaspase-8 concentration, we downregulated procaspase-8 in HeLa-CD95 cells using RNA interference (Figure 8C). The decrease in procaspase-8 level resulted in a significantly lower amount of p43-FLIP after CD95 stimulation. Subsequently, it led to the abolishment of CD95-mediated phosphorylation of IκB. Furthermore, the amount of active caspase-8, p18, was decreased, hence leading to reduced apoptosis. Therefore, by perturbing the ratio of procaspase-8 to c-FLIPL at the DISC, we have influenced the induction of apoptosis and NF-κB activation. The phase diagram shown in Figure 8A predicts that either increasing or decreasing the amount of c-FLIPL leads to a different signaling mode. To test this prediction, we modulated c-FLIPL in HeLa-CD95 cells and measured cell death. Figure 8D displays specific apoptosis of Hela-CD95 cells after CD95 stimulation at various concentrations around the threshold determined in Figure 6. In agreement with the phase diagram (Figure 8A), we observed that c-FLIPL overexpression resulted in a strong reduction of apoptosis. The downregulation of c-FLIPL, on the contrary, did not lead to a significant change of the death rate (Supplementary Figure S7). Thus, high amounts of c-FLIPL affect the level of programmed cell death after CD95 stimulation. Moreover, we could further confirm by western blot analysis that downregulation of c-FLIP led to a reduction of the levels of p43-FLIP and phosphorylated IκBα after CD95 stimulation (Supplementary Figure S8A). We wanted to rule out the possibility that the reduced amount of phosphorylated IκBα is because of increased caspase activity upon c-FLIPL downregulation, which could potentially block activation of NF-κB. Therefore, we performed the analysis in the presence of zVAD-fmk (Supplementary Figure S8B). The results were similar: stable downregulation of c-FLIPL led to a reduction of IκBα phosphorylation after CD95 stimulation. In addition, to control the specificity of c-FLIP downregulation and further confirm the requirement of cleavage of c-FLIPL to p43-FLIP, we performed a reconstitution experiment in HeLa-CD95–c-FLIP-deficient cells (Figure 8E). Cells reconstituted with WT c-FLIPL were able to generate p43-FLIP and increased IκBα phosphorylation upon CD95 stimulation. In contrast, cells reconstituted with the noncleavable mutant of c-FLIPL (D376E) did not show processing to p43-FLIP (Figure 8E; Supplementary Figure S9). Noticeably, as postulated by the model, this resulted in a strong reduction of the levels of IκBα phosphorylation upon CD95 stimulation. We also demonstrated that this effect does not result from a change of caspase activity of HeLa-CD95 cells reconstituted with c-FLIPL (D376E), as the extent of caspase-3 activation and PARP cleavage was comparable with that in c-FLIP-deficient cells (Figure 8E). Taken together, these data confirm our model postulates that cleavage of c-FLIPL to p43-FLIP is required for CD95-mediated NF-κB activation and that the DISC protein levels determine cell fate in a nonlinear manner, highlighting the role of signal processing within the DISC.
Discussion
In this study, we propose an integrated kinetic model of CD95-mediated apoptosis and NF-κB signaling. This was achieved by integrating mechanistic knowledge of DISC assembly and caspase activation with a simple scheme of NF-κB activation. The details of DISC formation have been mostly disregarded in earlier modeling studies of CD95 signaling. Typically, the only function of the DISC was to convert signals from CD95L into active caspase-8. If included, c-FLIP was assigned only an inhibitory function and p43-FLIP generation was neglected (Fussenegger et al, 2000; Bentele et al, 2004; Hua et al, 2005). As a recent study has indicated that the specific configuration of proteins at the CD95 DISC activates distinct signaling pathways (Budd et al, 2006), we built a mechanistic model with a high level of detail at the DISC and performed a systematic model reduction. The resulting model is simple yet fully compatible with the current paradigm of CD95 signaling. A large part of its components were observed experimentally, which increased the reliability of the parameter estimates. Furthermore, it could reproduce the experimentally observed dynamics and proved to be a powerful predictive tool.
