Abstract
A protease digestion strategy was incorporated into single-point SUPREX (stability of unpurified proteins from rates of H/D exchange), which is an H/D exchange- and mass spectrometry-based assay for the detection of protein-ligand binding. Single-point SUPREX is an abbreviated form of SUPREX in which protein-ligand binding interactions are detected by measuring the increase in a protein’s thermodynamic stability upon ligand binding. The new protease digestion protocol provides a noteworthy increase in the efficiency of single-point SUPREX because peptide masses can be determined with greater precision than intact protein masses in the MALDI readout of single-point SUPREX. The protocol was evaluated in test screens on two model protein systems, including cyclophilin A (CypA) and the minor allele variant of human alanine:glyoxylate aminotransferase (AGTmi). The test screening results obtained on both proteins revealed that the peptide readout of the single-point SUPREX-protease digestion protocol was more efficient than the intact protein readout of the original single-point SUPREX protocol at discriminating hits and non-hits. In addition to this improvement in screening efficiency, the protease digestion strategy described here is expected to significantly increase the generality of the single-point SUPREX assay.
Introduction
We recently reported a MALDI-MS-based technique for the high-throughput detection of protein-ligand binding interactions.1,2 The technique, hereafter referred to as single-point SUPREX, is an abbreviated version of SUPREX (stability of unpurified proteins from rates of H/D exchange), which is an H/D exchange- and mass spectrometry-based approach for evaluating the free energy difference between the folded and unfolded forms of a protein (i.e., the folding free energy).3,4 Since the folding free energy of proteins generally decreases (i.e., becomes more negative) upon ligand binding, the SUPREX methodology provides a useful means by which to detect and quantify the thermodynamic properties of protein-ligand binding interactions.4-13
In a recent application of single-point SUPREX, we screened an 880-member compound library for binding to cyclophilin A (CypA)2 and identified an important limitation of the single-point SUPREX protocol. The efficiency of the protocol was found to be highly dependent on the precision of the mass measurements and on the number of globally protected amide protons in the protein. In cases where the error associated with the protein mass measurement in the single-point SUPREX protocol is not small compared to the number of globally protected amide protons in the protein, the efficiency of the assay is low (i.e., it can be difficult to discriminate between hits and non-hits). For example, the efficiency of the CypA screen in our recent work2 was significantly reduced in comparison with the screening efficiency found for S-protein in our original proof-of-concept study.1 Since CypA and S-protein have similar numbers of globally protected amide proteins (as judged by the amplitudes of their SUPREX curves), the reduced efficiency was attributed to the errors associated with the MALDI mass measurements on CypA (i.e., ± ~6 Da) being larger than those on S-protein (i.e., ± 1.5 Da).
Described here is the introduction of a protease digestion strategy into the single-point SUPREX protocol. The single-point SUPREX-protease digestion protocol developed in this work is analogous to the protease digestion protocol we recently developed to expand the scope of the conventional SUPREX protocol.14 The strategy, which incorporates a protease digestion after the H/D exchange reaction in SUPREX, utilizes the SUPREX behavior of the detected peptides to report on the SUPREX behavior of the intact protein. Since peptide molecular weights can be determined with greater precision than intact protein molecular weights using MALDI-TOF mass spectrometry, we reasoned that the single-point SUPREX behavior of the protein could be determined with greater precision by reading out the peptide molecular weights.
As part of this work, the single-point SUPREX-digestion protocol was evaluated in the context of two model protein systems, CypA and the minor allele variant of human alanine:glyoxylate aminotransferase (AGTmi). CypA is a potential target for diagnostic imaging agents and pharmaceutical intervention because it is overexpressed in lung tumor cells and seems to be important for tumor growth.15,16 CypA was specifically chosen for this study to allow for a direct comparison to the results obtained in our previous screening experiment with CypA.2 AGTmi is a potential target for therapeutic intervention via pharmacological chaperones (i.e., compounds that bind and stabilize the native state of a protein) for patients with primary hyperoxaluria type I.17,18 AGTmi was chosen for this study because it represents a protein that is not amenable to screening using the original single-point SUPREX protocol due to its large size and relatively small number of globally protected amide protons.
Experimental Section
Materials
Human CypA and human AGTmi were expressed and purified as in our previous studies on these proteins.2,19 The following reagents were purchased from Sigma-Aldrich (St. Louis, MO): deuterium oxide (99.9% atom D), sodium deuteroxide (for pH adjustment; 40 wt% in D2O, 99.5% atom D), deuterium chloride (for pH adjustment; 35 wt% in D2O, 99% atom D), porcine pepsin, bovine pancreas insulin, pyridoxal 5′-phosphate (PLP), and sinapinic acid (SA). The internal standard ACTH clip (18-139) was from Bruker Daltonics (Billerica, MA). Guanidine hydrochloride (GdmCl) was purchased from EMD (Gibbstown, NJ), and trifluoroacetic acid (TFA) was purchased from Halocarbon (River Edge, NJ). Acetonitrile (ACN) was purchased from Fisher (Fair Lawn, NJ), and deuterated phosphoric acid was from Cambridge Isotope Laboratories (Andover, MA). Cyclosporin A (CsA) was from LKT Laboratories (St. Paul, MN).
Conventional SUPREX-Protease Digestion Protocol
CypA and AGTmi were initially subjected to SUPREX analyses using the conventional SUPREX-protease digestion protocol. These experiments were performed using a set of deuterated H/D exchange buffers containing 20 mM phosphate and deuterated GdmCl at concentrations between 0 and 4 M. The deuterated GdmCl was prepared using four cycles of dissolution in D2O and lyophilization. The analyses on CypA involved combining 1-μL aliquots of a 50 μM CypA stock solution with 0.5 μL of DMSO and initiating H/D exchange by mixing the resulting 1.5-μL aliquots with 3.5 μL of each deuterated H/D exchange buffer. After a five minute exchange time, the H/D exchange reaction in each deuterated H/D exchange buffer was quenched by adding 4 μL of the reaction mixture to 3 μL of aqueous HCl (pH ~ 0.8). A short pepsin digestion was performed by adding 3 μL of a 1 μM pepsin solution to the quenched H/D exchange reaction buffers. After a 1 min digestion, a 2-μL aliquot of the digested protein in each H/D exchange buffer was combined with 6 μL of matrix solution (saturated SA in 45% ACN/54.9% water/0.1% TFA), and 1 μL of the final matrix solution was spotted onto the MALDI target. The solvent was evaporated using a stream of nitrogen gas. Spectra were externally calibrated using the Peptide Calibration Standard II mix from Bruker.
