Abstract
Previous studies have led to a picture wherein the replication of DNA progresses at variable rates over different parts of the budding yeast genome. These prior experiments, focused on production of nascent DNA, have been interpreted to imply that the dynamics of replication fork progression are strongly affected by local chromatin structure/architecture, and by interaction with machineries controlling transcription, repair and epigenetic maintenance. Here, we adopted a complementary approach for assaying replication dynamics using whole genome time-resolved chromatin immunoprecipitation combined with microarray analysis of the GINS complex, an integral member of the replication fork. Surprisingly, our data show that this complex progresses at highly uniform rates regardless of genomic location, revealing that replication fork dynamics in yeast is simpler and more uniform than previously envisaged. In addition, we show how the synergistic use of experiment and modeling leads to novel biological insights. In particular, a parsimonious model allowed us to accurately simulate fork movement throughout the genome and also revealed a subtle phenomenon, which we interpret as arising from low-frequency fork arrest.
Keywords: cell cycle, ChIP-chip, DNA replication, replication fork, simulation
Introduction
In mitotic division, cells duplicate their DNA in S phase to ensure that the proper genetic material is passed on to their progeny. This process of DNA replication is initiated from several hundred specific sites, termed origins of replication, spaced across the genome. It is essential for replication to begin only after G1 and finish before the initiation of anaphase (Blow and Dutta, 2005; Machida et al, 2005). To ensure proper timing, the beginning stages of DNA replication are tightly coupled to the cell cycle through the activity of cyclin-dependent kinases (Nguyen et al, 2001; Masumoto et al, 2002; Sclafani and Holzen, 2007), which promote the accumulation of the pre-replication complex (pre-RC) at the origins and initiate replication. Replication fork movement occurs subsequent to the firing of origins on recruitment of the replicative helicase and the other fork-associated proteins as the cell enters S phase (Diffley, 2004). The replication machinery itself (polymerases, PCNA, etc.) trails behind the helicase, copying the newly unwound DNA in the wake of the replication fork.
One component of the pre-RC, the GINS complex, consists of a highly conserved set of paralogous proteins (Psf1, Psf2, Psf3 and Sld5; Kanemaki et al, 2003; Kubota et al, 2003; Takayama et al, 2003). In Xenopus egg extracts, GINS has been shown to localize at sites of unwound DNA (Pacek et al, 2006). In yeast, the complex associates with paused replication forks (Calzada et al, 2005), and directly interacts with several fork proteins (Gambus et al, 2006). Chromatin immunoprecipitation (ChIP) experiments show that the GINS complex moves away from specific autonomous replication sequences (ARSs) at the time of initiation (Takayama et al, 2003; Kanemaki and Labib, 2006). The GINS complex has been biochemically isolated with Cdc45 and Mcm2-7, which together are referred to as the replisome progression complex (RPC) or the CMG (Cdc45-MCM-GINS) (Gambus et al, 2006; Moyer et al, 2006); the CMG has been shown to have helicase activity in vitro (Moyer et al, 2006). Taken together, these data suggest that the GINS complex is an integral component of the replication fork and that its interaction with the genome correlates directly to the movement of the fork (reviewed in Labib and Gambus, 2007). Here, we used the GINS complex as a surrogate to measure features of the dynamics of replication—that is, to determine which origins in the genome are active, the timing of their firing and the rates of replication fork progression.
Previously, as many as 732 sites within the 16 chromosomes of the Saccharomyces cerevisiae genome have been reported as potential origins of replication (Nieduszynski et al, 2007) (www.oridb.org). Three separate studies monitoring the production of newly replicated DNA identified, respectively, 332, 260 and 444 origins in the yeast genome (Raghuraman et al, 2001; Yabuki et al, 2002; Feng et al, 2006). In addition, experiments using ChIP combined with microarray analysis (ChIP-chip) to search for the genomic localization of MCM and ORC proteins identified, respectively, 422 (Wyrick et al, 2001) and 529 sites (Xu et al, 2006). Thus, although it is likely that virtually all origins have been identified, exactly which are active during S phase remains ambiguous.
The timing of origin firing and the rates of fork progression have also been investigated by monitoring nascent DNA synthesis (Raghuraman et al, 2001; Yabuki et al, 2002). Origin firing was observed to occur as early as 14 min into the cell cycle and as late as 44 min (Raghuraman et al, 2001). A wide range of nucleotide incorporation rates (0.5–11 kb/min) were observed, with a mean of 2.9 kb/min (Raghuraman et al, 2001), whereas a second study reported a comparable mean rate of DNA duplication of 2.8±1.0 kb/min (Yabuki et al, 2002). In addition to these observations, replication has been inferred to progress asymmetrically from certain origins (Raghuraman et al, 2001). These data have been interpreted to mean that the dynamics of replication fork progression are strongly affected by local chromatin structure or architecture, and perhaps by interaction with the machineries controlling transcription, repair and epigenetic maintenance (Deshpande and Newlon, 1996; Rothstein et al, 2000; Raghuraman et al, 2001; Ivessa et al, 2003). In this study, we adopted a complementary approach for assaying replication dynamics, in which we followed GINS complexes as they traverse the genome during the cell cycle. These studies led us to a different view of replication fork dynamics wherein fork progression throughout the genome is symmetrical around origins, highly uniform in rate and little affected by genomic location.
