Abstract
Morphogen gradients infer cell fate as a function of cellular position. Experiments in Drosophila embryos have shown that the Bicoid (Bcd) gradient is precise and exhibits some degree of scaling. We present experimental results on the precision of Bcd target genes for embryos with a single, double or quadruple dose of bicoid demonstrating that precision is highest at mid-embryo and position dependent, rather than gene dependent. This confirms that the major contribution to precision is achieved already at the Bcd gradient formation. Modeling this dynamic process, we investigate precision for inter-embryo fluctuations in different parameters affecting gradient formation. Within our modeling framework, the observed precision can only be achieved by a transient Bcd profile. Studying different extensions of our modeling framework reveals that scaling is generally position dependent and decreases toward the posterior pole. Our measurements confirm this trend, indicating almost perfect scaling except for anterior most expression domains, which overcompensate fluctuations in embryo length.
Keywords: bicoid, Drosophila, morphogen gradients, precision, scaling
Introduction
Bicoid (Bcd) is a well studied morphogen involved in the patterning of the anterior–posterior (AP) axis of the Drosophila embryo (Driever and Nüsslein-Volhard, 1988a, 1988b). Zygotic downstream genes read out this gradient and their expression domains determine the basic body plan of the embryo along this axis. The positions of these domains are remarkably insensitive to fluctuations in the external environment (Houchmandzadeh et al, 2002; Crauk and Dostatni, 2005; Lucchetta et al, 2005) and their relative proportions are maintained across embryos of different sizes. The latter feature is referred to as scaling and occurs within a single species (Houchmandzadeh et al, 2002; Lott et al, 2007) and also across different species (Gregor et al, 2005, 2008; Lott et al, 2007).
Recent experiments also show that the Bcd gradient itself is rather precise and that its length scale correlates to some extent to the embryo size (Gregor et al, 2007a; He et al, 2008). These new findings suggest that the precision and scaling of Bcd target genes may, at least in part, be attributed to that of the morphogen gradient itself. Therefore, we focus here on single morphogen models that aim at explaining the precision and scaling of Bcd target genes at the level of the morphogen gradient. However, it is likely that other mechanisms (e.g. Bcd interactions with the staufen gene (Aegerter-Wilmsen et al, 2005), gap genes interactions (Jaeger et al, 2004a, 2004b, 2007; Jaeger and Reinitz, 2006; Manu et al, 2009a, 2009b), Bcd interactions with maternal Hunchback and the terminal system (Ochoa-Espinosa et al, 2009) or bistability (Lopes et al, 2008)) also contribute to further increase robustness in AP patterning.
In this work, we assess the spatial precision of expression domains for the gap genes Krüppel (Kr), Giant (Gt) and Hunchback (Hb), as well as the pair-rule gene Even-skipped (Eve) in more than 150 staining images of embryos with a single, double and quadruple dose of bcd. Our data indicate that the precision is maximal at mid-embryo as well as position dependent rather than gene dependent. This provides independent support for the conclusions drawn from direct measurements of Bcd (Gregor et al, 2007a; He et al, 2008) that the morphogen gradient itself is the main contributor to the precision of the target genes. It motivates our subsequent analytical investigation of noise propagation during Bcd gradient formation within a single morphogen modeling framework. Our analysis shows that fluctuations in the parameters affecting morphogen production, degradation, diffusion as well as nuclear trapping can give rise naturally to highest precision at mid-embryo, provided the Bcd gradient is decoded at pre-steady-state. Finally, we investigate the scaling of the morphogen gradient in a variety of models that couple the embryo size to parameters affecting gradient formation. To single out the most likely scenario, we measure scaling at the level of the target genes. We find that expression domain boundaries posterior to ∼40% embryo length (L) exhibit almost perfect scaling, whereas in the more anterior region we observe over-compensation to fluctuations in embryo size. This effect appears to be position dependent rather than gene dependent. Interestingly, it is in good agreement with a model that explicitly includes the nuclear trapping of Bcd. We conclude that the formation of the Bcd gradient itself is likely to be a main contributor to robust patterning along the AP axis and that pre-steady-state decoding and nuclear trapping are efficient means to increase robustness.