The conventional SUPREX-protease digestion protocol used to generate SUPREX data on AGTmi was identical to that described above for CypA with the following exceptions: 1) the AGTmi stock solution was 40 μM; 2) 2 μL of the protein stock solution was combined with 1 μL PBS (to substitute for the addition of PLP as a positive control in the screening experiments) and added to 7 μL of each deuterated H/D exchange buffer; 3) the stock pepsin concentration was 0.2 μM; 4) the digestion time was 30 sec; and 5) the internal standard was insulin.
As a part of this work, the conventional SUPREX-protease digestion protocol was used to evaluate the ΔGf and m-values associated with each AGTmi peptide fragment. To make these measurements, the conventional SUPREX-protease digestion protocol was carried out as described, except that a range of different H/D exchange times was used. The protocol was performed using five different H/D exchange times that included 0.5, 2, 5, 25, and 40 min. SUPREX curves were constructed from the data obtained at each H/D exchange time. The transition midpoint of each SUPREX curve was determined (see Data Collection and Analysis section), and Equation 1 was used to obtain m- and ΔGf values from the SUPREX data as previously described.4
| Equation 1 |
In Equation 1, R is the gas constant, T is the temperature in Kelvin, <kint> is the average intrinsic exchange rate of an unprotected amide proton, t is the H/D exchange time, m is defined as δΔGf/δ[Denaturant], ΔGf is the folding free energy of the protein in the absence of denaturant, n is the number of protein subunits involved in the folding reaction (n = 2 for AGTmi), and [P] is the protein concentration expressed in n-mer equivalents. The left side of the equality in Equation 1 will hereafter be referred to as ΔGapp.
Using Equation 1, plots of ΔGapp vs. C1/2SUPREX were generated for apo-AGTmi. These plots were fit using a linear regression routine, and the y-intercept and slope were taken to be the ΔGf and m-value, respectively. Values for <kint> were calculated using the relationship <kint> = 10pH-5 min-1.3
Single-Point SUPREX-Protease Digestion Protocol
The single-point SUPREX assay was performed by distributing the positive and negative controls used for each protein in alternating wells of a microtiter plate. Exchange reactions were performed in sets of 10 (i.e., 5 positive and 5 negative controls per set), and MALDI analysis of each set was performed before the exchange reaction for the subsequent set was initiated. For the screening protocol, all spectra were internally calibrated. The [M+H]+ signal from the ACTH clip (18-139) peptide standard from Bruker was used to calibrate CypA spectra, and the [M+H]+ ion signal from insulin was used to calibrate the AGTmi spectra.
The single-point SUPREX-protease digestion protocol is identical to the conventional SUPREX-protease digestion protocol described above except that instead of using a series of deuterated H/D exchange buffers containing a range of GdmCl concentrations, a single GdmCl concentration was used for the analysis of each protein. The GdmCl concentrations used for CypA and AGTmi were 2.5 and 2.0 M, respectively (see Figures 1B, 2B, and 2C). The only additional difference is that the AGTmi reactions were scaled down by a factor of 2 for the screening such that the final volume of the H/D exchange reactions was 5 μL instead of 10 μL. This helped to reduce protein consumption.
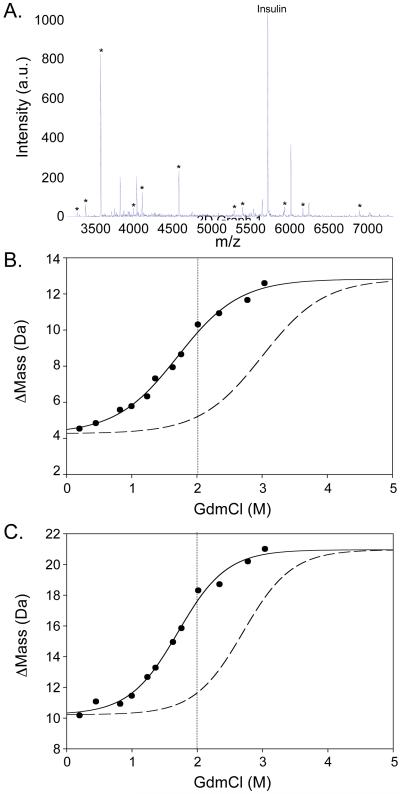
Figure 1.
A) A MALDI mass spectrum obtained for pepsin-digested CypA. The peptide ion signals marked with an asterisk were suitable for single-point SUPREX. B) A SUPREX curve obtained for a representative peptic fragment of CypA, fragment 2 (filled circles and solid line). The dashed curve represents the theoretical SUPREX curve expected for CypA fragment 2 in a SUPREX-protease digestion analysis performed in the presence of a hypothetical ligand with a Kd of 0.3 μM. The vertical dotted line indicates the concentration of GdmCl that was selected for the screening assay.
Figure 2.
A) A MALDI spectrum obtained for pepsin-digested AGTmi. The peptide ion signals marked with an asterisk were suitable for single-point SUPREX. B) and C) SUPREX curves obtained for AGTmi fragments 2 and 7, respectively (filled circles and solid line). These two fragments are representative of the two categories of SUPREX behavior observed for apo-AGTmi (see Results section). The dashed curves in B) and C) represent the SUPREX curves expected for AGTmi fragments 2 and 7 in SUPREX-protease digestion analyses performed in the presence of hypothetical ligands with a Kd of 10 μM. The vertical dotted lines indicate the concentration of GdmCl that was selected for the screening assay.