Results
In this study, we are particularly interested in the effect of chromosomal location on the dynamics of replication fork progression. To address this question on a genome-wide scale, we followed the spatial and temporal association of a protein that assembles at origins with the pre-RC and has been inferred to travel with the advancing replication forks. For this purpose, we chose Psf2, a component of the GINS complex (Takayama et al, 2003), and collected ChIP-chip (Ren et al, 2000; Tackett et al, 2005) data as a function of time throughout the cell cycle (shown for chromosome XVI and for the whole genome in Figure 1; Supplementary Figure S4).
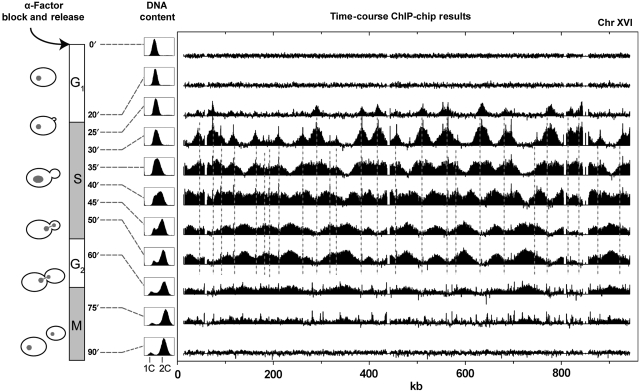
Figure 1.
Time-course ChIP-chip analysis of GINS complex binding to chromosome XVI through the cell cycle. S. cerevisiae cells (W303 strain, genomically tagged on Psf2—a component of the GINS complex) were synchronized in YPD media with α-factor. Starting from G1, ChIP-chip was performed at 11 time points through the cell cycle (see Materials and methods). The GINS complex was immunoisolated at each time point. Associated DNA was amplified and hybridized to yeast whole genome microarrays. Relative occupancies of the GINS complex for sites across chromosome XVI are shown for each time point. Locations of the initial binding sites are noted with dashes. Cell cycle state was determined by monitoring budding indices as well as DNA content by flow cytometry.
At early times (⩽20 min), the GINS complex does not exhibit any observable interaction with the genome as evidenced by the lack of peaks in the ChIP-chip signal (Figure 1) and lack of binding to other protein members of the RPC (data not shown). As the cell cycle progresses, the GINS complex begins to interact with specific chromosomal sites (indicated by the peaks and the dashed lines in Figure 1)—nearly all of which correspond to previously posited origins (Supplementary Table S1; Nieduszynski et al, 2007). The height of the peaks provides a measure of average occupancy, and their spreading indicates that some fraction of the origins within the cell population have fired. The data for chromosome XVI (Figure 1) indicate that 12 origins are occupied to some extent in the population, almost all of which have fired by 25 min. By 30 min the number of occupied origins increases to ∼28, after which time no additional origins appear to be populated. Bi-directional GINS progression from each of these origins (averaged over the population of cells) can be inferred from the spreading of the edges of the corresponding peaks. By 35 min, many of these edges have merged with those from adjacently spreading GINS and fewer locations on the chromosome remain wholly unoccupied. Spreading continues with time such that by 50 min GINS has progressed to most regions of the chromosome, and reduced occupancy is observed in the regions surrounding the origins. By 60 min the overall occupancy has fallen sharply across the chromosome indicating that the replication process for most of the population is nearing completion and the GINS complex has been released. The GINS re-associates with the origins at the beginning of the next cell cycle (data not shown). Thus, the Psf2 reporter provides an animated view of the GINS progression across an entire chromosome.