The large amount of CD95 sensitizes HeLa-CD95 cells for caspase and NF-κB activation and renders them a valuable tool for the study of p43/p41 and p43-FLIP generation at the DISC. We used live-cell imaging to show that CD95 induces apoptosis in HeLa-CD95 cells and that it is accompanied by translocation of NF-κB to the nucleus. However, we did not observe oscillatory dynamics as published by others (Nelson et al, 2004), which is likely to be a cell type-dependent phenomenon.
We provide new evidence for a direct link between CD95 stimulation and NF-κB activation. According to our study, c-FLIPL is cleaved to p43-FLIP, which interacts with the IKK complex leading to its activation. This pathway differs from p22-FLIP-mediated NF-κB activation, in which procaspase-8 cleaves c-FLIPL to p22-FLIP independently of CD95 stimulation (Golks et al, 2006a). We did not observe p22-FLIP formation in HeLa-CD95 cells on CD95 stimulation (data not shown). The RIP protein, another mediator of DR-induced NF-κB activation (Kataoka and Tschopp, 2004; Kreuz et al, 2004; Dohrman et al, 2005; Misra et al, 2005), was not detected to be associated with the IKK complex, which does not rule out that it is involved in the NF-κB pathway studied here.
In agreement with our findings, it was shown that Jurkat clones lacking IKKγ, FADD or caspase-8 abolished CD95-mediated upregulation of the NF-κB target gene IκBα (Kreuz et al, 2004). They also found that CD95-mediated NF-κB activity relies on a full-length procaspase-8 rather than on the processed form and upregulation of c-FLIPL and that c-FLIPS inhibits CD95-mediated NF-κB activity. Furthermore, c-FLIP-deficient T cells displayed diminished proliferation (Zhang and He, 2005). Furthermore, overexpression of c-FLIPL in Jurkat cells increased NF-κB activity upon TCR stimulation (Zhang and He, 2005), whereas downregulation of c-FLIPL was shown to either increase (Kreuz et al, 2004) or not influence CD95-induced NF-κB activity (Legembre et al, 2004). As suggested by our model analysis, these conflicting data might be explained by different levels of c-FLIP proteins in the different investigated cell systems. Data from c-FLIPL transgenic mice strongly support the role of c-FLIPL in proliferative pathways (Lens et al, 2002; Dohrman et al, 2005). We have found that c-FLIPL levels crucially determine the balance between apoptotic and NF-κB signaling by shaping the dynamics of DISC assembly. Although this finding is based on experiments performed in cell lines with limited physiological importance, we expect that the nonlinear dynamics of DISC assembly is a generic systems property of life/death decision making in CD95 signaling pathways. This is especially important for physiologically relevant cells, such as various cancer cells that are resistant towards death receptor-induced apoptosis. This hypothesis, however, needs careful experimental validation and will be subject to further investigation in our lab.
Our results support the emerging paradigm in CD95 signaling that the DISC can act as a potent signal processor deciding between life and death (Lavrik et al, 2007). Why then would the same receptor trigger two pathways with opposing phenotypes? Nuclear factor-κB is a transcription factor for the c-FLIP and the IAP family (Krammer et al, 2007). Hence, upregulation of these apoptosis inhibitors may maintain a threshold toward CD95-mediated apoptosis, thereby preventing unwanted apoptotic effects at low amounts of CD95L. In our modeling approach, we concentrated on early signaling events, which took place within a few hours after CD95 stimulation. We neglected gene expression induced by NF-κB as we did not observe upregulation of the c-FLIP isoforms at the protein level within the initial hours. It remains a challenge for future study to integrate a model of CD95-mediated signal transduction with a model of transcriptional regulation to capture the possible feedback from transcriptional regulation by NF-κB onto upstream CD95 signaling.
Materials and methods
Cell lines
HeLa-CD95 was generated by selection with G418, HeLa-CD95–c-FLIPL and HeLa-CD95–p65–mCherry by selection with G418 and puromycin (Sigma-Aldrich) according to standard protocols. HeLa, HeLa-CD95 and HeLa-CD95–c-FLIPL cells were maintained in DMEM (Life Technologies, Germany), 10 mM HEPES (Life Technologies), 50 μg/ml gentamycin (Life Technologies), 10% fetal calf serum (Life Technologies) in 5% CO2. G418 (0.5 mg/ml) was used to maintain HeLa-CD95 cells and a mix of G418 (0.25 mg/ml) and puromycin (0.2 μg/ml) was used to maintain HeLa-CD95–c-FLIPL and HeLa-CD95–p65–mCherry. Transfections were done using FuGene 6 (Roche, Switzerland).