The GdmCl concentrations used for the CypA and AGTmi screenings in this work were chosen based on theoretical SUPREX curves generated for hypothetical CypA- and AGTmi-ligand complexes with Kd values of 0.3 and 10 μM, respectively. The theoretical curves generated for the CypA- and AGTmi-ligand complexes were identical to the experimental SUPREX curves recorded for CypA and AGTmi in the absence of ligand, with the exception that the transition midpoints of the theoretical SUPREX curves were shifted to higher denaturant concentrations. The exact denaturant concentration shift for each SUPREX curve was determined by first calculating the expected binding free energy (i.e., ΔΔGf) for the hypothetical complexes using Equation 2.20
| Equation 2 |
In Equation 2, [L] is the concentration of free ligand in the H/D exchange buffer, n is the number of equivalent ligand binding sites in the protein, Kd is the dissociation constant of the hypothetical ligand, R is the ideal gas constant, and T is the temperature in K. In our calculations using Equation 2, n was set to 1 for CypA and 2 for AGTmi. The value for [L] was set to either 100 or 800 μM, according to the estimated free ligand concentrations in our CypA and AGTmi analyses, respectively. The resulting ΔΔGf values were used in Equation 3 to calculate the SUPREX curve transition midpoint shifts (i.e., ΔC1/2SUPREX values) expected for CypA and AGTmi upon binding to hypothetical ligands with Kd values of 0.3 and 10 uM, respectively.
| Equation 3 |
In Equation 3, ΔΔGf is the binding free energy defined by Equation 2, and m is defined as δΔGf/δ[Denaturant]. The m-value used in our CypA calculations, 3.7 kcal/(mol M), was taken from previously published SUPREX data on CypA,11 and the m-values used in our AGTmi calculations were derived from the experimental results in this work. Equation 3 is derived from Equation 1. Equation 3 is valid in cases where the m-value does not change upon ligand binding and the same H/D exchange time is used to analyze the protein in the absence and in the presence of ligand. These were the circumstances in this work.
A positive and negative control were analyzed for each model protein. A total of 30 replicate measurements were made on each control. The negative controls (i.e., the apo-protein samples) for the analyses using the single-point SUPREX-protease digestion protocol were prepared exactly as described above for the analyses using the conventional SUPREX-protease digestion protocol. The CypA positive controls (i.e., the CsA-bound samples) were prepared by combining 1 μL of the 50 μM CypA stock solution with 0.5 μL of 1 mM CsA in DMSO. The AGTmi positive controls (i.e., the PLP-bound samples) were prepared in a single stock solution by combining 2 parts of the 40 μM AGTmi stock solution with one part 8 mM PLP in PBS buffer and equilibrating the sample for 1 hr at room temperature. For the H/D exchange reactions, a 1.5-μL aliquot of the equilibrated protein solution was combined with 3.5 μL of the H/D exchange buffer.
Data Collection and Analysis
MALDI mass spectra were collected using a Bruker Daltonics (Billerica, MA) Ultraflex II TOF/TOF mass spectrometer with the following instrument parameters: an ion source 1 voltage of 25.0 kV, an ion source 2 voltage of 23.4 kV, a lens voltage of 6.5 kV, and a delay time of 100 ns. Gating was used to select peptides with molecular weights of greater than 1 kDa for CypA and less than 3 kDa for AGTmi. Mass spectra were a sum of 100 laser shots. A total of 5 (for the single-point SUPREX-protease digestion protocol) or 10 (for the conventional SUPREX-protease digestion protocol) mass spectra were collected for each data point.
The m/z value for each peptide peak was determined using an in-house Matlab (The MathWorks, Inc., Natick, MA) program that performed a 9-point (CypA) or 19-point (AGTmi) floating average smoothing of the data, a one-point calibration using the internal standard, and a center of mass calculation for the protein peak. To determine the extent of deuteration (i.e., the Δmass) for each peptide, the measured peptide masses from 5 or 10 replicate mass spectra were averaged, and this value was subtracted from the mass of the undeuterated peptide.
In the case of the conventional SUPREX-protease digestion protocol, the Δmass values were plotted vs. [GdmCl], and the data was fit to a four parameter sigmoidal equation using a nonlinear regression routine in SigmaPlot (SYSTAT Software, Inc., San Jose, CA) to determine the denaturant concentration at the transition midpoint. In the case of the single-point SUPREX-protease digestion protocol, the Δmass values were subjected to a three-point moving average smoothing. The smoothed Δmass values will hereafter be called Δmassav values. Z’-factors were calculated using Equation 4.21
| Equation 4 |
In Equation 4, σc+ is the standard deviation of the positive controls, σc- is the standard deviation of the negative controls, μc+ is the average Δmass value of the positive controls, and μc- is the average Δmass value of the negative controls. The use of the moving average allowed for the calculation of a Z’-factor for each Δmassav value. Ultimately, the Z’-factors determined for a given peptide were averaged to determine a single average Z’-factor for each peptide (see Tables 1 and 2).
Table 1. SUPREX Data and Screening Efficiency for CypA.
| Fragment number |
Molecular Weight (Da) |
C1/2SUPREX (M, GdmCl) |
SUPREX Curve Amplitude (Da) |
Average Standard Deviation (Da)a |
Z’-Factor | False Positives |
False Negatives |
|---|---|---|---|---|---|---|---|
| 1 | 1623 | 1.9 | 2.5 | 0.3 | 0.4 | 0 | 0 |
| 2 | 1865 | 1.9 | 3.8 | 0.3 | 0.4 | 0 | 0 |
| 3 | 2375 | 1.6 | 2.6 | 0.4 | 0.4 | 0 | 0 |
| 4 | 2427 | 1.6 | 1.6 | 0.3 | 0.2 | 0 | 0 |
| 5 | 2574 | 1.6 | 1.9 | 0.3 | 0.1 | 0 | 0 |
| 6 | 2671 | 1.7 | 1.5 | 0.4 | -0.2 | 0 | 1/30 (3%) |
| 7 | 2742 | 1.7 | 1.8 | 0.3 | -0.2 | 0 | 0 |
| 8 | 2839 | 1.6 | 3.9 | 0.3 | 0.4 | 0 | 0 |
| 9 | 3083 | 1.9 | 3.7 | 0.3 | 0.2 | 0 | 0 |
| 10 | 6046 | 2.1 | 5.8 | 0.7 | -0.9 | 3/30 (10%) | 0 |
This value is the average of the standard deviations of the mass measurements from each point in a representative SUPREX curve.