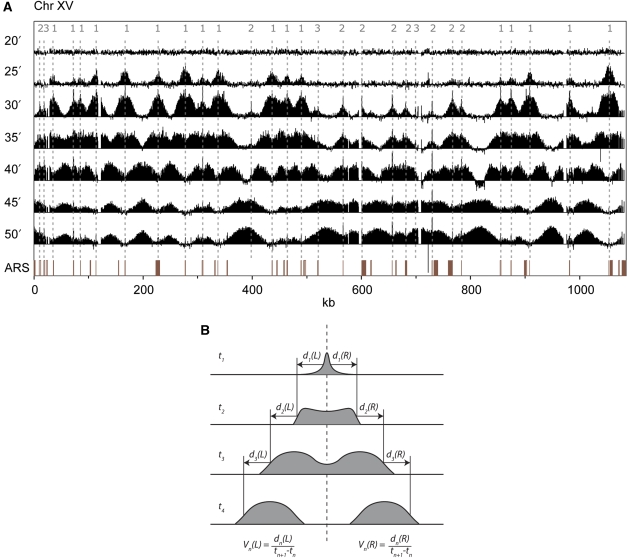
Similar data to that shown in Figure 1 were obtained for all 16 yeast chromosomes (Supplementary Figure S4). At the time resolution of the experiment (5 min), we discern three broad categories that describe the association of GINS with origins. The first includes those origins that fire in the interval 20–25 min. The second includes those that fire in the interval 25–30 min. The final category includes origins to which the GINS complex binds, but from which bidirectional spreading is not observed. Although others have reported similar early and late firing origins, our data reveal the majority of origins fire in a much narrower time window—that is, 15 versus 30 min (Yamashita et al, 1997; Poloumienko et al, 2001; Raghuraman et al, 2001; Yabuki et al, 2002). Applying this classification system to chromosome XV (Figure 2A), we identified a total of 29 origins, of which 17 are category 1, 9 are category 2 and 3 are category 3. In the whole genome, we identified 168, 135 and 24 origins, respectively, in categories 1, 2 and 3 (Supplementary Table S1, which also provides a detailed comparison with previous studies). Overall, we observe 303 origins that give rise to active GINS progression (i.e. categories 1 and 2). Although a small number of late firing origins may be obscured by proximal early firing origins, our data were determined with sufficiently high resolution and signal-to-noise to provide an accurate map of the majority of origins that are active in the cell cycle.
Figure 2.
Time-course ChIP-chip analysis of GINS complex binding to chromosome XV through S phase. (A) At the beginning of S phase (20 min time points) the GINS complex has not yet associated with any part of the chromosome. As S phase progresses, replication initiates at the indicated origins (25–30 min time points) whereupon the GINS complex moves away bi-directionally with the replication forks. Replication of a particular chromosomal region is finished when adjacent forks collide, giving rise to broad peaks in the inter-origin spaces. The locations of previously defined ARSs (excluding dubious origins as defined by Nieduszynski et al, 2007) are shown as red columns in the bottom panel. (B) Schematic of fork progression rate calculations.
Most of the 303 active origins fire within a short time window (∼10 min) (Figures 1, 2A; Supplementary Figure S4). We determined the progression rates for 278 spreading peaks emanating from 199 of these 303 origins by direct measurement of the positions of the peak edges as a function of time (Figure 2B). We obtain an average progression rate of 1.6±0.3 kb/min—a value that is considerably lower and more narrowly distributed than previous replication rate estimates (2.9 kb/min with a range between 0.5–11 kb/min (Raghuraman et al, 2001) or 2.8±1.0 kb/min (Yabuki et al, 2002). The bidirectional spreading that emanates from the origins progressed to the left and right at closely comparable rates.
From these data, we infer that this movement is largely symmetric and occurs at a highly characteristic rate, implying that, in contrast to previous inferences relating chromatin position to replication rate, there are few obvious chromosomal features that locally alter uniform GINS progression. To test the generality of this rather simple view of GINS progression throughout the genome, we used our measurements to generate an iterative model of this process. Our model (Materials and methods) uses a parsimonious set of assumptions: (1) the firing time of a given origin in a population of cells is normally distributed with a mean firing time specific to the origin; (2) the standard deviation of the firing times is constant for all origins in each simulated region; (3) the velocity of progression is the same for all forks (v=1.6 kb/min); and (4) GINS fall off the chromosomes when adjacent forks collide.
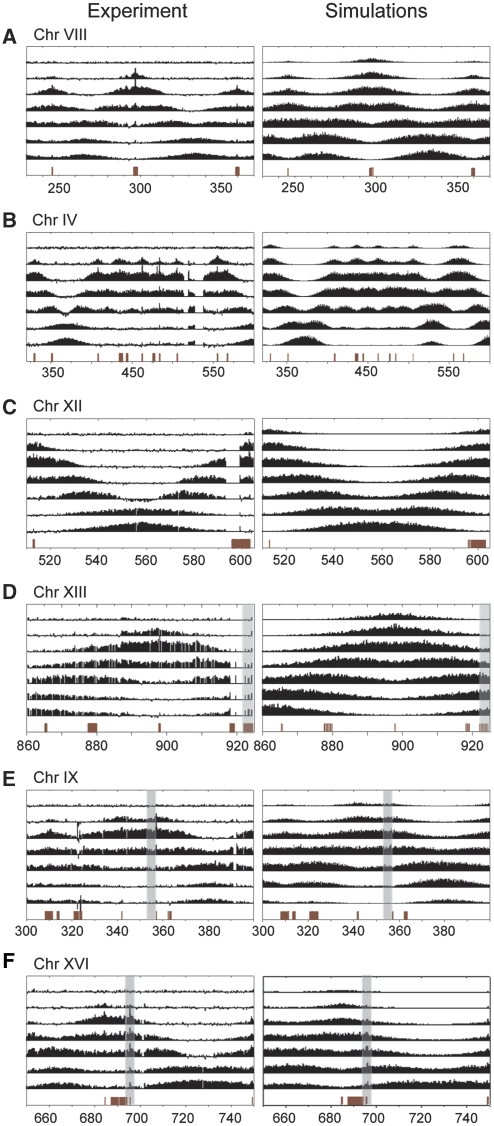
Examples comparing simulations based on our model with experimental data are provided in Figure 3. The top panel (Figure 3A) compares a region containing a single category 1 origin and two adjacent category 2 origins. The model accurately recapitulates the features exhibited in the experimental data. The same is true of the more complex region containing seven category 1 origins shown in Figure 3B. It also accounts for fork movement between the most widely spaced origins (∼100 kb apart) (Figure 3C). Such regions are completely filled in at our experimentally determined average rate of 1.6 kb/min, within the duration of S phase (∼30–35 min at room temperature). Figure 3D and E show that regions proximal to the telomeres and centromeres are as easily modeled as any other region of the genome. Category 3 origins were also readily modeled by allowing a small amount of binding without bidirectional spreading (Figure 3F). We conclude that our simple model successfully recapitulates GINS movement throughout the genome, indicating that this movement is largely uniform irrespective of location. By varying the assumed fork velocity, our model also allows us to estimate the accuracy of velocities inferred from the time-resolved ChIP-chip analysis. Velocity changes as small as 0.4 kb/min can readily be discerned (Supplementary Figure S8).