Results
The gap and pair-rule gene expression domains are precise
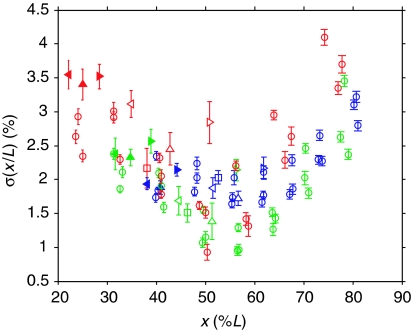
We studied staining images of 154 Drosophila melanogaster embryos at cleavage cycle 14 (see Methods in Bergmann et al, 2007). Using image processing tools described in ‘Materials and methods', we measured the relative positions x/L of the protein domains of Gt, Hb, Kr and Eve. We screened both wild-type embryos with two copies of bcd and mutant strains with one or four bcd copies resulting in shifted expression domains. We observed that in mid-embryo, the standard deviation of the relative domain localizations, σ(x/L), is between 1 and 2% and it tends to be higher (2–4%) toward the anterior and posterior poles (Figure 1).
Figure 1.
Measuring precision of Bcd target genes. Measure of imprecision σ(x/L) of the gap and pair-rule gene expression domains as a function of position x along the AP axis (see Supplementary Dataset S1). Errors (bars) were estimated by computing the standard deviation across 50 independent (semi-automated) markings of expression domain boundaries. For Gt (filled triangles) and Kr (empty triangles), we show results for their left (◃) and right (▹) boundaries, as well as for the center (▵) of their expression domain. The Hb (□) domain is characterized by the boundary where its concentration drops and the Eve stripes (○) by the position at which their intensity is maximal. Color code: 1xbcd (red), 2xbcd=wild type (green), 4xbcd (blue).
These results are in good agreement with previous results on the Hb domain (Houchmandzadeh et al, 2002). Importantly, they show that the precision of the target genes has the same magnitude and positional trend as that of Bcd according to Gregor et al (2007a). Moreover, our experimental analysis shows that precision is more position dependent than gene dependent. Indeed, when expression domains are shifted because of different bcd mRNA dosages, the precision of the domain seems to change according to the new domain position. Overall, all the investigated domains follow similar precision trends. This suggests that a major contribution to the precision of the Bcd target genes can be attributed to the morphogen gradient itself. We note that position dependent precision may also arise as an experimental artifact when analyzing embryos with different orientations and varying ages (times classes T5–T8 of nuclear cycle 14) or because of imperfect scaling (cf. Supplementary Text S3). However, estimating the maximal contributions of such effects we still find that higher precision at mid-embryo is statistically significant (cf. Supplementary Text S1; Supplementary Figure S4).
Modeling precision in a single morphogen framework
Given the aforementioned pieces of evidence for achieving precision at the level of the gradient formation, we consider a French-flag model (Wolpert, 1969) in which a single morphogen gradient induces expression domain boundaries in a concentration-dependent manner (see Materials and methods). During the first stages of embryonic development, 13 nuclear divisions occur about every 10 min. Gregor et al (2007b) have recently shown that nuclear concentrations stabilize at cycle 10. From a robustness perspective, this means that Bcd read-out is based on a similar number of molecules and so the associated stochastic noise does not change significantly from cycle 10 on. Therefore, we focus here on the noise propagated from the external Bcd gradient and investigate the fluctuations in domain localization when perturbing the various parameters characterizing the gradient formation (i.e. production, degradation, diffusion and nuclear trapping rates).