DNA constructs
The CD95–GFP fusion was made by fusing the entire coding sequence of CD95 5′ to mGFP (Snapp et al, 2003) with the linker TRDPPVAT in between. To generate cells stably expressing c-FLIPL, the coding sequence of c-FLIPL was cloned in the pIRESpuro2 vector (Clontech). p65–mCherry and pSilencer 3.1-H1 Neo vector were kindly provided by Dr Nathan Brady. The FLAG-IKKγ plasmid was a kind gift from Dr Ralf Marienfeld, FLAG-IKKα and -β plasmids were kind gifts from Professor Hiroyasu Nakano; MEKK1 plasmid was a kind gift from Professor Peter Angel and the luciferase reporter construct was provided by Dr Rüdiger Arnold (Arnold et al, 2001). The vector for c-FLIP downregulation was provided by Professor Martin Leverkus (Diessenbacher et al, 2008).
Cloning of c-FLIPL and c-FLIPL mutants
c-FLIP was cloned into the pcDNA3 expression vector (Invitrogen) using the KpnI and ApaI restriction sites. Several mutants were obtained through overlapping PCR. The mutated nucleotides are indicated in bold. The following primers were used:
c-FLIPL_for: 5′-CGGGGTACCATGTCTGCTGAAGTCATCC-3′
c-FLIPL_rev: 5′-GCCGGCCCTTATGTGTAGGAGAGGATAAG-3′
D196E_for: 5′-GTCTCAAGGAGCCTTCAAATAACTTCAGGCTCC-3′
D196E_rev: 5′-GAAGGCTCCTTGAGACTCTTTTGGATTGCTGC-3′
p43_rev: 5′-GCGGCCCCAATCCACCTCCAAGAGGCTGC-3′
c-FLIP-reconstitution_for: 5′-GGCGCTGGCACTAGGTATAGGAATGTTCTCCAAGCAG-3′
c-FLIP-reconstitution_rev: 5′-ATACCTAGTGCCAGCGCCTTGAACAGACTGCTTGTAC-3′
Antibodies and reagents
Anti-FADD mAb (IgG1) was purchased from Transduction Laboratories (Lexington, KY). Anti-IκBα, anti-phospho-IκBα, anti-IKKα/β/γ, anti-CD95 C20 polyclonal antibodies and anti-RIP monoclonal antibody were purchased from Santa Cruz Biotechnology (USA). Anti-caspase-8 monoclonal antibody C15 (mouse IgG2b) recognizes the p18 subunit of caspase-8 (Scaffidi et al, 1997). Anti-FLIP monoclonal antibody NF6 (mouse IgG1) recognizes the N-terminal part of c-FLIP (Scaffidi et al, 1999). Anti-FLAG monoclonal antibody M2 (mouse IgG1) was purchased from Sigma (Deisenhofen, Germany). As anti-CD95 antibody we always used anti-APO-1 antibody. Anti-APO-1 antibody is an agonistic monoclonal antibody (IgG3) recognizing an epitope on the extracellular part of CD95 (APO-1/Fas; Trauth et al, 1989). LZ-CD95L was prepared as described before (Walczak et al, 1999). Horseradish peroxidase-conjugated goat anti-mouse IgG1, -2a and -2b were purchased from Southern Biotechnology Associates (Echingen, Germany). All chemicals used were of analytical grade and purchased from Merck (Darmstadt, Germany) or Sigma. Renilla (pGL4.74) vector was from Promega (Mannheim, Germany).