Table 2. SUPREX Data and Screening Efficiency for AGTmi.
| Fragment number |
Molecular Weight (Da) |
C1/2SUPREX (M, GdmCl) |
SUPREX Curve Amplitude (Da) |
Average Standard Deviation (Da)a |
Z’-Factor | False Positives |
False Negatives |
|---|---|---|---|---|---|---|---|
| 1 | 3221 | 1.7 | 7.8 | 0.6 | 0.4 | 0 | 0 |
| 2 | 3334 | 1.7 | 8.4 | 0.6 | 0.5 | 0 | 0 |
| 3 | 3531 | 1.6 | 7.6 | 0.5 | 0.3 | 0 | 0 |
| 4 | 3965 | 1.7 | 7.3 | 0.7 | 0.0 | 0 | 1/30 (3%) |
| 5 | 4078 | 1.6 | 7.8 | 0.6 | 0.1 | 0 | 0 |
| 6 | 4564 | 1.3 | 8.0 | 1.0 | -0.3 | 0 | 1/30 (3%) |
| 7 | 5288 | 1.7 | 10.6 | 1.3 | 0.2 | 0 | 0 |
| 8 | 5404 | 1.5 | 8.6 | 1.3 | 0.3 | 0 | 0 |
| 9 | 5951 | 1.5 | 8.9 | 1.2 | 0.0 | 0 | 1/30 (3%) |
| 10 | 6197 | 1.4 | 10.1 | 1.3 | 0.1 | 1/30 (3%) | 0 |
| 11 | 6941 | 1.5 | 9.2 | 2.2 | -1.3 | 0 | 5/30 (17%) |
This value is the average of the standard deviations of the mass measurements from each point in a representative SUPREX curve.
Results
Selection of Screening Conditions
The pepsin digestion conditions used in this work yielded a number of peptide fragments for CypA and AGTmi in the 1 to 8 kDa size range (see Figures 1A and 2A). However, only 10 of the CypA peptide fragments and 11 of the AGTmi peptide fragments detected in the MALDI readouts were suitable for the single-point SUPREX protocol. An important prerequisite for a peptide fragment to be suitable for the single-point protocol is that it must yield a SUPREX curve in the conventional SUPREX-protease digestion protocol. Shown in Figures 1B, 2B, and 2C are SUPREX curves generated for representative CypA and AGTmi fragments using the conventional SUPREX-protease digestion protocol. Tables 1 and 2 summarize the CypA and AGTmi fragments that yielded SUPREX curves. As shown in Figures 1A and 2A, some peptide fragments of CypA and AGTmi yielded ion signals in the MALDI mass spectra but were not suitable for the single-point SUPREX protocol. These included fragments that did not have a denaturant dependence to their Δmass values using the conventional SUPREX-protease digestion protocol as well fragments with ion signals that were not well-resolved, had poor S/N ratios, and/or were not consistently detected in the MALDI readout of the conventional SUPREX-protease digestion protocol.
The transition midpoints of all 10 CypA fragments in Table 1 were within experimental error of each other (i.e., within a 0.5 M window). The average transition midpoint for the CypA fragments, 1.8 M, is also the same as the transition midpoint expected for the intact protein based on SUPREX data we have previously collected on CypA.11 The SUPREX analyses on the intact CypA in our previous work11 did not include a 5 min H/D exchange time. However, the ΔGf and m-values, 11.3 0.7 kcal/mol and 3.7 kcal/(mol M) (respectively), we previously reported for the purified CypA sample11 can be used in Equation 1 to calculate a 1.8 μM transition midpoint for the intact protein using a 5 min H/D exchange time in the SUPREX experiment. Such consistency between the SUPREX transition midpoint obtained on the intact protein using the conventional SUPREX protocol and on the peptide fragments using the conventional SUPREX-protease digestion protocol is expected for protein systems that fold and unfold in a concerted manner.14
The transition midpoints of all 11 AGTmi fragments listed in Table 2 were also within experimental error of each other (i.e., within a 0.5 M window). However, in contrast to the CypA results, the average transition midpoint for the AGTmi fragments, 1.6 M, is significantly different that that previously reported for the intact protein (i.e., 1.0 M) under similar conditions.19 Such a discrepancy is expected for protein systems that do not fold and unfold in a concerted manner (see Discussion section).14
The choice of the denaturant concentration and H/D exchange time used in the single-point SUPREX-protease digestion protocol is determined by the stringency of the selection and by the biophysical properties of the protein under study. The CypA and AGTmi assays in this work were designed to select ligands with dissociation constants (Kd values) less than 0.3 and 10 μM, respectively. These selection criteria were chosen to permit selection of the control ligands in this study, CsA and PLP, which have previously determined binding affinities in these ranges. For example, Kd values of 30-200 nM have been reported for the CypA-CsA binding interaction,22-25 and a Km value of 7.5 μM has been reported for the AGTmi-PLP binding interaction.26
Knowledge about the protein folding behavior of the target protein (i.e., ΔGf and m-values) is also needed to determine a useful denaturant concentration and H/D exchange time for the single-point SUPREX protocol. In particular, it is necessary to have a measure of the m-value associated with each peptide fragment to be used in the single-point SUPREX-protease digestion protocol (i.e., the peptide fragments that yielded SUPREX curves). In the case of proteins like CypA that fold and unfold in a concerted manner (see above), the ΔGf and m-values associated with all the peptide fragments that yield SUPREX curves will be similar to each other and similar to the ΔGf and m-values measured using the conventional SUPREX protocol to analyze the intact protein. The ΔGf and m-values for CypA used in these studies (-11.3 kcal/mol and 3.7 kcal/(mol M), respectively) were taken from earlier SUPREX studies on the intact protein.11
In the case of proteins like AGTmi which do not fold and unfold in a concerted manner, it is important to determine the m-value of each fragment using the conventional SUPREX-protease digestion protocol. Summarized in Table 3 are the ΔGf and m-values determined for the 11 AGTmi peptide fragments that yielded SUPREX curves using the conventional SUPREX-protease digestion protocol. The SUPREX curve transition midpoints used to generate the biophysical data in Table 3 are summarized in the Supporting Information. The 4.7 kcal/mol range of ΔGf values in Table 3 is consistent with the experimental error expected for replicate ΔGf value determinations by SUPREX (i.e., ± 10-15%). However, the 3.1 kcal/(mol M) range of m-values in Table 3 is greater than the experimental error expected for replicate m-value determinations by SUPREX (i.e., ± 20-30%). Inspection of the m-values in Table 3 reveals that the 11 peptide fragments in Table 3 can be divided into two groups based on the magnitude of their m-value. This grouping includes one group of peptides with m-values in the range of 3.2 - 4.2 kcal/(mol M) (i.e., Fragments 1-6 and 8) and one group of peptides with m-values in the range of 5.0 to 6.3 kcal/(mol M) (i.e., Fragments 7 and 9-11). The range of m-values in each group is consistent with the experimental error expected for replicate m-value determinations by SUPREX (i.e., ± 20-30%). These results suggest that there are at least two discrete categories of global unfolding/refolding reactions in apo-AGTmi.