We want to assess the variation in domain position Δx, which is induced by embryo-to-embryo fluctuations of magnitude Δq in a parameter q affecting the morphogen gradient formation. Δx can be estimated analytically to first order in Δq
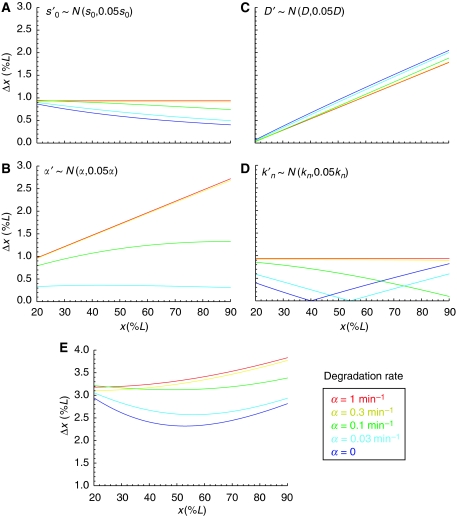
where we used that the threshold concentration is fixed, yielding dM=(∂M/∂x)dx+(∂M/∂q)dq=0. This estimate can be computed for any morphogen distribution M(x, t) with explicit dependence on q. Here, we consider the time-dependent solution of a model in which Bcd is produced at the anterior pole with production rate s0, diffuses according to a uniform diffusion constant D, is degraded at uniform rate α and is trapped and released by the nuclei in the embryo at rates kn, k−n, respectively (see Materials and methods). In Figure 2, we plotted the imprecision measure Δx for small fluctuations (5%) in each of these parameters (Figure 2A–D) as well as a combination of fluctuations in the production and diffusion rates (Figure 2E) (numerical simulations gave very similar results, data not shown). We choose ti=100 min as the time when patterning is initiated, but qualitatively our results only depend on the relation between ti and the Bcd decay time τ=1/α (for ti≪τ the gradient has not yet reached steady-state). We find that fluctuations in s0 (Figure 2A) give rise to decreasing Δx toward the posterior pole if the pre-steady-state gradient is read out (i.e. for small α). In contrast, for fluctuations in D or α, Δx increases toward the posterior pole (Figure 2B and C). Thus, in the case of Bcd pre-steady-state decoding (bluish curves) and fluctuations both in the production and degradation or diffusion rates (Figure 2E), maximal precision of the Bcd gradient around mid-embryo can arise naturally, as was indeed observed directly by Gregor et al (2007a) and indirectly by our analysis (cf. Figure 1). In contrast, for high decay rates (reddish curves) the gradient rapidly equilibrates such that fluctuations in the production rate give rise to uniform noise in Bcd concentrations, preventing the possibility of minimal noise in the central region. Interestingly, fluctuations in the nuclear trapping rate alone also yield higher precision around mid-embryo (Figure 2D) if the Bcd gradient is decoded early in pre-steady-state. However, the position with highest precision is sensitive to the decoding time, shifting more toward the posterior pole for later decoding.
Figure 2.
Modeling precision of nuclear Bcd. Predicted positional variability at t=100 min for various Bcd degradation rates α={1, 0.3, 0.1, 0.03, 0} min−1 and their corresponding diffusion constants D={167, 55, 17, 5, 3} μm2/s, ensuring λ=0.2L at t=150 min (numerical fit); 5% noise was added to the Bcd profile at the level of (A) the production rate, (B) the diffusion constant, (C) the degradation rate and (D) the nuclear trapping rate. In (E), a combination of noise was added to the Bcd profile (17% on the production- and 6% on the diffusion rates; all contributions are assumed to be independent). (L=1 for all embryos.)
Our modeling approach provides a proof of principle that maximal precision at mid-embryo can arise if the gradient is decoded before steady-state. However, we cannot rule out that other mechanisms may yield such a pattern of precision even for a steady-state Bcd gradient (e.g. in the wing disc, it was shown that cell-to-cell variability in the production, diffusion and degradation rates can yield higher precision around mid-field, Bollenbach et al, 2008).