Analysis of total cellular lysates
A total of 107 cells were either treated with indicated amounts of anti-CD95 antibody for indicated periods of time at 37°C or left untreated, washed twice in 1 × PBS and lysed subsequently in lysis buffer A (30 mM Tris–HCl (pH 7.5), 150 mM NaCl, 2 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (Sigma, Germany), protease inhibitor cocktail, 1% Triton X-100 (Serva, Germany) and 10% glycerol). If pre-treated with zVAD-fmk, cells were pre-incubated for 30 min at 37°C with the indicated concentrations of zVAD-fmk before stimulation. Total cellular lysates were analyzed using SDS–PAGE gels. Proteins were transferred to Hybond nitrocellulose membrane (Amersham Pharmacia Biotech, Germany), blocked with 5% non-fat dry milk in PBS/Tween (PBS plus 0.05% Tween 20) for 1 h, washed with PBS/Tween and incubated with the primary antibody in PBS/Tween overnight at 4°C. Blots were developed with a chemoluminescence method according to the manufacturer's protocol (Perkin Elmer Life Sciences, Germany).
Live-cell imaging
Cells were cultured in 8-well Lab-Tek chambered cover glasses (Nunc, Germany). During imaging, cells were maintained at 37°C in DMEM without phenol red and complemented with 20 mM HEPES buffer and 10% FCS. We used a Leica SP2 laser scanning confocal microscope (Leica Microsystems, Germany) with a × 63 oil immersion lens. For excitation, the laser wavelength was set to 488 nm for GFP and 594 nm for mCherry. The recording spectrum was 498–535 nm for GFP and 604–686 nm for mCherry.
IKKγ IP
A total 1.2 × 106 HeLa-CD95 cells were lysed in lysis buffer A (with or without stimulation, divided into two equal parts and the IP was performed by using 2 μg anti-IKKγ antibody or isotype control antibodies together with 30 μl of protein-A–Sepharose. Immunoprecipitations were performed overnight at 4°C; then beads were washed five times with 20 volumes of lysis buffer and subjected to western blot analysis as described above.
CD95 DISC IP
A total of 1 × 107 HeLa-CD95 cells were lysed in lysis buffer A, with or without stimulation, divided into two equal parts and the DISC IP was performed by using 5 μg anti-CD95 (anti-APO-1) antibody together with 30 μl of protein A–Sepharose. Immunoprecipitations were performed overnight at 4°C; then beads were washed five times with 20 volumes of lysis buffer and subjected to western blot analysis as described above.
NF6 and FLAG IPs
A total of 2 × 106 293T cells were transfected using the calcium phosphate method 1 day before lysis. Then cells were lysed in a volume of 1 ml for 30 min at 0°C. The FLIP IP was performed using 100 μl of NF6 hybridoma supernatant together with 30 μl of protein A–Sepharose. The FLAG IP was performed by using 4 μg anti-FLAG mAb together with 30 μl of protein A–Sepharose. Immunoprecipitations were performed for 1 h at RT or overnight at 4°C, then beads were washed five times with 20 volumes of lysis buffer and subjected to western blot analysis as described above.
Stable siRNA expression
We used stable expression of siRNA as published previously (Diessenbacher et al, 2008). The siRNA FLIP vector or control empty vector were transfected into the HeLa-CD95 cell line as described earlier (Diessenbacher et al, 2008). The downregulation of caspase-8 was done with a pSilencer 3.1-H1 Neo vector. The siRNA target sequence was 5′-gggtcatgctctatcagat-3′ as desribed by Wagner et al (2004). c-FLIPL-deficient cells were reconstituted with c-FLIPL-mutants using Lipofectamine transfection reagent according to the manufacturer's protocol. Silent mutations were introduced in WT c-FLIPL and c-FLIPL (D376E) to avoid silencing by siRNA (refer to cloning of c-FLIPL and c-FLIPL mutants). Only 15 ng DNA per well of a 6-well plate were transfected to obtain an approximately physiological level of c-FLIPL.
Quantification of western blot time series
The western blots were scanned and analyzed using ImageJ (http://rsb.info.nih.gov/ij/). The local background from bands was subtracted and their intensity was quantified. For normalization, the signal was divided by the anti-tubulin signal. For alignment of time, series from repeated experiments the signals were multiplied with a scalar to minimize the sum of mean-squared distances of measurements at the same time points. Finally, the average signal was computed and scaled.