Figure 3.
S-phase time-course data (experiment and simulations) for several genomic regions of interest. (A) ChrVIII coordinates 240–370 kb. A category 1 origin flanked by two category 2 origins. (B) Seven category 1 origins spaced unevenly between 320 and 600 kb of ChrIV. (C) ChrXII coordinates 510–600 kb. The gap in between the two noted origins is 97 kb, among the longest inter-origin distances observed in the yeast genome. (D) The right-hand telomeric region of ChrXIII. The telomeric region is shaded. (E) The centromeric region of ChrIX (shaded gray). (F) A category 3 origin (shaded gray) located at ∼695 kb (ARS1625) in ChrXVI.
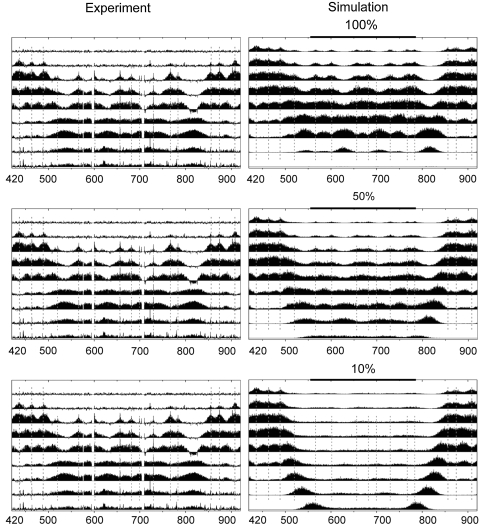
For simplicity, all the origins illustrated in Figure 3A–E are assumed to fire with unit efficiency. A refined model should include the possibility that different origins fire with different efficiencies. For example, replication is known to initiate at a reduced efficiency in many late firing origins (Yamashita et al, 1997; Poloumienko et al, 2001; Yabuki et al, 2002). In addition, there is a pattern of alternating groups of early and late firing origins across the genome (McCarroll and Fangman, 1988; Yamashita et al, 1997; Poloumienko et al, 2001; Raghuraman et al, 2001; Yabuki et al, 2002). We observe similar trends in this work (see Supplementary Table S1). As an example, we consider the cluster of late firing origins located on the right arm of chromosome XV between 500 and 800 kb (Figure 2A). We find that our model fails to accurately simulate this region if the firing efficiency for these late firing origins is too high (100%) or too low (10%). However, we can accurately simulate fork dynamics in this region if we assume that the efficiencies for these sites are decreased to 50% relative to the surrounding early firing origins (Figure 4). Thus, not only does the model provides an overall picture of the dynamic nature of GINS, but also a means to extract quantitative details concerning factors such as firing efficiency.
Figure 4.
Varying efficiencies applied a group of origins located in the right arm of ChrXV. The six category 1 origins on either end are given 100% efficiencies, whereas the efficiency applied to the cluster of seven category 2 origins in the middle of this region is indicated above each panel.
Indeed, the method is sufficiently sensitive to detect small heterogeneities in replication dynamics and in the process sheds new light on the phenomenon described as ‘replication pausing’—short duration stalling of forks at numerous specific sites in the genome (Deshpande and Newlon, 1996; Ivessa et al, 2003; Azvolinsky et al, 2006; Azvolinsky et al, 2009). For example, pausing intervals at tRNA genes have previously been estimated at ∼10 s (Deshpande and Newlon, 1996; Ivessa et al, 2003)—that is, ∼4 times longer than the interval required for unimpeded replication fork transit of a tRNA gene. Although such events would seem too subtle to detect with 5 min resolution (Azvolinsky et al, 2009), we observed sharp features at 267 out of 275 these tRNA genes (Supplementary Figures S11 and S12), indicating that our methodology is sensitive enough to detect small perturbations, and that GINS movement can indeed be hindered to some degree (Figure 5). However, rather than a brief pause of each fork as it passes through these sites, these features persist late into the cell cycle. Simulation of several different scenarios (Figure 6; Supplementary Figure S2) suggests that these features may represent infrequent long-term arrest events, occurring with a probability of <0.5% for any given fork passing through a tRNA gene (Supplementary Figure S3). This phenomenon was also observed at 81 of the 83 snoRNA and snRNA genes, and 95 of the 100 other most highly transcribed genes in the genome (Figure 5; Supplementary Figures S2 and S11), and appears to be independent from the direction of transcription.