In our analysis, we assumed the classical French-flag model (Wolpert, 1969) in which domain boundaries are determined from critical morphogen concentrations. This is a simplification and recent work by the Reinitz group (Jaeger et al, 2004a, 2004b, 2007; Jaeger and Reinitz, 2006; Surkova et al, 2008; Manu et al, 2009a, 2009b) showed that the gap gene expression domains are established by a highly dynamic process characterized by drifting domain boundaries. In our modeling, we neglected the gap gene dynamics and rather focused on how precision can be achieved already at the level of the Bcd gradient, as suggested by our data (as well as those reported in Gregor et al, 2007a; He et al, 2008). Thus, we assume that the positional information is transmitted relatively early to the gap genes, around cycle 10 (as suggested in Lucchetta et al, 2008). The precision and proper scaling of this read-out is then maintained (and possibly refined) by the gap genes independently of Bcd, because of their cross- and auto-regulation (Bergmann et al, 2007). Meanwhile, the Bcd gradient may continue to evolve and eventually decays at the onset of gastrulation (e.g. by activation of its PEST domain, Niessing et al, 1999).
Modeling scaling
We next sought to extend our modeling framework to accommodate also scaling. To quantify scaling analytically at the morphogen level, we define a scaling coefficient S(x, t) for a position x and time t
Perfect scaling corresponds to S=1. In this case, fluctuations in embryo length, dL/L, are exactly compensated by fluctuations in position, dx/x, implying perfectly conserved proportions. We use the terms hypo- and hyper-scaling to refer to S<1 and S>1, respectively. A position that hypo-scales does not compensate enough for a change in embryo size, meaning that in a bigger embryo the absolute position is not shifted enough posteriorward to keep the correct proportions. In contrast, hyper-scaling is the tendency to overcompensate for a change in embryo size.
Scaling is a property of the external gradient, which is then transmitted to the nuclear concentrations (cf. equation (7)). Assuming that the threshold concentration is fixed (implying dM=(∂M/∂x)dx+(∂M/∂L)dL=0) it follows from equation (2) that the scaling coefficient is
The above definition of scaling is generic and can be computed for any morphogen distribution M(x, t) with explicit dependence on L. Here, we consider a model in which the embryo length impacts the generation of the morphogen gradient through the nuclei density. Specifically, assuming that at decoding time all embryos have the same number of nuclei independent of their size (which agrees with the deterministic doubling of nuclei at each cycle), the nuclei density N depends on the embryo size like N∝L−n, where n∈[1;3] (n=3 corresponds to a uniform distribution of nuclei, whereas n=2 is true if nuclei are distributed on a shell with a fixed width, Gregor et al, 2007b). In this scenario, we find that scaling is time dependent and position dependent (Figure 3B; see Supplementary Figure S9 for the dependence of scaling on N/K and n). Specifically, anterior domain boundaries hyper-scale, in particular if decoding occurs relatively late, whereas posterior domains show very good scaling for a wide range of decoding times.
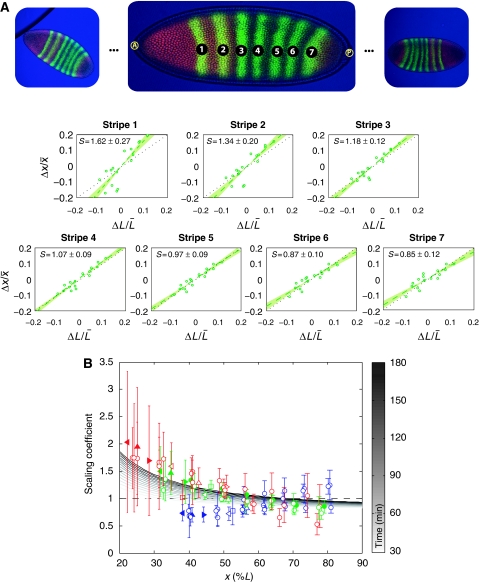
Figure 3.