NF-κB activation assay
The 24-well titer plates were seeded with 0.5 × 105 293T cells the day before transfection. Cells were transfected using the calcium phosphate method with various expression vectors, together with the NF-κB-driven luciferase reporter plasmid (500 ng) and a Renilla plasmid for normalization (100 ng). Cells were washed with PBS at 16 h after transfection and lysed for 20 min at RT in 50 μl lysis buffer (Passive lysis buffer, Promega) followed by centrifugation (10 000 × g, 20 min) to sediment insoluble materials. A volume of 5 μl of cell lysates were mixed with 50 μl of the luciferase assay mixture (470 μM Beetle Luciferin (Promega), 1.07 mM (MgCO3)4 Mg(OH)2·5 H2O, 20 mM N-Tris-(hydroxymethyl)-methylglycine, 2.67 mM MgSO4, 100 μM EDTA, 33.3 mM DTT, 270 μM CoA(OAc), 530 μM ATP). Relative light units were measured with a Berthold duoluminomat (Bad Wildbad, Germany). For the assays in HeLa-CD95 cells, 6-well titer plates were seeded with 0.5 × 105 HeLa-CD95 cells the day before transfection. Cells were transfected using Fugene with various expression vectors, together with the NF-κB-driven luciferase reporter plasmid (2 μg; Arnold et al, 2001) and Renilla luciferase plasmid (100 ng) used as normalization control. Subsequently, measurements were performed similar to those for 293T cells.
Cell death assay
Cells were plated and treated as indicated with anti-APO-1 antibody for 18 h and 1 μg/ml propidium iodide for 30 min. Cell death was assessed by propidium iodide uptake and quantified using a Cytomics FC 500 MPL flow cytometer (Beckman Coulter). Specific cell death was calculated as follows: (percentage of experimental cell death−percentage of spontaneous cell death)/(100−percentage of spontaneous cell death) × 100 and calibrated to 100% for the peak value.
Modeling and simulation
The molecular interactions depicted in Figure 3 were noted down as uni- or bimolecular reactions and subsequently translated into a system of coupled ordinary differential equations according to the law of mass action (Supplementary Table S1). The reduced model is available in SBML format as Supplementary information. We used the software MATLAB (MathWorks, USA) and SBtoolbox (http://www.sbtoolbox.org) for modeling, simulation and analysis. Unknown reaction constants, initial concentrations and data scaling parameters were estimated by maximizing the maximum likelihood criterion with MATLAB's lsqnonlin function (Supplementary Table S2; for details see Bentele et al (2004)). For every model, the estimation was repeated several thousand times from different, randomly chosen starting points to avoid convergence to local minima. Model fits were compared using the χ2 measure of goodness of fit. The normalized fit was obtained by dividing the χ2 value of the complete model by the χ2 value of the reduced model and subsequent multiplication by 100%.
Sensitivity analysis
The relative sensitivity of a variable xi at time t (in min) with respect to a parameter pj is given by
Time-averaged sensitivities were computed according to
We summed absolute values to rule out that positive and negative sensitivities cancel each other. For the pie chart, parameters were grouped into categories as displayed. All values, Sij, belonging to the same category were summed and subsequently compared.
Supplementary Material
Supplemetary tables S1–2, Supplementary figures S1–9
Acknowledgments
We acknowledge the support by the Helmholtz Alliance on Systems Biology (NW1SBCancer/UniHD) and the BMBF-funded ForSys grant VIROQUANT (0313923). JB received support from the Center for Modeling and Simulation in the Biosciences (BIOMS). We thank Nadine Sachs, Jennifer Hoetzel and Tatjana Schmidt for help with the experiments; Martin Leverkus for providing us with the siRNA FLIP vector; Eunice Hatada and Nathan Brady for critical reading of this paper; Wilhelm Sander Stiftung, HRJRJ-2/2008, SFB 405 and the Tumorzentrum Heidelberg/Mannheim for supporting our study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Arnold R, Liou J, Drexler HC, Weiss A, Kiefer F (2001) Caspase-mediated cleavage of hematopoietic progenitor kinase 1 (HPK1) converts an activator of NFkappaB into an inhibitor of NFkappaB. J Biol Chem 276: 14675–14684 [DOI] [PubMed] [Google Scholar]