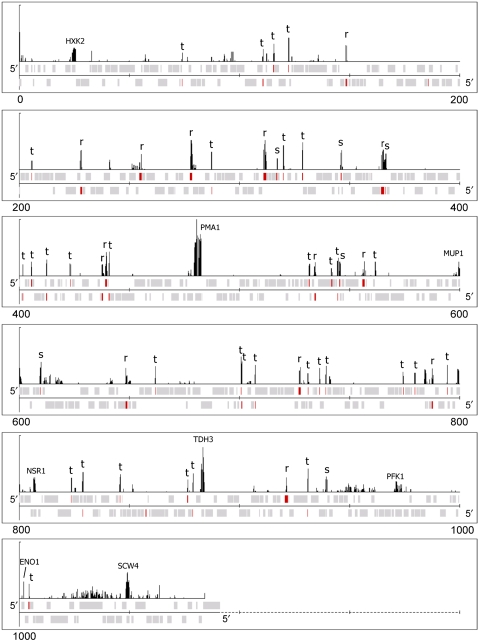
Figure 5.
Persistent features associated with GINS progression through highly transcribed genes. ChIP-chip data for the GINS with the entire ChrVII are shown for the 75 min time point. The baseline has been raised to emphasize peaks. Persistent features can be observed at all of the tRNA genes (t), snoRNA genes and snRNA genes (s), rRNA genes (r), as well as six out of the other nine most highly expressed genes on this chromosome (indicated by the gene name).
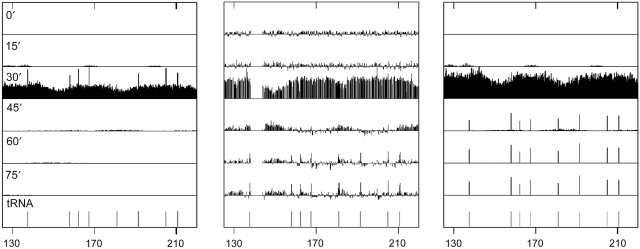
Figure 6.
The GINS complex undergoes long-term arrest at tRNA genes. (Center) ChIP-chip data for the association of the GINS complex with a region of chromosome VI (125–220 kb) containing eight tRNA genes. Data are shown in 15 min increments through the cell cycle. During late S phase sharp spikes are observed, which coincide with the position of the tRNA genes. We observe similar long-lived spikes that coincide with 267 of the 275 tRNA genes in the yeast genome. Two alternative models for explaining these spikes are presented: (left) in each cell, the replication forks are assumed to pause for 6 s at each tRNA gene; (right) the replication forks are assumed to have a 0.2% chance of long-term (t=∞) arrest at each tRNA gene. Additional model parameter conditions are shown in the Supplementary information.
Discussion
This study monitors the dynamic progression of the GINS complex along the genome during the cell cycle. Our data, along with previously determined interactions of the GINS complex, strongly suggest that GINS moves with the helicase at the replication fork. Our time-resolved ChIP-chip data reveal that GINS binds to active replication origins and spreads bi-directionally and symmetrically as S phase progresses. A similar approach has been used to monitor Polε localization on yeast chromosome III, albeit at lower time resolution (Hiraga et al, 2005). As monitored by GINS movement, the majority of origins appear to fire in the first ∼15 min of S phase. A small fraction (∼10%) of the origins to which GINS binds show no evidence of spreading (category 3 origins), although it remains possible that these peaks represent passively fired origins (Shirahige et al, 1998). Once an active origin fires, the GINS complex moves at an almost constant rate of 1.6±0.3 kb/min. Its movement through the inter-origin regions is consistent with that of a protein complex associated with a smoothly moving replication fork. This progression rate is considerably lower and more tightly distributed than those inferred from previous genome-wide measurements assayed through nascent DNA production (Raghuraman et al, 2001; Yabuki et al, 2002). It is of interest to consider potential sources of these discrepancies, such as those arising from the integration inherent to monitoring nascent DNA accumulation. Thus, for example, integration in regions flanking inefficient origins produces aberrant skewing of the measured times of replication. Such problems are avoided by our direct measurement of a specific fork-associated protein Psf2 (discussed in Supplementary Figure S1). However, it may be conjectured that the discrepancies discussed above can be explained by the possibility that the replication forks are not tightly coupled to the replicative polymerases with respect to their dynamics (Walter and Newport, 2000; Pacek et al, 2006). We do not consider this likely because it would require that the polymerases leave the origin considerably later than the fork, exposing large stretches of unpaired DNA.