Table 3. SUPREX-Derived Thermodynamic Parameters for Apo-AGTmi Peptide Fragments using the Conventional SUPREX-Protease Digestion Protocol.
| Fragment Number |
Molecular Weight (Da) |
ΔGf (kcal/mol)a |
m (kcal/mol/M)a |
|---|---|---|---|
| 1 | 3221 | -18.4 ± 0.4 | 4.2 ± 0.3 |
| 2 | 3334 | -18.1 ± 0.3 | 4.0 ± 0.2 |
| 3 | 3531 | -17.2 ± 0.4 | 3.7 ± 0.6 |
| 4 | 3965 | -16.6 ± 0.6 | 3.3 ± 0.4 |
| 5 | 4078 | -16.5 ± 0.2 | 3.2 ± 0.2 |
| 6 | 4564 | -16.3 ± 0.4 | 3.8 ± 0.3 |
| 7 | 5288 | -19.9 ± 0.9 | 5.1 ± 0.5 |
| 8 | 5404 | -17.1 ± 0.2 | 3.7 ± 0.2 |
| 9 | 5951 | -21.0 ± 1.0 | 6.3 ± 0.7 |
| 10 | 6197 | -19.0 ± 0.6 | 5.4 ± 0.6 |
| 11 | 6941 | -19.0 ± 1.0 | 5.0 ± 0.8 |
The error given is the fitting error obtained in a linear least squares analysis.
Ultimately, the ΔGf and m-values were used to predict the ligand-induced shift in the SUPREX curve transition midpoint (see Experimental Section). Figures 1 and 2 show the expected SUPREX curve transition midpoint shifts for the representative CypA and AGTmi peptide fragments. These shifts are those predicted for the representative CypA and AGTmi peptide fragments upon the binding of CypA and AGTmi to hypothetical ligands with Kd values of 0.3 and 10 μM, respectively. The denaturant concentration at which there is a maximum difference between the Δmass values expected for the unbound and bound forms of the protein is selected for the single-point SUPREX-protease digestion protocol. In the proof-of-principle studies described here on CypA and AGTmi, the selected denaturant concentrations were 2.5 and 2.0 M, respectively (see Figures 1B, 2B, and 2C).
The selected denaturant concentrations used in these proof-of-principle studies were based on using a 5 min H/D exchange time in the SUPREX protocol. In theory, longer or shorter H/D exchange times could have been used. The use of longer or shorter H/D exchange times would shift the SUPREX curve transition midpoints of the peptide fragments to lower or higher denaturant concentrations, respectively, as described previously.14 However, the expected transition midpoint shift upon ligand binding would be the same, irrespective of the H/D exchange time used. In practice, we found that the use of a relatively short (5 min) H/D exchange minimized Δmass values in the pre-transition baseline of the SUPREX curves we recorded. This helped to maximize the Δmass value difference for the unbound and bound forms of the protein.
Evaluation of the Single-Point SUPREX-Protease Digestion Assay
The single-point SUPREX-protease digestion assay was used in two model screening experiments, one to select for CsA binding to CypA and one to select for PLP binding to AGTmi. The conditions chosen for the screening experiments included denaturant concentrations of 2.5 and 2.0 M for CypA and AGTmi (respectively) and a 5 min H/D exchange time (see above). Each screen involved 30 replicate analyses of a positive control and a negative control. The Δmass values obtained in these replicate analyses of the positive and negative controls were recorded for the 10 suitable CypA fragments and the 11 suitable AGTmi fragments (see Tables 1 and 2).