Measuring scaling of Bcd target genes. (A) Scaling of the Eve stripes for wild-type bcd mRNA dosage, co-stained with Hb. For each stripe in each embryo (images on top), fluctuations in the domain position Δx/x̄ are plotted against fluctuations in embryo size ΔL/L̄. Scaling coefficients are then estimated by linear regression. Errors show 68% confidence intervals from the regression analysis. (B) Measured scaling coefficients of the gap and pair-rule gene expression domains as a function of position x along the AP axis (see Supplementary Dataset S2). Errors (bars) represent 68% confidence intervals from the linear regression. For Gt (filled triangles) and Kr (empty triangles), we show results for their left (◃) and right (▹) boundaries, as well as for the center (▵) of their expression domain. The Hb (□) domain is characterized by the boundary where its concentration drops and the Eve stripes (○) by the position at which their intensity is maximal (filled circles represent Eve co-stained with Hb, as in (A)). Color code: 1xbcd (red), 2xbcd=wild type (green), 4xbcd (blue). We also show scaling as predicted by our model (grayscale). Parameters are chosen such that the profile is closest to an exponential decay with length scale λ=0.2L at t=150 min (see Supplementary Figure S12 for temporal evolution).
We also investigated three other models that give rise to scaling by coupling the nuclear degradation, cytoplasmic degradation or the morphogen production rate to the embryo length (cf. Supplementary Text S2). We find that their scaling behavior is qualitatively similar (in the sense that S decreases toward the posterior pole) but more sensitive to the Bcd decoding time (Supplementary Figure S8).
The gap and pair-rule gene expression domains scale with embryo size
The model we investigated above predicts that scaling should be position dependent rather than gene dependent and close to perfect except for anterior most domains. To test these two predictions experimentally on our collection of staining images, we adapted the continuous definition of the scaling coefficient S(x, t) in equation (2) for discrete measurements as follows:
where 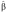 is the estimated slope from a linear regression x=α+βL of the domain positions xi (with mean value x̄) onto their respective embryo sizes Li (with mean value L̄) (see Figure 3A). In our data, the relative embryo size fluctuations were of the order of 10–15%. The results of our scaling analysis for Gt, Hb, Kr and Eve in embryos with single, double and quadruple bcd dosage are presented in Figure 3B. We observe that anterior domains indeed tend to hyper-scale, whereas mid-embryo and posterior domains show good scaling (P-value <0.00021, cf. Supplementary Text S1; Supplementary Figure S5). Moreover, the magnitude of the scaling coefficient depends mainly on the position of the respective domain (boundary) rather than the associated gene. This is in good agreement with the model discussed above, whereas in the other models we investigated (cf. Supplementary Text S2) it is difficult to obtain close to perfect scaling both at mid-embryo and toward the posterior pole.
is the estimated slope from a linear regression x=α+βL of the domain positions xi (with mean value x̄) onto their respective embryo sizes Li (with mean value L̄) (see Figure 3A). In our data, the relative embryo size fluctuations were of the order of 10–15%. The results of our scaling analysis for Gt, Hb, Kr and Eve in embryos with single, double and quadruple bcd dosage are presented in Figure 3B. We observe that anterior domains indeed tend to hyper-scale, whereas mid-embryo and posterior domains show good scaling (P-value <0.00021, cf. Supplementary Text S1; Supplementary Figure S5). Moreover, the magnitude of the scaling coefficient depends mainly on the position of the respective domain (boundary) rather than the associated gene. This is in good agreement with the model discussed above, whereas in the other models we investigated (cf. Supplementary Text S2) it is difficult to obtain close to perfect scaling both at mid-embryo and toward the posterior pole.
Perspectives
Precision and scaling are important to achieve robust pattern formation. Our new position-dependent measures provide a unified quantification of scaling for both functional descriptions of gradient profiles derived from models (equation (2)) and experimentally determined expression domains (equation (4)). These measures will be useful to address scaling also in different systems like the wing-disk. Importantly, our measures clearly identify perfect scaling (S=1) corresponding to the correct preservation of proportions, which is not necessarily equivalent to perfect correlation (see Supplementary Text S4 for a detailed discussion).