In this work, we also observe a large number of low-intensity persistent features at sites of high transcriptional activity (e.g. tRNA genes). We were able to accurately simulate these features by assuming they are the result of low probability arrest of replication forks at these sites. Previously, pausing of forks had been observed at such sights, where in certain cases the pause appeared to be coordinated with head-on collisions between the replication and transcriptional machineries (Deshpande and Newlon, 1996). Here, we do not observe any significant directional dependence of fork arrest at the tRNA genes, 40% of which transcribe in the same direction as the movement of the replication fork. Thus, the presently described features at highly transcribed genes do not appear to exclusively correlate with head-on collisions. The extremely low frequency of these events in wild-type cells suggests they are due to low probability stochastic occurrences during the replication process. It is hoped that future studies will resolve whether these persistent features indeed represent rare instances of fork arrest, or are the result of some alternative process. These may include, for example, the deposition of GINS complexes (or perhaps more specifically Psf2) once a pause has been resolved.
In this work, we have made extensive use of modeling to test a number of different hypotheses and assumptions. In particular, iterative modeling allowed us to infer that GINS progression is uniform and smooth throughout the genome. We have also shown the potential of simulations for estimating firing efficiencies. In the future, extending such firing efficiency simulations to the whole genome should allow us to make correlations with chromosomal features such as nucleosome occupancy. Such correlations may help in determining factors that govern the probability of replication initiation throughout the genome.
Materials and methods
Yeast strain construction and growth conditions
S. cerevisiae strains are from the W303 background. Strain MSY1 was made by tagging the PSF2 gene with a protein A (PrA) affinity tag at the C-terminal coding sequence through homologous recombination (Tackett et al, 2005). The BAR1 gene, which encodes for the α-factor protease, was replaced with the KANMX4 selection cassette, making the strain hypersensitive to α-factor block. Cells were grown in rich YEPD medium for synchronization.
Cell synchronization
MSY1 was grown in a 10 l Bioflo 410 fermenter (New Brunswick Scientific) to a density of ∼7 × 106/ml at 30°C. The mating pheromone α-factor was added to a final concentration of 50 nM, and the cells were incubated for another 3 h. Cell arrest in G1 was verified by the complete presence of the shmoo morphology. The cells were harvested and centrifuged at 5000 r.p.m. at 4°C and washed twice with ice-cold YPD. The culture was re-suspended as quickly as possible in fresh media at 25°C. Initial cell density for all time-course experiments was ∼1 × 107/ml. Budding indices for both types of time-course experiments (before and after formaldehyde incubation) are shown in the Supplementary Figure S9.
Time-course sampling
Time points were collected in two separate experiments—(a) every 15 min after release from the G1 block from 0 to 105 min and (b) every 5 min beginning 20 min after block (through S phase) from 20 to 50 min. For each time point to be used for ChIP, a 750 ml sample was cross-linked by incubation in 1% formaldehyde at room temperature for 20 min. The ChIP sample was centrifuged at 4500 r.p.m. for 4 min at 4°C, washed with ice-cold Tris pH 7.8 and centrifuged again at 3000 r.p.m. for 3 min at 4°C. The pellet was re-suspended in 500 μl of cold Lysis buffer (50 mM Hepes pH 7.5, 1.2% polyvinylpyrrolidine) and the cells were dripped into liquid nitrogen forming small pellets. The pellets were stored at −75°C until use, and then cryogenically ground three times at 30 Hz with a mixer mill (Retsch MM301). Ground samples were kept frozen until resuspended for IPs.
Flow cytometry
In all, 10 ml of cells from each time point were fixed in 70% ethanol for 1 h at room temperature, and prepared for FACS analysis using previously described methods (Haase and Lew, 1997). Cells were stained with 50 μg/ml propidium iodide, and DNA content was analyzed using a Facscalibur I (BD Biosciences). Data analysis was performed with FloJo 8.3.3 (http://www.flowjo.com/).
Chromatin immuno-precipitation
ChIP was performed as reported earlier (Tackett et al, 2005); 0.5 g of each frozen, ground time point was suspended in 1 ml of ChIP lysis buffer (20 mM Hepes pH 7.5, 150 mM NaCl, protease inhibitor cocktail (Roche)), and sonicated at 4°C, to an average size of ∼400 bp (Supplementary Figure S10). The IP sample was incubated for 1 h at 4°C with IgG-conjugated 3 μm dynabeads (Invitrogen 143-01), and eluted as reported earlier (Tackett et al, 2005). A PCR cleanup kit was used to purify the DNA samples (Qiagen 28104).