- Ashkenazi A, Dixit VM (1998) Death receptors: signaling and modulation. Science 281: 1305–1308 [DOI] [PubMed] [Google Scholar]
- Bagci EZ, Vodovotz Y, Billiar TR, Ermentrout GB, Bahar I (2006) Bistability in apoptosis: roles of Bax, Bcl-2, and mitochondrial permeability transition pores. Biophys J 90: 1546–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart BC, Legembre P, Pietras E, Bubici C, Franzoso G, Peter ME (2004) CD95 ligand induces motility and invasiveness of apoptosis-resistant tumor cells. EMBO J 23: 3175–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele M, Lavrik I, Ulrich M, Stosser S, Heermann DW, Kalthoff H, Krammer PH, Eils R (2004) Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis. J Cell Biol 166: 839–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd RC, Yeh WC, Tschopp J (2006) cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol 6: 196–204 [DOI] [PubMed] [Google Scholar]
- Chang DW, Xing Z, Pan Y, Algeciras-Schimnich A, Barnhart BC, Yaish-Ohad S, Peter ME, Yang X (2002) c-FLIP(L) is a dual function regulator for caspase-8 activation and CD95-mediated apoptosis. EMBO J 21: 3704–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary PM, Eby MT, Jasmin A, Kumar A, Liu L, Hood L (2000) Activation of the NF-kappaB pathway by caspase 8 and its homologs. Oncogene 19: 4451–4460 [DOI] [PubMed] [Google Scholar]
- Cheong R, Bergmann A, Werner SL, Regal J, Hoffmann A, Levchenko A (2006) Transient I kappa B kinase activity mediates temporal NF-kappa B dynamics in response to a wide range of tumor necrosis factor-alpha doses. J Biol Chem 281: 2945–2950 [DOI] [PubMed] [Google Scholar]
- Cheong R, Hoffmann A, Levchenko A (2008) Understanding NF-kappaB signaling via mathematical modeling. Mol Syst Biol 4: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diessenbacher P, Hupe M, Sprick MR, Kerstan A, Geserick P, Haas TL, Wachter T, Neumann M, Walczak H, Silke J, Leverkus M (2008) NF-kappaB inhibition reveals differential mechanisms of TNF versus TRAIL-induced apoptosis upstream or at the level of caspase-8 activation independent of cIAP2. J Invest Dermatol 128: 1134–1147 [DOI] [PubMed] [Google Scholar]
- Dohrman A, Kataoka T, Cuenin S, Russell JQ, Tschopp J, Budd RC (2005) Cellular FLIP (long form) regulates CD8(+) T cell activation through caspase-8-dependent NF-kappa B activation. J Immunol 174: 5270–5278 [DOI] [PubMed] [Google Scholar]
- Eissing T, Conzelmann H, Gilles ED, Allgower F, Bullinger E, Scheurich P (2004) Bistability analyses of a caspase activation model for receptor-induced apoptosis. J Biol Chem 279: 36892–36897 [DOI] [PubMed] [Google Scholar]
- Fischer U, Janicke RU, Schulze-Osthoff K (2003) Many cuts to ruin: a comprehensive update of caspase substrates. Cell Death Differ 10: 76–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussenegger M, Bailey JE, Varner J (2000) A mathematical model of caspase function in apoptosis. Nat Biotechnol 18: 768–774 [DOI] [PubMed] [Google Scholar]
- Golks A, Brenner D, Fritsch C, Krammer PH, Lavrik IN (2005) c-FLIPR, a new regulator of death receptor-induced apoptosis. J Biol Chem 280: 14507–14513 [DOI] [PubMed] [Google Scholar]
- Golks A, Brenner D, Krammer PH, Lavrik IN (2006a) The c-FLIP-NH2 terminus (p22-FLIP) induces NF-kappa B activation. J Exp Med 203: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golks A, Brenner D, Schmitz I, Watzl C, Krueger A, Krammer PH, Lavrik IN (2006b) The role of CAP3 in CD95 signaling: new insights into the mechanism of procaspase-8 activation. Cell Death Differ 13: 489–498 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2004) Signaling to NF-kappaB. Genes Dev 18: 2195–2224 [DOI] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF (1952) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117: 500–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Levchenko A, Scott ML, Baltimore D (2002) The I kappa B-NF-kappa B signaling module: temporal control and selective gene activation. Science 298: 1241–1245 [DOI] [PubMed] [Google Scholar]