Interestingly, our experimental analysis using embryos with different bcd dosages showed that both precision and scaling are more position dependent than gene dependent, suggesting that the morphogen gradient itself has an important function to set robust domain boundaries. As our study showed, gradient formation of a single morphogen can naturally ensure maximal robustness only in a limited patterning domain. For Bcd, this appears to be mid-embryo, where most of its targets are expressed. Outside of this domain, other systems and mechanisms may have evolved to cooperate in maintaining and potentially increasing robustness. Considering precision and scaling as position- (and possibly time-) dependent features will allow to better characterize and develop models for these systems.
Materials and methods
Image analysis
We use an interface developed in MATLAB (Supplementary Figure S1) to extract and analyze expression profiles from the staining images of different Bcd target genes, yielding quantitative information on the positions of each protein domain (Bergmann et al, 2007). A total of 154 staining images were analyzed using a semi-automated analysis tool where the position of the anterior and posterior poles were marked 50 times for each embryo. On the basis of this input, the software extracted a rectangular region from the image from which it generated protein concentration profiles and automatically determined the positional information of the gap and pair-rule gene expression domains. Thus, for each embryo, its total length and the domain localizations were characterized by a mean value and a standard deviation computed from the 50 markings. The right boundary of the anterior Hb expression domain was defined as the position at which the decline in the Hb concentration is the steepest. Gt and Kr domains were described similarly by their left and right boundaries, whereas the position with maximal staining intensity characterized their center. As the Eve stripes are not so broad, we only extracted the position at which the Eve intensity was maximal. (All the staining images are available for download at http://www.unil.ch/cbg/morphogen/Images_paper.zip)
We also developed a second, almost fully automated analysis tool that only requires human input for selecting which of the two poles is the anterior one (while their position was detected automatically). This tool was used to analyze the expression domain positions of a set of 119 embryos, which included 78 (51%) from the first analysis. Precision and scaling results turned out to be very similar for the two analysis approaches, suggesting that measurement errors and inter-embryo variation (e.g. because of different embryo orientations) can be safely neglected (Supplementary Figure S6). We also note that part of the observed fluctuations in embryo size may be induced by the fixation procedure. However, computing precision and scaling by considering embryos on different slides separately yielded similar results (Supplementary Figure S7), suggesting that fixation or other batch effects have no positional bias toward either of the embryo poles.
Modeling
We consider a model in which Bcd is produced at the anterior pole with production rate s0, diffuses according to a uniform diffusion constant D, is degraded at uniform rate α and is trapped and released by the nuclei in the embryo at rates kn, k−n, respectively. The corresponding coupled partial differential equations for the free morphogen concentration M(x, t) and the nuclear concentration Mn(x, t) read (Bergmann et al, 2007; Coppey et al, 2007; Gregor et al, 2007b):
where N is the nuclei density and s0δ(x) the source term localized at x=0 (anterior pole). We consider zero-flux (Neumann) boundary conditions at the posterior pole and that there is no morphogen at the initial time (t=0). Assuming that nucleo-cytoplasmic exchanges occur rapidly (∂Mn/∂t∣M≈0), as demonstrated by the experiments reported in Gregor et al (2007b), the following effective diffusion equation holds for the free morphogen (Bergmann et al, 2007; Coppey et al, 2007; Gregor et al, 2007b):
where D̃=D/(1+(N/K)), 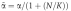 and
and 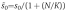 (with K=k−n/kn). At time t=150 min (corresponding to nuclear cycle 14), the morphogen profile is numerically fitted to an exponentially decaying gradient with length scale λ=0.2L. Thus, given a degradation rate, the diffusion constant is adjusted accordingly (see caption of Figure 2 for numerical values). We account for the presence of the nuclei by setting N/K=1 (i.e. to be specific, we assume that the probability that external Bcd is trapped equals the probability that nuclear Bcd is released; yet our qualitative results for precision are robust with respect to the exact choice of N/K).