As expected, the Δmass values recorded for each peptide fragment were generally lower for the positive controls than for the negative controls (see Figures 3 and 4 as well as Figures S1-6 in the Supplemental Information). However, the separation between the positive and negative controls in each screen varied for each peptide fragment, making some peptide fragments more ideal than others for the single-point SUPREX-protease digestion assay. Shown in Figures 3 and 4 are assay data obtained for CypA and AGTmi peptides with the highest and lowest Z’-factors in the single-point SUPREX-protease digestion assay. The dashed lines in these figures represent the cutoff values that would be used to select for hits when performing a single-point SUPREX assay. These cutoff values were determined by subtracting three standard deviations from the Δmassav values as in our previous work.2
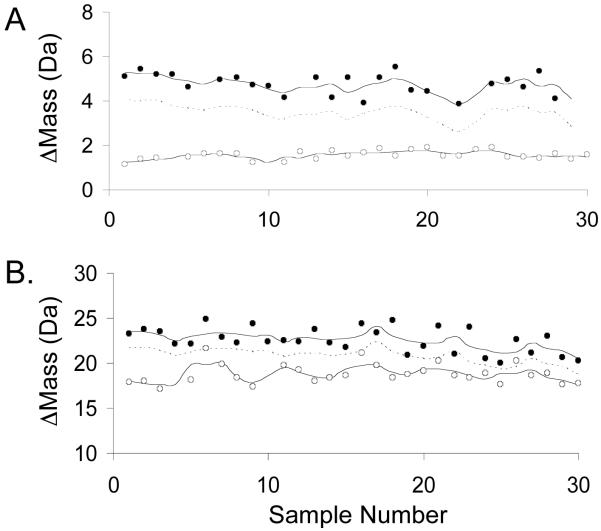
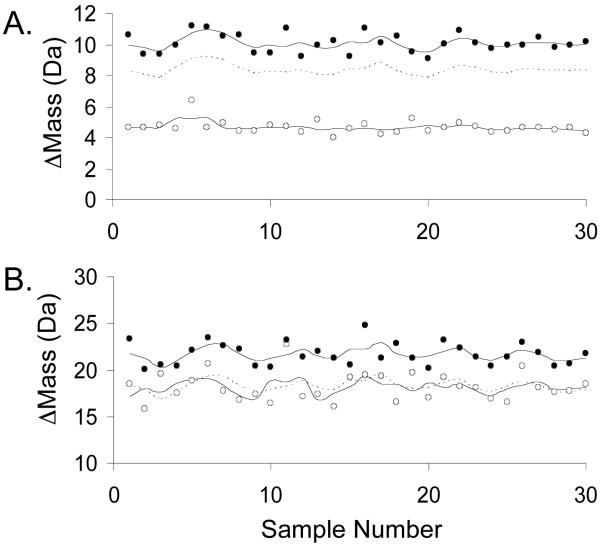
Figure 3.
Control screening data obtained for two CypA peptides with different Z’-factors. A) Data obtained using fragment 2 with a Z’-factor of 0.4. B) Data obtained using fragment 10 with a Z’-factor of -0.9. In each panel the data obtained on the negative and positive controls are represented with filled and open circles, respectively. The upper and lower solid lines represent the Δmassav of the negative and positive controls, respectively, and the dashed line represents the cutoff values, which are three standard deviations below the negative control Δmassav values.
Figure 4.
Control screening data obtained for two AGTmi peptides with different Z’-factors. A) Data obtained using fragment 2 with Z’-factor of 0.5. B) Data obtained using fragment 11 with a Z’-factor of -1.3. In each panel the data obtained on the negative and positive controls are represented with filled and open circles, respectively. The upper and lower solid lines represent the Δmassav of the negative and positive controls, respectively, and the dashed line represents the cutoff values, which are three standard deviations below the negative control Δmassav values.
One way to evaluate screening efficiency is to examine the rates of false positives and false negatives. Summarized in Tables 1 and 2 are the false positive and false negative rates obtained using the control Δmass values collected for CypA and AGTmi along with the calculated cutoff values described above. The false positive and false negative rates observed for most of the peptides were 0%, and all of the peptide fragments had false positive and false negative rates ≤ 10%, with the single exception of AGTmi fragment 11 (Table 2).
The screening efficiency of the selection assay was also evaluated using an analytical parameter called the Z’-factor. This parameter gives a measure of the separation between the positive and negative controls in an assay.21 Z’-factors must be at least zero for an assay to be viable, and a Z’-factor equal to zero is generally considered acceptable for a “yes/no” assay. However, Z’-factors greater than zero (0 < Z’ ≤ 1) are best for screening assays, with more efficient screening assays yielding larger values (up to Z’ = 1). The Z’-factors obtained for the CypA fragments varied from -0.9 to 0.4, with 7 of the 10 fragments yielding Z’-factors greater than 0. The Z’-factors obtained for the AGTmi fragments varied from -1.3 to 0.5, with 9 of the 11 fragments yielding Z’-factors of at least 0.
Discussion
The average standard deviations in Tables 1 and 2 are consistent with the expected mass accuracy, ± ~200 ppm, of the MALDI mass spectrometer used in this work. As expected, the average standard deviations of the larger fragments were larger than the average standard deviations of the smaller fragments. Interestingly, there was not a correlation between a peptide fragment’s molecular weight and its Z’-factor. This is due in large part to the absence of any correlation between the molecular weight of a peptide fragment and the amplitude of a peptide fragment’s SUPREX curve. The absence of such a correlation between amplitude and molecular weight is to be expected since the amplitude of a peptide’s SUPREX curve is not determined by the number of amide positions in the peptide (i.e., peptide size). Rather, the amplitude of a peptide’s SUPREX curve is determined by the number of amide positions in the peptide that are globally protected when the intact protein is folded in solution. This is related to the protein’s three-dimensional structure and the location of the peptide within that structure.
The Δmass values used to evaluate the Z’ factors in this work were not subject to a back-exchange correction. Deuterium losses clearly occurred during the digestion step and during the MALDI sample preparation and analysis. The Δmass values observed for the negative controls in this work were approximately 50-60% of those expected if all the amide protons in the peptide were exchanged. This degree of back exchange is consistent with that typically observed using the SUPREX with protease digestion protocol.14 Back-exchange correction factors could be applied to the Δmass values obtained for the positive and negative controls in this work. However, we found that this does not significantly impact Z’ factors in the single-point SUPREX protocol. While the Δmass-value difference between the positive and negative controls is increased upon application of a correction factor, there is a concomitant increase in the Δmass-value error that is propagated in the back-exchange-correction calculation. Thus, Z’ factors, which are a function of both the Δmass-value difference between the positive and negative controls and the Δmass-value error (see Equation 4), are not changed.