- Hu WH, Johnson H, Shu HB (2000) Activation of NF-kappaB by FADD, Casper, and caspase-8. J Biol Chem 275: 10838–10844 [DOI] [PubMed] [Google Scholar]
- Hua F, Cornejo MG, Cardone MH, Stokes CL, Lauffenburger DA (2005) Effects of Bcl-2 levels on Fas signaling-induced caspase-3 activation: molecular genetic tests of computational model predictions. J Immunol 175: 985–995 [DOI] [PubMed] [Google Scholar]
- Hughes MA, Harper N, Butterworth M, Cain K, Cohen GM, MacFarlane M (2009) Reconstitution of the death-inducing signaling complex reveals a substrate switch that determines CD95-mediated death or survival. Mol Cell 35: 265–279 [DOI] [PubMed] [Google Scholar]
- Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB (2005) A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310: 1646–1653 [DOI] [PubMed] [Google Scholar]
- Karin M, Lin A (2002) NF-kappaB at the crossroads of life and death. Nat Immunol 3: 221–227 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Budd RC, Holler N, Thome M, Martinon F, Irmler M, Burns K, Hahne M, Kennedy N, Kovacsovics M, Tschopp J (2000) The caspase-8 inhibitor FLIP promotes activation of NF-kappaB and Erk signaling pathways. Curr Biol 10: 640–648 [DOI] [PubMed] [Google Scholar]
- Kataoka T, Tschopp J (2004) N-terminal fragment of c-FLIP(L) processed by caspase 8 specifically interacts with TRAF2 and induces activation of the NF-kappaB signaling pathway. Mol Cell Biol 24: 2627–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME (1995) Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J 14: 5579–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer PH (2000) CD95's deadly mission in the immune system. Nature 407: 789–795 [DOI] [PubMed] [Google Scholar]
- Krammer PH, Arnold R, Lavrik IN (2007) Life and death in peripheral T cells. Nat Rev Immunol 7: 532–542 [DOI] [PubMed] [Google Scholar]
- Kreuz S, Siegmund D, Rumpf JJ, Samel D, Leverkus M, Janssen O, Hacker G, Dittrich-Breiholz O, Kracht M, Scheurich P, Wajant H (2004) NF kappa B activation by Fas is mediated through FADD, caspase-8, and RIP and is inhibited by FLIP. J Cell Biol 166: 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger A, Schmitz I, Baumann S, Krammer PH, Kirchhoff S (2001) Cellular flice-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the cd95 death-inducing signaling complex. J Biol Chem 276: 20633–20640 [DOI] [PubMed] [Google Scholar]
- Lavrik I, Krueger A, Schmitz I, Baumann S, Weyd H, Krammer PH, Kirchhoff S (2003) The active caspase-8 heterotetramer is formed at the CD95 DISC. Cell Death Differ 10: 144–145 [DOI] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Krammer PH (2005) Caspases: pharmacological manipulation of cell death. J Clin Invest 115: 2665–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Riess D, Bentele M, Eils R, Krammer PH (2007) Analysis of CD95 threshold signaling: triggering of CD95 (FAS/APO-1) at low concentrations primarily results in survival signaling. J Biol Chem 282: 13664–13671 [DOI] [PubMed] [Google Scholar]
- Legembre P, Barnhart BC, Zheng LX, Vijayan S, Straus SE, Puck J, Dale JK, Lenardo M, Peter ME (2004) Induction of apoptosis and activation of NF-kappa B by CD95 require different signalling thresholds. EMBO Rep 5: 1084–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legewie S, Bluthgen N, Herzel H (2006) Mathematical modeling identifies inhibitors of apoptosis as mediators of positive feedback and bistability. PLoS Comput Biol 2: e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lens SM, Kataoka T, Fortner KA, Tinel A, Ferrero I, MacDonald RH, Hahne M, Beermann F, Attinger A, Orbea HA, Budd RC, Tschopp J (2002) The caspase 8 inhibitor c-FLIP(L) modulates T-cell receptor-induced proliferation but not activation-induced cell death of lymphocytes. Mol Cell Biol 22: 5419–5433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grutter MG (2002) The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 277: 45162–45171 [DOI] [PubMed] [Google Scholar]