Real-time PCR
Real-time PCR was performed as sample validation before array hybridization (Supplementary Figure S13); 1 μl of undiluted sample was analyzed in 20 μl reaction volume with 900 nM forward (ARS306taqfor—5′-TCGTCTAAGTCCTTGTAATGTAAGGTAAGA-3′) and reverse primers (ARS306taqrev—5′-GCTTGGGTTTGTGACTTACTAACG-3′), and 250 nM probe (ARS306taqprobe—5′-FAM-TGCAAGCATCTTGTTTGTAACGCGATTG-TAMRA-3′). Samples were analyzed with a 7900HT Sequence Detection System (Applied Biosystems), for 45 cycles of denaturation at 94°C for 15 s, annealing at 43°C for 30 s and extension at 72°C for 30 s. Results are normalized to ACT1 levels (primer sequences are ACT1taqfor—5′-CTCCGTCTGGATTGGTGGTT-3′, ACTtaq1rev—5′-TGGACCACTTTCGTCGTATTCTT-3′, ACT1taqprobe—5′-FAM-TTGACTACCTTCCAACAA-TAMRA-3′).
Microarray experiments
The samples were prepared for hybridization to yeast 4x44k whole genome microarrays (Agilent) with average spatial resolution of ∼290 nt, and analyzed as described earlier (Ren et al, 2000; Tackett et al, 2005). The program SignalMap (Nimblegen) was used to visualize the data. The list of previously known origins was obtained from the OriDB website (Nieduszynski et al, 2007).
Calculating fork velocities
Fork velocities were estimated by measuring the distance between the peak edges in successive time points (Figure 2B), and dividing by 5 min. When edges began to merge with each other we ignored these data points. The rates are reported in kilobases per minute (kb/min).
Data normalization
The microarray results were returned as log2 ratios of IP fluorescence versus WCE fluorescence. For the purpose of normalization, the data were binned using a 5000 bp bin size. The distributions for the average intensities and the RMSD for these bins can be seen in Supplementary Figure S5A. The first normalization of the data was done by finding the intensity where 3% of the bins had a negative average intensity. This gives a robust zero level that is independent of occasional large negative outliers in the data. The centroid of the RMSD distribution was then found for each time point and the intensities were scaled in such a way that these centroids were set to 1 (Supplementary Figure S5B). The second normalization was performed by finding an intensity threshold that separated the signal from the noise, and by normalizing the centroid of the RMSD distribution of the noise (Supplementary Figure S5C). An additional correction was used for time points where the data were dominated by signal. This correction (1.5-fold for the 40 min time point) was based on the assumption that the number of forks in a wave is constant after firing of that origin has ceased and before the front of the wave has reached an adjacent wave of forks traveling in the opposite direction (see, for example the 35 and 40 min data in Figure 3C).
A small number of microarray time points exhibited signal saturation. This was corrected by re-scanning the same arrays at a lower gain, and adjusting the intensities of the saturated pixels according to average intensity ratio between the two scans (Dudley et al, 2002).
Simulations
Fork progression was modeled use the following set of assumptions: (i) the start of replication at origin i is normally distributed with an average start time ti and a standard deviation σ; (ii) replication progresses at a constant velocity, v=1.6 kb/min, for all replication forks and over the whole genome; (iii) each origin has an associated efficiency, ei; (iv) pausing might occur at a pause site j in a fraction (f) of the cells with a probability (Pj) and a duration (dj).
The mean start times, the standard deviation of the start times and origin efficiencies were determined from our data by minimizing the sum of the square differences between simulation results and experimental data. For each pair of adjacent origins, this minimization procedure used five slices of the data. These slices were chosen to encompass each of the origins and three equidistant regions between them, and the width of the slices was chosen to be 10% of the distance between the neighboring origins. The median was calculated for each of these slices, and used to calculate the sum of square differences. Each origin was assumed to have its own mean start time, ti, but the standard deviation of the start time was assumed to be constant in each simulation region. We also assumed that the GINS complex does not linger at the origin for any significant period of time, and after binding moves away at 1.6 kb/min. Finally, the GINS complexes fall off when adjacent forks collide. Each simulation was performed 10 000 times (to simulate 10 000 cells), using a random number generator to determine start times within the Gaussian distribution for a particular origin. Examples of the effects of changing the parameters used in the optimizations are presented in Supplementary Figures S6–S8.
Data availability
The complete experimental data can be found at http://prowl.rockefeller.edu/data/yeast_repl and has been deposited in MIAME compliant form in the Gene Expression Omnibus (accession number GSE19818).
Supplementary Material
Supplementary Discussion and data
Acknowledgments
We thank Bruz Marzolf for help with hybridizing and scanning of the arrays, and Conrad Nieduszynski for assistance with creating origin lists. We are grateful to Michael O'Donnell, Frederick Cross, Michael Rout, Andrew Murray, Bruce Stillman and Paul Nurse for insightful discussion and suggestions. We also thank all members of the Chait laboratory for their assistance. This work was supported by NIH grants U54 RR022220 (JA and BTC), P50 GM076547 (JA), and P41 RR00862 (BTC).
Footnotes
The authors declare that they have no conflict of interest.