It is noteworthy that addition of the protease digestion step to the single-point SUPREX protocol did not have a significant effect on the throughput of the protocol. In fact, the throughput of the control screenings using the single-point SUPREX-protease digestion protocol in this work was 1.8 min/ligand. This is slightly better than the throughput of 3 min/ligand that was achieved using the single-point SUPREX protocol in our previous work.2 However, a direct comparison of the screening rate in this work to that in our previous work is difficult because the screening was performed by one researcher instead of two, and different MALDI instruments were used in the two experiments. The MALDI instrument used in this work had a higher repetition rate laser and a faster sample positioning mechanism, and these differences in instrument performance are likely responsible for the improved rate.
Unlike many H/D exchange and protease digestion techniques, the single-point SUPREX-protease digestion strategy does not require extensive peptide mapping. The assay is designed to detect changes in the protein’s global stability upon ligand binding rather than to map the detailed conformational changes that result from ligand binding. This means that extensive peptide coverage is also not critical for the assay since the detection of ligand binding does not require the analysis of a peptide that is located at the binding site. The assay described here is similar to an approach recently reported by Chalmers and coworkers,27 which also relies on the read-out of selected peptides after proteolysis of the H/D exchanged protein in order to detect protein-ligand binding. However, the single-point SUPREX-protease digestion assay is unique with respect to its use of chemical denaturant. It is the chemical denaturant that makes the single-point SURPEX-protease digestion protocol especially sensitive to the global/subglobal changes in protein stability that result form ligand binding.
In the case of proteins with cooperative unfolding/refolding transitions (like CypA), all of the peptide fragments that yield a SUPREX curve in the protease digestion protocol report on the same cooperative unfolding/refolding reaction in the protein. Thus, any one or all of the peptide fragments with appropriately high Z’-factors could be used in the single-point SUPREX-protease digestion assay. In fact, the multiple peptides in the assay provide a built-in redundancy. For example, in the CypA screening, one could choose to monitor fragments 1,2,3, and 8, all of which have Z’-factors of 0.4. In effect, this would be similar to performing four replicate screenings.
In the case of proteins that do not have highly cooperative unfolding/refolding reactions (like AGTmi), at least one peptide with an appropriately high Z’-factor must be detected for each sub-global unfolding/refolding reaction in the protein that is to be probed. For example, in large, multidomain proteins, peptide fragments from globally protected regions in each of the different domains must be observed for the assay to be effective. If peptide fragments from globally protected regions of a particular domain are missing (e.g., due to signal suppression effects in the MALDI readout), then ligands targeting that particular domain will appear as false negatives. However, it is important to emphasize that extensive peptide coverage of a specific domain is still not needed for the single-point SUPREX-protease digestion protocol. As we have previously shown with the conventional SUPREX-protease digestion protocol, any peptide fragment derived from a globally protected region of the domain can be used to report on the biophysical properties of the entire domain (as long as the domain folds/unfolds in a concerted manner).14
A potentially complicating issue in the application of the single-point SUPREX-protease digestion protocol to proteins with multiple, non-concerted, sub-global unfolding/refolding reactions can arise if the ΔGf and/or m-values associated with one peptide fragment’s SUPREX behavior are significantly different than the ΔGf and/or m-values associated with another peptide fragment’s SUPREX behavior. Such variability can make it difficult to choose a single set of selection conditions (i.e., H/D exchange time and denaturant concentration) at which there is a maximum difference between the Δmass values expected for the unbound and bound forms of the protein in the single-point SUPREX-protease digestion assay. However, our results with AGTmi indicate that some such variability can be tolerated. For example, even though two classes of AGTmi peptide fragments were observed (i.e., one class with ΔGf and m-values close to 20 kcal/mol and 5.5 kcal/(mol M), respectively, and one class with ΔGf and m-values close to 17 kcal/mol and 3.7 kcal/(mol M)), a single set of conditions was effectively used to select for PLP binding to AGTmi. Moreover, our results suggest that PLP binding to AGTmi has a stabilizing effect on both of the concerted folding/refolding reactions detected in our experiments, as both classes of AGTmi peptides could be used to detect PLP binding.
The single-point SUPREX-protease digestion protocol described here is a significant improvement over the conventional single-point SUPREX protocol because it more efficiently discriminates between hits and non-hits in high-throughput screening experiments involving larger proteins (i.e., proteins with MW > ~10 kDa). For example, the results obtained in this work reveal that Z’-factors of 0.4 could be achieved using the single-point SUPREX-protease digestion protocol to screen ligands for CypA binding. Such Z’-factors are significantly higher than the best possible Z’-factors that were achieved using the conventional single-point SUPREX protocol on CypA (Z’ ~ 0.0).2
In the case of AGTmi, the protease digestion protocol was critical for the successful application of single-point SUPREX. Using data from the SUPREX analysis of intact AGTmi,19 the Z’-factor using the conventional single-point SUPREX protocol is expected to be approximately -0.2, indicating that screening the intact protein using the original protocol would not result in the efficient selection of ligands for AGTmi. In contrast, when a protease digestion step is incorporated into the protocol, AGTmi becomes amenable to screening using single-point SUPREX, and screening efficiencies comparable to those of CypA (using the protease digestion protocol) were achieved. We anticipate that this method will significantly increase the generality of the single-point SUPREX protocol for the analysis of large proteins like AGTmi.
Conclusions
We have shown that the efficiency of single-point SUPREX can be improved by incorporating a protease digestion step into the assay. The new single-point SUPREX-protease digestion assay was evaluated using two model proteins. Using the new strategy, Z’-factors of up to 0.5 were achieved, and false positive and false negative rates as low as 0% were achieved in screening experiments performed on the two model proteins. These Z’-factors and false positive and false negative rates were significantly improved over those that could be obtained using the single-point SUPREX assay without the protease digestion protocol. The protocol is expected to be applicable to a wide range of protein systems, including large proteins that are not easily detected in their intact form using MALDI-MS as well large, multidomain proteins that require the protease digestion protocol for successful analysis by SUPREX. Also, since multiple peptides can be monitored simultaneously, the assay has a built in redundancy that can be exploited to further improve the efficiency of a high-throughput screening experiment.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Michael J. Campa for providing the CypA used in this project. This work was funded by NIH grants to M.C.F. (R21 CA127064-01A1) and C.L.T. (R21 DK075291-01A1).