(with K=k−n/kn). At time t=150 min (corresponding to nuclear cycle 14), the morphogen profile is numerically fitted to an exponentially decaying gradient with length scale λ=0.2L. Thus, given a degradation rate, the diffusion constant is adjusted accordingly (see caption of Figure 2 for numerical values). We account for the presence of the nuclei by setting N/K=1 (i.e. to be specific, we assume that the probability that external Bcd is trapped equals the probability that nuclear Bcd is released; yet our qualitative results for precision are robust with respect to the exact choice of N/K).
According to the nuclear trapping model (equation (5)), the nuclear concentration cn, which determines the precision and scaling of Bcd target genes, is given by (see also Coppey et al, 2007)
where νn is the nuclear volume. Importantly, according to equation (7) the fluctuations in the nuclear concentrations cn are proportional to those in the external gradient, provided we can neglect variability in νn and K.
Supplementary Material
* Image analysis and control experiments:
We discuss the image analysis tools and limitations as well as the validity of our results.
* Scaling models
We discuss other models that could yield scaling of Bicoid as well as the advantages of our measure of scaling compared to correlations.
Dataset S1. Numerical values for the quantitative analysis of precision (relative position x/L, imprecision s(x/L), error, gene name, co-staining (for Eve domains), bcd dosage).
Dataset S2. Numerical values for the quantitative analysis of scaling (relative position x/L, scaling coefficient, 68% CI, gene name, co-staining (for Eve domains), bcd dosage).
Acknowledgments
We thank Naama Barkai, Ben-Zion Shilo and Zvi Tamari for careful reading of the manuscript, as well as all the members of the Bergmann Computational Biology Group and David Morton de Lachapelle for their support and comments during the whole project. In particular, we thank Toby Johnson, Karen Kapur and Sascha Dalessi for their help with the theoretical analyses and are very grateful to Zoltán Kutalik who implemented the fully automated version of the profile extraction tool. We are also grateful for financial support from the Giorgi-Cavaglieri Foundation, the Swiss National Science Foundation (Grant #3100AO-116323/1), SystemsX.ch (through the WingX project) and the European Framework Project 6 (through the AnEuploidy project).
Footnotes
The authors declare that they have no conflict of interest.
References
- Aegerter-Wilmsen T, Aegerter CM, Bisseling T (2005) Model for the robust establishment of precise proportions in the early Drosophila embryo. J Theor Biol 234: 13–19 [DOI] [PubMed] [Google Scholar]
- Bergmann S, Sandler O, Sberro H, Shnider S, Schejter E, Shilo B-Z, Barkai N (2007) Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol 5: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach T, Pantazis P, Kicheva A, Bökel C, González-Gaitán M, Jülicher F (2008) Precision of the Dpp gradient. Development 135: 1137–1146 [DOI] [PubMed] [Google Scholar]
- Coppey M, Berezhkovskii AM, Kim Y, Boettiger AN, Shvartsman SY (2007) Modeling the bicoid gradient: diffusion and reversible nuclear trapping of a stable protein. Dev Biol 312: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crauk O, Dostatni N (2005) Bicoid determines sharp and precise target gene expression in the Drosophila embryo. Curr Biol 15: 1888–1898 [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C (1988a) The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54: 95–104 [DOI] [PubMed] [Google Scholar]
- Driever W, Nüsslein-Volhard C (1988b) A gradient of bicoid protein in Drosophila embryos. Cell 54: 83–93 [DOI] [PubMed] [Google Scholar]