References
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA (2006) The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA (2009) Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A (2005) Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K (2005) Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19: 1905–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande A, Newlon C (1996) DNA replication fork pause sites dependent on transcription. Science 272: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Diffley JF (2004) Regulation of early events in chromosome replication. Curr Biol 14: 778–786 [DOI] [PubMed] [Google Scholar]
- Dudley AM, Aach J, Steffen MA, Church GM (2002) Measuring absolute expression with microarrays with a calibrated reference sample and an extended signal intensity range. Proc Natl Acad Sci USA 99: 7554–7559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Collingwood D, Boeck ME, Fox LA, Alvino GM, Fangman WL, Raghuraman MK, Brewer BJ (2006) Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol 8: 148–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Haase SB, Lew DJ (1997) Flow cytometric analysis of DNA content in budding yeast. Methods Enzymol 283: 322–332 [DOI] [PubMed] [Google Scholar]
- Hiraga S, Hagihara-Hayashi A, Ohya T, Sugino A (2005) DNA polymerases alpha, delta, and epsilon localize and function together at replication forks in Saccharomyces cerevisiae. Genes Cells 10: 297–309 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA (2003) The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. CTYP-3. Mol Cell 12: 1525–1536 [DOI] [PubMed] [Google Scholar]
- Kanemaki M, Labib K (2006) Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J 25: 1753–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Sanchez-Diaz A, Gambus A, Labib K (2003) Functional proteomic identification of DNA replication proteins by induced proteolysis in vivo. Nature 423: 720–725 [DOI] [PubMed] [Google Scholar]
- Kubota Y, Takase Y, Komori Y, Hashimoto Y, Arata T, Kamimura Y, Araki H, Takisawa H (2003) A novel ring-like complex of Xenopus proteins essential for the initiation of DNA replication. Genes Dev 17: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labib K, Gambus A (2007) A key role for the GINS complex at DNA replication forks. Trends Cell Biol 17: 271–278 [DOI] [PubMed] [Google Scholar]
- Machida YJ, Hamlin JL, Dutta A (2005) Right place, right time, and only once: replication initiation in metazoans. Cell 123: 13–24 [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H (2002) S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature 415: 651–655 [DOI] [PubMed] [Google Scholar]
- McCarroll RM, Fangman WL (1988) Time of replication of yeast centromeres and telomeres. Cell 54: 505–513 [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR (2006) Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA 103: 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ (2001) Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411: 1068–1073 [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Hiraga S-I, Ak P, Benham CJ, Donaldson AD (2007) OriDB: a DNA replication origin database. Nucleic Acids Res 35: 40–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC (2006) Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell 21: 581–587 [DOI] [PubMed] [Google Scholar]
- Poloumienko A, Dershowitz A, De J, Newlon CS (2001) Completion of replication map of Saccharomyces cerevisiae chromosome III. Mol Biol Cell 12: 3317–3327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL (2001) Replication dynamics of the yeast genome. Science 294: 115–121 [DOI] [PubMed] [Google Scholar]
- Ren B, Robert F, Wyrick JJ, Aparicio OM, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA (2000) Genome-wide location and function of DNA binding proteins. Science 290: 2306–2309 [DOI] [PubMed] [Google Scholar]
- Rothstein R, Michel B, Gangloff S (2000) Replication fork pausing and recombination or ‘gimme a break’. Genes Dev 14: 1–10 [PubMed] [Google Scholar]
- Sclafani RA, Holzen TM (2007) Cell cycle regulation of DNA replication. Annu Rev Genet 41: 237–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirahige K, Hori Y, Shiraishi K, Yamashita M, Takahashi K, Obuse C, Tsurimoto T, Yoshikawa H (1998) Regulation of DNA-replication origins during cell-cycle progression. Nature 395: 618–621 [DOI] [PubMed] [Google Scholar]
- Tackett AJ, Dilworth DJ, Davey MJ, O'Donnell M, Aitchison JD, Rout MP, Chait BT (2005) Proteomic and genomic characterization of chromatin complexes at a boundary. J Cell Biol 169: 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H (2003) GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev 17: 1153–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Newport J (2000) Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell 5: 617–627 [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Aparicio JG, Chen T, Barnett JD (2001) Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science 294: 2357–2360 [DOI] [PubMed] [Google Scholar]
- Xu W, Aparicio JG, Aparicio OM, Tavaré S (2006) Genome-wide mapping of ORC and Mcm2p binding sites on tiling arrays and identification of essential ARS consensus sequences in S. cerevisiae. BMC Genomics 7: 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuki N, Terashima H, Kitada K (2002) Mapping of early firing origins on a replication profile of budding yeast. Genes Cells 7: 781–789 [DOI] [PubMed] [Google Scholar]
- Yamashita M, Hori Y, Shinomiya T, Obuse C, Tsurimoto T, Yoshikawa H, Shirahige K (1997) The efficiency and timing of initiation of replication of multiple replicons of Saccharomyces cerevisiae chromosome VI. CTYP-3. Genes Cells 2: 655–665 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Discussion and data
Data Availability Statement
The complete experimental data can be found at http://prowl.rockefeller.edu/data/yeast_repl and has been deposited in MIAME compliant form in the Gene Expression Omnibus (accession number GSE19818).