- Misra RS, Jelley-Gibbs DM, Russell JQ, Huston G, Swain SL, Budd RC (2005) Effector CD4(+) T cells generate intermediate caspase activity and cleavage of caspase-8 substrates. J Immunol 174: 3999–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85: 817–827 [DOI] [PubMed] [Google Scholar]
- Nelson DE, Ihekwaba AEC, Elliott M, Johnson JR, Gibney CA, Foreman BE, Nelson G, See V, Horton CA, Spiller DG, Edwards SW, McDowell HP, Unitt JF, Sullivan E, Grimley R, Benson N, Broomhead D, Kell DB, White MRH (2004) Oscillations in NF-kappa B signaling control the dynamics of gene expression. Science 306: 704–708 [DOI] [PubMed] [Google Scholar]
- Park SG, Lee T, Kang HY, Park K, Cho KH, Jung GH (2006) The influence of the signal dynamics of activated form of IKK on NF-kappa B and anti-apoptotic gene expressions: a systems biology approach. FEBS Lett 580: 822–830 [DOI] [PubMed] [Google Scholar]
- Peter ME, Budd RC, Desbarats J, Hedrick SM, Hueber AO, Newell MK, Owen LB, Tschopp J, Wajant H, Wallach D, Wiltrout RH, Zornig M, Lynch DH (2007) The CD95 receptor: apoptosis revisited. Cell 129: 447–450 [DOI] [PubMed] [Google Scholar]
- Rehm M, Huber HJ, Dussmann H, Prehn JHM (2006) Systems analysis of effector caspase activation and its control by X-linked inhibitor of apoptosis protein. EMBO J 25: 4338–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Verveer PJ, Bastiaens PI (2007) Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol 9: 324–330 [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, Debatin KM, Krammer PH, Peter ME (1998) Two CD95 (APO-1/Fas) signaling pathways. EMBO J 17: 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi C, Medema JP, Krammer PH, Peter ME (1997) FLICE is predominantly expressed as two functionally active isoforms, caspase-8/a and caspase-8/b. J Biol Chem 272: 26953–26958 [DOI] [PubMed] [Google Scholar]
- Scaffidi C, Schmitz I, Krammer PH, Peter ME (1999) The role of c-FLIP in modulation of CD95-induced apoptosis. J Biol Chem 274: 1541–1548 [DOI] [PubMed] [Google Scholar]
- Snapp EL, Hegde RS, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J (2003) Formation of stacked ER cisternae by low affinity protein interactions. J Cell Biol 163: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprick M, Rieser E, Stahl H, Grosse-Wilde A, Weigand M, Walczak H (2002) Caspase-10 is recruited to and activated at the native TRAIL and CD95 death-inducing signalling complexes in a FADD-dependent manner but can not functionally substitute caspase-8. EMBO J 21: 4520–4530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H, Bidere N, Zheng L, Cubre A, Sakai K, Dale J, Salmena L, Hakem R, Straus S, Lenardo M (2005) Requirement for caspase-8 in NF-kappaB activation by antigen receptor. Science 307: 1465–1468 [DOI] [PubMed] [Google Scholar]
- Sun N, Zhao HY (2004) Genomic approaches in dissecting complex biological pathways. Pharmacogenomics 5: 163–179 [DOI] [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W, Debatin KM, Krammer PH (1989) Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 245: 301–305 [DOI] [PubMed] [Google Scholar]
- Wagner KW, Engels IH, Deveraux QL (2004) Caspase-2 can function upstream of bid cleavage in the TRAIL apoptosis pathway. J Biol Chem 279: 35047–35052 [DOI] [PubMed] [Google Scholar]
- Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, Chin W, Jones J, Woodward A, Le T, Smith C, Smolak P, Goodwin RG, Rauch CT, Schuh JC, Lynch DH (1999) Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 5: 157–163 [DOI] [PubMed] [Google Scholar]
- Xiong W, Ferrell JE (2003) A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature 426: 460–465 [DOI] [PubMed] [Google Scholar]
- Zhang N, He YW (2005) An essential role for c-FLIP in the efficient development of mature T lymphocytes. J Exp Med 202: 395–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemetary tables S1–2, Supplementary figures S1–9