- Gregor T, Bialek W, de Ruyter van Steveninck RR, Tank DW, Wieschaus EF (2005) Diffusion and scaling during early embryonic pattern formation. Proc Natl Acad Sci USA 102: 18403–18407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, McGregor AP, Wieschaus EF (2008) Shape and function of the Bicoid morphogen gradient in dipteran species with different sized embryos. Dev Biol 316: 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Tank DW, Wieschaus EF, Bialek W (2007a) Probing the limits to positional information. Cell 130: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor T, Wieschaus EF, McGregor AP, Bialek W, Tank DW (2007b) Stability and nuclear dynamics of the bicoid morphogen gradient. Cell 130: 141–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Wen Y, Deng J, Lin X, Lu LJ, Jiao R, Ma J (2008) Probing intrinsic properties of a robust morphogen gradient in Drosophila. Dev Cell 15: 558–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh B, Wieschaus E, Leibler S (2002) Establishment of developmental precision and proportions in the early Drosophila embryo. Nature 415: 798–802 [DOI] [PubMed] [Google Scholar]
- Jaeger J, Blagov M, Kosman D, Kozlov KN, Manu, Myasnikova E, Surkova S, Vanario-Alonso CE, Samsonova M, Sharp DH, Reinitz J (2004a) Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster. Genetics 167: 1721–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J, Reinitz J (2006) On the dynamic nature of positional information. Bioessays 28: 1102–1111 [DOI] [PubMed] [Google Scholar]
- Jaeger J, Sharp DH, Reinitz J (2007) Known maternal gradients are not sufficient for the establishment of gap domains in Drosophila melanogaster. Mech Dev 124: 108–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger J, Surkova S, Blagov M, Janssens H, Kosman D, Kozlov KN, Manu, Myasnikova E, Vanario-Alonso CE, Samsonova M, Sharp DH, Reinitz J (2004b) Dynamic control of positional information in the early Drosophila embryo. Nature 430: 368–371 [DOI] [PubMed] [Google Scholar]
- Lopes FJP, Vieira FMC, Holloway DM, Bisch PM, Spirov AV (2008) Spatial bistability generates hunchback expression sharpness in the Drosophila embryo. PLoS Comput Biol 4: e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott SE, Kreitman M, Palsson A, Alekseeva E, Ludwig MZ (2007) Canalization of segmentation and its evolution in Drosophila. Proc Natl Acad Sci USA 104: 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF (2005) Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature 434: 1134–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Vincent ME, Ismagilov RF (2008) A precise Bicoid gradient is nonessential during cycles 11–13 for precise patterning in the Drosophila blastoderm. PLoS ONE 3: e3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim A-R, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J (2009a) Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors. PLoS Comput Biol 5: e1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim A-R, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J (2009b) Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors. PLoS Biol 7: e1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Dostatni N, Jäckle H, Rivera-Pomar R (1999) Sequence interval within the PEST motif of Bicoid is important for translational repression of caudal mRNA in the anterior region of the Drosophila embryo. EMBO J 18: 1966–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochoa-Espinosa A, Yu D, Tsirigos A, Struffi P, Small S (2009) Anterior-posterior positional information in the absence of a strong Bicoid gradient. Proc Natl Acad Sci USA 106: 3823–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkova S, Kosman D, Kozlov K, Manu, Myasnikova E, Samsonova AA, Spirov A, Vanario-Alonso CE, Samsonova M, Reinitz J (2008) Characterization of the Drosophila segment determination morphome. Dev Biol 313: 844–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L (1969) Positional information and the spatial pattern of cellular differentiation. J Theor Biol 25: 1–47 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
* Image analysis and control experiments:
We discuss the image analysis tools and limitations as well as the validity of our results.
* Scaling models
We discuss other models that could yield scaling of Bicoid as well as the advantages of our measure of scaling compared to correlations.
Dataset S1. Numerical values for the quantitative analysis of precision (relative position x/L, imprecision s(x/L), error, gene name, co-staining (for Eve domains), bcd dosage).
Dataset S2. Numerical values for the quantitative analysis of scaling (relative position x/L, scaling coefficient, 68% CI, gene name, co-staining (for Eve domains), bcd dosage).