References
- 1.Powell KD, Fitzgerald MC. J. Comb. Chem. 2004;6:262–269. doi: 10.1021/cc034051e. [DOI] [PubMed] [Google Scholar]
- 2.Hopper ED, Roulhac PL, Campa MJ, Patz EF, Fitzgerald MC. J. Am. Soc. Mass Spectrom. 2008;19:1303–1311. doi: 10.1016/j.jasms.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghaemmaghami S, Fitzgerald MC, Oas TG. Proc. Natl. Acad. Sci. U. S. A. 2000;97(15):8296–8301. doi: 10.1073/pnas.140111397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Powell KD, Fitzgerald MC. Biochemistry. 2003;42:4962–4970. doi: 10.1021/bi034096w. [DOI] [PubMed] [Google Scholar]
- 5.Powell KD, Ghaemmaghami S, Wang MZ, Ma L, Oas TG, Fitzgerald MC. J. Am. Chem. Soc. 2002;124:10256–10257. doi: 10.1021/ja026574g. [DOI] [PubMed] [Google Scholar]
- 6.Ma LY, Fitzgerald MC. Chem. Biol. 2003;10(12):1205–1213. doi: 10.1016/j.chembiol.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 7.Roulhac PL, Powell KD, Dhungana S, Weaver KD, Mietzner TA, Crumbliss AL, Fitzgerald MC. Biochemistry. 2004;43(50):15767–15774. doi: 10.1021/bi0481848. [DOI] [PubMed] [Google Scholar]
- 8.Roulhac PL, Weaver KD, Adhikari P, Anderson DS, DeArmond PD, Mietzner TA, Crumbliss AL, Fitzgerald MC. Biochemistry. 2008;47(14):4298–4305. doi: 10.1021/bi701188x. [DOI] [PubMed] [Google Scholar]
- 9.Tang L, Hopper ED, Tong Y, Sadowsky JD, Peterson KJ, Gellman SH, Fitzgerald MC. Anal. Chem. 2007;79(15):5869–5877. doi: 10.1021/ac0700777. [DOI] [PubMed] [Google Scholar]
- 10.Tong Y, Wuebbens MM, Rajagopalan KV, Fitzgerald MC. Biochemistry. 2005;44(7):2595–601. doi: 10.1021/bi047762h. [DOI] [PubMed] [Google Scholar]
- 11.Wang MZ, Shetty JT, Howard BA, Campa MJ, Patz EF, Fitzgerald MC. Anal. Chem. 2004;76(15):4343–4348. doi: 10.1021/ac049536j. [DOI] [PubMed] [Google Scholar]
- 12.Weaver KD, Heymann JJ, Mehta A, Roulhac PL, Anderson DS, Nowalk AJ, Adhikari P, Mietzner TA, Fitzgerald MC, Crumbliss AL. Journal of Biological Inorganic Chemistry. 2008;13(6):887–898. doi: 10.1007/s00775-008-0376-5. [DOI] [PubMed] [Google Scholar]
- 13.Williams JC, Roulhac PL, Roy AG, Vallee RB, Fitzgerald MC, Hendrickson WA. Proc. Natl. Acad. Sci. U. S. A. 2007;104(24):10028–10033. doi: 10.1073/pnas.0703614104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang LJ, Roulhac PL, Fitzgerald MC. Anal. Chem. 2007;79(22):8728–8739. doi: 10.1021/ac071380a. [DOI] [PubMed] [Google Scholar]
- 15.Campa MJ, Wang MZ, Howard BA, Fitzgerald MC, Patz EF., Jr. Cancer Res. 2003;63:1652–1656. [PubMed] [Google Scholar]
- 16.Howard BA, Furumai R, Campa MJ, Rabbani ZN, Vujaskovik Z, Wang XF, Patz EF. Cancer Res. 2005;65(19):8853–8860. doi: 10.1158/0008-5472.CAN-05-1219. [DOI] [PubMed] [Google Scholar]
- 17.Danpure CJ. Am. J. Nephrol. 2005;25(3):303–310. doi: 10.1159/000086362. [DOI] [PubMed] [Google Scholar]
- 18.Lumb MJ, Danpure CJ. J. Biol. Chem. 2000;275(46):36415–36422. doi: 10.1074/jbc.M006693200. [DOI] [PubMed] [Google Scholar]
- 19.Hopper ED, Pittman AMC, Fitzgerald MC, Tucker CL. J. Biol. Chem. 2008;283(45):30493–30502. doi: 10.1074/jbc.M803525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schellman JA. Biopolymers. 1975;14:999–1018. doi: 10.1002/bip.1975.360140113. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TDY, Oldenburg KR. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Handschumacher RE, Harding MW, Rice J, Drugge RJ. Science. 1984;226(4674):544–547. doi: 10.1126/science.6238408. [DOI] [PubMed] [Google Scholar]
- 23.Harding MW, Handschumacher RE. Transplantation. 1988;46(2):S29–S35. doi: 10.1097/00007890-198808001-00006. [DOI] [PubMed] [Google Scholar]
- 24.Holzman TF, Egan DA, Edalji R, Simmer RL, Helfrich R, Taylor A, Burres NS. J. Biol. Chem. 1991;266(4):2474–2479. [PubMed] [Google Scholar]
- 25.Liu J, Albers MW, Chen CM, Schreiber SL, Walsh CT. Proc. Natl. Acad. Sci. U. S. A. 1990;87(6):2304–2308. doi: 10.1073/pnas.87.6.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coulter-Mackie MB, Lian Q, Wong SG. Prot. Expr. Purif. 2005;41(1):18–26. doi: 10.1016/j.pep.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. J. Biomol. Tech. 2007;18(4):194–204. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.